Abstract
Chromosomal inversions disrupt recombination in heterozygotes by both reducing crossing-over within inverted regions and increasing it elsewhere in the genome. The reduction of recombination in inverted regions facilitates the maintenance of hybridizing species, as outlined by various models of chromosomal speciation. We present a comprehensive comparison of the effects of inversions on recombination rates and on nucleotide divergence. Within an inversion differentiating Drosophila pseudoobscura and Drosophila persimilis, we detected one double recombinant among 9,739 progeny from F1 hybrids screened, consistent with published double-crossover frequencies observed within species. Despite similar rates of exchange within and between species, we found no sequence-based evidence of ongoing gene exchange between species within this inversion, but significant exchange was inferred within species. We also observed greater differentiation at regions near inversion breakpoints between species versus within species. Moreover, we observed strong “interchromosomal effect” (higher recombination in inversion heterozygotes between species) with up to 9-fold higher recombination rates along collinear segments of chromosome two in hybrids. Further, we observed that regions most susceptible to changes in recombination rates corresponded to regions with lower recombination rates in homokaryotypes. Finally, we showed that interspecies nucleotide divergence is lower in regions with greater increases in recombination rate, potentially resulting from greater interspecies exchange. Overall, we have identified several similarities and differences between inversions segregating within versus between species in their effects on recombination and divergence. We conclude that these differences are most likely due to lower frequency of heterokaryotypes and to fitness consequences from the accumulation of various incompatibilities between species. Additionally, we have identified possible effects of inversions on interspecies gene exchange that had not been considered previously.
Keywords: interchromosomal effect, recombination, inversions, Drosophila pseudoobscura, chromosomal speciation
Introduction
In the study of speciation, a crucial question is how species persist despite gene flow. One proposed solution is that chromosomal inversions partition the genome into regions protected from gene flow by reducing recombination over long stretches (Noor et al. 2001; Rieseberg 2001). Although reduced recombination in inversion heterozygotes has been documented repeatedly within species (Roberts 1976; Ishii and Charlesworth 1977), few studies estimate recombination rates in interspecies hybrids heterozygous for inversions. Further, by focusing on the importance of inversions in the reduction of recombination within inverted regions, recent work has largely overlooked the major global increases in recombination rate in inversion heterozygotes (but see Portin and Rantanen 1990), known as the “interchromosomal effect” (Schultz and Redfield 1951). Rather than determining the effects of the interchromosomal effect on global nucleotide variability, emphasis has instead been placed on identifying the mechanism of this process (Joyce and McKim 2010).
We examine here the extent to which inversions differentiating species affect recombination rates 1) within inverted regions, 2) at inversion boundaries, and 3) throughout the remainder of the genome. We focus our research on hybrids between Drosophila pseudoobscura and Drosophila persimilis, a classical model system for studying chromosomal inversions, hybridization, and speciation (Dobzhansky 1937). Drosophila pseudoobscura is found across Western North America extending north to Canada, east to Central Texas, and south to Central America. Drosophila persimilis maintains a smaller range restricted mainly to the western US and is contained within the range of D. pseudoobscura (Dobzhansky and Sturtevant 1938; Dobzhansky and Epling 1944). These species are morphologically identical; however, male hybrids are sterile (Dobzhansky 1936). They diverged approximately 0.5–0.85 Ma (Aquadro et al. 1991; Hey and Nielsen 2004; Leman et al. 2005); however, parts of the genome carry a signature of more recent hybridization (Machado et al. 2002; Hey and Nielsen 2004; Machado et al. 2007; Kulathinal et al. 2009). Despite this ongoing gene flow, these taxa were diagnosed as different species with three major paracentric inversions largely distinguishing them —two on the X chromosome and one on the second chromosome (Dobzhansky and Epling 1944). The D. pseudoobscura arrangement of the right arm of the X (hereafter XR) is also present in populations of D. persimilis exhibiting meiotic drive with skewed sex ratios, referred to as Sex Ratio (SR) D. persimilis (Sturtevant and Dobzhansky 1936). Additionally, both species segregate multiple arrangements among individuals on chromosome 3 (Dobzhansky and Sturtevant 1938; Powell 1992).
