Abstract
One-bead-one-compound (OBOC) libraries provide a powerful tool for drug discovery as well as biomedical research. However, screening a large number of beads/compounds (>1 million) and rank ordering the initial hits (which are covalently attached to a solid support) according to their potencies still post significant technical challenges. In this work, we have integrated some of the latest technical advances from our own as well as other laboratories to develop a general methodology for rapidly screening large OBOC libraries. The methodology has been applied to synthesize and screen a cyclic peptide library that features: (1) spatially segregated beads containing cyclic peptides on the surface layer and linear encoding peptides in their interior; (2) rapid on-bead screening of the library (>1 million) by a multi-stage procedure (magnetic bead sorting, enzyme-linked assay, and fluorescence based screening); (3) selective release of cyclic peptides from single positive beads for solution-phase determination of their binding affinities; and (4) hit identification by partial Edman degradation/mass spectrometry (PED/MS). Screening of the library against protein phosphatase calcineurin (Cn) identified a series of cyclic peptides that bind to the substrate-docking site for nuclear factor of activated T cells (NFAT) with KD values of ~1 μM. Further improvement of the affinity and specificity of these compounds may lead to a new class of immunosuppressive agents that are more selective and therefore less toxic than cyclosporine A and FK506.
Keywords: Calcineurin, combinatorial library, cyclic peptides, high-throughput screening, protein-protein interaction
INTRODUCTION
One-bead-one-compound (OBOC) libraries, in which each bead carries many copies of a unique compound, have become a powerful tool in biomedical research as well as the drug discovery process1. OBOC libraries of very high structural diversity can be readily synthesized by the split-and-pool method2. In principle, all of the library compounds, while still attached to individual beads, may be simultaneously screened against a target of interest (e.g., a protein receptor, an enzyme, or live cells), thus allowing the rapid identification of the most active compounds from the library. In practice, however, the potential of the OBOC libraries has not been fully realized due to a number of technical obstacles. First, screening millions of beads by the conventional method, which involves viewing the beads under a microscope and manually removing the “hits” (e.g., fluorescently labeled beads), is impractical for large-scale applications. Second, during on-bead screening, the signal strengths on beads (e.g., fluorescence intensity) do not always correlate with the potency of the ligands on the beads; in fact, false positive hits are common during on-bead screening due to the high ligand density on beads, which permits the simultaneous interaction of a macromolecular target with multiple ligand molecules on a bead3. Therefore, in order to identify the most active hit(s), one must resynthesize and test all of the initial hits individually. Depending on the number of initial hits, this process can be extremely time-consuming and expensive. Finally, screening of OBOC libraries requires post-screening hit identification, which remains an unresolved problem for large, nonpeptidic libraries.
Several recent technical advancements have removed some of the obstacles. Automated fluorescence-activated bead sorting has been employed to isolate the positive beads4, although it is still somewhat slow and requires a specially configured bead sorter, which is not available in most laboratories. Alternatively, with protein coated magnetic nanoparticles, hits can be rapidly separated from negative beads by magnetic seperation4c, 5. To identify the most active hit(s) without resynthesis, Hintersteiner et al. labeled the compound on each positive bead with a fluorescent dye through click chemistry, released it into solution, and determined its binding affinity to the target protein by fluorescence anisotropy4d. By using the material derived from a single 90-Gm TentaGel bead (which carries ~100 pmol of compounds), they were able to determine the KD value of each hit and rank order all of the hits according to their potencies. Alternatively, Kodadek and co-workers interfaced the on-bead screening with a microarray technique to quantitate the potency of hit compounds directly released from the positive beads5. Another notable development was the synthesis of OBOC libraries on spatially segregated beads that featured a reduced ligand density on the bead surface but a normal loading in the bead interior3, 6. The lower surface ligand density greatly reduced the amount of nonspecific binding and binding by weak ligands, while the normal ligand loading inside the bead (which is inaccessible to macromolecular targets such as proteins) provided enough material for hit identification.
In this work, we attempt to integrate the recent advancements into a single platform that allows a researcher to routinely synthesize and screen OBOC libraries of >1 million compounds to identify the most active compound(s) in a few weeks, without the use of any special equipment. The platform involves the following key features: (1) synthesis of OBOC libraries on spatially segregated beads that contain cyclic peptides on bead surface and linear encoding peptides in their interior; (2) rapid on-bead screening of a large number of beads (>1 million) by a multi-stage screening procedure (magnetic bead sorting, enzyme-linked assay, and fluorescence based screening); (3) in-solution determination of the binding affinity of hit compounds directly released from single beads (no hit resynthesis), and (4) high-throughput hit identification by partial Edman degradation/mass spectrometry (PED/MS)7.
We have applied this platform to identify cyclic peptides that inhibit the interaction between calcineurin (Cn) and its substrate, nuclear factor of activated T cell (NFAT). Cn is a protein serine/threonine phosphatase that plays a key role in immune response by activating T cells8. NFAT is a transcription factor which, upon dephosphorylation by Cn, translocates into the nucleus where it activates the expression of interleukin 2 (IL-2) and stimulates the immune response9. Naturally occurring small molecules such as cyclosporin A (CysA) and FK506, which inhibit the phosphatase activity of Cn, have been widely used as immunosuppressant drugs in post-allogeneic organ transplant. These compounds bind to cellular proteins cyclophilin and FK506-binding protein (FKBP), respectively, and the resulting binary complexes bind to Cn and sterically block the access of NFAT and other protein substrates to the Cn active site10. Although effective as immunosuppressants, these compounds also inhibit Cn phosphatase activity in other signaling pathways resulting in undesired side effects. For example, the use of FK506 and CysA for organ transplantation has been associated with increased incidence of diabetes11. Therefore, a compound that selectively inhibits the Cn-NFAT interaction (and prevents NFAT dephosphorylation) would act as an effective immunosuppressant but without affecting the other signaling functions of Cn (and thus less side effects). Previous studies have identified a 16-mer linear peptide VIVIT (MAPGHPVIVITGPHEE) that inhibits the Cn-NFAT interaction without affecting the Cn phosphatase activity12. Attachment of a cell penetrating peptide (R11) makes the peptide cell permeable and the resulting conjugate can cause immunosuppression in transplanted mice and prevent cardiac hypertrophy without affecting insulin secretion, a common side effect associated with FK506 treatment13. Since linear peptides generally have poor bioavailability (e.g., susceptibility to proteolytic degradation) and pharmacokinetic properties, we set out to develop cyclic peptide inhibitors against the Cn-NFAT interaction, which should have improved pharmacological properties. We show here that synthesis and screening of an OBOC cyclic peptide library against Cn by the high-throughput platform identified a family of cyclic peptides that bind to the NFAT-docking site with low micromolar affinities.
