Abstract
The constitutive DNase I hypersensitive site at the 5′ end of the chicken β-globin locus marks the boundary of the active chromatin domain in erythroid cells. The DNA sequence containing this site has the properties of an insulator, as shown by its ability in stable transformation experiments to block enhancer–promoter interaction when it lies between the two, but not when it lies outside, and to protect against position effects in Drosophila. We now show that the chicken insulator can protect a stably integrated gene, which is otherwise subject to great variability of expression, from chromatin-mediated repression in cell culture. When the integrated reporter gene is surrounded by insulator elements, stably transformed cell lines display consistent enhancer-dependent expression levels, in accord with the strength of the enhancer. In the absence of insulators, long-term nonselective propagation of cells carrying the integrated reporter gene results in gradual extinction of the reporter’s expression, with expression patterns from tandemly repeated inserted genes suggesting that the extinction of adjacent genes is coupled. We show that the uninsulated reporter genes, in addition to becoming transcriptionally inactive, lose several epigenetic hallmarks of active chromatin, including nuclease accessibility, DNA hypomethylation, and histone hyperacetylation during time in culture. Treatment with inhibitors of histone deacetylase or DNA methylation reverses the extinction of the uninsulated genes. Extinction is completely prevented by flanking the reporter construct with insulators. Furthermore, in contrast to the uninsulated reporter genes, chromatin over the insulated genes retains nuclease accessibility and histone hyperacetylation. However, there is no clear correlation between the presence of the insulators and the level of DNA methylation. This leads us to propose a model for the insulator’s ability to protect against extinction in the transformed cell lines and to function as a chromatin boundary for the chicken β-globin locus in normal erythroid cells.
Keywords: Insulator, transcription, DNA methylation, histone acetylation, silencing
Insulators are DNA sequences that can function as directional blocking elements in a number of environments. Two classes of behavior have been described. In the first, the insulator interferes with promoter–enhancer interactions if and only if it lies between them. In the second, insulators placed on either side of a stably transformed reporter gene serve to protect the gene against position effects, that is, against the influence of nearby strong endogenous enhancers or heterochromatic regions. The earliest described and best-characterized insulators are the scs/scs′ elements of the HSP70 heat-shock locus and the suppressor of Hairy-wing [su(Hw)] elements within the gypsy retrotransposon in Drosophila. Studies of Kellum and Schedl (1991) first showed that a stably integrated reporter eye color gene, surrounded by scs elements, is expressed in a position-independent manner. Geyer and Corces (1992) established the directional enhancer-blocking properties of su(Hw); this element was subsequently shown to provide insulation from position effects as well (Roseman et al. 1993).
There have been fewer examples of insulating activity in vertebrates (Chung et al. 1993; Zhong and Krangel 1997). Work in this laboratory has focused on a 1.2-kb DNA element from the 5′ end of the chicken β-globin locus, which has been found capable of directionally blocking enhancer activity in stable transformation assays. Experiments analogous to those with the scs element show that the chicken globin insulator is similarly capable of conferring position-independent expression on an eye color reporter gene in Drosophila (Chung et al. 1993).
These results raise the possibility that the chicken globin insulator might be capable of conferring position-independent behavior on genes stably transformed into vertebrate cells, which would make possible a more detailed examination of the mechanisms by which position effects are generated. In this paper we describe experiments in which a test gene expressing a cell-surface marker driven by a globin promoter and enhancer, and surrounded by insulator elements, is introduced into a pre-erythroid cell line. We show that the insulator is capable of protecting the reporter against position effects, in the form of both stimulatory and repressive signals that might be present in neighboring regions of the genome. In particular, the insulator elements can protect against the gradual silencing of expression found in most cell lines we observed. We show that this silencing is accompanied by DNA methylation at the gene’s promoter and by histone deacetylation in the region containing the gene, with inhibition of DNA methylation or of histone deacetylation reversing the silencing. Correlative with protection of gene activity, we observe that the presence of the insulators results in the maintenence over the reporter gene of histone hyperacetylation but not always of promoter hypomethylation. This suggests possible mechanisms for insulator action.
Results
Position effects
The chicken βA-globin gene, which carries a strong enhancer at its 3′ end (Fig. 1A), has been studied extensively in our laboratory. This gene was modified to serve as a probe of position effects in cell culture. We made reporter plasmids in which the globin coding region was replaced with a fragment of an interleukin 2 receptor cDNA [IL2R, tac fragment (Leonard et al. 1984)], so that the IL2R coding region was driven by the chicken βA-globin promoter and the strong β/ε enhancer. When expressed, the antigenic IL2R cell-surface marker allows detection and activity-dependent sorting, either by fluorescence cytometry (FACS; Fig. 1B) (Fordis et al. 1990) or magnetic bead sorting (MACS) (Giordano et al. 1991). For experiments in which the effects of the insulator were tested, we also made constructions that were flanked by the 1.2-kb chicken globin insulator element.
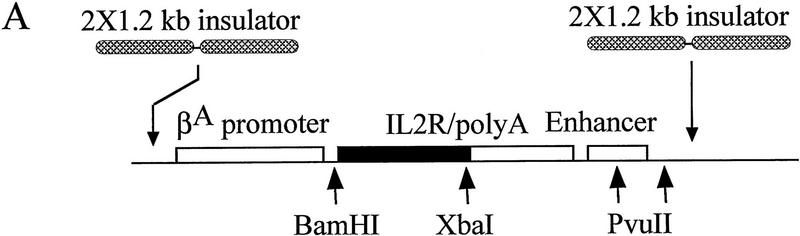
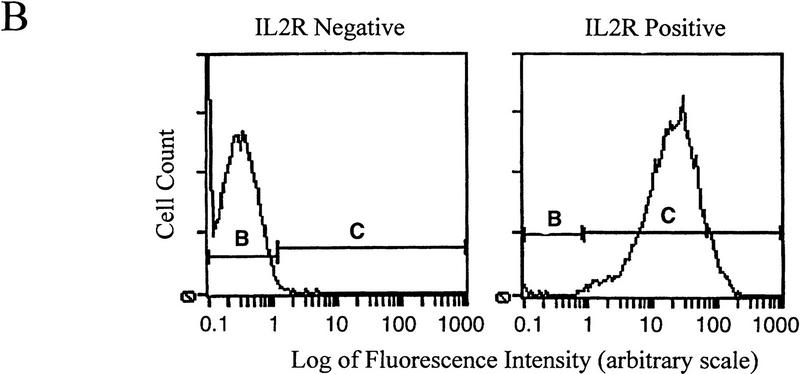
Figure 1.
(A) The IL2R cDNA and SV40 splice/polyadenylation signal are linked to the chicken βA-globin promoter and a 121-bp minimal β/ε enhancer element or enhancers mutated at either the NF–E2 site or at both GATA sites. These mutations decrease expression to 10% and 6%, respectively, of that of the wild-type (Reitman and Felsenfeld 1988) and reduce the probability of forming a hypersensitive site over the enhancer when the construct is stably integrated in 6C2 cells (Boyes and Felsenfeld 1996). Furthermore, the expression cassette was flanked with two copies of the 1.2-kb globin insulator. (B) FACS profiles of 6C2 cells untransfected with the IL2R construct (left) and following stable transfection and integration of one copy of the IL2R reporter with wild-type enhancer (right; these are line 005 cells, below). On these histograms, cell number is indicated on the y-axis, and the logarithm (10−1 through 103) of FITC fluorescence intensity on the x-axis. The horizontal bars denoted B and C define the ranges of IL2R negative and positive cells, respectively.
