Abstract
Formation of the dorsal–ventral axis of the Drosophila wing depends on activity of the LIM–homeodomain protein Apterous (Ap). Here we report that Ap activity levels are modulated by dLMO, the protein encoded by the Beadex (Bx) gene. Overexpression of dLMO in Bx mutants interferes with Apterous function. Conversely, Bx loss-of-function mutants fail to down-regulate Apterous activity at late stages of wing development. Biochemical analysis shows that dLMO protein competes for binding of Apterous to its cofactor Chip. These data suggest that Apterous activity depends on formation of a functional complex with Chip and that the relative levels of dLMO, Apterous, and Chip determine the level of Apterous activity. The dominant interference mechanism of dLMO action may serve as a model for the mechanism by which LMO oncogenes cause cancer when misexpressed in T cells.
Keywords: Beadex, LMO protein, LIM–homeodomain, Drosophila, wing development, Apterous
Axis formation in Drosophila limb development is controlled by localized expression of secreted signaling molecules. Hedgehog (Hh), Decapentaplegic (Dpp), and Wingless (Wg) form activity gradients that define the spatial domains of target gene expression in the developing legs and wings (Diaz-Benjumea and Cohen 1995; Zecca et al. 1995, 1996; Lecuit et al. 1996; Lecuit and Cohen 1997; Nellen et al. 1996; Neumann and Cohen 1996, 1997; Strigini and Cohen 1997). Both spatially restricted expression and appropriate levels of activation of the signaling pathways are critical for normal patterning of the limbs. Misexpression of genes at different levels of these regulatory hierarchies leads to pattern abnormalities. Misexpression of the signaling molecules can lead to axis duplication (Struhl and Basler 1993; Basler and Struhl 1994; Capdevila and Guerrero 1994; Diaz-Benjumea et al. 1994; Diaz-Benjumea and Cohen 1995; Felsenfeld and Kennison 1995; Ingham and Fietz 1995; Zecca et al. 1995). In addition overactivation (or underactivation) of a signaling pathway involved in proper spatial localization of downstream effector genes can also perturb normal limb development (e.g., Johnson et al. 1995; Axelrod et al. 1996; Neumann and Cohen 1996). Finally, misexpression of the effector genes themselves can lead to abnormalities in limb development (e.g., de Celis et al. 1996a; Grimm and Pflugfelder 1996; Gorfinkiel et al. 1997; Sturtevant et al. 1997).
These observations suggest that systematic misexpression of genes in the developing limbs might provide an effective way to screen for genes involved in the cell signaling processes that control limb development. The modular–misexpression system developed by Rørth (1996) was used to carry out a large-scale screen for genes that perturb wing development (Rørth et al. 1998). The system allows conditional misexpression of genes tagged by insertion of a P element that carries a GAL4 regulatable EP (enhancer and a basal promoter) oriented to direct expression of adjacent genomic sequences. When combined with a source of GAL4, the EP element will direct expression of any gene that happens to lie next to its insertion site. The screen identified EP insertions at hh, patched (ptc), and dpp, genes with known roles in limb patterning (Rørth et al. 1998) as well as a number of new loci that are implicated in wing patterning by virtue of their overexpression phenotypes.
Here we report the characterization of a gene identified by the EP screen that is involved in dorsal–ventral (DV) patterning of the wing. We present genetic and biochemical evidence that the product of the Beadex (Bx) gene regulates Apterous (Ap) activity levels. Gain-of-function mutants reduce Apterous activity. Conversely, loss-of-function mutants of Bx appear to increase Ap activity. Ap encodes a LIM–homeodomain protein that specifies dorsal cell fate (Cohen et al. 1992; Diaz-Benjumea and Cohen 1993; Williams et al. 1993; Blair et al. 1994). A LIM-binding protein called Chip has been identified as a possible cofactor for Ap (Morcillo et al. 1997). We show that Bx mutants overexpress a LIM-only protein, dLMO, that binds to Chip and thereby interferes with formation of a functional complex between Ap and Chip. We also show that Ap induces expression of its antagonist dLMO, which suggests that Ap and dLMO constitute a feedback mechanism and that the relative levels of dLMO, Chip, and Ap determine Ap activity levels in vivo. These findings suggest a molecular model for the mechanism of action of LMO oncogenes in causing leukemia and lymphoma (Fisch et al. 1992; McGuire et al. 1992).
Results
An antagonist of Ap function in dorsal cells
Two independently isolated EP insertions on the X chromosome produced phenotypes suggesting a role in DV axis formation when expressed in the developing wing. Adult wings from flies carrying EP1306 or EP1394 lack most of the normal wing margin and show irregular bits of ectopic margin associated with overgrowths in the dorsal surface of the wing when the EP lines are expressed under control of optomoter blind–gal4 (omb–gal4) (not shown). A milder version of this phenotype is observed when EP1394 is expressed in a narrow stripe of cells in the center of the wing blade under control of ptc–gal4 (Fig. 1). The dorsal surface of the wing is extensively overgrown and contains an ectopic wing margin along the edge of the overgrowth (Fig. 1B, red arrow). Comparable effects are observed with both EP lines.
Figure 1.
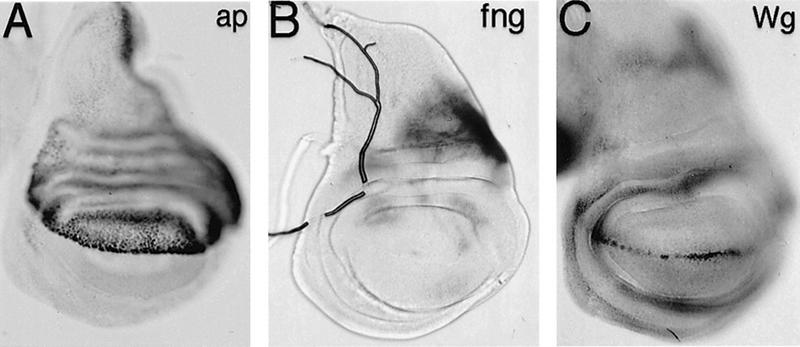
Misexpression of EP1306 interferes with Ap function. (A) Cuticle preparation of a wild-type wing colored blue to highlight the region of ptc–gal4 expression in the region between veins 3 and 4. The wing is viewed from the ventral side, with the anterior margin to the top. (B) Cuticle preparation of a ptc–gal4; EP1394 wing. The wing is distorted because of the extensive overgrowth of the dorsal compartment. In this example dorsal (d) is to the top and ventral (v) to the bottom, in effect a 90° rotation perpendicular to the wing in A. The anterior margin is viewed edge on. The red arrow indicates an ectopic wing margin running along the edge of the overgrowth in the dorsal compartment. The ectopic margin consists of posterior structures and has both dorsal and ventral elements. Ectopic anterior margins are usu ally incomplete. The topology of these wings is best understood in terms of Wg expression (see D). Comparable effects are seen in all cases for EP1394 and EP1306. (C,D) Wg expression in wild-type (C) and ptc–gal4; EP1306 (D) wing imaginal discs visualized by antibody staining. Wg expression is interrupted at the DV boundary of the ptc–gal4; EP1306 disc and an ectopic stripe of Wg runs along the edge of the ptc–gal4 domain (red arrow). Note the relative overgrowth of the dorsal compartment. (E,F) ap–lacZ expression in wild-type (E) and ptc–gal4; EP1306 (F) wing discs visualized using anti-β-galactosidase. Note that the normally sharp boundary between Ap-expressing dorsal cells and ventral cells is disturbed in the ptc–gal4 domain (arrow). This suggests a defect in Ap function in cells expressing EP1306, because Ap activity is important in generating the DV compartment boundary (Diaz-Benjumea and Cohen 1993; Blair et al. 1994). (G,H) fng–lacZ expression in wild-type (G) and ptc–gal4; EP1306 (H) wing discs visualized by histochemical activity staining. fng–lacZ is not expressed in dorsal cells in which EP1306 is overexpressed. We verified that Ap regulates fng–lacZ by ectopic expression of Ap in the ptc–gal4 stripe in the ventral compartment in ptc–gal4; UAS–Ap flies (data not shown).
