Abstract
The ESCRT apparatus has multiple Ubiquitin-binding domains and participates in a wide variety of cellular processes. Many of these ESCRT-dependent processes are keenly regulated by Ub, which serves as a lysosomal sorting signal for membrane proteins targeted into multivesicular bodies and which may serve as a mediator of viral budding from the cell surface. Hints that both ESCRTs and Ub work together in the processes such as cytokinesis, transcription, and autophagy are beginning to emerge. Here we explore the relationship between ESCRTs and Ub in MVB sorting and viral budding.
Keywords: ESCRTs, Ubiquitin, Endosome, Lysosome, Vesicle formation
I. ESCRT-MEDIATED SORTING OF UBIQUITINATED PROTEINS INTO MULTIVESICULAR BODIES
Pathways into the endosomes
The first recognized function of the Endosomal Sorting Complex Required for Transport (ESCRT) apparatus was the biogenesis of multivesicular bodies/endosomes (MVBs) (1). ESCRTs have a clear role in forming and pinching off intralumenal vesicles (ILVs) that bud into the endosomal lumen as well as capturing and sorting ubiquitinated membrane protein cargo into those vesicles, leading to their lysosomal degradation (2). The ESCRT apparatus is comprised of four distinct multimeric complexes (0-III), which interact with each other as well as several accessory factors that support disassembly and recycling of the complexes for future rounds of ILV biogenesis. ESCRT-0, -I, and –II all contain ubiquitin-binding domains (UBDs), implying they serve a critical role in gathering ubiquitinated membrane cargo at endosomes for eventual sorting into intralumenal MVB vesicles.
A wealth of data now supports the view that the covalent attachment of ubiquitin (Ub) serves as a necessary and sufficient sorting signal for entry into the degradative MVB/lysosomal pathway for a wide variety of proteins (3-5). These include a range of tyrosine kinase receptors, G-coupled protein receptors, channels and transporters, which are down regulated in response to various stimuli and misfolded cell surface proteins that are recognized as a quality control process by ubiquitin ligases (5). Where tested, Ub-dependent trafficking of membrane proteins into the MVB pathway relies on the ESCRT system; supporting the consensus view that ESCRT-dependent endosomal sorting has a near exclusive relationship with its ubiquitinated membrane protein cargo (Ub-cargo). A number of ESCRT-independent MVB sorting pathways have been described both in vivo and in vitro (6-9); however, the sorting signals that direct proteins into these pathways are only beginning to be understood. Importantly, these alternate pathways are independent of Ub, supporting the notion that the ESCRT pathway mediates the sorting of most, if not all, Ub-cargo into MVBs.
In contrast, the attachment of Ub to membrane proteins may not be the only signal that mediates their sorting into the lysosomal lumen in an ESCRT-dependent manner. For example, the ESCRT-I subunit Tsg101 is required for the lysosomal degradation of the T-Cell co-receptor CD4 when induced by the cytosolic HIV-1 protein, Nef. Yet neither Nef nor CD4 require lysine ubiquitination (10). Further, the loss of the cytosolic lysine resides on the opioid receptor, DOR, slows but does not block its sorting into ILVs, despite its dependence on Hrs (ESCRT-0) and the Vps4 ATPase, an ESCRT accessory factor that controls activity of ESCRT-III (11). Similarly, attenuating ubiquitination of the yeast protein Sna3 appears to only slow its sorting into ILVs along the ESCRT-dependent MVB pathway (12). Finally, the unliganded Transferrin Receptor-2 (TfR2) undergoes lysosomal degradation independent of its sole cytosolic lysine in an ESCRT-dependent manner (13). However, drawing firm conclusions from all of these experiments is difficult for several reasons. First, the ubiquitination of residues other than lysine can occur (14, 15). Additionally, the Ub signal maybe carried by other proteins with which these proteins associate. Indeed, DOR associates with Dysbindin, a known target of at least one E3 ligase, and Sna3 associates with the HECT-domain E3 ligase Rsp5, which undergoes autoubiquitination (12, 16). On the other hand, alternate interactions between cargo and ESCRTs could drive this Ub-independent, ESCRT-mediated sorting. This possibility has been nicely demonstrated by the use of cargo proteins engineered to bypass the need for ubiquitination through direct contact with the ESCRTs via a binding site to HD-PTP, a Bro1/ALIX homolog (17). For the above example cargos, they may associate with the ESCRT apparatus indirectly through their interactions with components that directly interact with the ESCRTs since Nef, Dysbindin and Rsp5 all associate with ESCRT components (16, 18, 19). Except for TfR2, all of the example proteins above do undergo ubiquitination that may provide for efficient MVB sorting under physiological conditions, leaving the seeming Ub-independent route as a backup mechanism. It will be important to verify these apparent Ub-independent, ESCRT-dependent sorting mechanisms and determine their wider physiological meaning.
Recent studies have expanded the range of ESCRT-dependent cargos to include cytosolic proteins, positioning the ESCRT-MVB pathway as a mediator of signal transduction events and a contributor to a subset of autophagy pathways. The transcription regulatory protein Jun is degraded by the ESCRT/MVB pathway once ubiquitinated (20). The ESCRT/MVB pathway is also necessary to sequester GSK3β into ILVs. This sequestration serves as a way to stimulate the Wnt pathway by effectively stabilizing β-catenin against the combined actions of phosphorylation and Ub-dependent degradation by GSK3β and SCF-βTrCP (21). In addition, the ESCRT/MVB pathway may mediate the autophagic sequestration of a broad range of cytosolic proteins into ILVs in an Hsc70-dependent process (22). How these cargos interact with the ESCRT machinery has yet to be determined, but Ub may serve as a potential link. For instance, GSK3β stays bound to its substrates (23), which are targets of the Ub-ligase SCF-βTrCP, potentially providing an Ub-sorting tag for recognition by the Wnt receptor/GSK3β complex. Similarly, Hsc70 associates with the Ub-ligase CHIP that promotes Ub-dependent lysosomal degradation of membrane proteins (22, 24, 25). Additionally, Hsc70 itself is ubiquitinated and associates with Hrs (24). Together, these recent data indicate ESCRTs may have a wide variety of proteins they can sort into MVBs and it will be instructive to resolve the role that Ub plays in these functions.
ESCRTs as Receptors for Ubiquitinated membrane protein cargo
At the endosome, the ESCRT apparatus couples the sorting of Ub-cargo with the formation of ILVs. Many biochemical and structural studies support a direct role for ESCRTs in recognizing Ub-cargo and follow from early bioinformatic studies connecting ESCRTs and Ub-interacting proteins (4, 26, 27). We now know that the ESCRT apparatus is filled with plethora of Ub-binding domains (UBDs), with the prediction that there may yet be more. Some of the ESCRTs have UBDs that may allow them to act as Ub-Sorting Receptors that recognize Ub attached to a variety of cargo proteins and deliver them into ILVs. However, not all UBDs may serve to bind and capture Ub-cargo. Further, the functions of Ub-binding and ubiquitination events that occur during the MVB sorting process have yet to be fully resolved.
The ESCRT-0 complex, composed of yeast Vps27 and Hse1 or mammalian Hrs and STAM1/2, has the best evidence for being an Ub-cargo receptor. The ESCRT-0 heterodimer has no less that 5 UBDs, housed in both α-helical Ubiquitin-Interaction Motifs (UIM) and the N-terminal VHS domains, which have weak affinities for Ub on the order of 100-500µM typical of other UBDs. Loss of the UIM domains within yeast ESCRT-0 causes defects in Ub-cargo sorting while leaving other ESCRT-0 functions intact, indicating that the primary function of the Ub-UIM interaction is dedicated to the recognition of Ub-cargo (28). Similarly, combined loss of Ub-binding by the Hse1 VHS-domain and the Vps27 UIM-domains dramatically blocks Ub-cargo sorting while still preserving other aspects of ESCRT-0 function (29). Studies in mammalian cells also emphasize a role for the dUIM domain of Hrs (a UIM variant that has two Ub-binding surfaces). Loss of this domain disrupts the ability of Hrs to associate with ubiquitinated proteins, to sequester Ub-cargo on endosomes, and to alter endosomal morphology when overexpressed (30-32). Therefore, the multiple UBDs within ESCRT-0 have a role in recognizing the attachment of Ub on cargo proteins.