RESULTS AND DISCUSSION
Design and Synthesis of Cyclic Peptide Library
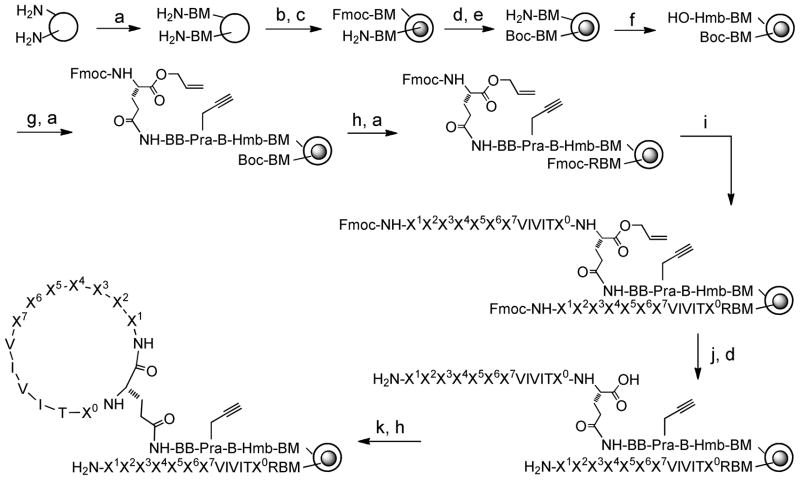
Selective dephosphorylation of NFAT by Cn critically depends on the interaction between a substrate-docking site on the Cn catalytic domain and a conserved PxIxIT motif in NFAT, which is located N-terminal to the phosphorylation sites in NFAT’s regulatory domain14. Interfering with the Cn-NFAT interaction by mutating the conserved sequence impairs NFAT’s activation capabilities14. Thus, targeting the NFAT-binding site of Cn (rather than the enzyme’s catalytic site) represents an alternative strategy for more specific inhibition of Cn-NFAT signaling and selective suppression of T cell activation15. Previous screening of a consensus PxIxIT sequence-based combinatorial peptide library identified the VIVIT peptide that exhibits a 50-fold higher affinity for Cn than the native sequence (KD = 500 nM)15. In order to maximize the probability of obtaining a cyclic peptide ligand with high affinity and selectivity for Cn, we designed a biased peptide library in which all members of the library contained the VIVIT motif as the core structure (Scheme 1). Seven random residues (X1–X7) were added to the N-terminal side of the VIVIT motif; it was anticipated that some of the random sequences would make additional interactions with the Cn surface and enhance the binding affinity (and potentially selectivity) of the VIVIT motif. For positions X1–X5, we used a 25-amino acid set that was selected based on their structural diversity, metabolic stability, and commercial availability16. It includes ten proteinogenic α-L-amino acids [Ala, Arg, Asp, Gln, Gly, His, Ile, Ser, Trp, and Tyr], five non-proteinogenic α-L-amino acids [L-4-fluorophenylalanine (Fpa), L-homoproline (Pip), L-norleucine (Nle), L-ornithine (Orn), and L-phenylglycine (Phg)], and ten α-D-amino acids [D-2-naphthylalanine (D-Nal), D-Ala, D-Asn, D-Glu, D-Leu, D-Lys, D-Phe, D-Pro, D-Thr and D-Val]. Structural analysis of the Cn-VIVIT peptide complex showed that the bound peptide adopted anti-parallel β-sheet conformation at the VIVIT core region17. Since it has previously been shown that the D-Pro-L-Pro motif promotes the formation of a β-turn structure,18 we biased the X6 and X7 positions toward prolyl residues. At the X6 position, 50% of the library members contained a D-Pro, while the other 50% had Gly, Ala, D-Ala, Pip or His. Similarly, the X7 position had 50% L-Pro and the other half were Gly, Ala, D-Ala, Pip, or Nα-methylalanine (Mal). Therefore, a significant fraction of our library members (~25%) is expected to adopt an antiparallel β-sheet conformation, which should facilitate their interaction with the NFAT-docking site on Cn. Because we did not know what ring size would be optimal for the cyclic peptides, we generated five different ring sizes by inserting GAA, AAA, GA, AA, G, A or no amino acid at the X0 position and by deleting the X1 residue from 10% of the library members. The theoretical diversity of the resulting library is ~1010.
Scheme 1.
Design and Synthesis of Cyclic Peptide Library against Cn.
Reagents and conditions: a) standard Fmoc/HBTU chemistry; b) soak in water; c) 0.5 equiv. Fmoc-OSu in Et2O/CH2Cl2; d) Di-tert-butyl dicarbonate; e) piperidine; f) 4-hydroxymethylbenzoic acid (Hmb)/HBTU/HOBt; g) Fmoc-β-Ala-OH/DIC; h)TFA; i) split-and-pool synthesis by Fmoc/HATU chemistry; j) Pd(PPh3)4; and k) PyBOP, HOBt.
The library was synthesized on TentaGel microbeads with a diameter of ~130 Gm (~106 beads/g, ~300 pmol peptides/bead). To facilitate hit identification, each resin bead was spatially segregated into outer and inner layers19, with a cyclic peptide displayed on the bead surface (thus accessible to a macromolecular target) and the corresponding linear peptide confined to the bead interior to serve as an encoding tag (inaccessible to the macromolecular target)16, 20. To permit affinity measurement of any initial hits in solution without resynthesis of the hits, we incorporated an L-propargylglycine (Pra) residue into the linker region of the cyclic peptides (Scheme 1). Its side-chain alkyne group was later used for selective labeling with a fluorescent probe through the click chemistry21. To spatially segregate the beads, resin bearing the dipeptide β-Ala-Met (β-Ala for flexibility and Met for peptide release by CNBr) was soaked in water, drained, and quickly suspended in 1:1 (v/v) DCM/Et2O containing 0.5 equivalent of N-(9-fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu). Because the organic solvent is immiscible with water, only peptides on the bead surface were exposed to and reacted with the activated ester. The beads were washed with DMF and the remaining free N-terminal amines inside the beads (~0.5 equivalent) were protected with a Boc group. After removal of the Fmoc group, a p-hydroxymethylbenzoic acid (Hmb) linker, which is installed for later peptide cleavage (vide infra), a β-Ala, L-Pra, two additional β-Ala, and Fmoc-L-Glu-OAll (All = α-allyl ester) were sequentially coupled to the peptides in the outer layer. The glutamate serves as a convenient anchor for attachment of the cyclic peptide to the solid support via its side chain, while allowing later peptide cyclization at its α-carboxyl group. The N-terminal Boc group was then removed from the inner peptides by treatment with trifluoroacetic acid (TFA) and an arginine residue was added to provide a fixed positive charge to facilitate later sequencing by PED/MS. Next, the VIVIT motif and the random region were synthesized by the split-and-pool method2a to generate the OBOC library. Finally, the α-allyl group on the C-terminal glutamate and the N-terminal Fmoc group were removed and the surface peptides were cyclized by treatment with benzotriazol-1-yl-oxy-tris(pyrrolidino)phosphonium hexafluorophosphate (PyBOP). Due to the absence of a C-terminal Glu, the inner peptides cannot undergo cyclization and thus remain in the linear form. Side-chain deprotection with TFA rendered the library ready for screening.