The presence of the ectopic wing margin and the overgrowth of the dorsal compartment suggested that EP1394 misexpression might cause ectopic Wg expression. Wg is normally expressed in a stripe along the DV boundary of the wild-type wing imaginal disc (Fig. 1C), where it acts locally to induce nearby cells to adopt wing margin identity (Phillips and Whittle 1993; Couso et al. 1994). Ectopic Wg expression has been shown to induce ectopic wing margin formation and overgrowth of the surrounding tissue (Diaz-Benjumea and Cohen 1995; Kim et al. 1995; de Celis et al. 1996b; Doherty et al. 1996; Neumann and Cohen 1997). In ptc–gal4; EP1306 discs, Wg is ectopically expressed in cells parallel to the ptc–gal4 stripe in the dorsal compartment (Fig. 1D, red arrow). This ectopic Wg stripe corresponds to the ectopic wing margin shown in Figure 1B.
The observation that EP1306 expression induces ectopic Wg in dorsal cells but not in ventral cells suggested involvement of the ap and fringe (fng) genes. ap is expressed in dorsal cells (Fig. 1E), in which it is thought to induce fng expression (Irvine and Wieschaus 1994). In ptc–gal4; EP1306 discs, an ap reporter gene (Fig. 1F) and Ap protein (not shown) continue to be expressed normally in dorsal cells; however, fng–lacZ expression is lost in the dorsal compartment where ptc–gal4 is expressed (arrow Fig. 1H). Loss of fng explains the abnormal expression of Wg in EP1306-expressing wing discs. Wg is induced at the interface, where cells that express fng meet cells that do not (Kim et al. 1995). Loss of fng in cells expressing EP1306 will lead to loss of Wg at the DV boundary and to ectopic Wg along the new boundary of fng expression in the dorsal compartment. Removing fng activity from a large patch of dorsal cells in somatic mosaic clones produces a comparable effect (Kim et al. 1995). The similarity in these phenotypes suggests that the effects of overexpressing EP1306 can be explained by the observed loss of fng expression in dorsal cells. These observations suggest that Ap function is compromised in cells that overexpress EP1306.
EP1306 directs expression of the Bx gene
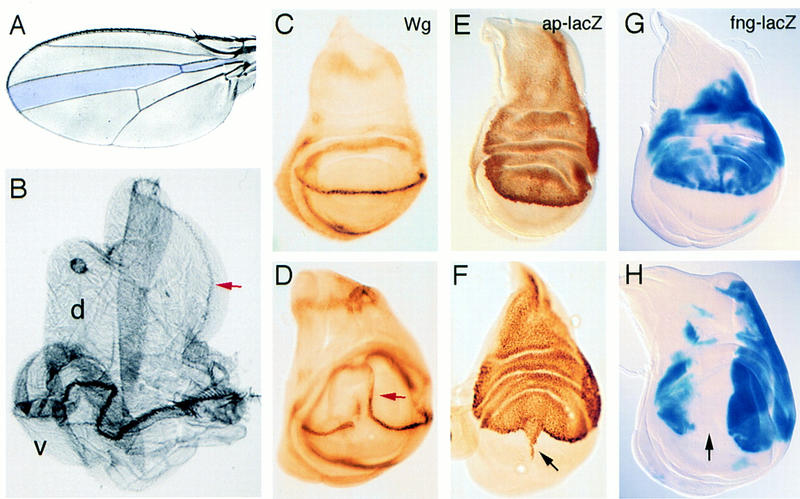
EP1306 and EP1394 were mapped to cytological position 17C1-4 by in situ hybridization to polytene chromosomes. This corresponds to the location of the Bx gene. Bx alleles are dominant mutations that are thought to increase gene activity (Lifschytz and Green 1979; Mattox and Davidson 1984). The severity of the Bx mutant phenotype is enhanced by introducing an extra copy of the wild-type gene and reduced by removing a copy. In addition, increasing the number of copies of the normal Bx gene (by tandem duplication) produces the same wing defects as are found in the dominant Bx mutant (cited in Lifschytz and Green 1979). This suggests that the Bx mutant phenotype is caused by overexpression of the normal gene product. We observe that sequences adjacent to the insertion sites of EP1306 and EP1394 are overexpressed in Bx1 mutant wing discs (Fig. 2), suggesting that these EP elements direct expression of the Bx gene. To confirm that Bx mutants produce comparable effects to overexpression of EP1306, we examined Wg, ap, and fng expression in Bx1 and Bx2 mutant wing discs. ap–lacZ expression is normal, whereas fng–lacZ expression is reduced in dorsal cells of the wing pouch (Fig. 3A,B). Wg expression is reduced and irregular at the margin (Fig. 3C).
Figure 2.
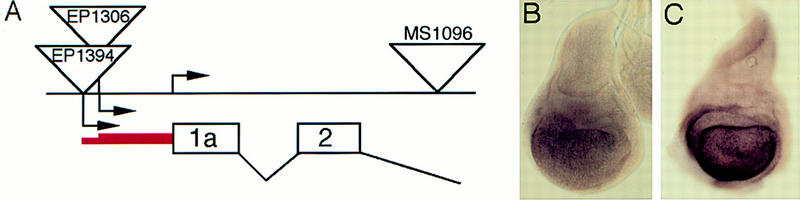
EP1306 directs expression of the Bx–dLMO transcript. (A) Molecular map of EP1306, EP1394, and MS1096 insertions at the dLMO locus. (B,C) In situ hybridization to wild-type (B) and Bx1 (C) wing discs using an antisense RNA probe prepared from the dLMO cDNA. The dLMO transcript is overexpressed in Bx mutant wing discs. The discs in B and C were processed in parallel to allow direct comparison of staining intensity. To verify that overexpression of dLMO is the cause of the EP1306 and Bx mutant phenotypes, a newly isolated dLMO cDNA was modified to introduced an epitope tag, cloned into pUAST and expressed under ptc–gal4 control. The resulting phenotypes were identical to those produced by ptc–gal4; EP1306 (data not shown).