ESCRT-0 is also structurally well-suited for its role as an Ub-cargo receptor. Hydrodynamic studies indicate that ESCRT-0 is flexible allowing it to interact with a wide variety of proteins that display Ub-sorting tags in different contexts (33). The multiple UBDs within ESCRT-0 could provide conformational plasticity for recognizing diverse of Ub-cargos. In addition, ESCRT-0 can associate with other UBD-containing proteins such as Eps15b, broadening the range of Ub-cargo substrates with which it could interact (34). Finally, higher order structures of ESCRT-0 have been observed suggesting it may be multimeric on membranes and thus provide a strong Ub-binding platform to sequester a variety of Ub-cargo (35, 36).
ESCRT-0 is optimally situated to arbitrate between the recycling and degradation of cargo. It is localized to clathrin patches on endosomes that contain both recycling cargo proteins and proteins destined for degradation (37, 38). Loss of the ESCRT-0 subunit Vps27 causes defects in the recycling of the TGN protein Vps10 back from endosomes (39). Similarly, recycling of some but not all proteins is perturbed in mammalian cells by the loss of ESCRT-0 (40-42). Hrs directly interacts with machinery that promotes recycling such as retromer components Snx1 and Vps35 (43, 44), and overexpressing mutant Hrs lacking its Snx1-binding sites shunts more EGFR towards lysosomal degradation compared to wildtype Hrs (43). In addition, Hrs directly interacts with SCAMP3 and α-actinin, both of which promote recycling from endosomes (42, 45). ESCRT-0 likely exploits distinct sets of protein interacts to switch between ushering proteins for recycling versus promoting their degradation. The degradative pathway is fostered by PTAP motifs within Hrs/Vps27 that bind the Ub variant E2 (UEV)-domain of TSG101/Vps23 of ESCRT-I, which would help convey cargo farther along the pathway towards MVb intralumenal vesicles (46, 47). Interestingly, disrupting the ESCRT-I-binding motifs within Vps27 or the PTAP-binding (ESCRT-0) site within the TSG101 UEV-domain does not have a major effect on sorting of MVB cargo suggesting that ESCRT-0 has additional ways of networking with ESCRT-I and –II (48-50). The signals that toggle ESCRT-0 between interactions with ESCRT-I for degradation or interactions with alternate machinery for recycling are not known.
ESCRT-0 is thought to convey Ub-cargo to the ESCRT -I and -II complexes. Both human and yeast ESCRT -I and -II can form a supercomplex bridged by the Vps28 C-terminal domain (51, 52). These ESCRTs also contain UBDs: ESCRT-I binds Ub via the UEV-domain in Vps23/TSG101 and the yeast ESCRT-I subunit Mvb12 binds Ub using its C-terminal domain, which is positioned near the Vps23 UEV-domain in the overall ESCRT-I structure (50, 53, 54). The ESCRT-II Vps36/EAP45 subunit also binds Ub; EAP45 binds via its N-terminal GLUE-domain and the yeast Vps36 binds via an NZF-domain that is inserted into the GLUE domain (55, 56). The idea that cargo sorting may happen sequentially and involve a “hand-off” of cargo from ESCRT-0 was supported by finding multiple UBDs within ESCRT-I/II. Initial experiments indicated that these UBDs contribute at some level to the function of ESCRTs (53, 56). However, recent studies of mutant yeast strains with all three of the UBDs within ESCRT-I/II inactivated showed only modest defects in sorting Ub-cargo (50). If Ub-cargo is handed off to ESCRT-I/II, then these results indicate there must be additional, yet undiscovered, UBDs within this complex. There may also be other proteins that associate with ESCRT-I or ESCRT-II that contain the critical UBDs, such as γ2 subunit of AP2, the V-domain within Alix, or the Ub-binding UBAP1 protein predicted to be a novel component of ESCRT-I (57-60) (61).
Alternatively, the UBDs within ESCRTs I/II may serve a purpose other than binding Ub-cargo. Interestingly, combined loss of ESCRT-I/II UBDs only cause a dramatic sorting defects when combined with mutations that perturb the overall organization of the ESCRT apparatus (50). These data imply that Ub-binding may be important not for just cargo sorting but also for the integrity or assembly of the ESCRT apparatus. Interestingly, a similar result was found with ESCRT-0. When some of the UBDs within the Vps27/Hse1 complex were inactivated, dramatic defects in sorting Ub-cargo were seen, but other ESCRT-0 functions (eg maintaining endosome morphology and sorting of lumenal vacuolar hydrolases) were intact (28, 29). However, loss of all the UBDs within ESCRT-0 caused a severe loss-of-function, indistinguishable from complete loss of the complex (29). One possibility is that Ub-cargo itself participates directly in the organization of the ESCRT apparatus. Thus, rather that act as a passive substrate, Ub-cargo concentrated within endosomal subdomains helps assemble the ESCRT apparatus around it to effect cargo sorting and vesicle formation. Another proposed role for ESCRT UBDs is that rather than binding Ub-cargo, they instead bind other ubiquitinated proteins, in particular the ESCRT components that can become ubiquitinated (62). These ubiquitination events are driven either by direct binding of ESCRTs with Ub-ligases, or indirectly by association of ESCRT UBDs with auto-ubiquitinated ligases. Potentially, ubiquitination on ESCRTs could induce intra- or intermolecular interactions with the UBDs to promote their activity. However, blocking ESCRT ubiquitination by mutation of lysine residues or by creating ESCRT fusion proteins tied to the catalytic domains of deubiquitinating proteases neither stimulates nor inhibits sorting of Ub-cargo signifying that ESCRT ubiquitination is not used as a critical mechanism at the stage of MVB sorting (63). Thus, the exact role of the ESCRT-I/II UBDs in MVB sorting-- whether they interact with Ub-cargo or other ubiquitinated proteins, and what function such interactions provide-- remain open questions. Importantly, ESCRTs have a number of other cellular functions, and it may instead be these processes wherein particular UBDs have evolved well-defined and unique roles.
Modular solution for sorting subsets of Ub-cargo
Deletion of any of the ESCRT complexes in yeast results in a dramatic phenotype including accumulation of aberrant endosomal compartments and loss of intralumenal vesicle formation, supporting the idea that ESCRTs act collectively and concertedly. However, one of the themes echoed in the other reviews on ESCRTs in Traffic, is that they are far more modular and individual complexes can play major roles in a given process while others do not. Similarly, it may be that instead of a single ESCRT-0 complex used for Ub-cargo sorting, a variety of Ub-binding proteins could serve an “ESCRT-0 like” function to collect Ub-cargo at endosomal sub-domains and convey it into the rest of the ESCRT apparatus. A more modular arrangement (Fig. 1) would specify that other proteins execute an “ESCRT-0” function, by associating with Ub-cargo at clathrin and PtdIns-3P enriched endosomal subdomains and helping position the rest of the ESCRT apparatus. These ESCRT-0 like proteins share common molecular features in that they bind Ub, clathrin, PtdIns-3P and share structural domains such as the N-terminal VHS domain. Several lines of evidence support this general scheme. First, many organisms do not have a cononical Hrs/STAM but do have other ESCRT-0 like complexes such as a Tom1 ortholog (64-66). The ESCRT-0-like proteins also have the capacity to bind to ESCRT-I, providing the ability to interface with the rest of the ESCRT apparatus to convey Ub-cargo to ILVs (67-69). Functional parallels are beginning to emerge from cells that express multiple ESCRT-0 like proteins to suggest both overlapping and unique functions. For instance, although GGA proteins are known to transport both ubiquitinated and nonubiquitinated proteins from the trans-Golgi Network to endosomes, they also localize to endosomes and influence transport of ubiquitinated proteins from the cell surface into MVBs (63, 69, 70). TOM1 and TOM1L1 and their interacting partner Tollip, when overexpressed together, sequester ubiquitinated proteins on endosomal subdomains that are largely distinct from the subdomains which contain the endosomal marker protein EEA1, mirroring a phenotype shared by overexpression of Hrs (71). In addition, some of these alternate ESCRT-0 proteins have a clear role in the degradation of known MVB cargos; Tollip is required for deliver ubiquitinated IL-1β receptors to the lysosome for degradation and Tom1L1 is required for efficient EGFR degradation (72, 73). Just how well these alternative complexes can function as ESCRT-0 complexes, how they might functionally couple to the rest of the ESCRT apparatus, or whether they function just prior to Hrs/STAM ESCRT-0 as a type of “pre-ESCRT” remains to be elucidated.
Figure 1.