Library Screening
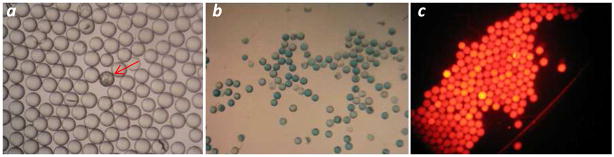
Conventional on-bead screening involved labeling a target of interest (e.g., a protein) with a biotin or fluorescent group and manually isolating the resulting colored beads under a dissecting or fluorescence microscope2a, 22. While this method is effective for small amounts of resin (<100 mg or 200,000 beads), the task becomes increasingly more challenging as the number of beads increases. The human eye also lacks the ability to make quantitative comparison of the color (or fluorescence) intensities on different beads. Fluorescence-based bead sorting has also been used to isolate positive beads after labeling the target with a fluorescent group4. However, sorting requires specialized equipment and is still relatively slow. By using a COPAS bead sorter (Union Biometrica, MA), Hu et al. recently reported a sorting rate of 30–40 beads per second4c. At this rate, screening 1.0 g of a library on the popular 90-μm TentaGel resin (~2.86 × 106 beads) would take 20–27 h to finish; for 1.0 g of library on 35-μm TentaGel resin (4.55 × 107 beads), the sorting process would take 13–18 days. In light of this, we designed a three-stage screening protocol by adapting a recently reported magnetic sorting technique4b, 5 to our system. During the first screening step, the cyclic peptide library (1.0 g, ~1 million beads) was incubated with the biotinylated Cn (100 nM). After extensive washing, the library beads were mixed with 6–7 × 107 streptavidin-coated magnetic beads (2.8 Gm in diameter) and sorted by applying a magnetic field. This was readily accomplished by suspending the beads in a 50-mL conical tube and placing a magnet next to wall of the tube. The positive beads, due to the presence of magnetic particles on them, were attracted to the wall, while the nonbinding beads settled to the bottom of the tube. After removing the nonbinding beads form the bottom, the positive beads (~1000 beads) were retrieved by removing the magnetic field. Figure 1a shows a small portion of the library immediately before the sorting step; a single positive bead is clearly visible (darker colored) due to the presence of magnetic particles on its surface. During the secondary screening step, the ~1000 hits were incubated again with the biotinylated Cn (150 nM), followed by the addition of a streptavidin-alkaline phosphatase (SA-AP) conjugate. Binding of Cn to a bead recruits SA-AP to the bead surface which, upon the subsequent addition of 5-bromo-4-chloro-3-indolyl phosphate (BCIP), produces a turquoise colored precipitate on the positive bead (Figure 1b). The ~300 most intensely colored beads were manually isolated by using a micropipette and exhaustively washed to remove the turquoise dye and any remaining protein. The resulting beads were subjected to a third round of screening, during which the ~300 beads were incubated with 2 μM Cn labeled with Texas-red. It should be noted that the choice of Texas-red as the fluorescent label was crucial, as TentaGel beads have strong background fluorescence at shorter wavelengths23. As expected, all of the ~300 beads showed strong red fluorescence (Figure 1c), while the control beads (negative beads from the magnetic sorting step) showed essentially no color (not shown). A total of 113 most intensely colored beads were selected for further analysis.
Figure 1.
Library screening against Cn. (a) A portion of the library beads after incubation with biotinylated Cn and streptavidin-coated magnetic beads (the positive bead is indicated with an arrow); (b) library beads after incubation with biotinylated Cn, SA-AP, and BCIP; and (c) library beads after incubation with Texas red-labeled Cn. Panels a and b were viewed under a dissecting microscope, whereas panel c was viewed under a fluorescence microscope.
Fluorescent Labeling and Selective Release of Cyclic Peptides
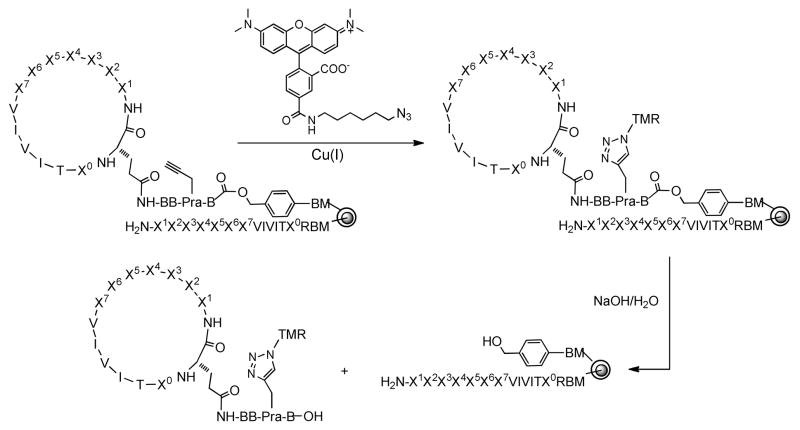
The 113 hits were still too many for individual resynthesis and affinity measurement. To determine which bead(s) contains the most potent Cn ligand(s), we adopted a strategy recently reported by Auer and coworkers to rank order all initial hits according to their potencies without compound resynthesis4d. It involves on-bead labeling of each hit compound with a fluorescent probe, releasing the labeled compound into solution, and determining its binding affinity to the target protein by fluorescence anisotropy (FA)24. Since our cyclic peptide library was prepared on spatially segregated beads, it gave us a unique opportunity to selectively label only the cyclic peptides with a fluorescent dye and release it from the resin for solution-phase binding analysis. Thus, treatment of the 113 beads with tetramethylrhodamine (TMR) azide in the presence of Cu(I) selectively labeled the cyclic peptides at the Pra residue imbedded in the linker region (Scheme 2). The linear encoding peptide was not affected because it does not contain the Pra residue. After exhaustive washing to remove any excessive dye molecule, the beads were placed into individual microcentrifuge tubes (one bead/tube) and treated with a NaOH solution. The cyclic peptide, which was attached to the resin via an ester linkage, was selectively released, while the linear peptide remained attached to the resin. Neutralization with HCl provided ~26 μL of a dye-labeled cyclic peptide stock solution (~1 μM), ready for solution phase binding analysis.
Scheme 2.
On-Bead Labeling and Selective Release of Cyclic Peptides
Solution-Phase Affinity Measurement and Rank Ordering of the Hits
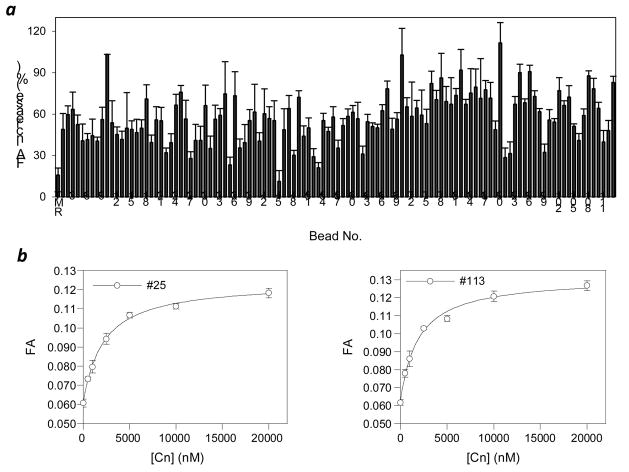
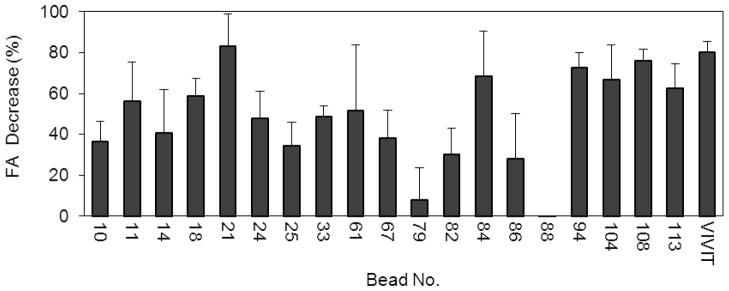
Binding of the TMR-labeled cyclic peptides to Cn was analyzed by measuring the FA values of the labeled peptides, which should increase upon binding to a macromolecular receptor24. Because of the large number of samples, we carried out the binding studies in two stages. During the first stage, FA analysis was performed at a fixed concentration of both Cn (5 μM) and the TMR-labeled cyclic peptide (100 nM) and the percentage of FA increase (relative to the control without Cn) was measured. Out of the 113 peptides, 44 (39%) had an FA increase of ≥60%, while the control with free TMR (not attached to any peptide) had an increase of less than 20% (Figure 2a). Beads that gave both high absolute FA values and high percentage increases upon the addition of Cn (39 beads) were subjected to peptide sequencing by PED/MS (vide infra) and unambiguous sequences were obtained for 19 of the beads (Table 1). Next, for each of the 19 compounds, a full titration curve was generated by incubating the TMR-labeled peptide (50 nM) with varying concentrations of Cn (0–20 μM) and measuring their FA increases. The dissociation constant (KD) for each cyclic peptide was obtained by curve fitting. Figure 2b shows the binding curves of two representative cyclic peptides derived from beads No. 25 and No. 113, having KD values of 1.9 and 1.5 μM, respectively. The KD values for the other 17 cyclic peptides were similarly determined and were all in the low μM range (Table 1 and Figure S1). The KD values allowed us to rank order the 19 hits according to their binding affinities to Cn. The most potent cyclic peptide (from bead No. 10) had a KD value of 0.92 μM, while the least active peptide (from bead No. 79) had a KD value of 20 μM.