To examine how inversions affects recombination rates within inverted regions, we focused on the largest (XR) inversion because we expect higher potential for double-crossover events and gene flow between the two arrangements (Navarro et al. 1997). We evaluated the double-crossover rate in female hybrids between D. pseudoobscura and D. persimilis across this ∼12.5 Mb inversion through a direct assay. We further compared divergence of different karyotypes between-species and within-species for the XR-chromosome inversion using published genome sequences of D. pseudoobscura (Richards et al. 2005) and D. persimilis (Clark et al. 2007), and an assembled genome sequence we generated for D. persimilis possessing the SR arrangement.
We also examined the effects of inversion heterozygosity on recombination rates elsewhere in the genome. To this end, we quantified recombination at the boundaries of inversions by estimating the amount of recombination between markers outside, but near, the breakpoints of each inversion. These regions have higher rates of divergence, similar to markers inside inverted regions (Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009; McGaugh et al. forthcoming), suggesting that they experience less recombination than neighboring regions. Finally, we calculate the effect inversions have on enhancing recombination rates throughout the rest of the genome by comparing recombination rates in hybrids to a published recombination map of D. persimilis (Stevison and Noor 2010).
Expected Rates of Exchange within Inverted Regions Based on Within-Species Inversion Polymorphisms
The major prerequisite of modern chromosomal speciation models (for recent review, see Faria and Navarro 2010) is that crossover products are rarely or not recovered between inverted segments. In Drosophila, single crossovers between heterokaryotypes of a paracentric inversion result in nonviable gametic products, which are subsequently shunted to the polar bodies prior to oviposition (Sturtevant and Beadle 1936; Carson 1946; Coyne et al. 1993; Navarro et al. 1997). Therefore, any exchange between heterokaryotype regions is likely due to either gene conversion or double-crossover events. Due to the strong effect of interference acting over distances as long as 8–10 Mb in Drosophila (Weinstein 1918; Foss et al. 1993; Fitzpatrick et al. 2009; Stevison and Noor 2010), only inversions with large recombinational distances (>20 cM) are expected to achieve an observable rate of double-crossover (Navarro et al. 1997).
Empirical analysis of exchange in inversion heterozygotes has followed two approaches: controlled crosses to measure the rate of double-crossover and sequence analysis of targeted regions within an inversion to infer historical exchange. Empirical estimates between segregating inversions in Drosophila have observed an average of 10−4 double crossovers within a single generation using phenotypic mutants located near the center and ends of inversions spanning 30–80 cM (Levine 1956; Ishii and Charlesworth 1977). This double-crossover rate is lower than recent empirical studies using microsatellite markers within inversions spanning 7.7–11.7 Mb along the O chromosome in D. subobscura, which found one double-crossover among 391 individuals in one inversion heterokaryotype cross (∼10−3) and none in two other crosses of inversion heterokaryotypes (Pegueroles et al. 2010). In comparison, the double-crossover rate across similar regions in homokaryotypes is approximately 5–10% (Gruneberg 1935; Spurway and Philip 1952; Novitski and Braver 1954; Robbins 1974; Pegueroles et al. 2010), showing that double-recombination products are not produced as frequently as expected in inversion heterozygotes. Sequencing approaches have enhanced our understanding of differentiation within inverted regions by being able to detect double-crossover events or historical gene flux due to conversion within an inverted region (Rozas and Aguade 1994; Wesley and Eanes 1994; Hasson and Eanes 1996; Betran et al. 1997; Laayouni et al. 2003; Schaeffer and Anderson 2005; Nobrega et al. 2008; White et al. 2009). These studies confirm predictions by theoretical models for flux rates to be highest near the center of inverted regions (Navarro et al. 1997; Kirkpatrick and Barton 2006; Feder and Nosil 2009).