Figure 3.
Bx interferes with Ap function. (A) ap–lacZ expression in a Bx1 mutant wing disc. Ap protein expression is also normal in Bx1 discs (not shown). (B) fng–lacZ expression in a Bx1 mutant wing disc. (C) Wg protein expression in a Bx2 mutant wing disc. Comparable effects on Wg, ap, and fng expression were observed in Bx1 and Bx2 mutant discs.
Genetic interactions between Bx, ap, and chip
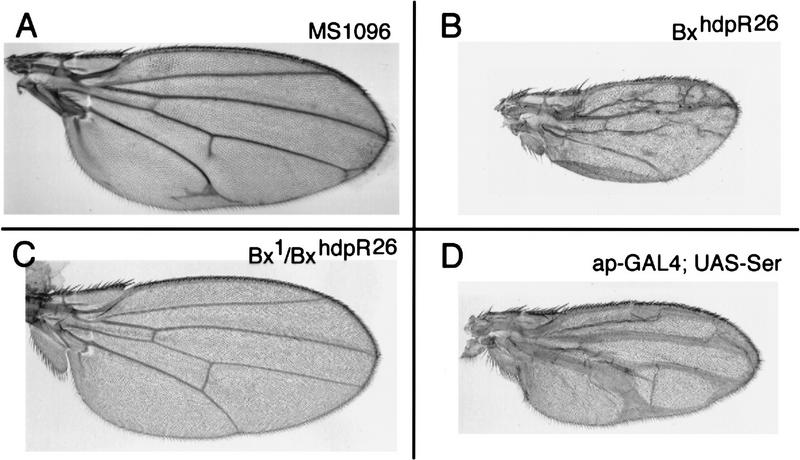
Adult wings of homozygous Bx1 flies show severe scalloping of the wing margin and transformation of dorsal to ventral fate in the alula at the posterior margin of the wing (Fig. 4A,B). The abnormalities in Bx wings resemble those produced by reducing ap function (Butterworth and King 1965; Wilson 1981). Consistent with the suggestion that Bx mutants reduce Ap function, we observed genetic interaction between Bx and ap, fng, and chip mutants. Flies heterozygous for Bx1 and a wild-type copy of the gene show mild notching (Fig. 4C). The severity of this weak Bx1 phenotype can be enhanced by simultaneously removing one copy of the ap gene (Fig. 4D), by removing one copy of fng (Fig. 4E), or by removing one copy of chip (Fig. 4F), a gene proposed to act as a cofactor for Ap (Morcillo et al. 1997). The strong Bx1 phenotype can be completely suppressed by increasing the level of ap in dorsal cells (in flies of genotype Bx1/Y; ap–gal4; UAS–ap; Fig. 4G). Increasing the level of fng expression using ap–gal4; UAS–fng also suppresses the wing margin defects but causes other defects in the internal organization of the wing (Fig. 4H). Taken together, these genetic interactions suggest that the defects caused by overexpressing Bx are due to reduced Ap activity.
Figure 4.
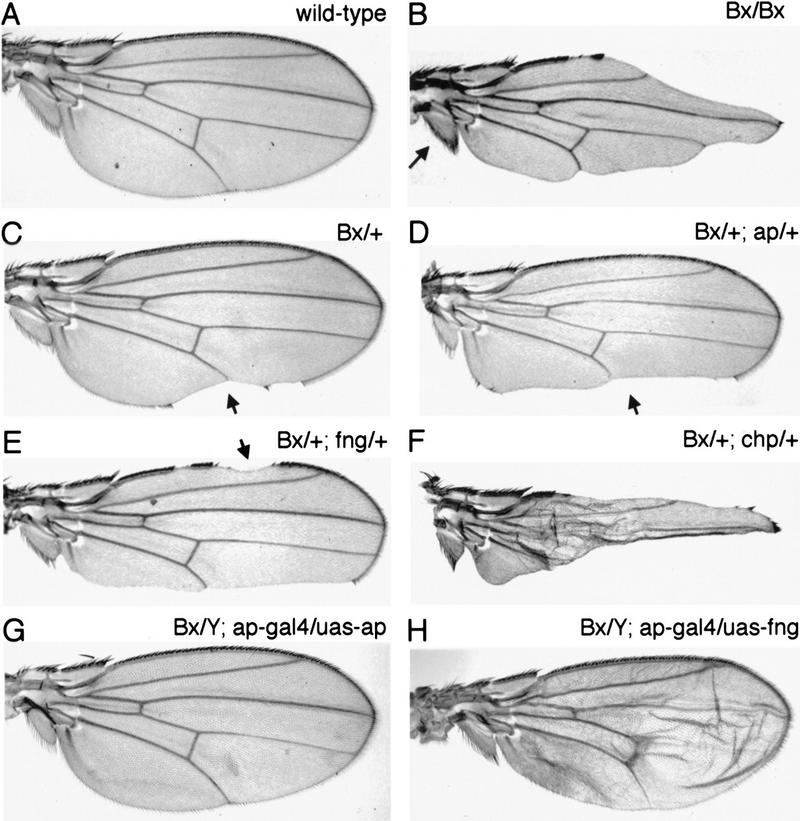
Genetic interactions between Bx and ap. (A) Cuticle preparation of a wild-type wing. (B) Bx1 homozygous mutant wing. Detailed analysis of the Bx1 phenotype suggests a close functional relationship to ap. There is extensive scalloping of the wing margin. In addition, bristles are found on both the dorsal and ventral surfaces of the alula (arrow, the defect in not visible at this magnification). In wild-type wings the alula has bristles only on the ventral surface. The alterations in Bx1 suggest a partial transformation of dorsal structures toward ventral identity, suggesting that ap activity might be reduced. (C) Bx1/+ heterozygous mutant wings typically produce small notches at the posterior margin (arrow). (D) Bx1/+; apUGO35/+ mutant wing. apUGO35 is a null allele (Cohen et al. 1992). Note that notching is more extensive than in the Bx1/+ wing. apUGO35 is completely recessive and does not cause a visible phenotype when heterozygous. (E) Bx1/+; fng80/+ mutant wing. Notching of the margin is more extensive than in the Bx1/+ wing, and also occurs anteriorly. fng80 does not produce a phenotype when heterozygous (Irvine and Wieschaus 1994). (F) Bx1/+; chipe55/+ mutant wing. Removing one copy of chip strongly enhances the loss of wing tissue. chipe55 does not produce a phenotype when heterozygous (Morcillo et al. 1997). (G) Bx1/Y; ap–gal4; UAS–apterous wing. Both the scalloping and DV fate transformation in the alula are rescued. (H) Bx1/Y; ap–gal4; UAS–fng wing. The margin scalloping is fully rescued, but DV fate transformation of the alula is not, suggesting that this represents a defect due to underexpression of another Ap target gene.