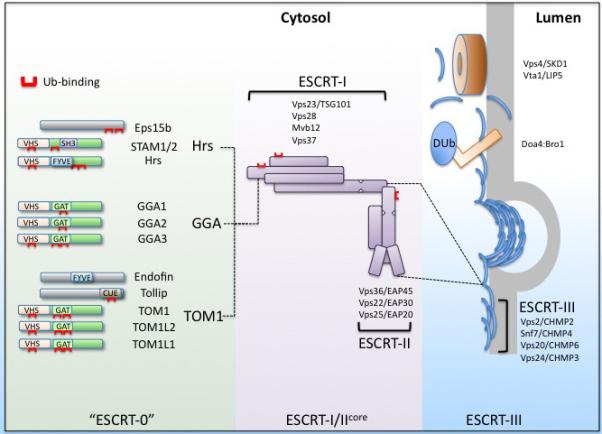
Modular arrangement of ESCRTs for sorting Ubiquitinated cargo into Intralumenal endocytic vesicles. The ESCRT apparatus contains several Ub-binding domains (Red). These are distributed amongst ESCRT-0 and a wider set of proteins that might serve a similar parallel function as ESCRT-0. Together, these proteins recognize ubiquitinated membrane cargo and deliver it to the ESCRT-I/II supercomplex which coordinates cargo sorting and incorporation into intralumenal vesicles with the ESCRT-III apparatus used for completing intralumenal vesicle biogenesis. The individual components of the ESCRT-I and II are listed along with their mammalian counterparts. For simplicity, the various isoforms of particular mammalian ESCRT subunits are not designated. Once cargo is sorted and vesicles are formed, a polymer of ESCRT-III subunits forms and facilitates scission of intralumenal vesicles. The Doa4 deubiquitinating enzyme is recruited to the ESCRT-III complex by interacting with Bro1. In mammalian cells, AMSH and USP8 can also interact with ESCRT-III, although that interaction is not dependent on being bridged by the Bro1 homolog, Alix. The Vps4 AAA ATPase complex is required for de-polymerizing the ESCRT-III complex.
ESCRTs and Ubiquitination machinery
ESCRTs associate with both Ub ligases and peptidases. This association could mediate modification of cargo or the ESCRTs, thereby controlling cargo degradation or regulating ESCRT activity. One clear case of cargo modification is the role of yeast Doa4/Ubp4. Doa4/Ubp4 is a deubiquitinating enzyme that associates with the Snf7 ESCRT-III subunit via Bro1, a homolog of Alix (74, 75). Doa4 is required to remove Ub from cargo just prior to its entry into ILVs; loss of Doa4 leads to accumulation and degradation of Ub in the vacuole and a wide array of phenotypes that result from the general depletion of Ub. Interestingly, the process of cargo deubiquitination may be regulated and, in turn, provide a mechanism to balance Ub levels in the cell and thus cross-regulate numerous functions (76). For instance, Doa4 and Bro1 associate with Rfu1, an inhibitor of Doa4 activity (77). When Rfu1 is deleted, Doa4 becomes more active and generates more unconjugated Ub to cause promiscuous ubiquitination of cellular proteins, which inturn becomes toxic when other aspects of the Ub system are compromised. The prominent role of Doa4 in deubiquitinating cargo at the end of their sorting reaction poses a series of interesting puzzles. One is how cargo is trapped during the sorting reaction so that removal of Ub would no longer allow cargo to escape from incorporation into ILVs. A second mystery is how might Doa4 be coupled with the process of vesicle budding allowing Doa4 enough time to efficiently remove Ub. Recently, binding of ESCRT-III Snf7 by the N-terminal Bro1-domain of Bro1 has been shown to inhibit Vps4-ATPase mediated disassembly of ESCRT-III, potentially providing a mechanism where a Bro1-Doa4 complex might retard subsequent vesicle formation/scission until sufficient deubiquitination is completed (78).
It has been harder to pin down a mammalian enzyme that is functionally equivalent to Doa4 to recycle off Ub from cargo just prior to its incorporation into ILVs. Two enzymes, AMSH and UBPY/USP8, are good candidates. However, these enzymes do not appear to have a simple role in modifying cargo at a single step of the sorting process. In some cases, loss of AMSH and USP8 functions accelerate cargo degradation, suggesting they intervene early in the trafficking of the receptor to strip away the attached Ub sorting signals (79-81). Yet in other instances, loss of these enzymes blocks cargo degradation, (81-88) signifying a more general role in line with the effects observed upon loss of Doa4 in budding yeast that perturbs cargo ubiquitination. The broad defects observed in doa4Δ yeast mutants are also observed upon loss of AMSH in organisms such as fission yeast and plants suggesting that both AMSH and USP8 help maintain the pool of free ubiquitin by recycling it from MVB cargo (89, 90). The idea that these DUbs have multiple roles within the MVB pathway is supported by their protein interaction patterns, with both DUbs able to interact “early” in the pathway with ESCRT-0 via the SH3 domain in STAM as well as “late” in the sorting process by binding ESCRT-III via MIT-MIM interactions (82, 85, 86, 91-95). In addition to these possible roles, it has been proposed that these DUbs may control the levels of ESCRT components, since loss of UBPY/USP8 results in lower steady state levels of ESCRT-0 (87, 96). However, in other instances ubiquitination does not alter ESCRT levels, providing the possibility that ubiquitination of ESCRTs may provide a mode of regulation that is potentially mediated through ESCRT UBDs directing intra- and intermolecular interactions with ubiquitinated ESCRT subunits (62, 82, 97-101). To resolve multiple roles for these DUbs, future studies will be obliged to carefully dissect the contributions of various DUb interactions on the fate of specific Ub-cargo on a case-by-case basis.
ESCRTs also associate with Ub ligases including Nedd4-family HECT-type ligases such as AIP4, Rsp5, Nedd4 and a host of Ring ligases including POSH, Tal, and Mahagouin (99-112). Investigating the function of these interactions has lead to models similar to those explaining the association of deubiquitinating enzymes with ESCRTs. Many of the HECT-type ubiquitin ligases have clear roles in the modifying cargo (113). In yeast, the Rsp5 ligase associates with ESCRT-0 to modify cargo and ensure efficient sorting into MVBs (19). Ligase association has also been found to cause ubiquitination of the ESCRTs themselves, and this can correlate with both inhibitory and stimulatory effects of the ligase on MVB sorting. In some cases, ubiquitination controls the levels of ESCRT components by inducing their proteasomal degradation (107, 114). However, in other instances ubiquitination does not alter ESCRT levels, providing the possibility that ubiquitination of ESCRTs may provide a mode of regulation (82, 100, 102-106, 115, 116). The likelihood that ubiquitination of ESCRTs is part of an essential mechanism used for MVB sorting has been diminished by recent studies where ESCRT-dependent MVB sorting was found to operate even when ESCRT ubiquitination was blocked by either removal of all ubiquitinated lysines or attaching DUb activity to multiple places within the ESCRT apparatus (63). Nevertheless, whether ubiquitination can inhibit ESCRTs within particular regulatory pathways or whether it fosters functions of ESCRTs other than MVB sorting are distinct possibilities.
What do ESCRTs recognize as a MVB sorting signal?