Figure 2.
Evaluation of the binding affinity of the 113 hits to Cn by fluorescence anisotropy. (a) Comparison of all 113 hits at a fixed concentration of Cn (5 μM) and TMR-labeled cyclic peptide (100 nM). The percentage of FA increase was relative to the anisotropy value of the corresponding peptide in the absence of Cn. (b) Plots of FA values as a function of the Cn concentration for cyclic peptides derived from beads No. 25 and 113. The curves were fitted to the data as described under Experimental Procedures. All measurements were performed in triplicates.
Table 1.
Cyclic Peptides Identified from Library Screening against Cn
| Bead No. | Peptide Sequences |
KD (μM)
|
|
|---|---|---|---|
| Single-bead | Resynthesized | ||
| 10 | Tyr-His-Ile-Gly-D-Leu-Gly-Pro-Val-Ile-Val-Ile-Thr-Ala-Ala-Glu | 0.92 ± 0.37 | 8.4 ± 2.8 |
| 113 | Nle-D-Pro-Pip-Arg-D-Ala-D-Ala-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Glu | 1.6 ± 0.3 | |
| 25 | Nle-Gly-Tyr-D-Ala-Arg-D-Pro-Pro-Val-Ile-Val-Ile-Thr-Ala-Ala-Glu | 1.9 ± 0.3 | 7.6 ± 4.8 |
| 67 | Gly-Orn-Phg-Gly-Ala-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Ala-Glu | 2.1 ± 0.9 | |
| 82 | D-Leu-Arg-Arg-Ile-Gln-D-Pro-Pro-Val-Ile-Val-Ile-Thr-Ala-Ala-Glu | 2.4 ± 0.6 | 1.6 ± 0.3 |
| 88 | D-Lys-D-Thr-Ser-Arg-Arg-D-Pro-Pro-Val-Ile-Val-Ile-Thr-Gly-Glu | 2.6 ± 1.8 | |
| 108 | Tyr-Nle-Orn-Arg-Gln-Ala-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Ala-Glu | 2.8 ± 0.8 | |
| 84 | Arg-Arg-D-Leu-Ile-Arg-Pip-Pro-Val-Ile-Val-Ile-Thr-Ala-Ala-Glu | 3.0 ± 0.7 | 0.74 ± 0.35 |
| 104 | D-Nal-Arg-D-Val-D-Asn-Gln-His-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Glu | 3.0 ± 0.8 | |
| 33 | D-Ala-D-Leu-D-Asn-D-Lys-Phg-D-Ala-Pro-Val-Ile-Val-Ile-Thr-Glu | 3.3 ± 0.6 | |
| 24 | D-Lys-Gln-D-Phe-Gln-D-Leu-His-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Ala-Glu | 3.4 ± 1.0 | |
| 18 | D-Lys-D-Val-Arg-Pip-D-Glu-D-Pro-Pro-Val-Ile-Val-Ile-Thr-Ala-Ala-Glu | 3.5 ± 1.0 | |
| 86 | Gly-Asp-Arg-Ala-Arg-Gly-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Glu | 3.8 ± 1.4 | 30 ± 20 |
| 14 | D-Nal-Phg-His-His-His-Pro-Val-Ile-Val-Ile-Thr-Ala-Ala-Glu | 3.8 ± 0.9 | |
| 94 | D-Lys-Gln-Arg-Ala-D-Thr-His-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Ala-Glu | 4.4 ± 0.8 | |
| 21 | D-Glu-D-Val-D-Pro-D-Pro-D-Ala-D-Pro-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Ala-Glu | 4.8 ± 1.2 | |
| 61 | Ala-Orn-D-Nal-Ser-D-Glu-Ala-Pro-Val-Ile-Val-Ile-Thr-Glu | 5.4 ± 3.0 | 16 ± 5 |
| 11 | Ser-D-Pro-His-Gly-Pip-Pro-Val-Ile-Val-Ile-Thr-Gly-Ala-Ala-Glu | 5.7 ± 2.2 | |
| 79 | D-Thr-D-Ala-Arg-Nle-Arg-Gly-Pro-Val-Ile-Val-Ile-Thr-Glu | 20 ± 12 | |
Hit Identification by PED/MS
Others and we have previously reported PED/MS as a high-throughput peptide (or peptoid) sequencing technique and have applied it to sequence >30000 peptides/year in our laboratory7, 25. In essence, a bead selected from an OBOC library is subjected to Edman degradation in the presence of a small amount of a competing electrophile (e.g., Fmoc-OSu). Repetition of the PED cycle converts a homogeneous peptide population into a series of progressively shorter fragments, which are then analyzed by MALDI-TOF mass spectrometry. Its high-throughput capability is due to the fact that all of the hits isolated from an OBOC library (or even beads from different libraries if they contain different signature motifs such as different linker sequences) may be pooled into a single reactor for PED treatment, greatly reducing the amount of effort for sequencing a large number of hits. Unfortunately, PED of the 39 hits as a pool would result in the loss of their individual identity and is thus not compatible with the current platform. To maintain the identity of each hit while achieving relatively high throughput, we adapted the PED reaction to the 96-well plate format. Thus, the 39 beads that carried the most active hits were retrieved from the sample plates that contained the 113 cyclic peptide stock solutions, placed into individual wells (one bead/well) of an AcroPrep 96-well filter plate, and subjected to seven cycles of PED (Figure S2). After that, the linear peptide and its degradation products were released from each bead by treatment with CNBr and analyzed by MALDI-TOF MS to determine the sequence of the hit peptide.