Bx (dLMO) protein competes for formation of a complex between Ap and Chip
BLAST searches with the DNA sequence flanking the EP1306 insert showed that the EP element is located near the 5′ end of the dLMO gene (Fig. 2A). dLMO had been cloned previously by homology to the human oncogene encoding the LIM domain protein LMO-2 (Zhu et al. 1995). The EP elements EP1394 and EP1306 are located 280 and 235 residues, respectively, from the 5′ end of exon 1a of dLMO and direct expression of dLMO. To integrate the genetic and molecular nomenclature we will refer to the protein encoded by the Bx locus as the dLMO protein.
LIM domains are thought to mediate protein interactions and are found in a variety of different types of proteins, often in combination with other recognized protein domains, as in the LIM–homeodomain (HD) proteins (for review, see Dawid et al. 1995). dLMO belongs to a class of LIM domain proteins that have two LIM domains and no other recognizable motifs (hence, the designation LMO, for LIM only). In view of the effects of dLMO on Ap function, we asked whether dLMO can interact with Ap and Chip proteins. ap encodes a LIM–HD protein (Cohen et al. 1992). chip encodes a member of the Ldb (LIM-domain binding) family of proteins (Morcillo et al. 1997). Ldb proteins have been shown to bind to the LIM domains of LIM–HD proteins like Ap (Agulnik et al. 1996; Morcillo et al. 1997). The Xenopus Ldb1 protein binds and activates the LIM–HD protein XLim1 in neuraxis induction (Agulnik et al. 1996). Likewise, CLIM1, another Ldb protein, binds and promotes transactivation by LIM3 (Bach et al. 1997). This is thought to occur by alleviating intramolecular repression, perhaps by preventing the endogenous LIM domains of LIM–HD proteins from interfering with homeodomain function (Sanchez-Garcia et al. 1993). LDB proteins also bind to the LIM domains of nuclear LMO proteins of the type encoded by Bx (Agulnik et al. 1996).
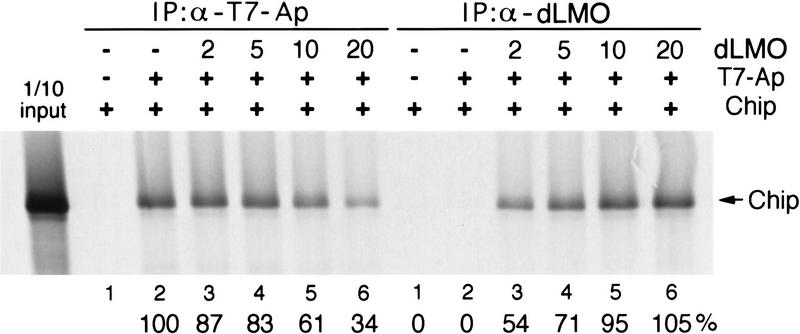
Genetic interactions between chip and ap suggest that as for Ldb1 and XLim1, Chip binding might activate Ap function (Morcillo et al. 1997). When overexpressed, Bx appears to interfere with Ap function without affecting either Chip or Ap protein expression (not shown). This raises the possibility that dLMO might interfere with binding between Ap and Chip. This was tested using a coimmunoprecipitation assay in which the binding between constant amounts of Chip and Ap proteins was challenged by increasing concentrations of Bx protein (Fig. 5). Chip protein can be immunoprecipitated with T7–epitope-tagged Ap protein and anti-T7 antibody, showing that Ap and Chip proteins bind in vitro (Fig. 5, sample 2, left). Binding between Chip and Ap was challenged by adding increasing amounts of in vitro-translated dLMO protein. The binding reactions (samples 1–6) were split in equal parts and immunoprecipitated with anti-T7 or with antibody to dLMO [Fig. 5, T7 immunoprecipitations (IPs) are on the left; dLMO IPs on the right]. We observed a dose-dependent decrease in the amount of Chip immunoprecipitating with Ap as the amount of dLMO protein was increased (Fig. 5, left, samples 3–6) and a corresponding increase in the amount of Chip immunoprecipitating with dLMO (Fig. 5, right, samples 3–6). These observations indicate that dLMO can bind to Chip in vitro and can compete for binding between Chip and Ap in a concentration-dependent manner. As a further test, the LIM domains of Ap were expressed as a GST fusion protein and tested for binding to full-length dLMO, Chip, and Ap proteins. Ap binds to itself and to Chip but not to dLMO in the GST–pull-down assay (data not shown). This suggests that dLMO interferes with formation of the active Ap–Chip complex by competing with Ap for binding to Chip.
Figure 5.
dLMO competes for binding between Ap and Chip. Coimmunoprecipitation of Chip by binding to Ap or to dLMO. Binding reactions (samples 1–6) contained equal amounts of 35S-labeled Chip protein synthesized in vitro. Samples 2–5 contain 5 μl of Ap lysate; samples 3–6 contain increasing amounts of Bx lysate (μl). After binding the samples were split into equal parts and immunoprecipitated with anti-T7 or anti-Bx. Quantitation of the relative amount of Chip precipitated is indicated below the lanes.
Bx activity in normal wing development
Bx loss-of-function mutants have been described previously under the name heldup (hdp) because of the abnormal posture of the wings (Lifschytz and Green 1979). Unfortunately, the original hdp mutants are no longer available. To study the normal function of dLMO in wing development new mutants were generated by imprecise excision of the MS1096 P element. MS1096 is inserted in the second intron of the Bx–dLMO transcription unit (Fig. 2A) and produces a weak phenotype consisting of venation defects (Fig. 6A). New Bxhdp mutants were recovered in P-element excision screens on the basis of their adult wing phenotypes. The wings of the new Bxhdp mutants are reduced in size and show abnormalities in vein pattern (Fig. 6B). In addition the wing posture is abnormal, as described for the original hdp mutants (not shown).
Figure 6.
Bx loss-of-function mutant phenotypes. (A) Cuticle preparation of an MS1096 mutant wing. (B) BxhdpR26 wing. hdpR26 is a lack-of-function Bx allele generated by mobilization of the MS1096 P element. Note the small size of the wing and the abnormal thickening of the wing veins. (C) Bx1/BxhdpR26 wing. The Bx1 phenotype is completely suppressed (cf. Fig. 4C for Bx1/+). (D) ap–gal4; UAS–Ser wing.
hdp mutants behave as dominant suppressors of the Bx gain-of-function phenotype (hdp-a; Lifschytz and Green 1979). The MS1096 excision mutants completely suppress the Bx phenotype in heterozygous females (Fig. 6C), suggesting that they are loss-of-function mutants. This was confirmed by examining dLMO protein, which is expressed at much reduced levels in wing discs of the excision mutant hdpR26 (Fig. 7A,B). In wild-type discs dLMO protein is nuclear and is expressed at higher levels in the dorsal compartment of the mature third-instar disc than in the ventral compartment. This expression pattern mirrors that of the MS1096 GAL4 enhancer trap line (Capdevila and Guerrero 1994; data not shown). In early- to mid-third-instar discs both MS1096 and dLMO protein are restricted to the dorsal compartment. The observation that dLMO is initially expressed in dorsal cells and maintained at elevated levels in the dorsal compartment suggested that dLMO might be regulated by Ap. To test this possibility we forced ectopic Ap expression using dpp–gal4 to direct UAS–Ap in a stripe of cells along the anterior–posterior (AP) boundary of the wing disc. dLMO is induced in Ap-expressing ventral cells to the same elevated level typical of cells in the dorsal compartment (Fig. 8). Thus, Ap induces expression of dLMO in dorsal cells. The transition from exclusively dorsal expression to dorsal and ventral expression suggests that dLMO expression is initiated by Ap but comes under an additional control mechanism as the disc matures.