Cargo that undergoes MVB sorting is typically modified by multiple Ubs and short chains of Ub linked via K63 (117-120). At least some of the key Ub-ligases that modify cargo favor formation of K63 chains (121) and some of the Dubs involved in MVB sorting favor dismantling K63 chains over others (79, 122, 123). Critically, cells unable to make K63 chains, because they carry a mutant K63R Ub as their sole source of Ub, show severe MVB sorting defects for a number of cargos (70, 124). In addition, ESCRTs bind much better to polyubiquitin chains over mono-Ub (125). These data support the idea that K63 polyubiquitin chain may be the requisite sorting signal for ESCRT-dependent sorting into MVBs. However, at the molecular level it is not clear that ESCRTs uniquely favor recognition of K63 chains. All UBDs of the ESCRTs have been defined at the structural level in the context of binding a single Ub (126). Moreover, for well-characterized complexes such as ESCRT-0, the increased affinity for K63 chains over mono-Ub is explained by an increase in avidity by the concatermization of mono-Ub binding sites rather than a preference for a unique topology a K63 chain of Ub would provide (29). Interestingly, the multiple Ub-binding sites within ESCRT-0 provide little cooperativity that might be observed if polyubiquitin chains were to lie down simultaneously across multiple UBDs. Further, ESCRT-0 shows high affinity for diUb with either K63 or K48 linkages over mono-Ub but no discernable difference for linkage specify (29). This is in contrast to other UBDs that do have distinct bias for poly-Ub linkages such as Rap80 that has a strong bias for K63-linked chains and the UBAN motif of NEMO that favors linear Ub chains (127, 128). This emphasizes that the main function of multiple UBDs may be to foster recognition of a variety of cargos that present Ub in different contexts. One possible exception may be the NZF domain of yeast Vps36, which shares features of a subset of NZF domains that greatly favor K63-linked chains (56, 129). However, how well the Vps36 NZF domain favors K63 or whether Ub-binding via this domain even contributes to cargo recognition remain unclear as there are conflicting reports on its binding activity in vitro and no in vivo data specifically addressing this possibility (129, 130). K63-linked chains are clearly important for aspects of MVB sorting in vivo (131). However, they are not strictly required as a sorting signal since fusion of mutant Ub lacking K63 onto cargo is sufficient to restore MVB sorting in yeast strains that lack the ability to make K63 polybuqiutin chains (63). In addition, deletion of Ubp2 (a DUb that forms a complex with Rsp5 and favors dismantling K63 polyubiquitin chains) allows Rsp5 to make extended K63-linked chains, yet this effect diminishes MVB sorting, suggesting even an inhibitory role for K63 chains (122). With the increased binding K63 chains have towards ESCRTs, it makes sense that K63 chain formation would foster more efficient sorting of cargoes either by mediating more avid binding to ESCRTs or serving as a buffer against counteracting endosomal DUbs. Long Ub chains or “over-ubiquitination” of cargo may confound the sorting process by diminishing the ability of cargo to disengage from the ESCRTs in order to conclude the sorting process.
II. UBIQUITIN AND ESCRT-DEPENDENT VIRAL BUDDING
Contribution of Ub to ESCRT-dependent Viral Release
ESCRTs also mediate the release of wide variety of retroviruses in a process focused on the detachment of viral particles from the cell surface (132). Budding is mediated by the viral Gag protein, composed of domains that mediate membrane association, self-assembly with other Gag proteins, and the so-called Late domain required for the final scission steps that occur. Late domains amongst viruses are remarkably interchangeable and work when placed in a variety of postions within the Gag protein. ESCRTs bind motifs within the late domain and disruption of this interaction causes virus particles to accumulate at the cell surface, unable to undergo the final stages of membrane fission. Ub is heavily implicated in this process; however, a clear and unified role for Ub and its recognition by the ESCRTs has yet to fully become apparent. A direct role for ESCRTs to foster scission of viral particles and release from the cell surface is through their recognition of “late-domains” housed within viral Gag proteins. A core feature of this process is the ultimate recruitment of ESCRT-III to the site of viral budding (133, 134) reflected by the universal requirement for the Vps4 AAA-ATPase, a key regulator of ESCRT-III. The late domains themselves can interact with a variety of ESCRTs that in turn are thought to initiate a cascade of protein:protein interactions that eventually recruit ESCRT-III. Viruses have the remarkable ability to use a variety of interaction sites within the ESCRT apparatus to effect their budding. For instance, viral late domains contain peptide motifs such as PTAP, which allow binding to the TSG101 UEV-domain within ESCRT-I, or YPxL/LxxLY motifs that bind the V-domain of ALIX (135-138). Often viruses contain multiple ESCRT binding motifs presumably to provide redundant pathways for successful ESCRT-III recruitment. Viral late domains show remarkable flexibility and interchangeability in their ability to catalyze budding (139). This presents a formidable problem when interpreting the results from experiments that use chimeric viral Gag proteins since these experiments readily reveal what is sufficient for viral budding rather than what may be truly required for the budding of a specific natural virus.
Multiple observations imply an intimate role for Ub in ESCRT-dependent viral budding. Ub is a constituent of virus-like particles (VLP) formed by a variety of viral Gag proteins (140, 141). Treatment of infected host cells with proteasome inhibitors, which might deplete pools of available Ub, can block viral egress and fusion of Ub to Gag overcomes this inhibition (142). Decreased viral budding is also seen when mutant forms of Ub, which lack the ability to form K63-linked chains or residues important for binding a host of Ub-binding domains, are overexpressed (143). In addition, PPxY motifs, which interact with the WW domains within variety of Nedd4-family HECT-domain Ub ligases, can also provide both necessary and sufficient late domain function for viruses such as murine leukemia virus (MLV) and Rous sarcoma virus (RSV) (144-146). Overexpression of HECT-domain ligases stimulates release of such viruses, but only if the HECT-domain is enzymaticaly active (147, 148). Similarly, over-expression of proteins that can compete for occupancy of the ligase WW-domains inhibits virus release (108). Gag proteins carrying PPxY motifs also undergo ubiquitination (149). In addition, HIV-1 and feline immune deficiency virus (FIV), which do not use PPxY motifs that directly bind HECT-domain ligases, are also stimulated by over-expression of Ub-ligases, especially when their direct connection with ESCRT-I is severed (116, 150-152). Like the PPxY motif, the HIV-1 late domain motifs can also mediate ubiquitination of Gag proteins in some circumstances (150). Similarly, the residual budding of MLV lacking its PPxY motif can be enhance with overexpression of the Itch HECT-domain Ub ligase, which also correlates with increased levels of ubiquitinated Gag (153). Finally, support for a functional relationship between Nedd4 and Gag ubiquitination can be found in how interferon-induced expression of ISG15 interferes with viral budding. Part of this inhibition is manifested in both inhibition of Gag ubiquitintion and Nedd4 ligase activity, although certainly other ISG15-dependent processes surely contribute to virus suppression (154-156). The universal effect of Ub-ligases on different Gag budding systems stress the importance of a undefined target being ubiquitinated rather than mere direct physical connection with a ligase. Clearly ESCRT-dependent viral budding process involves Ub somehow, leaving the larger goal of finding the physiological mechanism(s) for how Ub contributes to this process.
What is Getting Ubiquitinated?
There is strong evidence that the target of ubiquitination can be the viral Gag protein itself. This supports previous models that postulated that ubiquitination of Gag could work co-operatively with other late domain binding proteins. Indeed, the UEV domain of TSG101, which is required for budding of many viruses, has increased affinity for PTAP-containing peptides that also contain an Ub moiety (137). Also, the PTAP binding motif and Ub can work synergistically when present on certain Gag proteins (60). There are many studies showing a positive correlation between Gag ubiquitination and the capacity to support budding (140, 149, 151, 153, 157, 158). More directly, loss of lysines within Gag proteins diminishes budding, although these lysines need not be confined to the late domain itself (159, 160). Importantly, for RSV Gag protein, the requirement for lysines is independent of their position, strongly indicating that they function to accept Ub rather than serve a structural role for the Gag protein itself. Similarly, budding of HTLV can be enhanced by the addition of non-native lysine residues (159). As proof of principle, it has been shown that fusion of Ub directly to HTLV Gag can largely suppress the need for its native TSG101- and Ub-ligase- binding late domain altogether (57). Also, an engineered Gag derived from Prototypic Foamy virus can form virus-like particles when fused to Ub, and does so more efficiently than when fused to PTAP (Tsg101-binding) or PPxY (HECT-ligase binding) motifs (60).