Despite our understanding of the rates of double-crossovers in inversions differentiating populations of Drosophila, we do not have a strong understanding of how inversions that differentiate species might differ in rates of exchange from inversions segregating within species. There are three reasons to expect exchange rates might differ in interspecies hybrids: 1) inversion heterozygotes segregating within populations are more abundant than those between species, which are dependent on the rate of premating isolation, 2) mechanistically, levels of sequence divergence influence rates of crossover, predicting lower levels of exchange when sequence divergence is higher between homologous chromosomes (Modrich and Lahue 1996), and 3) inversion heterozygotes between species will likely suffer some fitness consequences because of alleles conferring differential adaptation or incompatibilities within them, allowing selection to influence the detectable rate of historical exchange (Noor et al. 2001; Kirkpatrick and Barton 2006). Although this latter difference may not influence the rate observed in a single-generation cross, it may impact the observed exchange rate in a sequence-based analysis. For this reason, we used direct cross analysis to calculate double-crossover rate in a single generation despite a lack of evidence of ongoing exchange in a recent sequence-based test of migration within the inverted region of XR (Stevison 2011). Furthermore, by comparing both SR D. persimilis and D. pseudoobscura sequences (both carrying the ancestral XR arrangement) to the D. persimilis sequence, which carries the derived XR arrangement, we examine how exchange within the XR inversion varies in intraspecific versus interspecific comparisons.
Recombination Suppression of Inversions Extends beyond Breakpoints
Markers outside inverted regions, but near the breakpoints, tend to have heightened divergence, similar to markers found within inversions (Machado et al. 2007; Noor et al. 2007; Kulathinal et al. 2009). This effect is consistent with the observation of persistent recombination suppression in inversion heterozygotes outside the inversion, near boundary regions (hereafter inversion boundaries) (Dobzhansky and Epling 1948; Spurway and Philip 1952; Maynard Smith J and Maynard Smith S 1954; Ortiz-Barrientos et al. 2006; Kulathinal et al. 2009; Pegueroles et al. 2010). This pattern is not unique to Drosophila (White and Morley 1955; Roberts 1976; Stump et al. 2007; Strasburg et al. 2009), and similar effects of extended divergence at centromeric boundaries suggest that this pattern is not unique to inversion boundaries (Carneiro et al. 2010). Our systematic analysis of recombination rate at inversion boundaries aims to refine the previous estimates from Kulathinal et al. (2009) for the extent of recombination suppression at inversion boundaries in hybrids between D. pseudoobscura and D. persimilis.
Inversions as Global Recombination Modifiers
In addition to their proposed effects on species maintenance, chromosomal inversions increase recombination rates significantly throughout the rest of the genome (Sturtevant 1919; Schultz and Redfield 1951). Inversions alter crossover rates on the same chromosome (intrachromosomal effect) and on other chromosomes (interchromosomal effect). Due to multiple inversions in this system, we cannot tease apart intrachromosomal effect and interchromosomal effect in this study, so for simplicity, we refer to them collectively hereafter as ICE. Despite renewed interest in inversions and their importance in speciation (see recent reviews: Hoffmann and Rieseberg 2008; Brown and O'Neill 2010; Faria and Navarro 2010; Jackson 2011), advances in fine-scale recombination mapping technology, and the observation of extensive variation in recombination rate from humans to yeast (Stephan and Langley 1998; Gerton et al. 2000; Cirulli et al. 2007; Coop and Przeworski 2007; Kulathinal et al. 2008; Rockman and Kruglyak 2009; Stevison and Noor 2010), there has been relatively little research done on the role of ICE in contributing to nucleotide variation. Here, we map recombination rates on the second chromosome in single-generation hybrids (heterozygous for three inversions, see Introduction) and compare these measures with the fine-scale homokaryotype recombination map on the same chromosome recently published in D. persimilis (Stevison and Noor 2010).