Figure 7.
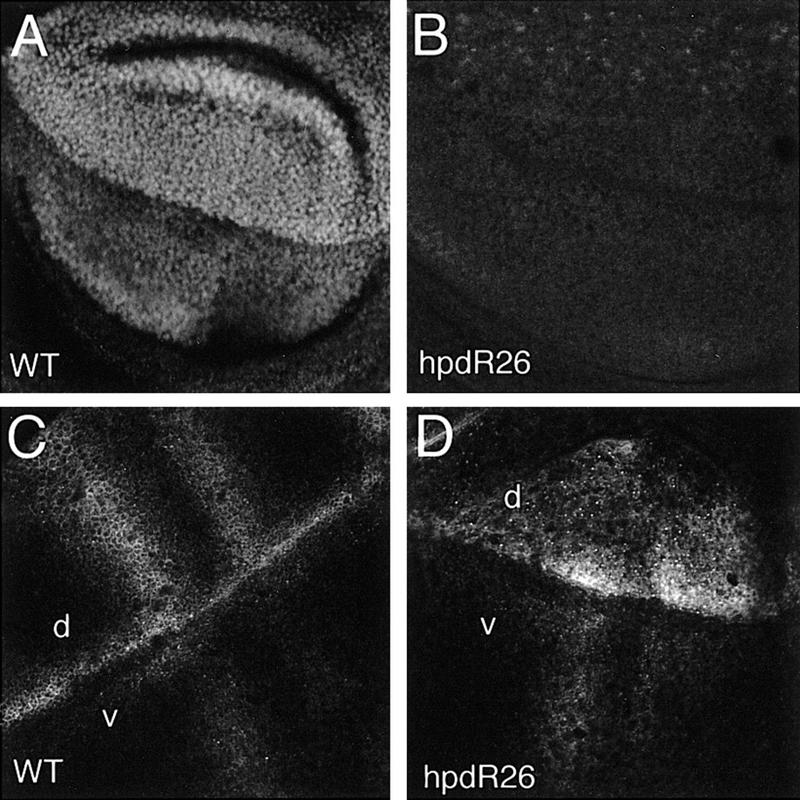
Reduced dLMO activity changes Ser expression. (A) dLMO protein expression in the wing pouch of a wild-type third-instar imaginal disc. Expression is higher in the dorsal compartment and near the AP boundary. dLMO protein is nuclear. Previous work using antibody to a vertebrate ortholog suggested that dLMO protein is cytoplasmic in the embryo (Zhu et al. 1995). We have not attempted to resolve this discrepancy. This image is at lower magnification than B–D. (B) BxhdpR26 wing disc. dLMO protein is expressed at reduced levels compared to wild-type. BxhdpR590 produces a similar wing phenotype but shows less severely reduced dLMO expression. (C) Ser protein in a wild-type third-instar wing disc. At this stage Ser expression has resolved to stripes flanking to the DV boundary and along the presumptive wing veins. (d) Dorsal compartment; (v) ventral compartment. (D) Ser expression in a BxhdpR26 loss-of-function mutant wing disc. Note the small size of the dorsal compartment and the relatively elevated level of Ser expression dorsally.
Figure 8.
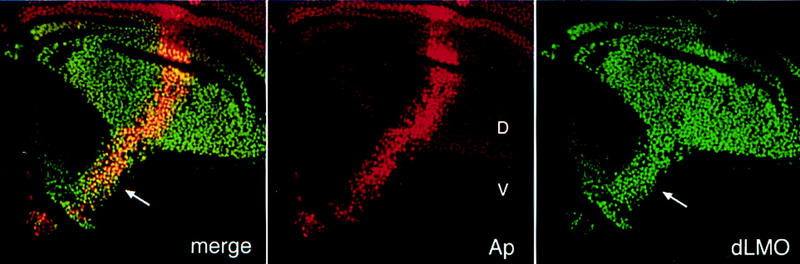
Ap induces dLMO expression. Wing disc expressing Ap in a stripe of cells along the AP boundary (genotype dpp–gal4 + UAS–ap). Ap protein is shown in red; dLMO protein in green. Ap directs dLMO expression at dorsal levels when misexpressed in ventral cells (arrow). Note that increased levels of Ap in the dorsal compartment do not obviously increase the level of dLMO protein in dorsal cells.
To ask whether the wing abnormalities caused by reducing dLMO levels might be due to an effect on Ap activity, we examined the effects of the Bxhdp excision mutants on Ap target gene expression. In early third-instar fng–lacZ and Serrate (Ser) are expressed evenly throughout the dorsal compartment of the wing disc and are thought to be regulated by Ap (Irvine and Wieschaus 1994; Diaz-Benjumea and Cohen 1995). In Bxhdp mutant discs, the size of the dorsal compartment is considerably reduced, consistent with the small wing phenotype (Fig. 7C,D). fng–lacZ expression is not affected in Bxhdp discs (not shown). Ser expression is elevated in the dorsal compartment and does not resolve normally into stripes along the DV boundary and wing veins (Fig. 7C,D). Ser expression in the ventral compartment appears normal. The stripes of Ser expression along the DV boundary and wing veins are both dorsal and ventral (Thomas et al. 1991) and are under different regulation than the early dorsal-specific domain (de Celis and Bray 1997; Micchelli et al. 1997). The abnormal pattern of Ser in the dorsal compartment of the Bxhdp may be due to superimposition of the early and late expression patterns. We suggest that this reflects a failure to down-regulate Ap activity as the disc matures.
To ask whether elevated Ser levels might contribute to the defects observed in Bxhdp mutant wings, we overexpressed Ser in the dorsal compartment of an otherwise wild-type disc using ap–gal4 to direct UAS–Ser expression (Fig. 6D). The resulting wings are small and show thickened veins but do not show the abnormalities in vein pattern observed in the Bxhdp mutant wings. Overexpression of fng using ap–gal4 in a wild-type background produces no phenotype (data not shown). These observations suggest that Ser overexpression contributes to the abnormalities observed in Bxhdp mutant wings but that there are likely to be additional factors.