While it has been demonstrated that ubiquitination of Gag can contribute to viral budding in some cases, other data confound the interpretation that this represents a universal and necessary aspect of ESCRT-dependent budding. The amount of ubiquitinated Gag protein is low, which might indicate that its does not play a stoichiometric role (140). Ubiquitination of Gag proteins can actually increase (albeit modestly) when their ESCRT binding motifs are compromised and their budding is blocked (161, 162). Targeting Ub-ligases to Gags results in their ubiquitination but does not always stimulate budding (158). When the major ubiquitination site of HTLV Gag is lost, budding is only slightly diminished, while intriguingly increasing the requirement for the Nedd4-family Ub ligases (163). Finally, an engineered Gag derived from Prototypic Foamy virus unable to undergo N-terminal or lysyl- ubiquitination supports formation of viral like particles either through a PTAP or a PPxY pathway. In addition, both of these pathways were stimulated by the overexpression of Nedd4-family ubiquitin ligase (60, 164). Interpreting these experiments to mean that Gag ubiquitination is not important comes with many caveats, however. While the level of Gag ubiquitination is only a few percent of the total Gag protein (140), it is quite comparable to the level achieved by Ub-cargo that undergoes MVB sorting and may simply reflect multiple rounds of ubiquitination/deubiquitination are occurring. Also, the place, timing and topology of ubiquitination may play a central role in this process and these qualities may not be resolved by examining steady-state levels of ubiquitinated Gag. Indeed, different ligases, which induce comparable steady state Ub-Gag levels but which produce different poly-ubiquitin chain topologies, show dramatic differences in their ability to promote HIV-1 budding (158). The fusion of Ub directly to Gag may allow it to bypass the spatio-temporal regulation that cell mediated ubiquitination is subject to, allowing it to robustly substitute as a late domain. Finally, the specter of non-amino linked ubiquitination through C, S, and T residues endures. Still, taken together, these results suggest that while Gag ubiquitination can contribute to budding, it may not be the only protein that can accept the Ub. Certainly other proteins at the site of budding can undergo ubiquitination. These include the ESCRT proteins themselves such as ESCRT-1 and Alix (116, 165). Also, the Nedd4-family of Ub-ligases catalyze their own ubiquitination (60). These or other proteins might serve as cryptic Ub acceptors that become functional when preferred substrates such as the Gag proteins themselves, are not available for ubiquitination. This may explain why HTLV-1 lacking its Gag ubiquitination site needs elevated Ub-ligase activity to restore efficient budding, since this might promote ubiquitination of less preferred cryptic sites (163). This stochastic but not precise mode of ubiquitination, as previously proposed, nicely accounts for the potential role(s) of Ub in viral budding (60, 164). Alternatively, ubiquitination of Gag proteins could be a red herring, with the role of Ub playing a more precise role in the modification of discrete ESCRT components to trigger their ability to effect viral budding (116) .
What is Recognizes the Ubiquitin in Viral Budding?
The larger question looms as to the function Ub actually provides to the process of viral budding. So will understanding what ultimately recognizes the attached Ub. A variety of ESCRTs contain UBDs, but only a handful of ESCRTs are strictly required for viral budding. Fusion of ESCRT-0 and ESCRT-II subunits to Gag fosters VLP budding (47, 166, 167). However, depletion of ESCRT-0 or ESCRT-II does not hamper budding of virus particles (60, 168, 169) making it unlikely that they play a critical direct role. Three components that are required for viral budding are ESCRT-I, Alix, and Nedd4-family ligases as their depletion severely hampers budding of viruses whose late domains interact with these factors (135, 170-173). All three of these components also bind Ub non-covalently, potentially providing the key UBD that recognizes either Ub-Gag or other ubiquitinated protein at the site of budding (53, 174, 175). The first steps to decipher which components actually play a role in Ub-dependent virus budding have used experimental contexts that heavily rely on some aspect of ubiquitination. These include the Nedd4-stimulated budding of HIV-1 lacking its TSG101-binding PTAP motif (116, 165), budding of Gag fused to Ub (57, 60), and budding of PPxY containing Gags that rely heavily on interacting with Nedd4-family ligases (60, 173). In these systems, depletion of ESCRT-I TSG101 and/or Alix significantly diminishes viral release, with additive effects when co-depletion of Alix and TSG101 were employed. Since Alix and ESCRT-I could each define interaction pathways that lead to Ub-independent recruitment of later acting ESCRT-III, the key experiment is to determine whether their UBDs play roles in these budding reactions. Budding of a Ub-fused EIAV Gag lacking its normal Alix binding site was found to be dependent on the Ub-binding activity of the TSG101 UEV domain, indicating this UBD may play a role in Ub-dependent viral budding. However, TSG101 with the same inactivating mutations within its UBD is capable of supporting Nedd4-stimulated budding of mutant HIV-1 (116). In addition, Gag-Ub fusions carrying Q62/E64 mutations within the Ub moiety that block binding to the TSG101 UEV domain only modestly diminish budding activity, in contrast to fusions with mutations of I44 or L8 that target the hydrophobic surface on Ub that is required for a number of UBD interactions (60, 174). These data imply that other UBDs play a role in Ub-dependent viral budding; indeed, there is no shortage of candidate UBDs either within the ESCRTs or their associated partners. Alix stands as a particularly attractive candidate, although the structural basis of its Ub-binding activity has not been revealed yet. Moreover, ESCRTs such as ESCRT-0 or ESCRT-II might even perform an auxiliary role in Ub recognition that may only be revealed when other ESCRT UBDs are inactivated. Interestingly, overexpression of Ub mutated at F4 diminishes VLP production of HIV-1 Gag (143). In addition, Gag-Ub fusions containing the F4A mutation show less budding that those fused to WT Ub, and show level of inhibition comparable to fusion with Ub lacking its TSG101 interface (60). To date, an UBD that relies on F4 of Ub has not been defined and it could be that this factor in combination with TSG101 constitutes what is authentically required for Ub-dependent viral budding.
Figure 2.
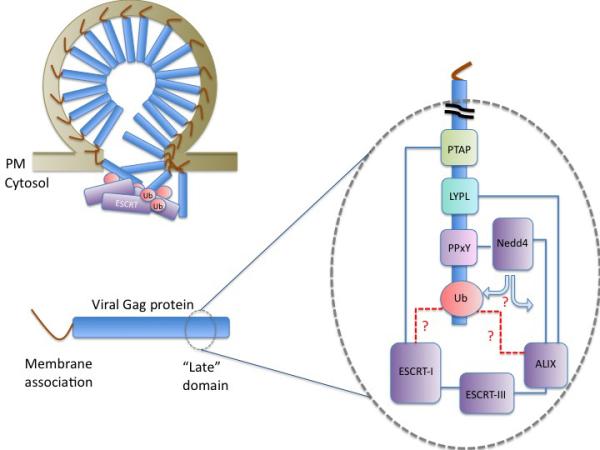
Viral Gag proteins contain late domains that interact with different cellular host proteins to promote viral egress. The schematic depicts a Gag protein with an N-terminal membrane association domain and a late domain containing various interaction sites. PTAP-motifs recruit ESCRT-I through interactions with the UEV-domain within Tsg101, LYPL/LxxLY-motifs recruit ALIX through interactions with its V-domain, and PPxY-motifs recruit Nedd4-family of Ub ligases. Gag proteins are ubiquitinated, which may promote association with ESCRTs; however, other cellular proteins may be functional targets of ubiquitination as well. Ubiquitin may be involved as a recruitment pathway by allowing TSG101, ALIX, or another ESCRT to bind Gag proteins. Alternatively, Ub may regulate aspects of ESCRT function. Intriguingly, the function of late domains along with the ESCRT pathway can be circumvented with a Gag containing a domain that allows for higher order oligomerization (176-178). These data indicate that the ESCRT dependent process may provide a similar function. Pictured at the tope left is a stochastic model whereby several alternate ubiquitinated proteins could interact with any number of Ub-binding ESCRTs or ESCRT-associated proteins to allow for viral budding.