The IL2R vectors were introduced into the chromatin of the chicken early erythroid 6C2 cell line by stable cotransfection with a hygromycin resistance gene as a selectable marker. Previous work has shown that the chicken β/ε enhancer assembles a nuclease accessible chromatin structure when inserted with the chicken βA-globin gene and promoter into 6C2 cells (Boyes and Felsenfeld 1996). Initially, cells were grown in the continued presence of hygromycin, and single-copy integrants were analyzed by FACS for IL2R expression. Expression was consistent in most wild-type enhancer lines in the control constructions without insulators (Fig. 2A). However, two such uninsulated lines, 124 and 134, of six single copy lines tested, showed a decreased, but still detectable, amount of IL2R surface marker. This variability of expression among lines is evidence of a moderate position effect on the wild-type enhancer. Much greater variability of expression was seen when the NF-E2 or GATA sites in the enhancer were mutated: Activities of two of four uninsulated single-copy lines of each mutant (204, 239, 409, and 413) approached that of the wild-type enhancer lines, whereas others had low activity (Fig. 2A).
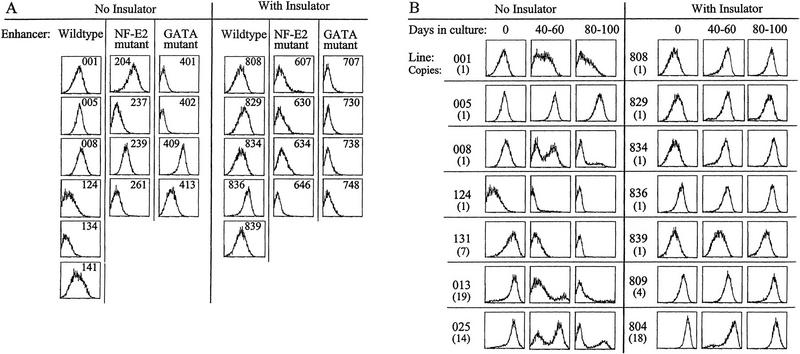
Figure 2.
FACS analyses for position effects (A) Cell lines carrying single copies of the IL2R constructs with or without flanking insulators were grown under selection for the hygromycin resistance marker and analyzed by FACS for IL2R cell surface activity. The most left-shifted histogram peaks, such as line 707, fall at the fluorescence intensity position of control, untransfected 6C2 cells analyzed under the same conditions (see Fig. 1B). (B) Wild-type enhancer lines, in single- and multiple-copy, both with and without insulators, were grown for up to 100 days in culture without hygromycin. Cells were analyzed during the time course for IL2R activity. Of the uninsulated lines tested, only one (005) remained active, whereas all lines with insulators retained full activity throughout passage. The rightward peak shifts of lines 013, 025, 809, and 804, reflect increased IL2R mRNA levels caused by multiple gene copies. When RNA was prepared from sorted cells, IL2R message levels were found to be unchanged in the active fractions relative to the initial, 100% active state, whereas message was undetectable in the inactive fractions (data not shown).
We asked whether this variability of expression could be prevented by introduction of the insulator element. When the constructs described above were flanked with two copies of the 1.2-kb β-globin insulator on either side, the level of expression for each type of enhancer construct was uniform in the single-copy integrants examined. Furthermore, the levels of expression obtained were commensurate with the wild-type and mutant enhancer strengths seen earlier in transient transfection into chick red blood cells, that is, double GATA mutant < NF-E2 mutant < wild type (Reitman and Felsenfeld 1988) (Fig. 2A, right). Thus, the insulator suppressed position effects on these reporters.
Effects of the insulator on extinction of expression
A different pattern of expression emerged when cell lines carrying the uninsulated wild-type enhancer were grown for extended time without hygromycin selection. Most clones without insulators, with single- as well as multiple-copy inserts, evolved into two populations of cells as revealed by biphasic FACS profiles (Fig. 2B, left). In these lines, a portion of the cells expressed as much IL2R as under initial selective conditions, whereas an increasing fraction became inactive for IL2R. For most lines, expression of IL2R was extinct in >90% of the cells of each population by 80 days of growth, with only one uninsulated line (005) maintaining high expression throughout long-term growth.
Again, we asked what effect the insulator had on this behavior. When we repeated the IL2R expression studies with cell lines carrying the wild-type enhancer flanked by insulator elements, we found stable, high levels of IL2R expression for all lines for the duration of the experiment (Fig. 2B, right). The insulator therefore protected these wild-type enhancer constructs from variegated silencing. This occurred in every cell line carrying the insulated transgene, suggesting that the insulator protects against position effects in a way that is independent of the site of integration of the reporter. In principle, this might occur if the insulator targeted the construct to integrate within specific active chromatin domains. To test this possibility, we performed experiments with constructs carrying both copies of the double 1.2-kb insulator element on the 5′ side of the IL2R reporter, so that the transgene was now adjacent to the same arrangement of two double 1.2-kb insulators but was completely unprotected on the 3′ side. In this configuration, the IL2R reporter was not protected against transcriptional inactivation (data not shown). This is not consistent with a mechanism in which the insulator elements might be capable of guiding the IL2R gene to an active chromatin domain but, rather, with a model in which inactivation can occur when the insulator is not present on both sides of the reporter gene, wherever it is integrated.
Coordinated activity of adjacent genes
In some of the uninsulated, wild-type lines (001 and 008), loss of IL2R expression did not occur if the cells were grown in the continued presence of hygromycin and suggested that hygromycin genes were integrated next to IL2R genes in the genome, as Southern blotting of restriction-digested DNA of these lines confirmed. Furthermore, when cells of the 001 or 008 lines that had grown for some time without hygromycin selection were sorted by MACS and replated, the IL2R-inactive cells were now hygromycin sensitive, whereas the IL2R-positive fractions remained hygromycin resistant (data not shown). This suggested that the activity of the two adjacent genes was coupled. The insulated cell line 836, which had at least one copy of the hygromycin resistance gene integrated adjacent to the insulated IL2R gene, behaved differently. Hygromycin resistance of these cells, all of which continued to express IL2R, was greatly reduced when hygromycin was added back after 80 days of nonselective culture (data not shown). Thus, the insulator decoupled inactivation of hygromycin marker from the neighboring IL2R transgene. This is also not consistent (see above) with models in which the insulator directs the integration site.
Furthermore, the biphasic expression profiles obtained for multicopy uninsulated integrants also suggested a coordination of adjacent genes. Line 025, for example, contains an estimated 14 copies of the gene in a tandem array, and line 013, 19 copies. Although we cannot be certain that the behavior at each gene copy is the same within any given cell, we observe that the total IL2R activity in the active cell population is significantly higher than in the single-copy genes, and is copy-number dependent, suggesting that multiple copies of the gene must be expressed simultaneously. Independent inactivation of each individual gene in the cluster would be expected to give a single peak of declining activity. It could not give rise to the kind of bimodal distribution we observe for lines 013 and 025, with very active and completely inactive cells present simultaneously. Therefore, as with the IL2R/hygromycin linkage described above, we conclude that inactivation is a coupled event, with the entire complement of repeated IL2R genes tending to become inactivated in a given cell all at once.
Changes in chromatin structure
We have shown that the insulator can prevent transcriptional silencing and position effects. Next, we wished to study the uninsulated lines as they inactivated, with the expectation that a better understanding of what caused the silencing might shed light on how the insulator functions to prevent it.
Inactivation was not caused by spontaneous mutations to the gene or its regulatory sequences, because inactivation was not permanent but, as will be shown below, was partially reversible (see Fig. 4A, below). Moreover, transient transfections of analogous reporters with the βA-promoter and β/ε enhancer driving the chloramphenicol acetyltransferase gene showed no loss of activity in the IL2R-inactive cells (data not shown), so that the complement of trans-acting factors required for transcription appeared uncompromised. Inactivation is therefore an epigenetic process; we thought it likely to be mediated through the transgene’s chromatin environment.