Materials and Methods
Single-Generation Estimate of Inversion Crossover Rate in Hybrids
Genome lines of D. pseudoobscura, Mesa-Verde, CO 2-25 (MV2-25, San Diego stock number #14011-0121.94) and D. persimilis, Mount Saint Helena 3 (MSH3, San Diego stock number #14011-0111.49), were crossed to generate heterozygous F1 hybrid females. These females were backcrossed to MV2-25 males to generate approximately 10,000 progeny to screen for crossovers along the XR inversion.
Using microsatellite or indel markers at the breakpoints and center of the XR inversion (spanning 12.5 Mb), we tested for double crossovers in F1 hybrids between D. pseudoobscura and D. persimilis. The rate of gene conversion is often assumed in recombination models to be uniform across an inversion, whereas the rate of double-crossover is expected to be highest near the center, due to the role of crossover interference (Navarro et al. 1997; Andolfatto et al. 2001). Therefore, if two crossover events occur within the inversion loop created at meiosis, they are more likely to span the center of the inversion than segments closer to the inversion breakpoints (Navarro et al. 1997). Chromosome arm XR differs by a single inversion between D. pseudoobscura and D. persimilis, and the breakpoints of this inversion have been mapped (Noor et al. 2007; Bhutkar et al. 2008). The following markers were used to assay interspecies double-crossover rate inside the inversion—DPS X063 (XR_group6: 12,588,339, center of inversion), centromeric breakpoint (XR_group6: 6,219,093), and telomeric breakpoint (XR_group8: 7,199,440). Primer sequences are included in supplementary file, Supplementary Material online. All individuals were genotyped at the center of the inversion and at least one of the breakpoint markers. Double crossovers were defined as individuals with an allele in the center of the inversion that was different from the alleles at the two breakpoints, indicating exchange within the inverted segment.
Calculation of Relative Node Depth between Non-SR D. persimilis, SR D. persimilis, and D. pseudoobscura
A total of four XR-chromosome sequences were used for this study—one D. miranda, one D. pseudoobscura, and two D. persimilis. Drosophila miranda (Mt. St. Helena 22) and SR D. persimilis (Mt. St. Helena 39) sequences were generated as Illumina 76 bp paired-end reads (NCBI SRP006823). DNA for the SR sequence was isolated from a single male fly from the F2 generation of a female captured in 1997 from Mount St. Helena, California, and lab strain male (X chromosome was wild caught), and amplified for genome sequencing using Qiagen’s Whole Genome Amplification service (REPLI-g Service, Single Tube (100 μg), Cat. no. 805923 Hilden, Germany). Drosophila miranda genome data was isolated from 15 flies from an inbred stock. SR D. persimilis reads were aligned to the D. pseudoobscura reference genome v2.9 using bwa-0.5.5 (file: dpse-all-chromosome-r2.9, alignment settings -e 4 -t 7) (Li and Durbin 2009). Consensus assemblies for D. persimilis were generated using the alignments output by bwa and Samtools 0.1.6 pileup option (pileup settings -s -c -T 0.9 -N 1) (Li et al. 2009). Drosophila miranda sequence reads were aligned using the same pileup process as SR D. persimilis and filtered through custom Python scripts that exclude indels with <70% of reads supporting them, 5 bp flanking putative indels, bases with less than 3 read coverage, and bases with a quality score (Phred scale) less than 30. Annotated alignments between the released genomes of D. persimilis (MSH3, Clark et al. 2007) and D. pseudoobscura (MV2-25, Richards et al. 2005) were produced previously from Kulathinal et al. (2009). The D. persimilis, SR D. persimilis, and D. miranda sequences were consolidated into one alignment file using D. pseudoobscura as a reference (available from the Dryad repository at doi:10.5061/dryad.7q0nq). Five megabases of both the centromeric and telomeric ends of this chromosome were excluded due to low diversity in these regions (Andolfatto and Wall 2003). This input file was then divided into 200-Kb windows and processed with a series of custom Perl scripts to both excise introns and correct out of frame coding sequences caused by indels in alignment file. Due to genetic similarities between each of the sequences and D. pseudoobscura, gene annotations for each were assumed to be the same as the annotated D. pseudoobscura sequence.