References
- 1.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95(6):847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 2.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143(5):682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 5.Hislop JN, von Zastrow M. Role of ubiquitination in endocytic trafficking of G-protein-coupled receptors. Traffic. 2011;12(2):137–148. doi: 10.1111/j.1600-0854.2010.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuffers S, Sem Wegner C, Stenmark H, Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10(7):925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- 7.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 8.Theos AC, Truschel ST, Tenza D, Hurbain I, Harper DC, Berson JF, Thomas PC, Raposo G, Marks MS. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev Cell. 2006;10(3):343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White IJ, Bailey LM, Aghakhani MR, Moss SE, Futter CE. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25(1):1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.daSilva LL, Sougrat R, Burgos PV, Janvier K, Mattera R, Bonifacino JS. Human immunodeficiency virus type 1 Nef protein targets CD4 to the multivesicular body pathway. J Virol. 2009;83(13):6578–6590. doi: 10.1128/JVI.00548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry AG, White IJ, Marsh M, von Zastrow M, Hislop JN. The role of ubiquitination in lysosomal trafficking of delta-opioid receptors. Traffic. 2010;12(2):170–184. doi: 10.1111/j.1600-0854.2010.01145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oestreich AJ, Aboian M, Lee J, Azmi I, Payne J, Issaka R, Davies BA, Katzmann DJ. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell. 2007;18(2):707–720. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Wang J, Meyers KR, Enns CA. Transferrin-directed internalization and cycling of transferrin receptor 2. Traffic. 2009;10(10):1488–1501. doi: 10.1111/j.1600-0854.2009.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309(5731):127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Herr RA, Rabelink M, Hoeben RC, Wiertz EJ, Hansen TH. Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol. 2009;187(5):655–668. doi: 10.1083/jcb.200908036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marley A, von Zastrow M. Dysbindin promotes the post-endocytic sorting of G protein-coupled receptors to lysosomes. PLoS One. 2010;5(2):e9325. doi: 10.1371/journal.pone.0009325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyotte A, Mironov A, McKenzie E, Woodman P. The Bro1-related protein HD-PTP/PTPN23 is required for endosomal cargo sorting and multivesicular body morphogenesis. Proc Natl Acad Sci U S A. 2008;105(17):6308–6313. doi: 10.1073/pnas.0707601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa LJ, Chen N, Lopes A, Aguiar RS, Tanuri A, Plemenitas A, Peterlin BM. Interactions between Nef and AIP1 proliferate multivesicular bodies and facilitate egress of HIV-1. Retrovirology. 2006;3:33. doi: 10.1186/1742-4690-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J, Kee Y, Huibregtse JM, Piper RC. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol Biol Cell. 2007;18(1):324–335. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda H, Kerppola TK. Lysosomal localization of ubiquitinated Jun requires multiple determinants in a lysine-27-linked polyubiquitin conjugate. Mol Biol Cell. 2008;19(11):4588–4601. doi: 10.1091/mbc.E08-05-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143(7):1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105(6):721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 24.Pridgeon JW, Webber EA, Sha D, Li L, Chin LS. Proteomic analysis reveals Hrs ubiquitin-interacting motif-mediated ubiquitin signaling in multiple cellular processes. FEBS J. 2009;276(1):118–131. doi: 10.1111/j.1742-4658.2008.06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26(6):347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 27.Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4(7):534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- 29.Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29(6):1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol. 2006;13(3):272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- 31.Urbe S, Sachse M, Row PE, Preisinger C, Barr FA, Strous G, Klumperman J, Clague MJ. The UIM domain of Hrs couples receptor sorting to vesicle formation. J Cell Sci. 2003;116(Pt 20):4169–4179. doi: 10.1242/jcs.00723. [DOI] [PubMed] [Google Scholar]
- 32.Raiborg C, Bache KG, Gillooly DJ, Madshus IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4(5):394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 33.Ren X, Kloer DP, Kim YC, Ghirlando R, Saidi LF, Hummer G, Hurley JH. Hybrid structural model of the complete human ESCRT-0 complex. Structure. 2009;17(3):406–416. doi: 10.1016/j.str.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roxrud I, Raiborg C, Pedersen NM, Stang E, Stenmark H. An endosomally localized isoform of Eps15 interacts with Hrs to mediate degradation of epidermal growth factor receptor. J Cell Biol. 2008;180(6):1205–1218. doi: 10.1083/jcb.200708115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayers JR, Fyfe I, Schuh AL, Chapman ER, Edwardson JM, Audhya A. ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J Biol Chem. doi: 10.1074/jbc.M110.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pullan L, Mullapudi S, Huang Z, Baldwin PR, Chin C, Sun W, Tsujimoto S, Kolodziej SJ, Stoops JK, Lee JC, Waxham MN, Bean AJ, Penczek PA. The endosome-associated protein Hrs is hexameric and controls cargo sorting as a “master molecule”. Structure. 2006;14(4):661–671. doi: 10.1016/j.str.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Sachse M, Urbe S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13(4):1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prekeris R, Klumperman J, Chen YA, Scheller RH. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J Cell Biol. 1998;143(4):957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131(3):603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanyaloglu AC, McCullagh E, von Zastrow M. Essential role of Hrs in a recycling mechanism mediating functional resensitization of cell signaling. EMBO J. 2005;24(13):2265–2283. doi: 10.1038/sj.emboj.7600688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang SH, Zhao L, Sun ZP, Li XZ, Geng Z, Zhang KD, Chao MV, Chen ZY. Essential role of Hrs in endocytic recycling of full-length TrkB receptor but not its isoform TrkB.T1. J Biol Chem. 2009;284(22):15126–15136. doi: 10.1074/jbc.M809763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Mol Biol Cell. 2005;16(5):2470–2482. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chin LS, Raynor MC, Wei X, Chen HQ, Li L. Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J Biol Chem. 2001;276(10):7069–7078. doi: 10.1074/jbc.M004129200. [DOI] [PubMed] [Google Scholar]
- 44.Popoff V, Mardones GA, Bai SK, Chambon V, Tenza D, Burgos PV, Shi A, Benaroch P, Urbe S, Lamaze C, Grant BD, Raposo G, Johannes L. Analysis of articulation between clathrin and retromer in retrograde sorting on early endosomes. Traffic. 2009;10(12):1868–1880. doi: 10.1111/j.1600-0854.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- 45.Aoh QL, Castle AM, Hubbard CH, Katsumata O, Castle JD. SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol Biol Cell. 2009;20(6):1816–1832. doi: 10.1091/mbc.E08-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci U S A. 2003;100(13):7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162(3):425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163(2):237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren X, Hurley JH. Structural basis for endosomal recruitment of ESCRT-I by ESCRT-0 in yeast. EMBO J. 2011;30(11):2130–2139. doi: 10.1038/emboj.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol. 2009;185(2):213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14(6):902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Emr SD, Williams RL. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 2007;26(2):600–612. doi: 10.1038/sj.emboj.7601501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106(2):145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 54.Kostelansky MS, Schluter C, Tam YY, Lee S, Ghirlando R, Beach B, Conibear E, Hurley JH. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell. 2007;129(3):485–498. doi: 10.1016/j.cell.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280(20):19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 56.Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23(7):1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joshi A, Munshi U, Ablan SD, Nagashima K, Freed EO. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic. 2008;9(11):1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doring T, Gotthardt K, Stieler J, Prange R. gamma2-Adaptin is functioning in the late endosomal sorting pathway and interacts with ESCRT-I and -III subunits. Biochim Biophys Acta. 2010;1803(11):1252–1264. doi: 10.1016/j.bbamcr.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 59.Sangsuriya P, Rojtinnakorn J, Senapin S, Flegel TW. Identification and characterization of Alix/AIP1 interacting proteins from the black tiger shrimp, Penaeus monodon. J Fish Dis. 2010;33(7):571–581. doi: 10.1111/j.1365-2761.2010.01156.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhadina M, Bieniasz PD. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog. 2010;6(10):e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Souza RF, Aravind L. UMA and MABP domains throw light on receptor endocytosis and selection of endosomal cargoes. Bioinformatics. 2010;26(12):1477–1480. doi: 10.1093/bioinformatics/btq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haglund K, Stenmark H. Working out coupled monoubiquitination. Nat Cell Biol. 2006;8(11):1218–1219. doi: 10.1038/ncb1106-1218. [DOI] [PubMed] [Google Scholar]
- 63.Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J Cell Biol. 2011;192(2):229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leung KF, Dacks JB, Field MC. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic. 2008;9(10):1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 65.Blanc C, Charette SJ, Mattei S, Aubry L, Smith EW, Cosson P, Letourneur F. Dictyostelium Tom1 participates to an ancestral ESCRT-0 complex. Traffic. 2009;10(2):161–171. doi: 10.1111/j.1600-0854.2008.00855.x. [DOI] [PubMed] [Google Scholar]