PED/MS analysis of the 39 positive beads resulted in 19 complete sequences (Table 1). For the other 20 beads, complete sequence assignment was not possible due to poor MS spectra (missing peaks). Inspection of the selected sequences reveals that they all contain a Pro at the X7 position. This is expected, as the NFAT-docking site has previously been shown to recognize a consensus sequence of PxIxIT. There was also a preference for a Gly immediately C-terminal to the VIVIT motif (X0); all five peptides that had a tripeptide as the X0 “residue” contained a Gly at this position. Note that the linear VIVIT ligand also has a Gly at the same position. These results validate our screening procedure. No strong sequence preference was observed at positions X1–X5, though there were substantially more basic than acidic residues in this region. The selected peptides also had four out of the five possible ring sizes, suggesting that there is substantial flexibility in the ring size. Finally, all of the selected cyclic peptides are less potent than the original linear VIVIT peptide (KD = 0.5 μM). It is possible that peptide cyclization locks the peptides into conformations that are less favorable for Cn binding. More probably, our library design may have restricted the cyclic peptides into a less favorable sequence space. Our recent SAR studies of the linear VIVIT peptide suggested that the sequence C-terminal to VIVIT is more critical than the N-terminal sequence for binding to Cn (TL and DP, unpublished result). Synthesis and screening of a redesigned cyclic peptide library with variable sequences C-terminal to VIVIT may provide more potent cyclic peptide ligands against Cn.
Cyclic Peptides Bind to the NFAT-Docking Site on Cn
To determine whether the selected cyclic peptides bind to the NFAT-docking site on Cn, we performed a competition experiment. Each of the TMR-labeled cyclic peptides (50 nM) was incubated with Cn (10 μM) in the presence of the linear VIVIT peptide (100 μM) and the amount of FA was measured and compared to that of the control (no linear VIVIT peptide added). The addition of the competing VIVIT peptide decreased the FA values of all cyclic peptides (typically 30–80%), with the exception of peptide No. 88, which had very low fluorescence signal (Figure 3). We therefore conclude that the cyclic peptides all bound to the same site (or overlapping sites) as the linear VIVIT peptide, i.e., the NFAT-docking site, which is consistent with our observation that Pro is exclusively selected at the X7 position.
Figure 3.
Histograms showing the competition between the TMR-labeled cyclic peptides (50 nM) and linear VIVIT peptide (100 μM) for binding to Cn (10 μM).
Binding Affinities of Resynthesized Hits
To test the validity of our library screening, seven of the selected sequences, which ranked 1st, 2nd, 3rd, 5th, 8th, 13th, and 18th on the basis of their Cn-binding affinity, were chosen for resynthesis and binding analysis (Table 1). The synthesis of peptide No. 113 failed at the cyclization step and therefore it was not included in the subsequent analysis. The other six peptides were labeled with TMR, purified by HPLC, and tested for binding to Cn by FA (Figure S3). There is a general agreement between the binding affinities derived from the single-bead analysis and those determined with purified peptides, in that all of the selected peptides bound Cn with KD values in the low μM range and that the more highly ranked peptides are generally more potent ligands (e.g., peptides No. 10, 25, 82), while the lower-ranking peptides are poorer binders (e.g., peptides No. 86 and 61). However, for a given peptide, the KD values derived from the two assays typically differ by ~3-fold (up to 9-fold for peptide No. 10). Some of the discrepancy was caused by the poor quality of the binding data derived from the single-bead analysis (e.g., peptides No. 61, 84, and 86) (Figure S1). Since only ~50 nM peptide could be used in the binding assay (due to limited material from a single bead), the amount of total fluorescence was low and the FA measurement was less reliable. Impurities in the cyclic peptide samples, which may include the corresponding linear peptide and its truncation fragments (generated during library synthesis for the purpose of differentiating mass-degenerate amino acids7), may also interfere with the interaction between a cyclic peptide and Cn (Figure S4). Nevertheless, the single-bead assay provides an approximate KD value for each hit compound, allowing the researcher to focus any subsequent efforts on the most potent hits.
Improved PED/MS Method
The low success rate (~50%) of hit identification by PED/MS prompted us to investigate the origin of the problem and to improve the success rate. In our previously reported procedure7, which was designed for degradation of multiple beads in a custom designed reaction vessel, beads to be sequenced were suspended in 160 μL of 2:1 (v/v) pyridine/water, to which an equal volume of 30–80:1 (mol/mol) phenylisothiocyanate (PITC)/Fmoc-OSu in pyridine was added. When the same procedure was carried out in 96-well plates, we noticed that the amount of peptide chain termination at each residue (which is determined by the rate of reaction with Fmoc-OSu) was uneven and sequence dependent. While insufficient chain termination at a particular residue results in a missing peak at that position, excessive termination at a position(s) (especially when it is near the N-terminus) depletes the peptide leading to weak or loss of signals in the C-terminal region. We reasoned that the amount of chain termination might be related to how quickly PITC and Fmoc-OSu diffuse into a bead during different PED cycles. Since PITC reacts rapidly with water whereas Fmoc-OSu does not, slow diffusion of the pyridine/PITC/Fmoc-OSu solution into a bead equilibrated in the aqueous environment (2:1 pyridine/water) would result in excessive reaction between PITC and water, lower the PITC/Fmoc-OSu ratio inside the bead, and produce excessive chain termination. This problem would be aggravated for PED reactions in 96-well plates, which do not allow adequate mixing of the two solutions. With this insight, we modified the PED procedure by first quickly mixing the pyridine/PITC/Fmoc-OSu and pyridine/water solutions and immediately adding the mixture to a bead(s) that had been dried by vacuum suction. This allows both PITC and Fmoc-OSu to rapidly diffuse into a bead and react with the peptide N-terminal amine so that the extent of peptide chain termination is more directly controlled by the PITC/Fmoc-OSu ratio. We have found that this simple modification leads to more uniform peak heights in the MS spectra and substantially higher success rates for PED/MS sequencing in both the conventional and the 96-well formats. For example, the modified procedure now routinely gives success rates of ≥80% for sequencing beads derived from the Cn library in the 96-well plate format.
SUMMARY
We have integrated some of the latest technologies associated with OBOC libraries to develop a robust platform, which allows a researcher to synthesize, screen, and identify the most active compound(s) from a >1 million-member OBOC library in a few weeks. This can be accomplished without any special equipment such as dedicated robotic systems and thus be performed in any chemical or biochemical laboratory. Application of this platform to a focused cyclic peptide library identified low micromolar ligands against the NFAT-binding site on Cn. To our knowledge, these represent the first examples of cyclic peptide inhibitors against the Cn-NFAT interaction.
EXPERIMENTAL PROCEDURES
Materials
Reagents for peptide synthesis were purchased from Peptides International (Louisville, KY), NovaBiochem (La Jolla, CA), Anaspec (San Jose, CA), Chem-Impex International Inc. (Wood Dale, IL), or AAPPTec (Louisville, KY). Sephadex G-25 column was from GE Healthcare (Piscataway, NJ). PITC (in 1-mL sealed ampoules) were purchased from Sigma-Aldrich (Saint Louis, MO) and a freshly opened ampoule was used in each experiment. Dynabeads M-280 streptavidin, 6-carboxyfluorescein succinimidyl ester (6-FAM, SE), N-hydroxysuccinimidyl ester of Texas Red-X, and tetramethylrhodamine azide were from Invitrogen (Carlsbad, CA). All solvents and other chemical reagents were purchased from Aldrich and were of analytical grade.