Custom Perl scripts (available from the Dryad repository at doi:10.5061/dryad.7q0nq) were written to calculate average pairwise sequence difference (π), defined as the number of mismatches divided by the total number of bases, along third codon positions of nondegenerate codons (C4) between 200-kb windows of each of the annotated D. pseudoobscura and D. persimilis sequences. C4 sites were used because they have been shown to have less evolutionary constraints than other coding regions (de Procé et al. 2009). Relative node depth (RND; Feder et al. 2005), a measure of species divergence that uses an outgroup to account for differences in mutation rate, was then calculated for these windows by dividing the pairwise differences of each sequence pair by the average of the pairwise sequence differences between each of the sequences and D. miranda (Machado et al. 2007).
High-Throughput Genotyping to Analyze Recombination Rate Changes throughout the Genome Outside Inversions
A subset of 480 individuals from the cross described in “Single-generation estimate of inversion crossover rate in hybrids” was genotyped to assay recombination rate changes relative to the within-species rate outside of the inversion due to the interchromosomal effect and at inversion boundaries (see supplementary table 1, Supplementary Material online). This cross design did not account for direction of the cross because a previous study in this system showed no difference in ICE between F1 hybrids backcrossed to either D. persimilis or D. pseudoobscura (Ortiz-Barrientos et al. 2006).
DNA was isolated from the 480 backcross offspring individually at the Genomic Sciences Lab at North Carolina State University for subsequent genotyping with 96 single nucleotide polymorphisms (SNPs) (see methods and scripts described in Stevison and Noor (2010)). A subset of 42 markers corresponds to positions on the second chromosome of the within-species recombination map generated previously for D. persimilis (Stevison and Noor 2010). These markers were designed to assess the pervasiveness of the interchromosomal effect on this chromosome due to inversion heterozygosity in the F1 females of this cross. This chromosome was targeted due to the fine-scale nature of the existing within-species map (markers apx. 150–200 kb apart) relative to other chromosomes.
Another subset of 48 markers (see supplementary table 1, Supplementary Material online) was designed to further fine-map recombination reduction at inversion boundaries as in Kulathinal et al. (2009). These markers correspond to 2–3 Mb outside the breakpoints of each of the three major chromosomal inversions differentiating D. pseudoobscura and D. persimilis (six boundary regions total). Of these 48 markers designed for inversion boundaries, there were six on each XL boundary, seven on each XR boundary, and six on each Chromosome 2 (C2) boundary. Additionally, there were two markers within the XR inversion (plus two additional markers designed as genotyping replicates at the center of the inversion, see below) and four within both the XL and C2 inversions.
Finally, six markers were designed to duplicate the genotyping markers used in the inversion recombination survey of XR, so as to include this subset of individuals in the data for the original cross (see Single-Generation Estimate of Inversion Crossover Rate in Hybrids).
SNP markers were screened in all offspring using the Illumina BeadXpress platform (Fan et al. 2003) (Illumina, Inc. San Diego, CA) at the Genomic Analysis Facility within the Duke University Center for Human Genome Variation. The output consisted of raw genotypes at all markers for all individuals. Eleven total markers were not useful for analysis for various reasons, such as polymorphism within strains or monomorphism between strains. Furthermore, eight individuals did not amplify at any of the markers, and two individuals were removed from two different subsets due to greater than 10% missing data in that subset of markers. One individual was removed prior to assembly of the C2 map, and another individual was removed from the XL boundary map prior to assembly.