Thus, both gain-of-function and loss-of-function Bx mutant phenotypes can be attributed to abnormal regulation of Ap activity. We conclude that Ap induces dLMO expression in the wing disc and that dLMO then functions as part of a feedback system to regulate the level of Ap activity.
Discussion
Ap activity depends on the relative levels of Ap, dLMO, and Chip proteins
Analysis of the LIM–HD proteins has suggested that LIM domains may act as intramolecular negative regulatory domains that block activity of the homeodomain (Sanchez-Garcia et al. 1993; Agulnik et al. 1996). Deleting or mutating the LIM domains activates the homeodomain in LIM–HD proteins. Binding of the LDB protein Ldb1 activates Xlim1, apparently by binding to its LIM domains. This suggests that complex formation between LDB proteins and LIM–HD proteins is necessary to activate LIM–HD proteins (Agulnik et al. 1996). The finding that chip, a Drosophila relative of Ldb1, shows genetic interaction with ap and can bind to Ap protein in yeast (Morcillo et al. 1997) and in vitro (Fig. 5) suggests a similar functional relationship between these proteins in Drosophila wing development. Our finding that overexpression of dLMO can functionally inactivate Ap in vivo and that dLMO can interfere with the formation of a complex between Ap and Chip in vitro provides strong support for the proposal that Ap must bind Chip to be activated (represented schematically in Fig. 9).
Figure 9.
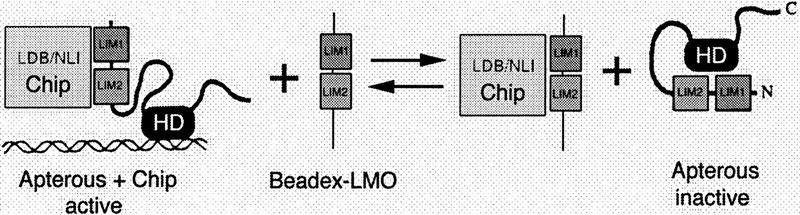
Model for Ap regulation by dLMO. Schematic depiction of complex formation between Chip, Ap, and dLMO proteins. Ap forms an inactive protein when the amino-terminal LIM domains are free. Following the scheme of Dawid et al. (1995), this is depicted as being due to intramolecular binding between LIM and homeodomain regions, but other possibilities are equally plausible. Chip can bind to the LIM domains of Ap or dLMO. Our data suggest that dLMO and Ap compete for binding to Chip and that varying the relative levels of Ap and dLMO will shift the equilibrium between active Ap–Chip complexes and inactive free Ap, by favoring formation of dLMO–Chip complexes. Although Chip is depicted as a monovalent protein here, its ortholog NLI dimerizes and can bind LIM domains as either a monomer or a dimer (Jurata and Gill 1997). This raises the possibility that higher-order complexes involving Ap, Chip, and dLMO might be of functional significance.
Biochemical studies have shown that the Chip ortholog NLI (nuclear LIM interactor), binds LIM domains through a short domain in the carboxy-terminal portion of the protein (Jurata and Gill 1997). In addition NLI forms homodimers via an amino-terminal domain. Although dimerization of NLI is not required for LIM-domain binding, higher-order complexes can form, consistent with the possibility that the functional complex might be composed of two NLI/LIM–HD dimers. Whether the active Ap–Chip complex is a dimer or a tetramer, overexpression of dLMO would increase the proportion of Chip bound by dLMO and thereby reduce the pool of Chip protein available for binding to Ap. Conversely, mutants that reduce dLMO levels might allow formation of more than normal levels of functional Ap–Chip complex.
In this context it is striking that the Bx mutant phenotype can be completely suppressed in vivo by overexpressing Ap. Increasing the concentration of Ap presumably shifts the balance of competition for Chip toward formation of functional Chip–Ap complexes. Likewise, reducing the amount of either Ap or Chip in vivo would shift the balance toward formation of dLMO–Chip complexes and thus enhance the severity of the Bx mutant phenotype as shown in Figure 4. We note that the genetic interaction between chip and Bx is stronger than between ap and Bx, suggesting that the endogenous level of Chip may be more limiting than that of Ap.
Ap activity is modulated during wing development
Selector genes such as ap are often thought of as simple binary switches. Our results suggest that the situation is more complex and that Ap activity levels are modulated during wing development. Ap expression is restricted to dorsal cells. We have identified dLMO as a new target for regulation by Ap and have shown that dLMO functions to modulate Ap activity. These observations suggest that Ap, Chip, and dLMO are components of a regulatory feedback loop that controls Ap activity levels. We have presented evidence that the balance in the levels of these three proteins is important for determining the level of Ap activity in vivo. We note that this regulation occurs at the level of protein activity, not at the level of gene expression, and suggest that this may reflect a requirement to fine-tune activity levels regionally as the wing develops. Genetic analysis suggests that Chip may have additional functions (Morcillo et al. 1997). Given that dLMO is expressed in regions where Ap is not known to function, it is likely that dLMO may have other functions as well. Chip and dLMO may regulate the activity of LIM–HD proteins in other developmental contexts and in postembryonic homeostatic processes.
A possible molecular mechanism for the oncogenic activity of vertebrate LMO genes
Gain-of-function mutations that cause misexpression of vertebrate LMO proteins have been implicated in cancers of the lymphoid system. Genes encoding the LMO1 and LMO2 proteins were identified by chromosomal translocations associated with leukemia (McGuire et al. 1989; Boehm et al. 1991). LMO1 and LMO2 have been shown to induce leukemia and lymphoma when misexpressed in T cells in transgenic mice (Fisch et al. 1992; McGuire et al. 1992). We have shown here that overexpression of the Drosophila LMO protein causes a dominant interfering activity that reduces activity of the LIM–HD protein Ap. We present evidence that this occurs by competitive inhibition of formation of a functional complex between the LDB protein, Chip, and Ap. We suggest that the molecular mechanism by which LMO proteins cause lymphoid cancers might be similar. It is possible that an as yet unidentified LIM–HD protein is required for proper differentiation or maintenance of the differentiated state in T cells and that loss of its function through overexpression of an LMO protein can lead to a failure in control of cell proliferation.
Materials and methods
Fly strains
The EP collection consists of 2300 independent P-element insertions available through the Berkeley Drosophila Genome Project (Rørth et al. 1998). ap–lacZ is described in Cohen et al. (1992); fng–lacZ is described in Irvine and Wieschaus (1994).
Molecular analysis of EP1306, EP1394, and MS1096
DNAs flanking EP1306, EP1394, and MS1096 were cloned by plasmid rescue. Sequence analysis showed that EP1306 and EP1394 are located 235 and 280 bp upstream from exon 1 of the dLMO gene (Zhu et al. 1995) and that MS1096 is located in intron 2 as indicated in Figure 2. cDNA clones were isolated from an imaginal disc cDNA library using the flanking DNA as probe and used to make antisense RNA probes for in situ hybridization to imaginal discs. Sequence analysis of our dLMO cDNA clone and of the genomic flank (accession no.) indicated that the dLMO open reading frame (ORF) differs in the amino-terminal region from that published for dLMO, due to a frameshift in the published sequence of the dLMO cDNA (x83012; Zhu et al. 1995).