Expression, Purification, and Labeling of Cn
The catalytic domain of human Cn was expressed as a glutathione S-transferase fusion protein (GST-Cn) in Escherichia coli BL21 cells and purified on a glutathione-Sepharose column as previously described26. To label GST-Cn with biotin, 500 μL of GST-Cn (100 μM) in 100 mM NaHCO3 (pH 8.5) was mixed with 3 molar equivalents of N-hydroxysuccinimidyl biotin dissolved in DMSO (10 mg/mL). The mixture was incubated for 30 min at room temperature and any remaining biotin ester was quenched by treatment with 10 μL of 1 M Tris buffer (pH 8.5) for 5 min. The reaction mixture was passed through a Sephadex G-25 column and eluted with 30 mM HEPES (pH 7.4), 150 mM NaCl to remove any free biotin. Protein concentration was determined by the Bradford assay and UV absorbance at 280 nm. The extinction coefficient for GST-Cn was calculated to be 87,000 M−1 cm−1 using Protoparam27. Both methods gave similar concentration values. Labeling of the protein with Texas-Red was similarly carried out but with the following modifications. The reaction mixture was kept in the dark and the stoichiometry of labeling was determined by comparing the absorbance at 280 and 595 nm (absorption maximum for Texas-Red; ε = 80,000 M−1 cm−1).
Library Synthesis
The peptide library was synthesized on 1.0 g of TentaGel S NH2 resin (130 μm, 0.2 mmol/g, ~300 pmol/bead). All of the manipulations were performed at room temperature unless otherwise noted. The linker sequence (BM) was synthesized with 5 equiv of Fmoc-amino acids, using HBTU/HOBt/DIEA as the coupling reagents. The coupling reaction was typically allowed to proceed for 1.5 h and the beads were washed with DMF (3×) and DCM (3×). The Fmoc group was removed by treatment twice with 20% piperidine in DMF (5 + 15 min) and the beads were exhaustively washed with DMF (6×). To spatially segregate the beads into outer and inner layers, the resin was washed with DMF and water, and soaked in water overnight. The resin was drained and suspended in a solution of Fmoc-OSu (0.50 equiv) in 10 mL of 1:1 (v/v) DCM/diethyl ether. The mixture was incubated on a rotary shaker for 30 min at room temperature. The beads were washed with 1:1 DCM/diethyl ether (3×) and DMF (8×) to remove water from the beads and then treated with 50 equivalents of Di-tert-butyl dicarbonate and DIEA with catalytic amount of 4-dimethylaminopyridine (DMAP) in DCM (1 h). Next, 2 equivalents of 4-hydroxymethylbenzoic acid was added to the resin with HBTU/HOBt/DIEA (2:2:4 equiv) after the removal of Fmoc group. Fmoc-β-Ala-OH (5 equiv.) was coupled to the Hmb linker by N, N′-diisopropylcarbodiimide (DIC)/DMAP (5.5:0.1 equiv.), and the coupling was repeated twice to drive the reaction to completion. Then, L-Pra, two β-Ala, and Fmoc-L-glutamic acid α-allyl ester were sequentially coupled by standard Fmoc/HBTU chemistry. After that, the Boc protecting group was removed by TFA/water/triisopropylsilane (95:2.5:2.5), followed by the coupling of Fmoc-Arg. Next, the resin was split into 8 equal portions and sequences VIVIT, VIVIT, VIVITG, VIVITA, VIVITGA, VIVITAA, VIVITGAA, or VIVITAAA was added by standard Fmoc/HBTU chemistry. The resin was combined and the random region was synthesized by the split-and-pool method2a using 5 equivalents of Fmoc-amino acids and HATU as the coupling agent. The coupling reaction was repeated once to ensure complete reaction at each step. To differentiate isobaric amino acids during PED/MS sequencing, 4% (mol/mol) of CD3CO2D was added to the coupling reactions of D-Ala, D-Leu, D-Lys and Orn, while 4% CH3CD2CO2D was added to the Nle reactions.7 The allyl group on the C-terminal Glu residue was removed by overnight treatment with tetrakis(triphenylphosphine)palladium, triphenylphosphine, formic acid and diethylamine (1, 3, 10, 10 equiv, respectively) in anhydrous THF at room temperature. Finally, after the removal of the N-terminal Fmoc group, the surface peptides were cyclized by treating the resin with PyBOP/HOBt/NMM (5, 5, 10 equiv, respectively) in DMF for 3 h. Side-chain deprotection was achieved by treatment with reagent K (TFA/thioanisole/water/phenol/1,2-ethanedithiol, 82.5:5:5:5:2.5 v/v) for 2 h. The resulting library was washed with DCM, DMF, 5% N, N-diisopropylethylamine in DMF, 1:1 (v:v) DCM/diethyl ether, DMF, and DCM extensively and stored at −20 °C.
Library Screening
One gram of the library (~106 beads) was screened in three stages. The library resin was first swollen in DCM, washed extensively with DMF, doubly distilled H2O, and HBST-gelatin buffer (30 mM HEPES, pH 7.4, 150 mM NaCl, 0.05% Tween 20 and 0.1% gelatin), and incubated overnight at 4 °C with a blocking buffer (3% BSA in HBST-gelatin buffer). The library resin was incubated with the blocking buffer containing 100 nM biotinylated Cn for 6 h at 4 °C in Poly-Prep chromatography column (10mL, BioRad). Any unbound protein was washed away three times with HBST-gelatin buffer. Next, 100 μL of M280 streptavidin-coated dynabeads (6–7 × 107 beads) was added to the resin in the blocking buffer and incubated for 20 min at room temperature with gentle rotation. The library resins were washed nine times with HBST-gelatin buffer gently in the column to remove unbound magnetic particles and moved to a 50 mL falcon tube. A Dynal MPC-1 magnetic particle concentrator (Invitrogen) was then applied to collect all the hits. All of the positive beads (~1000) were transferred into a BioSpin column (0.8 mL, BioRad) and incubated with 0.8 mL of the blocking buffer containing streptavidin-alkaline phosphatase conjugate (1 μg/mL final concentration) at 4 °C for 10 min. The beads were quickly washed with 1 mL of the blocking buffer (3×) and 1 mL of a staining buffer (30 mM Tris, pH 8.5, 100 mM NaCl, 5 mM MgCl2, 20 μM ZnCl2) (3×). Then, 1 mL of the staining buffer and 100 GL of a BCIP stock solution (5 mg/mL) were added to the beads and intense turquoise color developed on positive beads in 15 min. The turquoise colored beads were manually removed under a dissecting microscope, and subjected to a third stage of screening after extensive washing with HBST buffer, ddH2O, and 8 M guanidine hydrochloride to remove the bound proteins. The resulting beads were incubated overnight with 5 μM Texas-red labeled Cn in blocking buffer at 4 °C. The library beads were viewed under an Olympus SZX12 microscope equipped with a fluorescence illuminator (Olympus America, Center Valley, PA) and the most intensely colored beads were manually collected as positive hits.