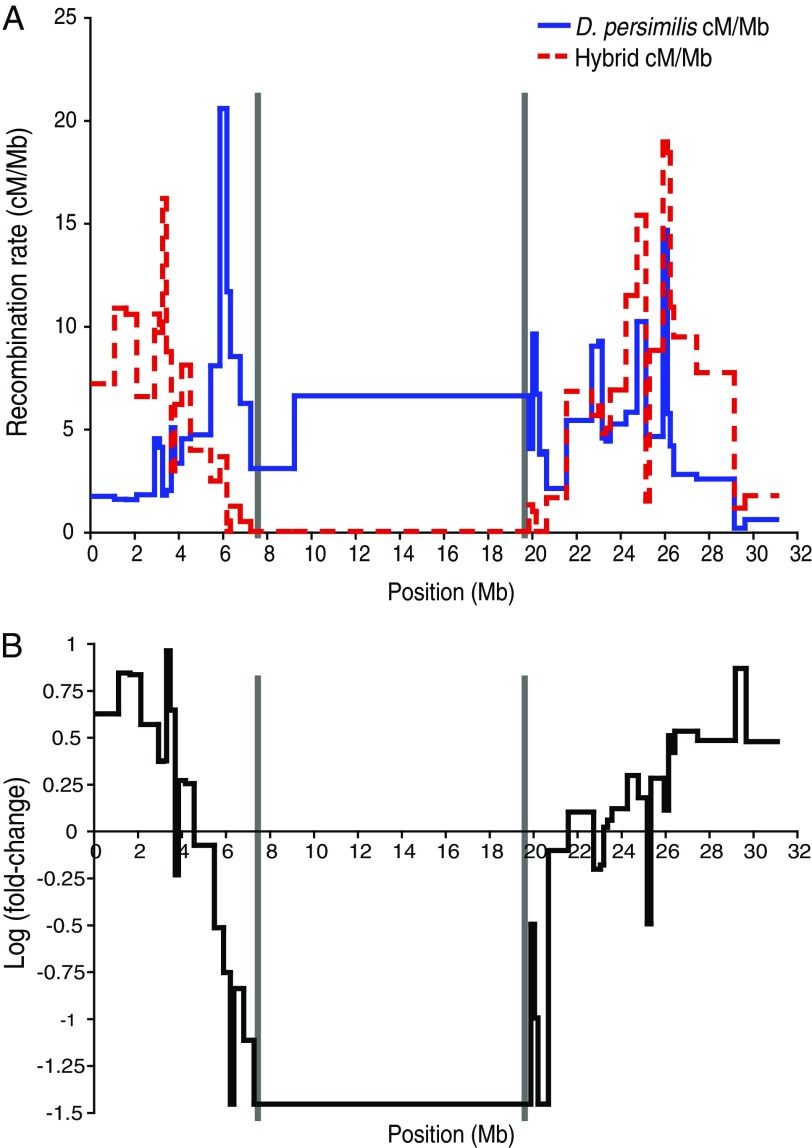
The raw data was processed to assess crossovers at each interval via scripts from Stevison and Noor (2010). For C2, a Kosambi cM/Mb value was calculated and compared with the published rate in D. persimilis (Stevison and Noor 2010) to calculate a fold change difference between the hybrid map and the pure species map (fig. 2). Previous studies comparing recombination rates between D. pseudoobscura and D. persimilis have noted very tight correspondence (Ortiz-Barrientos et al. 2006; Stevison and Noor 2010); therefore, we limited our pure species comparison with the D. persimilis recombination rates. To examine how variation in interspecies divergence may be predicted by changes in recombination due to inversions, we calculated for each recombination interval interspecies sequence divergence. For consistency with our previous measures of divergence (see “Calculation of RND between non-SR D. persimilis, SR D. persimilis, and D. pseudoobscura”), we defined divergence as the number of mismatches between D. persimilis and D. pseudoobscura divided by the total number of bases, along third codon positions of nondegenerate codons (C4) based on the annotated D. pseudoobscura and D. persimilis sequences.
FIg. 2.—
Inter-chromosomal effect. (A) A comparative plot of recombination rate along the second chromosome between the published D. persimilis map and the map of recombination rate in between species hybrids. (B) A plot of the fold change difference in recombination rates.