To produce UAS–Bx, three copies of the flu epitope tag were introduced at the amino terminus of the Bx protein by PCR (primers: CATATGTATCCCTATGACGTCCCCGATTATGCCTACCCTTACGATGTACCTGACTACGCGTATCCGTACGACGTTCCGGACTATGCTATGATGACTATGGAC and atggaattcCTCCTCCACCGCCGCCCATTCCTA). The PCR product was cloned into pCRScript (Stratagene) and recloned into pUAST (Brand and Perrimon 1993). UAS–ap was prepared by cloning a full-length ap cDNA (B8; Cohen et al. 1992) into pUAST.
Bx loss-of-function mutants
MS1096 is inserted in the dLMO gene in intron 2. The MS1096 wing venation phenotype can be reverted to wild-type excision of the P element indicating that MS1096 is an insertional mutant (not shown). New Bx loss-of-function mutants were generated by imprecise excision of the MS1096 P element. Sixty-two independent excision alleles were recovered. All produce similar phenotypes, though with different severity. Excision mutants derived by mobilization of EP1394 were recovered and produce comparable phenotypes. Complementation mapping of three alleles placed the mutations in the smallest genetic interval known to contain Bx—on the basis of failure to complement Df(1)fuE5 and on complementation of Df(1)fuB10 and Df(1)osUE19 (Eberl et al 1992). The excision alleles also fail to complement maggot3E, suggesting the maggot alleles may be lethal mutations of Bx. We refer to these alleles collectively as BxhdpR# to indicate that they have the properties previously described for revertants of Bx, known as hdp-a alleles.
Antibodies
Mouse anti-dLMO: The dLMO ORF was amplified by PCR and cloned into pGEX 2TK (Pharmacia, primers cgcggattcATGATGACTATGGAC and atggaattcCTCCTCCACCGCCGCCCATTCCTA). Fusion protein was purified on glutathione–agarose and used to immunize BALB/C mice. Polyclonal serum was used for histochemistry and IP. Control experiments with preimmune serum showed no nuclear staining or IP of dLMO (not shown). Mouse anti-Ser was from Thomas et al. (1991), mouse anti-Wg was from Brook and Cohen (1996), and rabbit anti-β-galactosidase was from Cappell; guinea pig anti-Ap was provided by Juan Botas (Baylor College of Medicine, Houston, TX).
IP
The Ap ORF was cloned into pET23A to introduce a T7 epitope tag at the amino terminus. The Bx ORF was in pSK isolated from a λ–Zap cDNA library. The Chip ORF was excised from a yeast expression plasmid provided by Patrick Morcillo (Morcillo et al. 1997) and recloned into pKS. Proteins were produced using a coupled transcription–translation kit (Promega). For IP Chip was labeled with [35S]methionine, and T7–Ap and dLMO were not labeled. Five microliters of 35S-labeled Chip lysate was incubated with 0 or 5 μl of Ap lysate and with 0–20 μl of dLMO lysate; the final volume of the binding reactions was adjusted to 30 μl with unprogrammed lysate. Binding reactions were assembled on ice and allowed to incubate at room temperature for 30 min. The reactions were diluted to 200 μl with 50 mm HEPES (pH 7.5), 150 mm NaCl, 2 mm MgCl2, 10% glycerol; 1% NP-40 with 1 mm PMSF, and 1 μg/ml each of aprotinin, pepstatin, and leupeptin (IP buffer), centrifuged for 5 min at 4°C to pellet insoluble material. Equal portions were incubated with 2 μg of mouse monoclonal antibody to the T7 epitope tag (Novagen) or with 2 μl of dLMO antiserum for 90 min on ice. Sixty microliters of a 1:1 suspension of protein A–agarose beads in IP buffer was added, and samples were incubated with gentle rocking at 4°C for 30 min, washed three times with IP buffer, and analyzed on a 10% SDS–polyacrylamide gel.
GST pull-downs
The LIM domains of Ap were amplified by PCR and cloned into pGEX2T (primers: atagaattcGACGACTGCTCCGGC; taactcgagACTGGATGAGGCGGTATC, lowercase letters indicate sequences added for cloning using EcoRI and XhoI). GST and GST–AP–LIM proteins were expressed in bacteria and purified on glutathione–agarose beads. The yield of GST–AP–LIM was very low, compared to GST. Fifty microliters of a 1:1 suspension of GST or GST–AP–LIM beads in IP buffer was mixed with 4 μl of [35S]methionine-labeled in vitro-translated protein in 200 μl of IP buffer, incubated for 1 hr at 4°C, washed three times in IP buffer, and analyzed on a 10% SDS–polyacrylamide gel.
Acknowledgments
We thank Katrin Weigmann for her participation in screening the EP collection, Eyrun Hjorleifsdottir for help in initial characterization of EP1306 and EP1394, and Ann-Mari Voie and Anna Cyrklaff for technical help. We thank K. Irvine, P. Martín, B. Royer-Pokora, P. Morcillo, D. Dorsett, A. Hilliker, N. Perrimon, and J. Botas for fly strains, DNA samples, and antibodies. M.M. is a fellow of the Human Frontiers Science Foundation. The sequence data described in this paper have been submitted to the GenBank data library under accession nos. AJ010387 (Drosophila melanogaster mRNA for beadex/dLMO protein), AJ010388 (Drosophila melanogaster genomic DNA flank of p-element EP1394), and AJ010389 (Drosophila melanogaster genomic DNA flank of p-element EP1306).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL scohen@embl-heidelberg.de; FAX 49 6221 387 166.