Figure 4.
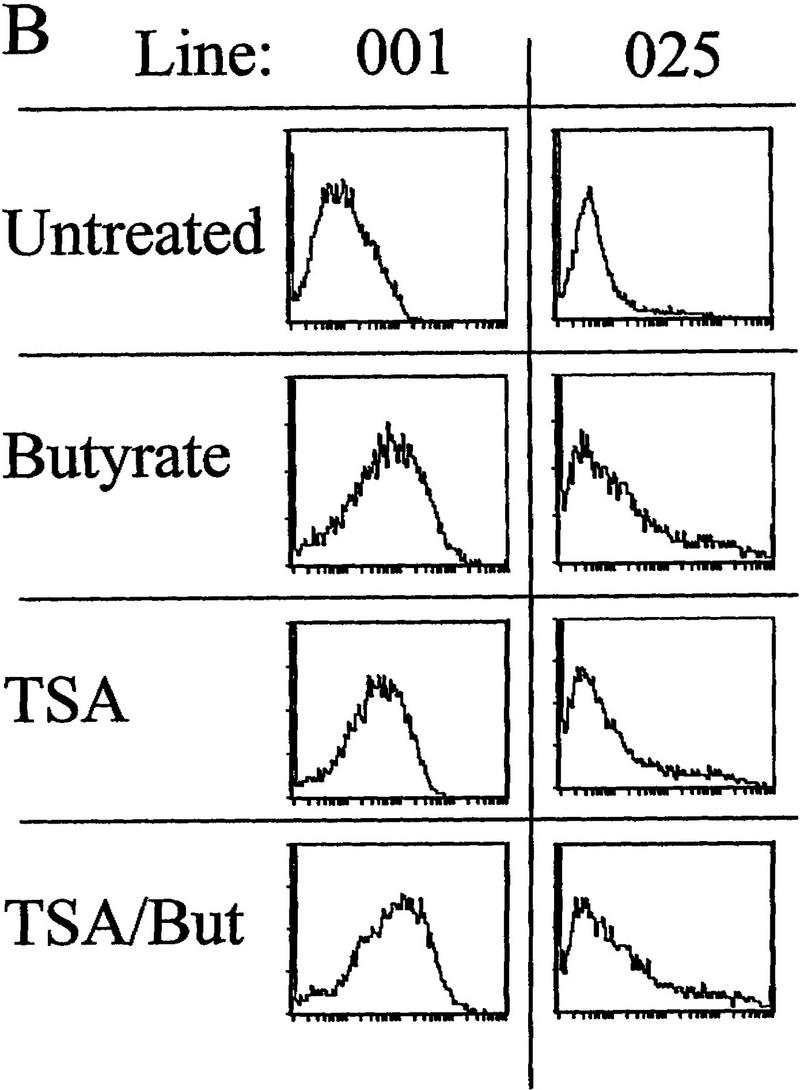
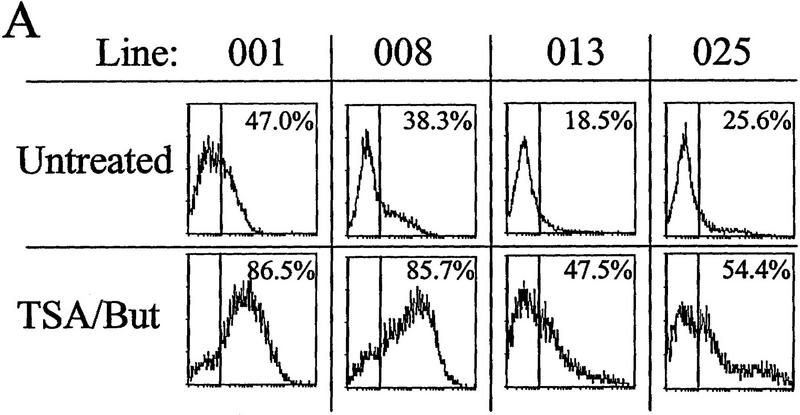
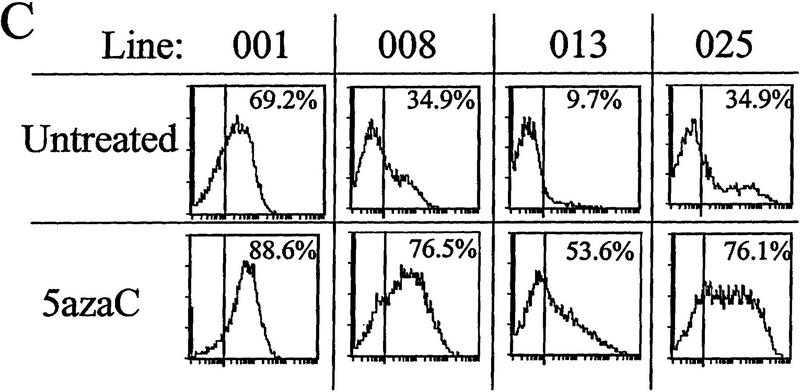
The effects of inhibitors of histone deacetylation and DNA methylation on transcription. (A) Histone deacetylase inhibitors reverse extinction of expression. Cells were sorted with magnetic beads, and the inactive fractions were replated and grown in either untreated medium for 3 days (top), or in medium plus 5 ng/ml TSA and 50 mm Na butyrate for 24 hr followed by washing and 2 days of recovery in untreated medium (bottom). On the third day, cells were analyzed by FACS for IL2R expression. The vertical line in each FACS panel marks the cutoff observed between inactive and active cells, and the number in the upper right corner of each indicates the percentage of total cells that fluoresce at intensities higher than that cutoff. (B) Inactive cells were treated with butyrate or TSA separately (same conditions as in A). (C) Inactive cells grown for 2 days untreated or in the presence of 50 μm 5-azaC, then assayed for IL2R by FACS. Percentages of cells in the active fluorescent range are denoted as in A.