- 66.Herman EK, Walker G, van der Giezen M, Dacks JB. Multivesicular bodies in the enigmatic amoeboflagellate Breviata anathema and the evolution of ESCRT 0. J Cell Sci. 124(Pt 4):613–621. doi: 10.1242/jcs.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puertollano R. Interactions of TOM1L1 with the multivesicular body sorting machinery. J Biol Chem. 2005;280(10):9258–9264. doi: 10.1074/jbc.M412481200. [DOI] [PubMed] [Google Scholar]
- 68.Yanagida-Ishizaki Y, Takei T, Ishizaki R, Imakagura H, Takahashi S, Shin HW, Katoh Y, Nakayama K. Recruitment of Tom1L1/Srcasm to endosomes and the midbody by Tsg101. Cell Struct Funct. 2008;33(1):91–100. doi: 10.1247/csf.07037. [DOI] [PubMed] [Google Scholar]
- 69.Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6(3):244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- 70.Lauwers E, Jacob C, Andre B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 2009;185(3):493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh Y, Shiba Y, Mitsuhashi H, Yanagida Y, Takatsu H, Nakayama K. Tollip and Tom1 form a complex and recruit ubiquitin-conjugated proteins onto early endosomes. J Biol Chem. 2004;279(23):24435–24443. doi: 10.1074/jbc.M400059200. [DOI] [PubMed] [Google Scholar]
- 72.Liu NS, Loo LS, Loh E, Seet LF, Hong W. Participation of Tom1L1 in EGF-stimulated endocytosis of EGF receptor. EMBO J. 2009;28(22):3485–3499. doi: 10.1038/emboj.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brissoni B, Agostini L, Kropf M, Martinon F, Swoboda V, Lippens S, Everett H, Aebi N, Janssens S, Meylan E, Felberbaum-Corti M, Hirling H, Gruenberg J, Tschopp J, Burns K. Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr Biol. 2006;16(22):2265–2270. doi: 10.1016/j.cub.2006.09.062. [DOI] [PubMed] [Google Scholar]
- 74.Amerik A, Sindhi N, Hochstrasser M. A conserved late endosome-targeting signal required for Doa4 deubiquitylating enzyme function. J Cell Biol. 2006;175(5):825–835. doi: 10.1083/jcb.200605134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richter C, West M, Odorizzi G. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 2007;26(10):2454–2464. doi: 10.1038/sj.emboj.7601692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Groothuis TA, Dantuma NP, Neefjes J, Salomons FA. Ubiquitin crosstalk connecting cellular processes. Cell Div. 2006;1:21. doi: 10.1186/1747-1028-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kimura Y, Yashiroda H, Kudo T, Koitabashi S, Murata S, Kakizuka A, Tanaka K. An inhibitor of a deubiquitinating enzyme regulates ubiquitin homeostasis. Cell. 2009;137(3):549–559. doi: 10.1016/j.cell.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 78.Wemmer M, Azmi I, West M, Davies B, Katzmann D, Odorizzi G. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J Cell Biol. 192(2):295–306. doi: 10.1083/jcb.201007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166(4):487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mukai A, Yamamoto-Hino M, Awano W, Watanabe W, Komada M, Goto S. Balanced ubiquitylation and deubiquitylation of Frizzled regulate cellular responsiveness to Wg/Wnt. EMBO J. 2010;29(13):2114–2125. doi: 10.1038/emboj.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bowers K, Piper SC, Edeling MA, Gray SR, Owen DJ, Lehner PJ, Luzio JP. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J Biol Chem. 2006;281(8):5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- 82.Berlin I, Higginbotham KM, Dise RS, Sierra MI, Nash PD. The deubiquitinating enzyme USP8 promotes trafficking and degradation of the chemokine receptor 4 at the sorting endosome. J Biol Chem. 285(48):37895–37908. doi: 10.1074/jbc.M110.129411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hasdemir B, Murphy JE, Cottrell GS, Bunnett NW. Endosomal deubiquitinating enzymes control ubiquitination and down-regulation of protease-activated receptor 2. J Biol Chem. 2009;284(41):28453–28466. doi: 10.1074/jbc.M109.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hislop JN, Henry AG, Marchese A, von Zastrow M. Ubiquitination regulates proteolytic processing of G protein-coupled receptors after their sorting to lysosomes. J Biol Chem. 2009;284(29):19361–19370. doi: 10.1074/jbc.M109.001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma YM, Boucrot E, Villen J, Affar el B, Gygi SP, Gottlinger HG, Kirchhausen T. Targeting of AMSH to endosomes is required for epidermal growth factor receptor degradation. J Biol Chem. 2007;282(13):9805–9812. doi: 10.1074/jbc.M611635200. [DOI] [PubMed] [Google Scholar]
- 86.Row PE, Liu H, Hayes S, Welchman R, Charalabous P, Hofmann K, Clague MJ, Sanderson CM, Urbe S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J Biol Chem. 2007;282(42):30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 87.Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281(18):12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- 88.Sierra MI, Wright MH, Nash PD. AMSH interacts with ESCRT-0 to regulate the stability and trafficking of CXCR4. J Biol Chem. 2010;285(18):13990–14004. doi: 10.1074/jbc.M109.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Isono E, Katsiarimpa A, Muller IK, Anzenberger F, Stierhof YD, Geldner N, Chory J, Schwechheimer C. The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell. 2010;22(6):1826–1837. doi: 10.1105/tpc.110.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwaki T, Onishi M, Ikeuchi M, Kita A, Sugiura R, Giga-Hama Y, Fukui Y, Takegawa K. Essential roles of class E Vps proteins for sorting into multivesicular bodies in Schizosaccharomyces pombe. Microbiology. 2007;153(Pt 8):2753–2764. doi: 10.1099/mic.0.2007/006072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agromayor M, Martin-Serrano J. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J Biol Chem. 2006;281(32):23083–23091. doi: 10.1074/jbc.M513803200. [DOI] [PubMed] [Google Scholar]
- 92.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Activation of the endosome-associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body-sorting machinery. Curr Biol. 2006;16(2):160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 93.Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88(3):333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 94.Kaneko T, Kumasaka T, Ganbe T, Sato T, Miyazawa K, Kitamura N, Tanaka N. Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain. J Biol Chem. 2003;278(48):48162–48168. doi: 10.1074/jbc.M306677200. [DOI] [PubMed] [Google Scholar]
- 95.Kato M, Miyazawa K, Kitamura N. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J Biol Chem. 2000;275(48):37481–37487. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- 96.Niendorf S, Oksche A, Kisser A, Lohler J, Prinz M, Schorle H, Feller S, Lewitzky M, Horak I, Knobeloch KP. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27(13):5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alwan HA, van Leeuwen JE. UBPY-mediated epidermal growth factor receptor (EGFR) de-ubiquitination promotes EGFR degradation. J Biol Chem. 2007;282(3):1658–1669. doi: 10.1074/jbc.M604711200. [DOI] [PubMed] [Google Scholar]
- 98.Angers A, Ramjaun AR, McPherson PS. The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans-Golgi network and endosomal system. J Biol Chem. 2004;279(12):11471–11479. doi: 10.1074/jbc.M309934200. [DOI] [PubMed] [Google Scholar]
- 99.Berlin I, Schwartz H, Nash PD. Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8.STAM complex. J Biol Chem. 285(45):34909–34921. doi: 10.1074/jbc.M109.016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Malik R, Marchese A. Arrestin-2 interacts with the endosomal sorting complex required for transport machinery to modulate endosomal sorting of CXCR4. Mol Biol Cell. 2010;21(14):2529–2541. doi: 10.1091/mbc.E10-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marchese A, Raiborg C, Santini F, Keen JH, Stenmark H, Benovic JL. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev Cell. 2003;5(5):709–722. doi: 10.1016/s1534-5807(03)00321-6. [DOI] [PubMed] [Google Scholar]
- 102.Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y, Roubini E, Cohen M, Wides R, Bacharach E, Schubert U, et al. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 2004;18(14):1737–1752. doi: 10.1101/gad.294904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bhandari D, Trejo J, Benovic JL, Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem. 2007;282(51):36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- 104.Jiao J, Sun K, Walker WP, Bagher P, Cota CD, Gunn TM. Abnormal regulation of TSG101 in mice with spongiform neurodegeneration. Biochim Biophys Acta. 2009;1792(10):1027–1035. doi: 10.1016/j.bbadis.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Katz M, Shtiegman K, Tal-Or P, Yakir L, Mosesson Y, Harari D, Machluf Y, Asao H, Jovin T, Sugamura K, Yarden Y. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 2002;3(10):740–751. doi: 10.1034/j.1600-0854.2002.31006.x. [DOI] [PubMed] [Google Scholar]
- 106.Kim BY, Olzmann JA, Barsh GS, Chin LS, Li L. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol Biol Cell. 2007;18(4):1129–1142. doi: 10.1091/mbc.E06-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim GH, Park E, Kong YY, Han JK. Novel function of POSH, a JNK scaffold, as an E3 ubiquitin ligase for the Hrs stability on early endosomes. Cell Signal. 2006;18(4):553–563. doi: 10.1016/j.cellsig.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 108.Rauch S, Martin-Serrano J. Multiple Interactions Between The ESCRT Machinery And Arrestin-Related Proteins: implications in PPXY-dependent budding. J Virol. 2010 doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol. 84(16):8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsuda M, Seong KH, Aigaki T. POSH, a scaffold protein for JNK signaling, binds to ALG-2 and ALIX in Drosophila. FEBS Lett. 2006;580(13):3296–3300. doi: 10.1016/j.febslet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 111.Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, Vidal M, Komander D, Serrano L, Lehner P, Sanderson CM. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19(10):1905–1911. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Medina G, Pincetic A, Ehrlich LS, Zhang Y, Tang Y, Leis J, Carter CA. Tsg101 can replace Nedd4 function in ASV Gag release but not membrane targeting. Virology. 2008;377(1):30–38. doi: 10.1016/j.virol.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10(6):398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 114.McDonald B, Martin-Serrano J. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol Biol Cell. 2008;19(2):754–763. doi: 10.1091/mbc.E07-09-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou R, Kabra R, Olson DR, Piper RC, Snyder PM. Hrs controls sorting of the epithelial Na+ channel between endosomal degradation and recycling pathways. J Biol Chem. 285(40):30523–30530. doi: 10.1074/jbc.M110.150755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chung H, Morita E, Von Schwedler U, Muller B, Krausslich H, Sundquist W. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82(10):4884. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21(6):737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 118.Roth AF, Davis NG. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J Biol Chem. 2000;275(11):8143–8153. doi: 10.1074/jbc.275.11.8143. [DOI] [PubMed] [Google Scholar]