On-Bead Labeling and Peptide Release
The positive beads derived from library screening were pooled, washed with water and DMF, and soaked in 60 μL of 1:1 (v/v) water/DMF mixture. The labeling reaction was initiated by the addition of 20 μL of freshly prepared ascorbic acid and copper sulfate solutions (each at 5 mg/ml in water) and 5 μL of tetramethylrhodamine azide in anhydrous DMSO (10 mM). The reaction was allowed to proceed at room temperature overnight in the dark. The reaction was terminated by extensive washing of the beads with water/DMF and the beads were transferred into individual microcentrifuge tubes (one bead/tube). Cyclic peptide release was carried out by the addition of 5 μL of 1 M NaOH solution into each tube and incubation for 1 h at room temperature in the dark. The solution was neutralized by the addition of 5.5 μL of 1 M HCl, transferred to a new tube, evaporated to dryness in a vacuum concentrator, and dissolved in 26 μL of double distilled water to generate a stock solution of ~1 μM (the concentration was estimated by measuring the absorbance of the rhodamine group). The beads containing linear coding peptides were kept in the original tubes and stored for later PED/MS sequencing.
Fluorescence Anisotropy
A primary FA experiment was performed by incubating 100 nM TAMRA-labeled cyclic peptide with 5 μM GST-Cn in 20 mM HEPES (pH 7.4), 150 mM NaCl, 2 mM magnesium acetate, and 0.1% BSA for 2 h at room temperature. The full FA titration experiment was similarly performed by incubating 50 nM the labeled cyclic peptide with varying concentrations (0 –20 μM) of GST-Cn. The FA competition assay was performed by incubating 50 nM labeled cyclic peptide with 10 μM GST-Cn for 1.5 h, followed by the addition of 100 μM the linear VIVIT peptide and incubation for another 1.5 h. The FA values were measured on a Molecular Devices Spectramax M5 spectrofluorimeter, with excitation and emission wavelengths at 545 and 585 nm, respectively. Equilibrium dissociation constants (KD) were determined by plotting the fluorescence anisotropy of TAMRA-labeled cyclic peptides as a function Cn concentration. The titration curves were fitted to the following equation, which assumes a 1:1 binding stoichiometry:
where Y is the measured anisotropy at a given concentration x of Cn; L is the cyclic peptide concentration; Qb/Qf is the correction fact for dye-protein interaction; Amax is the maximum anisotropy when all the peptides are bound to Cn, while Amin is the minimum anisotropy when all the peptides are free; Kd is the dissociation constant.
PED/MS Peptide Sequencing
Beads containing encoding linear peptides were placed into individual wells of an AcroPrep 96-well filter plate (Pall Corporation, PN5030) with one bead per well. To each well was added a freshly mixed solution containing 25 μL of pyridine/water (v/v 2:1) plus 0.1% triethylamine and 25 μL of Fmoc-OSU (2 μmol) and PITC (100 μmol) in dry pyridine. The reaction was allowed to proceed for 6 min and drained by a universal vacuum manifold system designed for 96-well plates (United Chemical Technologies, Inc.). The bead was washed five times with DCM and once with TFA, and incubated with 100 μL of TFA (2 × 6 min). The bead was washed with DCM and pyridine and PED cycle was repeated for n times, where n is the number of residues to be sequenced. At the last PED cycle, the N-terminal Fmoc group was removed by treatment with 20% piperidine in DMF. For MALDI-TOF analysis, the bead was treated with 100 μL of TFA containing ammonium iodide (1.0 mg) and dimethylsulfide (10 μL) for 20 min to reduce any oxidized Met. The bead was washed with water and transferred into a microcentrifuge tube and treated overnight with 20 μL of CNBr in 70% TFA (40 mg/mL) in the dark. The solvents were evaporated under vacuum to dryness and the peptides released from the bead were dissolved in 5 μL of 0.1% TFA in water. One μL of the peptide solution was mixed with 2 μL of saturated 4-hydroxy-α-cyanocinnamic acid in acetonitrile/0.1% TFA (1:1) and 1 μL of the mixture was spotted onto a MALDI sample plate. Mass spectrometry was performed on a Bruker Microflex MALDI-TOF instrument. The data obtained were analyzed by either Moverz software (Proteometrics LLC, Winnipeg, Canada) or Bruker Daltonics flexAnalysis 2.4 (Bruker Daltonic GmbH, Germany).
Supplementary Material
Acknowledgments
We thank Dr. Anjana Rao for providing the GST-Cn plasmid. This work was supported by a grant from the National Institutes of Health (GM062820).
Footnotes
Supporting Information. Additional cyclic peptide-Cn binding data and PED/MS conditions. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Lam KS, Lebl M, Krchnak V. The “One-Bead-One-Compound” Combinatorial Library Method. Chem Rev. 1997;97(2):411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]; (b) Lam KS, Liu RW, Miyamoto S, Lehman AL, Tuscano JM. Applications of one-bead one-compound combinatorial libraries and chemical microarrays in signal transduction research. Acc Chem Res. 2003;36(6):370–377. doi: 10.1021/ar0201299. [DOI] [PubMed] [Google Scholar]; (c) Meldal M. ‘One bead two compound libraries’ for detecting chemical and biochemical conversions. Curr Opin Chem Biol. 2004;8(3):238–244. doi: 10.1016/j.cbpa.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.(a) Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354(6348):82–4. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]; (b) Furka A, Sebestyen F, Asgedom M, Dibo G. General-Method for Rapid Synthesis of Multicomponent Peptide Mixtures. Int J Pept Prot Res. 1991;37(6):487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]; (c) Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and Use of Synthetic Peptide Combinatorial Libraries for Basic Research and Drug Discovery. Nature. 1991;354(6348):84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, Tan PH, Zhang Y, Pei D. On-bead screening of combinatorial libraries: reduction of nonspecific binding by decreasing surface ligand density. J Comb Chem. 2009;11(4):604–11. doi: 10.1021/cc9000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Needles MC, Jones DG, Tate EH, Heinkel GL, Kochersperger LM, Dower WJ, Barrett RW, Gallop MA. Generation and Screening of an Oligonucleotide-Encoded Synthetic Peptide Library. Proc Natl Acad Sci U S A. 1993;90:10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Muller K, Gombert FO, Manning U, Grossmuller F, Graff P, Zaegel H, Zuber JF, Freuler F, Tschopp C, Baumann G. Rapid identification of phosphopeptide ligands for SH2 domains - Screening of peptide libraries by fluorescence-activated bead sorting. J Biol Chem. 1996;271(28):16500–16505. [PubMed] [Google Scholar]; (c) Hu BH, Jones MR, Messersmith PB. Method for screening and MALDI-TOF MS sequencing of encoded combinatorial libraries. Anal Chem. 2007;79(19):7275–85. doi: 10.1021/ac070418g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Hintersteiner M, Kimmerlin T, Kalthoff F, Stoeckli M, Garavel G, Seifert JM, Meisner NC, Uhl V, Buehler C, Weidemann T, Auer M. Single Bead Labeling Method for Combining Confocal Fluorescence On-Bead Screening and Solution Validation of Tagged One-Bead One-Compound Libraries. Chem Biol. 2009;16(7):724–735. doi: 10.1016/j.chembiol.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Seamless Bead to Microarray Screening: Rapid Identification of the Highest Affinity Protein Ligands from Large Combinatorial Libraries. Chem Biol. 2010;17(1):38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Peng L, Liu R, Xu B, Lam KS. Applications of topologically segregated bilayer beads in ‘one-bead one-compound’ combinatorial libraries. J Pept Res. 2005;65(1):130–8. doi: 10.1111/j.1399-3011.2005.00192.x. [DOI] [PubMed] [Google Scholar]