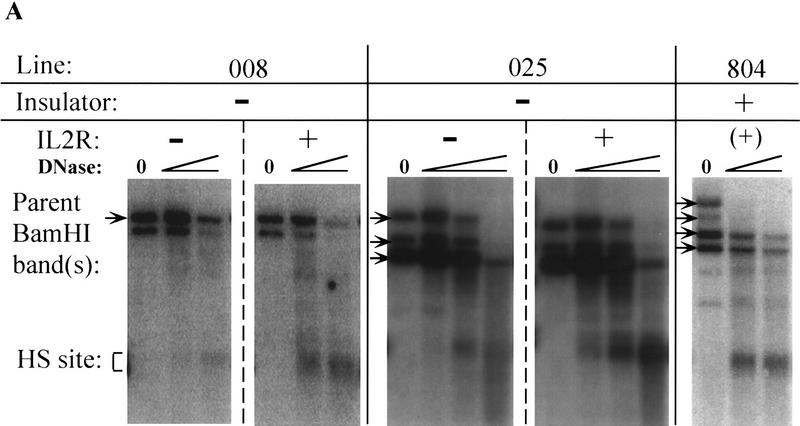
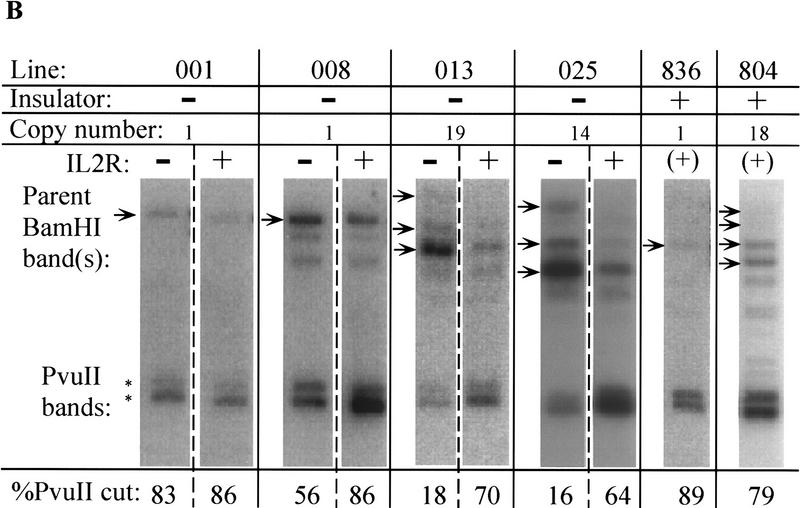
Recent studies have found differences in chromatin condensation between active and inactive cells taken from transgenic mice exhibiting variegation of expression (Festenstein et al. 1996; Garrick et al. 1996). To examine the chromatin structure around the enhancer in our expressing and nonexpressing cells, the uninsulated lines displaying biphasic inactivation were sorted by MACS at about the midpoint of inactivation, ∼50 days following removal of selection. Nuclei were prepared from the active and inactive IL2R fractions and digested with either DNase I or the restriction enzyme PvuII. In both a single-copy (008) and a multicopy (025) uninsulated line, DNase I hypersensitivity was much stronger in the IL2R active (+) cell populations compared with inactive (−), and hypersensitivity was also strong in a multicopy insulated line (804; this line was not sorted prior to preparation of nuclei and digestion because all cells are active) (Fig. 3A). For a more quantitative comparison, nuclei were also digested with the restriction enzyme PvuII. It has been shown that the accessibility of a PvuII site within the enhancer is reduced in integrated constructs carrying a mutated enhancer (Boyes and Felsenfeld 1996). In the IL2R-active fractions of sorted uninsulated lines as well as unsorted, completely active insulated lines, PvuII was accessible at between 64% and 89% of sites. PvuII cutting was, in general, decreased in the inactive uninsulated cell fractions, albeit to a varying degree depending on the line (Fig. 3B). However, the enhancer was accessible in cells of the active fractions, showing that an open chromatin conformation at the enhancer is correlated with expression.
Figure 3.
Nuclease accessibility in IL2R active and inactive cells. (A) A single-copy (008) and a multiple-copy (025) cell line at ∼50 days following release from hygromycin selection were sorted by MACS into IL2R inactive (−) and active (+) fractions. Nuclei from the sorted cells were digested with increasing concentrations of DNase I, as were nuclei from line 804, which carries multiple copies of the insulated construct and is fully active. DNA samples were analyzed by digesting with BamHI, electrophoresis and Southern blotting, and probing by hybridization to a radiolabeled DNA corresponding to the coding sequence of the IL2R cDNA (BamHI–XbaI fragment—Fig. 1). The arrays of parent BamHI bands in lines 025 and 804 are indicative of multiple integrants. Line 008 has one BamHI parent band at single-copy intensity; the second band below it hybridizes to probe at less than single-copy intensity, and on the basis of the sizes of several restriction digestion products, appears to be a separately integrated fragment of the IL2R construct that has lost the promoter and 3′ portion of the gene. (B) Single- and multiple-copy uninsulated lines were sorted by MACS as in A and nuclei were digested with PvuII, as were nuclei from unsorted, active single and multiple-copy insulated lines. PvuII produces a doublet of digested bands (asterisks) as a result of the two adjacent sites, one inside the enhancer fragment and one just outside it. Percent digestion was calculated by measuring band intensities on a PhophorImager and taking the ratio of the sum of the digested band doublet to the total of the parent (arrow) and digested bands. As a control for the extent of nuclease digestion, blots were stripped of labeled IL2R probe and reprobed with sequence specific to the chicken βA gene-coding region (not in the IL2R reporter plasmids) such that cutting was detected at the endogenous β/ε enhancer, whose accessibility to DNase I and PvuII was equivalent for all lines examined (data not shown). For most cell lines, sorting and nuclear isolation followed by DNase I or PvuII digestion was repeated two or three times. When repeated, the extent of digestion by the two nucleases was always reproducible.
Effects of inhibition of DNA methylation and histone deacetylation
The DNA of animal cells is subject to chemical modification by methylation at the 5 carbon position of cytidine bases at CpG dinucleotides. Methylation is not genomically uniform, as unmethylated CpGs are found preferentially in transcriptionally active chromatin (Naveh-Many and Cedar 1981). Conversely, hypermethylation is associated with transcriptional repression (for review, see Holliday 1987), for example, in X chromosome inactivation (Riggs 1984), during globin expression in red blood cell development, (McGhee and Ginder 1997; Ginder and McGhee 1981), and in the heritable repression of nonessential genes in cultured cells (Antequera et al. 1990). Moreover, there is increasing evidence of the importance of histone acetylation in the regulation of expression from chromatin templates (Wolffe 1996; Chen 1997; Pazin and Kadonaga 1997). To examine the roles of DNA methylation and histone acetylation in the uninsulated transgene activity, we again sorted biphasic cultures by MACS, and replated only the IL2R-inactive, unbound cells and tested the effects of inhibitors of deacetylase and DNA methylation. For the histone deacetylase inhibition experiment, the IL2R-negative cells were grown in either untreated medium for three days or in medium plus the histone deacetylase inhibitors trichostatin A (TSA; 5 ng/ml) and sodium butyrate (50 mm) for 1 day, followed by 2 days of recovery, and then analyzed by FACS. We observed that treatment with TSA/butyrate produced a significant reactivation of IL2R expression in all lines (Fig. 4A). This confirms the observation of Townes and coworkers (Chen et al. 1997) in other cell lines that extinction of expression can be reversed by use of histone deacetylase inhibitors. Treatment of our cultures with butyrate or TSA showed separately that each chemical stimulated equivalent reactivation, with no further activation when the two were combined (Fig. 4B). The deacetylase inhibitors did not stimulate transcription of these reporter constructs following transient transfection, nor did they provide further stimulation of IL2R expression in already active cells prior to long-term growth and transcriptional silencing (data not shown). Therefore, the ability of the inactivated IL2R transgene to reactivate preferentially in the presence of TSA or butyrate suggested histone deacetylation as a causal factor in the inactivation process.
Alternatively, the IL2R-negative cells were also replated following sorting into medium with or without the drug 5-azacytidine (5-azaC), a cytidine analog that inhibits methylation when incorporated into DNA. Following treatment with 5-azaC for 48 hr prior to FACS analysis, all four lines became active with respect to IL2R expression (Fig. 4C). As with TSA and butyrate, the degree of reactivation by 5-azaC varied among the four lines and, in fact, reactivation under the two conditions was quite similar for each line.