- 119.Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16(19):5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Springael JY, Galan JM, Haguenauer-Tsapis R, Andre B. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci. 1999;112(Pt 9):1375–1383. doi: 10.1242/jcs.112.9.1375. [DOI] [PubMed] [Google Scholar]
- 121.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29(12):3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kee Y, Munoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281(48):36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- 123.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455(7211):358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 124.Paiva S, Vieira N, Nondier I, Haguenauer-Tsapis R, Casal M, Urban-Grimal D. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J Biol Chem. 2009;284(29):19228–19236. doi: 10.1074/jbc.M109.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barriere H, Nemes C, Du K, Lukacs GL. Plasticity of polyubiquitin recognition as lysosomal targeting signals by the endosomal sorting machinery. Mol Biol Cell. 2007;18(10):3952–3965. doi: 10.1091/mbc.E07-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hurley J, Stenmark H. Molecular Mechanisms of Ubiquitin-Dependent Membrane Traffic. Ann Rev Biophys. 2011;40 doi: 10.1146/annurev-biophys-042910-155404. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28(16):2461–2468. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136(6):1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 129.Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol. 2009;16(12):1328–1330. doi: 10.1038/nsmb.1731. [DOI] [PubMed] [Google Scholar]
- 130.Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J. 2009;28(24):3903–3909. doi: 10.1038/emboj.2009.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20(4):196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 132.Goff SP. Host factors exploited by retroviruses. Nat Rev Micro. 2007;5(4):253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- 133.Baumgärtel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Kräusslich H-G, Bräuchle C, Müller B, Lamb DC. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011 doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 134.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13(4):394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fisher RD, Chung H-Y, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128(5):841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 136.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14(3):194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pornillos O, Alam SL, Rich RL, Myszka DG, Davis DR, Sundquist WI. Structure and functional interactions of the Tsg101 UEV domain. The EMBO Journal. 2002;21(10):2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pornillos O, Garrus JE, Sundquist WI. Mechanisms of enveloped RNA virus budding. Trends in Cell Biology. 2002;12(12):569–579. doi: 10.1016/s0962-8924(02)02402-9. [DOI] [PubMed] [Google Scholar]
- 139.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344(1):55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 140.Ott DE, Coren LV, Copeland TD, Kane BP, Johnson DG, Sowder RC, Yoshinaka Y, Oroszlan S, Arthur LO, Henderson LE. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J Virol. 1998;72(4):2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Putterman D, Pepinsky RB, Vogt VM. Ubiquitin in avian leukosis virus particles. Virology. 1990;176(2):633–637. doi: 10.1016/0042-6822(90)90035-p. [DOI] [PubMed] [Google Scholar]
- 142.Patnaik A, Chau V, Li F, Montelaro RC, Wills JW. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J Virol. 2002;76(6):2641–2647. doi: 10.1128/JVI.76.6.2641-2647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Strack B, Calistri A, Göttlinger HG. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J Virol. 2002;76(11):5472–5479. doi: 10.1128/JVI.76.11.5472-5479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68(10):6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69(9):5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yuan B, Li X, Goff SP. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18(17):4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168(1):89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Segura-Morales C. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. Journal of Biological Chemistry. 2005;280(29):27004. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 149.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci U S A. 2000;97(24):13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Usami Y, Popov S, Popova E, Gottlinger H. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82(10):4898. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Calistri A, Del Vecchio C, Salata C, Celestino M, Celegato M, Gottlinger H, Palu G, Parolin C. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J Cell Physiol. 2009;218(1):175–182. doi: 10.1002/jcp.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Votteler J, Iavnilovitch E, Fingrut O, Shemesh V, Taglicht D, Erez O, Sorgel S, Walther T, Bannert N, Schubert U, Reiss Y. Exploring the functional interaction between POSH and ALIX and the relevance to HIV-1 release. BMC Biochem. 2009;10:12. doi: 10.1186/1471-2091-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jadwin JA, Rudd V, Sette P, Challa S, Bouamr F. Late domain-independent rescue of a release-deficient Moloney murine leukemia virus by the ubiquitin ligase itch. J Virol. 2010;84(2):704–715. doi: 10.1128/JVI.01319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol. 2010;84(9):4725–4736. doi: 10.1128/JVI.02478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A. 2008;105(10):3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Malakhova OA, Zhang DE. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283(14):8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Blot V, Perugi F, Gay B, Prevost MC, Briant L, Tangy F, Abriel H, Staub O, Dokhelar MC, Pique C. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci. 2004;117(Pt 11):2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 158.Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Göttlinger H. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 2010;6(9) doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Spidel JL, Craven RC, Wilson CB, Patnaik A, Wang H, Mansky LM, Wills JW. Lysines close to the Rous sarcoma virus late domain critical for budding. J Virol. 2004;78(19):10606–10616. doi: 10.1128/JVI.78.19.10606-10616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gottwein E, Jäger S, Habermann A, Kräusslich H-G. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect. J Virol. 2006;80(13):6267–6275. doi: 10.1128/JVI.02177-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. J Virol. 2004;78(11):5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gottwein E, Kräusslich H-G. Analysis of human immunodeficiency virus type 1 Gag ubiquitination. J Virol. 2005;79(14):9134–9144. doi: 10.1128/JVI.79.14.9134-9144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J Virol. 2007;81(18):9769–9777. doi: 10.1128/JVI.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhadina M, McClure MO, Johnson MC, Bieniasz PD. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(50):20031–20036. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol. 2010;84(16):8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pincetic A, Medina G, Carter C, Leis J. Avian sarcoma virus and human immunodeficiency virus, type 1 use different subsets of ESCRT proteins to facilitate the budding process. J Biol Chem. 2008;283(44):29822–29830. doi: 10.1074/jbc.M804157200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bouamr F, Houck-Loomis BR, De Los Santos M, Casaday RJ, Johnson MC, Goff SP. The C-terminal portion of the Hrs protein interacts with Tsg101 and interferes with human immunodeficiency virus type 1 Gag particle production. J Virol. 2007;81(6):2909–2922. doi: 10.1128/JVI.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Langelier C, von Schwedler UK, Fisher RD, De Domenico I, White PL, Hill CP, Kaplan J, Ward D, Sundquist WI. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol. 2006;80(19):9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Janvier K, Pelchen–Matthews A, Renaud J-B, Caillet M, Marsh M, Berlioz-Torrent C. The ESCRT-0 Component HRS is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation. PLoS Pathog. 2011;7(2):e1001265. doi: 10.1371/journal.ppat.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stuchell MD, Garrus JE, Müller B, Stray KM, Ghaffarian S, McKinnon R, Kräusslich H-G, Morham SG, Sundquist WI. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem. 2004;279(34):36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 171.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD, Yaravoy A. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Garrus J, von Schwedler U, Pornillos O, Morham S, Zavitz K, Wang H, Wettstein D, Stray K, Côté M, Rich R. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 173.Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem. 2005;280(29):27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 174.Joshi A, Munshi U, Ablan S, Nagashima K, Freed E. Functional replacement of a retroviral late domain by ubiquitin fusion. Traffic. 2008;9(11):1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.French M, Kretzmann B, Hicke L. Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem. 2009;284(18):12071–12079. doi: 10.1074/jbc.M901106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Accola MA, Strack B, Gottlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74(12):5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5(6):e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Popova E, Popov S, Gottlinger HG. Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol. 84(13):6590–6597. doi: 10.1128/JVI.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