References
- Agulnik AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Axelrod JD, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between wingless and Notch signaling pathways mediated by Dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and OTX homeodomain proteins. Genes & Dev. 1997;11:1370–1380. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Blair SS, Brower DL, Thomas JB, Zavortink M. The role of apterous in the control of dorsoventral compartmentalization and PS integrin gene expression in the developing wing of Drosophila. Development. 1994;120:1805–1815. doi: 10.1242/dev.120.7.1805. [DOI] [PubMed] [Google Scholar]
- Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: Distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brook WJ, Cohen SM. Antagonistic interactions between Wingless and Decapentaplegic responsible for dorsal-ventral pattern in the Drosophila leg. Science. 1996;273:1373–1377. doi: 10.1126/science.273.5280.1373. [DOI] [PubMed] [Google Scholar]
- Butterworth FM, King RC. The developmental genetics of apterous mutants of Drosophila melanogaster. Genetics. 1965;52:1153–1174. doi: 10.1093/genetics/52.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdevila J, Guerrero I. Targeted expression of the signalling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B, McGuffin ME, Pfeifle C, Segal D, Cohen SM. apterous: A gene required for imaginal disc development in Drosophila encodes a member of the LIM family of developmental regulatory proteins. Genes & Dev. 1992;6:715–729. doi: 10.1101/gad.6.5.715. [DOI] [PubMed] [Google Scholar]
- Couso JP, Bishop S, Martinez-Arias A. The wingless signalling pathway and the patterning of the wing margin in Drosophila. Development. 1994;120:621–636. doi: 10.1242/dev.120.3.621. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Toyama R, Taira M. LIM domain proteins. C R Acad Sci Ser III Life Sci. 1995;318:295–306. [PubMed] [Google Scholar]
- de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996a;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Garcia-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996b;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen SM. Interaction between dorsal and ventral cells in the imaginal disc directs wing development in Drosophila. Cell. 1993;75:741–752. doi: 10.1016/0092-8674(93)90494-b. [DOI] [PubMed] [Google Scholar]
- ————— Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development. 1995;121:4215–4225. doi: 10.1242/dev.121.12.4215. [DOI] [PubMed] [Google Scholar]
- Diaz-Benjumea FJ, Cohen B, Cohen SM. Cell interactions between compartments establishes the proximal-distal axis of Drosophila limbs. Nature. 1994;372:175–179. doi: 10.1038/372175a0. [DOI] [PubMed] [Google Scholar]
- Doherty D, Fenger G, Younger-Shepherd S, Jan L-Y, Jan Y-N. Dorsal and ventral cells respond differently to the Notch ligands Delta and Serrate during Drosophila wing development. Genes & Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- Eberl DF, Perkins LA, Engelstein M, Hilliker AJ, Perrimon N. Genetic and developmental analysis of polytene section-17 of the X-Chromosome of Drosophila melanogaster. Genetics. 1992;130:569–583. doi: 10.1093/genetics/130.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld AL, Kennison JA. Positional signaling by hedgehog in Drosophila imaginal disc development. Development. 1995;121:1–10. doi: 10.1242/dev.121.1.1. [DOI] [PubMed] [Google Scholar]
- Fisch P, Boehm T, Lavenir I, Larson T, Arno J, Forster A, Rabbitts TH. T-cell acute lymphoblastic lymphoma induced in transgenic mice by the RBTN1 and RBTN2 LIM-domain genes. Oncogene. 1992;7:2389–2397. [PubMed] [Google Scholar]
- Gorfinkiel N, Morata G, Guerrero I. The homeobox gene Distal-less induces ventral appendage development in Drosophila. Genes & Dev. 1997;11:2259–2271. doi: 10.1101/gad.11.17.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S, Pflugfelder G. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Fietz MJ. Quantitative effects of hedgehog and decapentaplegic activity on the patterning of the Drosophila wing. Curr Biol. 1995;5:432–440. doi: 10.1016/s0960-9822(95)00084-4. [DOI] [PubMed] [Google Scholar]
- Irvine K, Wieschaus E. fringe, a boundary specific signalling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Grenier JK, Scott MP. patched overexpression alters wing disc size and pattern: Transcription and post-transcriptional effects on hedgehog targets. Development. 1995;121:4161–4170. doi: 10.1242/dev.121.12.4161. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. Functional analysis of the nuclear LIM domains interactor NLI. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Irvine KD, Carroll SB. Cell recognition, signal induction and symmetrical gene activation at the dorsal/ventral boundary of the developing Drosophila wing. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Cohen SM. Proximal-distal axis formation in the Drosophila leg. Nature. 1997;388:139–145. doi: 10.1038/40563. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook WJ, Ng M, Calleja M, Sun H, Cohen SM. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Lifschytz E, Green MM. Genetic identification of dominant overproducing mutations: The Beadex gene. Mol & Gen Genet. 1979;171:153–159. doi: 10.1007/BF00270001. [DOI] [PubMed] [Google Scholar]
- Mattox WW, Davidson N. Isolation and characterization of the Beadex locus of Drosophila melanogaster: A putative cis-acting negative regulatory element for the heldup-a gene. Mol Cell Biol. 1984;4:1343–1353. doi: 10.1128/mcb.4.7.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire EA, Hockett RD, Pollock KM, Bartholdi MF, O’Brien SJ, Korsmeyer SJ. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989;9:2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire EA, Rintoul CE, Sclar GM, Korsmeyer SJ. Thymic overexpression of Ttg-1 in transgenic mice results in T-cell acute lymphoblastic leukemia/lymphoma. Mol Cell Biol. 1992;12:4186–4196. doi: 10.1128/mcb.12.9.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes & Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a Dpp morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development. 1996;122:1781–1789. doi: 10.1242/dev.122.6.1781. [DOI] [PubMed] [Google Scholar]
- ————— Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- Phillips R, Whittle JRS. wingless expression mediates determination of peripheral nervous system elements in late stages of Drosophila wing disc development. Development. 1993;118:427–438. doi: 10.1242/dev.118.2.427. [DOI] [PubMed] [Google Scholar]
- Rørth P. A modular misexpression screen in Drosophila detecting tissue specific phenotypes. Proc Natl Acad Sci. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P, Szabo K, Bailey A, Laverty T, Rehm J, Rubin GM, Weigmann K, Milán M, Benes V, Ansorge W, Cohen SM. Systematic gain-of-function genetics in Drosophila. Development. 1998;125:1049–1057. doi: 10.1242/dev.125.6.1049. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Osada H, Forster A, Rabbitts TH. The cysteine-rich LIM domains inhibit DNA binding by the associated homeodomain in isl-1. EMBO J. 1993;12:4243–4250. doi: 10.1002/j.1460-2075.1993.tb06108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strigini M, Cohen SM. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development. 1997;124:4697–4705. doi: 10.1242/dev.124.22.4697. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Biehs B, Marin E, Bier EA. The spalt gene links the A/P compartment boundary to a linear adult structure in the Drosophila wing. Development. 1997;124:21–32. doi: 10.1242/dev.124.1.21. [DOI] [PubMed] [Google Scholar]
- Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with complex expression patterns in embryos and wing discs. Development. 1991;111:749–761. doi: 10.1242/dev.111.3.749. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Carroll SB. Pattern formation in a secondary field: A hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete subregions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Wilson TG. Expression of phenotypes in a temperature-sensitive allele of the apterous mutation in Drosophila melanogaster. Dev Biol. 1981;85:425–433. doi: 10.1016/0012-1606(81)90274-8. [DOI] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Sequential organizing activities of engrailed, hedgehog and decapentaplegic in the Drosophila wing. Development. 1995;121:2265–2278. doi: 10.1242/dev.121.8.2265. [DOI] [PubMed] [Google Scholar]
- ————— Direct and long-range action of a Wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zhu T, Bodem J, Keppel E, Paro R, Royer-Pokora B. A single ancestral gene of the human LIM domain oncogene family LMO in Drosophila: characterization of the Drosophila Dlmo gene. Oncogene. 1995;11:1283–1290. [PubMed] [Google Scholar]