Chromatin immunoprecipitation before and after long-term growth in culture
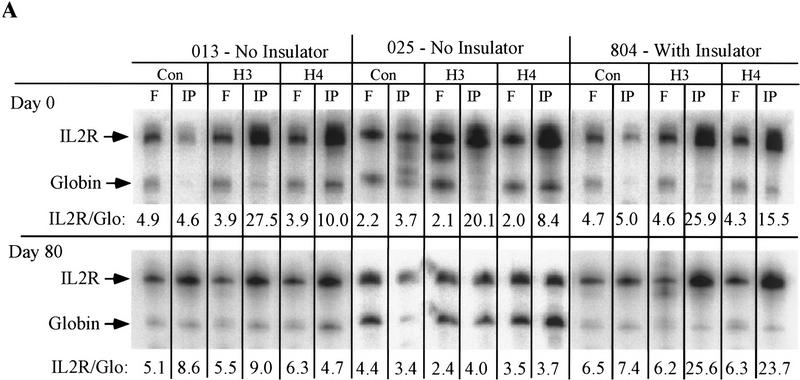
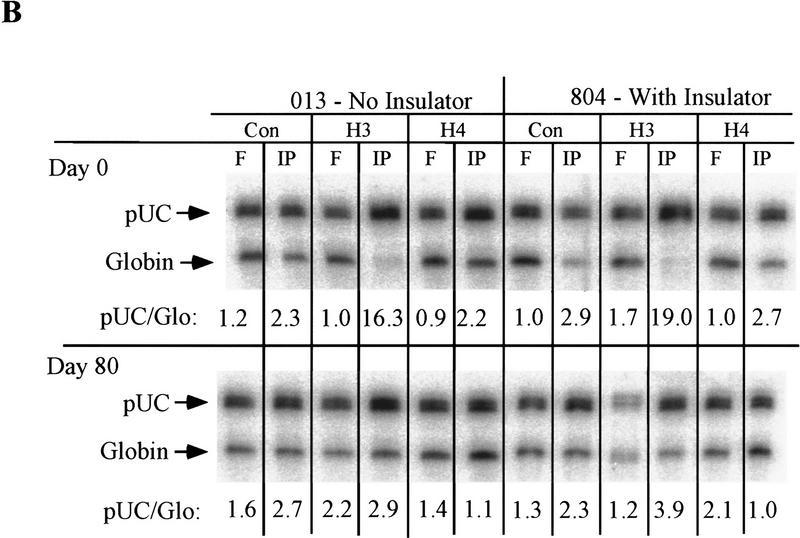
To confirm that inactivation was associated with changes in histone acetylation, we used antibodies against acetylated amino-terminal tails of histones H3 or H4 to immunoprecipitate chromatin (Hebbes et al. 1988) from uninsulated (013 and 025) and insulated (804) lines before and after removal from hygromycin selection. We used PCR to compare the amounts of IL2R and globin DNA (as a control) in the immunoprecipitated and free material. The IL2R sequences were over-represented in the DNA immunoprecipitated from these three lines grown in hygromycin, when IL2R transcription was fully active in all lines. However, after long-term nonselective growth, the preferential immunoprecipitation of IL2R DNA was seen in the insulated line but not the inactive, uninsulated line (Fig. 5A). Thus, histone acetylation, initially established in both lines, was lost in the uninsulated lines 013 and 025 during transcriptional inactivation but not in the insulated, continuously active line 804. As an additional control, PCR was also performed on these immunoprecipitated samples by use of primers directed to vector backbone sequences. This sequence is of pUC19 origin, present in both the uninsulated and insulated constructs, lying adjacent to and outside of the insulators with respect to the promoter/cDNA/enhancer cassette in the insulated construct. As shown (Fig. 5B), histones associated with these plasmid sequences adjacent to both the uninsulated and insulated transgenes are detectably hyperacetylated early in culture, in particular, more so at H3 than H4, but revert to acetylation levels indistinguishable from bulk chromatin after the 80-day period in culture. This result further establishes our conclusion that the insulator sequences cause neither the initial integration of the transgene into a more active chromatin site than would be available to the uninsulated transgene nor a dominant, bidirectional establishment of an active domain following integration.
Figure 5.
Histone acetylation assay on the uninsulated lines 013 and 025 and insulated line 804. Chromatin was immunoprecipitated with either no primary antibody (Con) as a control or with monoclonal antibody directed against acetylated histone H3 or H4 tails. (A) DNA from immunoprecipitated (IP) or free (F) histone complexes was subjected to PCR with primers specific to IL2R and globin sequences. The abundance of IL2R DNA detected in the fractions relative to the globin internal control was determined for three separate PCR reactions and given as an average below each lane, with standard deviations of 20% or less for each. IL2R to globin ratios between 4 and 6 are characteristic of genomic DNA; ratios substantially higher than this indicate relative hyperacetylation of histones bound to the IL2R sequence. (B) To examine the acetylation level of histones bound to transfected DNA outside the insulator sequences, the same immunoprecipitated DNA samples were amplified with a pair of primers directed to vector sequences and the globin control primers. All lines were assayed before release of hygromycin selection (top), when 013, 025, and 804 are all fully active, and after 80 days of nonselective growth (bottom), after which lines 013, 025 were nearly completely inactive while line 804 retained activity (see Fig. 2B).
Promoter methylation in active and inactive cells
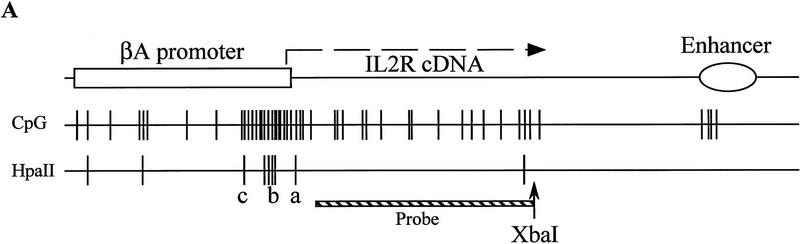
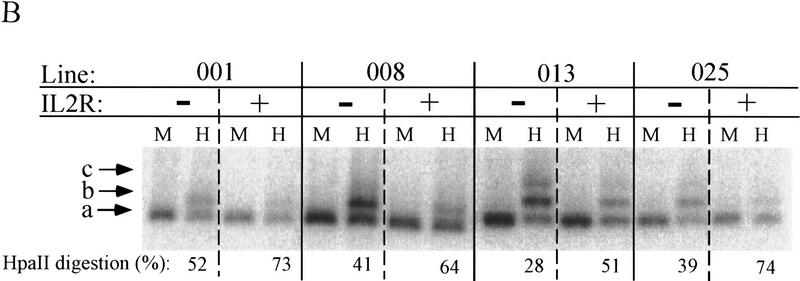
Because the βA-globin promoter contains a high density of potentially methylatable CpG dinucleotides clustered around the start site of transcription (Fig. 6A), we have made use of the ability of the restriction endonuclease HpaII to distinguish internally methylated from unmethylated CCGG sequences to assay for methylation differences over the transgene promoter in active and inactive cells. As in the nuclease accessibility studies above, cells of lines 001, 008, 013, and 025 were sorted by magnetic beads at between 40 and 50 days of nonselective growth. DNA was purified from these cells and digested with XbaI and MspI or HpaII. Cutting at the CCGG site marked “a” produces the smallest detectable band; appearance of the next higher band (actually an unresolved array of fragments generated by cutting at one or more of the cluster of four CCGG sites at “b”) is evidence of methylation at the a site. Similarly, the third band arises when cutting at the a and all four b sites is blocked by methylation. The overall extents of methylation varied somewhat among the lines studied, but within each line blocking of HpaII cutting by methylation at the a and b sites was greater in the IL2R-inactive fractions than in the active cells (Fig. 6B).
Figure 6.