- 7.(a) Chait BT, Wang R, Beavis RC, Kent SBH. Peptide ladder sequencing. Science. 1993;262:89–92. doi: 10.1126/science.8211132. [DOI] [PubMed] [Google Scholar]; (b) Wang P, Arabaci G, Pei D. Rapid sequencing of library-derived peptides by partial Edman degradation and mass spectrometry. J Comb Chem. 2001;3:251–254. doi: 10.1021/cc000102l. [DOI] [PubMed] [Google Scholar]; (c) Sweeney MC, Pei D. An improved method for rapid sequencing of support-bound peptides by partial edman degradation and mass spectrometry. J Comb Chem. 2003;5(3):218–22. doi: 10.1021/cc020113+. [DOI] [PubMed] [Google Scholar]; (d) Thakkar A, Wavreille AS, Pei D. Traceless capping agent for peptide sequencing by partial edman degradation and mass spectrometry. Anal Chem. 2006;78(16):5935–9. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- 8.(a) Aramburu J, Rao A, Klee CB. Calcineurin: From structure to function. Curr Topics Cell Regulation. 2000;3636:237–295. doi: 10.1016/s0070-2137(01)80011-x. [DOI] [PubMed] [Google Scholar]; (b) Rusnak F, Mertz P. Calcineurin: Form and function. Physiol Rev. 2000;80(4):1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]; (c) Li HM, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21(2):91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Ann Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]; (b) Crabtree GR. Generic signals and specific outcomes: Signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96(5):611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Farmer JD, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin Is a Common Target of Cyclophilin-Cyclosporine-a and Fkbp-Fk506 Complexes. Cell. 1991;66(4):807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 11.Odocha O, Mccauley J, Scantlebury V, Shapiro R, Carroll P, Jordan M, Vivas C, Fung JJ, Starzl TE. Posttransplant Diabetes-Mellitus in African-Americans after Renal-Transplantation under Fk-506 Immunosuppression. Transplantation Proc. 1993;25(4):2433–2434. [PMC free article] [PubMed] [Google Scholar]
- 12.Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285(5436):2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 13.(a) Noguchi H, Matsushita M, Okitsu T, Moriwaki A, Tomizawa K, Kang SY, Li ST, Kobayashi N, Matsumoto S, Tanaka K, Tanaka N, Matsui H. A new cell-permeable peptide allows successful allogeneic islet transplantation in mice. Nat Med. 2004;10(3L):305–309. doi: 10.1038/nm994. [DOI] [PubMed] [Google Scholar]; (b) Kuriyama M, Matsushita M, Tateishi A, Moriwaki A, Tomizawa K, Ishino K, Sano S, Matsui H. A cell-permeable NFAT inhibitor peptide prevents pressure-overload cardiac hypertrophy. Chem Biol Drug Design. 2006;67(3):238–243. doi: 10.1111/j.1747-0285.2006.00360.x. [DOI] [PubMed] [Google Scholar]
- 14.Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan PG. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1(5):627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 15.Roehrl MHA, Kang SH, Aramburu J, Wagner G, Rao A, Hogan PG. Selective inhibition of calcineurin-NFAT signaling by blocking protein-protein interaction with small organic molecules. Proc Natl Acad Sci U S A. 2004;101(20):7554–7559. doi: 10.1073/pnas.0401835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Joo SH, Voorhees JL, Brooks CL, Pei D. Synthesis and screening of a cyclic peptide library: discovery of small-molecule ligands against human prolactin receptor. Bioorg Med Chem. 2009;17(3):1026–33. doi: 10.1016/j.bmc.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Li HM, Rao AJ, Hogan PG. Structural delineation of the calcineurin-NFAT interaction and its parallels to PP1 targeting interactions. J Mol Biol. 2004;342(5):1659–1674. doi: 10.1016/j.jmb.2004.07.068. [DOI] [PubMed] [Google Scholar]; (b) Li HM, Zhang L, Rao A, Harrison SC, Hogan PG. Structure of calcineurin in complex with PVIVIT peptide: Portrait of a low-affinity signalling interaction. J Mol Biol. 2007;369(5):1296–1306. doi: 10.1016/j.jmb.2007.04.032. [DOI] [PubMed] [Google Scholar]; (c) Takeuchi K, Roehrl MHA, Sun ZYJ, Wagner G. Structure of the calcineurin-NFAT complex: Defining a T cell activation switch using solution NMR and crystal coordinates. Structure. 2007;15(5):587–597. doi: 10.1016/j.str.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Favre M, Moehle K, Jiang LY, Pfeiffer B, Robinson JA. Structural mimicry of canonical conformations in antibody hypervariable loops using cyclic peptides containing a heterochiral diproline template. J Am Chem Soc. 1999;121(12):2679–2685. [Google Scholar]; (b) Jiang LJ, Moehle K, Dhanapal B, Obrecht D, Robinson JA. Combinatorial biomimetic chemistry: Parallel synthesis of a small library of beta-hairpin mimetics based on loop III from human platelet-derived growth factor B. Helv Chim Acta. 2000;83(12):3097–3112. [Google Scholar]
- 19.Liu R, Marik J, Lam KS. A novel peptide-based encoding system for “one-bead one-compound” peptidomimetic and small molecule combinatorial libraries. J Am Chem Soc. 2002;124(26):7678–80. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 20.Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. High-throughput sequence determination of cyclic peptide library members by partial Edman degradation/mass spectrometry. J Am Chem Soc. 2006;128(39):13000–9. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- 21.(a) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (b) Tornoe CW, Christensen C, Meldal M. Peptidotriazoles on solid phase: [1,2,3]-triazoles by regiospecific copper(I)-catalyzed 1,3-dipolar cycloadditions of terminal alkynes to azides. J Org Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 22.(a) Kodadek T, Bachhawat-Sikder K. Optimized protocols for the isolation of specific protein-binding peptides or peptoids from combinatorial libraries displayed on beads. Molecular Biosystems. 2006;2(1):25–35. doi: 10.1039/b514349g. [DOI] [PubMed] [Google Scholar]; (b) Pei DH, Wavreille AS. Reverse interactomics: decoding protein-protein interactions with combinatorial peptide libraries. Mol Biosys. 2007;3(8):536–541. doi: 10.1039/b706041f. [DOI] [PubMed] [Google Scholar]
- 23.Olivos HI, Bachhawat-Sikder K, Kodadek T. Quantum dots as a visual aid for screening bead-bound combinatorial libraries. Chembiochem. 2003;4(11):1242–1245. doi: 10.1002/cbic.200300712. [DOI] [PubMed] [Google Scholar]
- 24.Kask P, Palo K, Fay N, Brand L, Mets U, Ullmann D, Jungmann J, Pschorr J, Gall K. Two-dimensional fluorescence intensity distribution analysis: Theory and applications. Biophys J. 2000;78(4):1703–1713. doi: 10.1016/S0006-3495(00)76722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakkar A, Cohen AS, Connolly MD, Zuckermann RN, Pei D. High-Throughput Sequencing of Peptoids and Peptide-Peptoid Hybrids by Partial Edman Degradation and Mass Spectrometry. J Comb Chem. 2009;11(2):294–302. doi: 10.1021/cc8001734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang SH, Li HM, Rao AJ, Hogan PG. Inhibition of the calcineurin-NFAT interaction by small organic molecules reflects binding at an allosteric site. J Biol Chem. 2005;280(45):37698–37706. doi: 10.1074/jbc.M502247200. [DOI] [PubMed] [Google Scholar]
- 27.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–52. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.