Restriction digestion analysis to determine the extent of DNA methylation over the transgene promoter. (A) Under a map of the reporter construct are illustrated the incidence of individual CpG dinucleotides and CCGG MspI–HpaII sites occuring in the sequence of the βA promoter, IL2R cDNA, and β/ε enhancer, showing the relatively CG-rich character of the promoter region. Indirect end labeling by digestion with XbaI and probing with IL2R cDNA detects a 867-bp fragment indicative of cutting at the site marked a. Cutting at any of a closely spaced quartet of sites at b produces indistinguishable indirect end-labeled bands of 961–1032 bp, and c gives a 1145-bp fragment. Very faint bands of higher molecular weight were only rarely seen. Digests were carried out overnight on 5 μg of DNA sample with 100 units of MspI or HpaII. Digestion by MspI always went to completion. The extent of HpaII digestion at the a site was quantitated for each HpaII lane as the percentage of the intensity of the a band relative to the sum of the a, b, and c bands in the lane by PhosphoImager analysis. (B) DNA from sorted cells carrying uninsulated IL2R transgenes was digested with XbaI and MspI (M) or HpaII (H). These cells had been passaged 40–50 days in culture and dispalyed biphasic populations of IL2R activity in FACS analyses (see Fig. 2). (C) The same digests were performed on insulated IL2R lines following continuous passage in culture for >80 days, at which point all were still completely positive for IL2R activity.
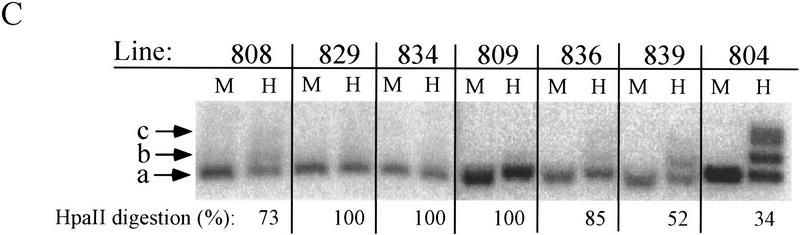
Given this correlation between methylation and transcriptional activity, we examined whether the promoters of the insulator-flanked genes were maintained in an undermethylated state. We isolated DNA from the insulated lines analyzed by FACS (see Fig. 2) that had been grown in culture for 80–100 days. These cells were not sorted by magnetic beads as all were completely active. Again, the promoter methylation state was assessed by digestion with XbaI and HpaII, or MspI. Although digestion was complete in three lines (829, 834, and 809), variable digestion was observed among four others (Fig. 6C). The a and b sites in line 804 were noticeably more methylated than in even inactive fractions of the uninsulated lines in Figure 5B. This was despite the fact that the 804 line retained its initial, copy-number-dependent level of IL2R activity as judged by FACS (remaining greater than one log, or 10-fold, brighter than the single-copy integrant lines; Fig. 2A), remained nuclease accessible at nearly all integrated gene copies (Fig. 3), and continued to be associated with acetylated histone H3 and H4 (Fig. 5A). Therefore, although we observe an overall tendency toward a reduced level of methylation in the insulated lines as in the active fractions of the uninsulated lines, there is no stringent requirement for a methyl-free promoter in the continuously active, insulator-flanked genes.
Discussion
The nuclear environment imposes position effects on gene expression when a gene finds itself, either through rearrangement or insertion as a transgene, close to a strong endogenous enhancer or a heterochromatic region. A number of DNA sequences have been identified as insulators against these position effects (Kellum and Schedl 1991; Roseman et al. 1993; Hagstrom et al. 1996). Among these sequences, the chicken β-globin insulator, at the 5′ end of the chicken β-globin locus, blocks an enhancer from activating a promoter when it lies between them, but lacks enhancer or promoter activity of its own (Chung et al. 1993, 1997). Here we show this element’s insulator activity in cell lines in two ways. First, transformed cell lines display enhancer-dependent expression levels when the IL2R reporter gene, driven by the chicken βA-globin promoter and wild-type or mutant β/ε enhancers, is surrounded by insulator elements. Furthermore, in the case of the wild-type enhancer and in the absence of insulators, long-term propagation of the transformed cells in culture without hygromycin selection results in gradual extinction of expression. This repression is reversible by inhibitors of histone deacetylase and of cytosine methylation and can be prevented entirely by flanking the reporter gene with insulators.
The biphasic decay of expression resembled that observed by others following removal of an enhancer stimulus from a transgene reporter (Walters et al. 1996), with the difference that the reporter system used here—a different transgene, enhancer, promoter, and cell line—was apparently more subject to silencing, because silencing occurred even in the enhancer’s presence. In this regard, the loss of expression observed here is consistent with what has been seen elsewhere with virally transduced reporter constructs (Chen et al. 1997). In light of these differences in behavior, it appears likely that the extent to which repression occurs can vary with the choice of reporter and cell type.
As in earlier studies (Walters et al. 1995; Chung et al. 1997), the insulator did not provide direct transcriptional stimulation per se. The initial activity of the wild-type enhancer integrants was the same with and without the insulators, and the mutant-enhancer integrants were on the whole less active with the insulator than without. Thus the insulator’s transcriptionally neutral role may be contrasted with the direct establishment of a transcriptionally active chromatin domain by locus control regions and other strong regulatory elements (Festenstein et al. 1996; Martin et al. 1996; Jenuwein et al. 1997). In contrast to Drosophila insulators such as scs′ or gypsy, whose cognate binding proteins BEAF (Zhao et al. 1995) or the product of su(Hw) (Roseman et al. 1993) have been identified, we know little about trans-acting proteins operating at the chicken globin insulator. Whatever these factors are, however, they appear to be less specialized than their Drosophila counterparts, as the globin insulator provides universal activity in all species and cell types tested thus far. For this reason, the chicken globin insulator has been used in the establishment of transgenic mouse strains that produce exogenous protein with the desired tissue specificity and robust expression. These include the introduction of a chimeric human progesterone receptor/yeast GAL4 transcription factor into murine liver (Wang et al. 1997) and the secretion of proteins in the milk of lactating female mice (H. Meade, pers. comm.). On the basis of the results reported here, it seems likely that the insulator will also be useful in assuring long term, predictable expression of stably integrated genes in cell culture.
Possible mechanisms of insulator action
We were interested in determining the role of chromatin structure in the extinction of expression observed when cell lines were cultured for 40–100 days in the absence of hygromycin selection. Confirming earlier work (Chen et al. 1997), we showed that brief incubation of silenced cells with inhibitors of histone deacetylation resulted in considerable reactivation. Because TSA and butyrate have been shown to affect the acetylation of certain nonhistone regulatory proteins (Cuisset et al. 1997; Imhof et al. 1997) the possibility should not be discounted that reactivation reflects the altered acetylation of one of these proteins rather than of the histones. However, we have shown that lower acetylation levels of the histones covering the IL2R reporter gene are indeed correlated with extinction of expression. Furthermore, we find that the histone deacetylase inhibitors have a stimulatory effect on this gene only in the context of the chromatin-mediated repression observed with long-term growth in culture, but not on transiently transfected templates carrying the same promoter and enhancer. We also note that these templates are active when transfected into cell populations in which expression from the stably integrated reporter has been extinguished, suggesting that extinction is not the result of inactivation of transcription factors.
A second clue to the mechanisms of inactivation and insulator action comes from the extinction experiments carried out under nonselective conditions, examining tandemly integrated genes—either a single copy of IL2R next to the hygromycin gene, or multiple copies of IL2R next to each other. In the absence of insulators, loss of IL2R activity in the IL2R/hygromycin pair was correlated with loss of hygromycin resistance: Cells expressing IL2R were also resistant. When the corresponding experiment was done with cells carrying IL2R surrounded by insulators (and the adjacent hygromycin gene external to the insulators), all cells expressed IL2R but were much more sensitive to hygromycin, showing that the expression of the two genes had been decoupled. In the case of the tandemly integrated IL2R genes, extinction of expression in each cell occurred in an all-or-none fashion, that is, there were two separate cell populations, one expressing many copies of the gene and the other none. Such behavior is not consistent with independent inactivation of individual gene copies in the cluster. Both kinds of experiments with tandemly integrated genes support a model in which there is coordinated activity or repression among adjacent sequences. In the case of the IL2R/hygromycin pair, the insulator blocks this coupling.
We suggest that IL2R repression in the absence of insulators takes place as a result of the coordinated imposition of a repressive chromatin structure over the transgene. The loss of IL2R expression is accompanied by three hallmarks of transcriptionally inactive chromatin: DNA hypermethylation, histone deacetylation, and a nuclease-insensitive chromatin structure. Recent studies of MeCP2, a murine protein that binds DNA at single methylated CpG pairs, show that it interacts indirectly with histone deacetylases (Jones et al. 1998; Nan et al. 1998). Although repression by histone deacetylation proceeds in other systems independently of DNA methylation (as recently observed, for example, via Rb/E2F complexes (Brehm et al. 1998)), the demonstration of a physical interaction between MeCP2 and histone deacetylase supports mechanisms of methylation-dependent repression via a progression of events beginning with DNA methylation leading to binding of methylated DNA specific proteins, local deacetylation of histones, and thus to gene inactivation (Kass et al. 1997). On the basis of this model, our data suggest that inactivation of the uninsulated IL2R reporter reflects a gradual increase in its methylation during repeated rounds of replication. At some point in this process, a threshold is reached allowing sufficient occupancy either of methyl-CpG-specific binding proteins or of histone deacetylase complexes, to result in coordinated deacetylation of the region. This would produce the bimodal expression patterns evident in the FACS analyses.
In contrast, flanking the IL2R reporter with insulators prevents transcriptional inactivation, allowing the IL2R gene to maintain a transcriptionally permissive chromatin environment. The insulator does not, however, protect against widespread promoter methylation in all cell lines, because the CpGs detected in the MspI–HpaII assay are subject to methylation at the a site in line 839 and the a as well as the four b sites in line 804 after long-term culture. We note that the MspI–HpaII assay detects only those CpG dinucleotides in CCGG sites and may overlook other CpGs, perhaps more critical sites whose methylation the insulator could conceivably serve to prevent in particular. However, given that the globin insulator also prevents position effect variegation (PEV) in Drosophila, where DNA methylation is not operant, its seems likely that the insulator protects against transcriptional silencing and histone deacetylation more directly. We suggest that the insulator functions as a barrier to a step in the inactivation process downstream of CpG methylation, such as the formation of methyl-C-binding protein/histone deacetylase complexes (Jones et al. 1998; Nan et al. 1998). Alternatively, the insulators could exclude recruitment of histone deacetylases to the protected domain, or they could sequester histone acetylases within it in sufficient amounts to overcome deacetylation reactions (Fig. 7). In any case, just as deacetylation can occur independently of DNA methylation, our results show that it is also possible when the insulators are present to have methylation without deacetylation or gene inactivation.
Figure 7.
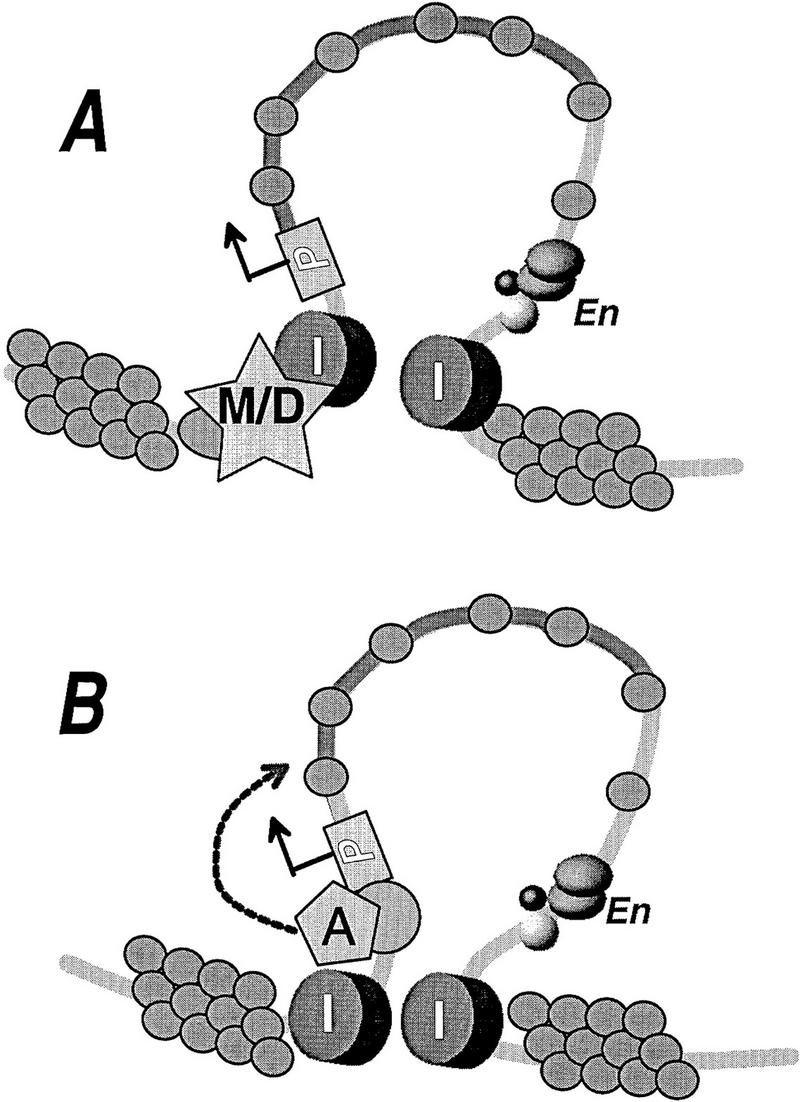
Two possible models of insulator action in protecting the reporter gene against formation of heterochromatin. In the first model (A), the insulator elements exclude methyl-CpG-binding proteins, histone deacetylases, or any other component required for formation of the silenced domain. Alternatively (B), the insulator elements are modeled as boundaries within which histone acetylase recruitment is preserved directly, thus favoring an acetylated, active domain over invading condensed chromatin. (I) Insulators; (P) promoter; (en) enhancer; (M/D) meCpG-binding protein/histone deacetylase complex; (A) histone acetylase.
The properties of the insulator described here are consistent with the role that Crane-Robinson and coworkers have proposed for it in the chicken β-globin locus (Hebbes et al. 1994). The element we have used is located at the 5′ end of the locus, where it appears to mark the boundary of the active chromatin domain. This locus has been carefully mapped in erythroid cells both by DNase I sensitivity and histone acetylation measurements (Hebbes et al. 1994). In that study, the boundary between the DNase-sensitive, hyperacetylated chromatin of the active β-globin locus and the insensitive, normally acetylated chromatin upstream was shown to occur just 5′ of the insulator region. We have now shown that the insulator can preserve the state of histone hyperacetylation over an exogenous gene, and we suggest that this may be part of its function in helping to maintain a boundary at its site in the chicken β-globin locus.
Materials and methods
The IL2R reporter vectors were made from analogous CAT reporter vectors under the control of the chicken βA-globin promoter and β/ε enhancer, described previously as the 120-bp EP fragment, along with a 4-bp transversion mutant to the NF-E2 site or a pair of such mutations to the two GATA sites [mutants 10 and 20/24, respectively (Reitman and Felsenfeld 1988)]. The CAT coding sequence was replaced with that of the Tac subunit of the human IL2R gene from pCMV–IL2R (Giordano et al. 1991). For the insulator-flanked analogous vectors, the βA promoter-IL2R gene–β/ε enhancer cassette was placed between the two paired copies of the 1.2-kb chicken globin insulators in pJC13-1 (Chung et al. 1993) from which the γ-neo and mouse LCR HS2 sequences had been removed (Fig. 1). Plasmids and cell line series are named as follows, all without and with flanking insulators, respectively: 000/100 and 800, wild-type enhancer; 200 and 600, NF-E2 mutation; 400 and 700, double GATA mutation. 6C2 cells were grown and transfected by electroporation of 1 μg of hygromycin resistance gene from pREP7 (Invitrogen) and 1 μg of the IL2R construct as described (Boyes and Felsenfeld 1996). Single-copy integrant clones were identified as such by digesting with enzymes that cut only once within the IL2R construct, probing Southern blots with IL2R sequence, and confirming the appearance of a single band. Copy number for multicopy lines was measured by comparing band intensity. The presence of the hygromycin resistance marker fragment adjacent to the IL2R reporter was tested by digesting with BamHI or XbaI (giving the parent IL2R band) and with rarely cutting restriction enzymes that cut in the hygro but not the IL2R fragment and probing with the IL2R BamHI–XbaI DNA. For most single-copy lines tested including 001, 008, and 836, this digestion detected the hygromycin fragment next to the IL2R reporter, in the transcriptionally downstream direction (data not shown).
For FACS analysis, harvested cells were washed in Hank’s balanced salt solution plus 0.1% bovine serum albumin and 0.1% sodium azide (HBSS) and reacted with 1 μg of mouse anti-IL2R monoclonal antibody (Upstate Biotechnology) for 30 min on ice, washed twice with HBSS, and reacted with 750 ng of FITC-conjugated goat anti-mouse IgG (Boeringer Mannheim) for 30 min on ice and washed. Labeled cells were analyzed by FAST systems (Gaithersburg, MD). For subsequent structural studies, separation was carried out by MACS, with the same anti-IL2R antibody as used for FACS bound to ferromagnetic bead-conjugated anti-mouse antibody (Dynal).
Nuclei for chromatin structural studies were prepared by lysing the cells in 10 mm Tris, 10 mm NaCl, 3 mm MgCl2, and 0.4% NP-40. Nuclei were pelleted, resuspended in the same buffer, and digested with either increasing concentrations of DNase I or with PvuII. Restriction enzyme digests were performed with 2 units of enzyme per 100 μl of nuclei for 30 min at 37°C. This time point was found to allow complete digestion of accessible sites, with digestion for longer times producing no further cutting (Boyes and Felsenfeld 1996). DNA was purified from these digestions, digested with BamHI, and probed by Southern blotting with a fragment of the IL2R cDNA.
Immunoprecipitation was carried out as described (Hebbes et al. 1988), with the exceptions that they were performed with one-tenth less antibody to nuclear DNA and immunobound material was precipitated with recombinant protein G–agarose beads. Monoclonal antibodies against acetylated Tetrahymena H3 and H4 tail peptides (Lin et al. 1989) were obtained from Upstate Biotechnology. Precipitation of ∼0.5% total DNA was routinely obtained under these conditions. We note that this is less fractional immunoprecipitation than seen in other studies (O’Neill and Turner 1995; A.L. Clayton, pers. comm.). As a result, the unbound chromatin fractions should not be considered to be completely depleted of hyperacetylated histone chromatin but are essentially bulk cellular chromatin, from which samplings, but not all, of hyperacetylated histone-bound DNA have been removed by immunoprecipitation. PCR was carried out (Pikaart et al. 1992) for 25 cycles by use of primer pairs that amplify a 64-bp product from within the IL2R cDNA or a 60-bp product from plasmid vector sequences, and a 53-bp product from the endogenous βA-globin first intron. These conditions give linearity with respect to input DNA. One primer of each pair was 5′ end labeled with 32P and detected by electrophoresis on TBE–urea gels and exposed to a PhosphorImager.
Acknowledgments
We thank Dr. Catherine Smith for guidance in the use of the IL2R marker and Dr. Alison Clayton for advice on histone immunoprecipitation. We thank Drs. David Clark, Michael Grunstein, Robert Martin, and Marc Reitman for thoughtful review of the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL GXF@vger.niddk.nih.gov; FAX (301) 496-0201.
References
- Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–604. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Chen WY, Bailey EC, McCune SL, Dong J-Y, Townes TM. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Chung JH, Bell AC, Felsenfeld G. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci. 1997;94:575–580. doi: 10.1073/pnas.94.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuisset L, Tichonicky L, Jaffray P, Delpech M. The effects of sodium butyrate on transcription are mediated through activation of a protein phosphatase. J Biol Chem. 1997;272:24148–24153. doi: 10.1074/jbc.272.39.24148. [DOI] [PubMed] [Google Scholar]
- Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Milou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- Fordis CM, Helmly B, Novotny E, Holter W, Barker JL, Howard BH. Transient analysis for antiproliferative gene activity. Proc Natl Acad Sci. 1990;87:1169–1173. doi: 10.1073/pnas.87.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Sutherland H, Robertson G, Whitelaw E. Variegated expression of a globin transgene correlates with chromatin accessibility but not methylation status. Nucleic Acids Res. 1996;24:4902–4909. doi: 10.1093/nar/24.24.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes & Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Ginder GD, McGhee JD. DNA methylation in the chicken adult β-globin gene: A relationship with gene expression. In: Stamatoyannopoulos G, Nienhuis AW, editors. Organization and expression of globin genes. New York, NY: Alan R. Liss; 1981. pp. 191–201. [Google Scholar]
- Giordano T, Howard T, Coleman J, Sakamoto K, Howard BH. Isolation of a population of transiently transfected quiescent and senescent cells by magnetic affinity cell sorting. Exp Cell Res. 1991;192:193–197. doi: 10.1016/0014-4827(91)90175-t. [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes & Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:686–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Forrester WC, Fernandez-Herrero LA, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–190. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kass SU, Pruss D, Wolffe AP. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Depper JM, Crabtree GR, Rudikoff S, Pumphrey J, Robb RJ, Kronke M, Svetlik PB, Peffer NJ, Waldmann TA, et al. Molecular cloning and expression of cDNAs for the human interleukin-2 receptor. Nature. 1984;311:626–631. doi: 10.1038/311626a0. [DOI] [PubMed] [Google Scholar]
- Lin R, Leone JW, Cook RG, Allis CD. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DIK, Fiering S, Groudine M. Regulation of β-globin gene expression: Straightening out the locus. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Ginder GD. Specific DNA methylation sites in the vicinity of the chicken β-globin genes. Nature. 1979;280:419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T, Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci. 1981;78:4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Pikaart M, Feng J, Villeponteau B. The polyoma enhancer activates chromatin accessibility on integration into the HPRT gene. Mol Cell Biol. 1992;12:5785–5792. doi: 10.1128/mcb.12.12.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Felsenfeld G. Mutational analysis of the chicken beta-globin enhancer reveals two positive-acting domains. Proc Natl Acad Sci. 1988;85:6267–6271. doi: 10.1073/pnas.85.17.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, DNA methylation, and differentiation revisited. In: Razin A, Cedar H, Riggs AD, editors. DNA methylation. New York, NY: Springer-Verlag; 1984. pp. 269–278. [Google Scholar]
- Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DIK. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters MC, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin DIK. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes & Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- Wang Y, DeMayo FJ, Tsai SY, O’Malley BW. Ligand-inducible and liver-specific target gene expression in transgenic mice. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Histone deacetylase: A regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- Zhao, K., C.M. Hart, and U. Laemmli. 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 879–889. [DOI] [PubMed]
- Zhong XP, Krangel MS. An enhacer-blocking element between α and δ gene segments within the human T cell receptor α/δ locus. Proc Natl Acad Sci. 1997;94:5219–5224. doi: 10.1073/pnas.94.10.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]