Results
Single-Generation Estimate of Inversion Crossover Rate in Hybrids
Of 9,739 individuals screened, one sample was confirmed as a double recombinant across the XR inversion, with the D. pseudoobscura allele at the center of the inversion and the D. persimilis allele just inside both breakpoints. Confirmation consisted of first repeating the initial genotyping at all three markers spanning the XR inversion. Next, a gene conversion event was ruled out by confirming the D. pseudoobscura allele at a second marker (DPSX051, chromosome scaffold XR_group6: position 12953124) near the center of the inversion, ∼360 kb from the original genotyping marker. This exchange is much larger than gene conversion tract lengths in Drosophila (Hilliker et al. 1994; Schaeffer and Anderson 2005). Finally, the sample was confirmed via Sanger sequencing at markers nearby the three genotyping markers, again showing a mismatch between the allele at the center of the inversion relative to the inversion breakpoints. Hence, we estimate a rate of 0.01% or ∼10−4 double-crossover events across this 12.5 Mb inversion in a single generation, though this figure may underestimate the total double-crossover rate because only a single position within the inversion was surveyed.
Comparisons between Intraspecific and Interspecific Inversion Difference of XR Chromosomes
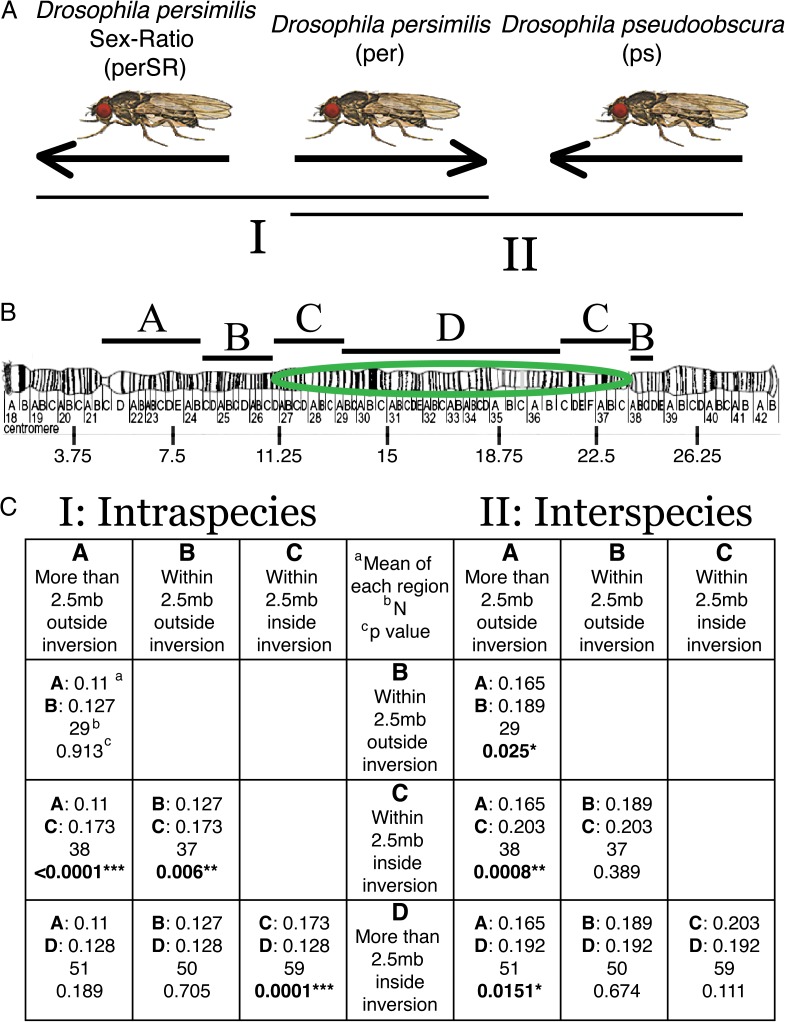
We calculated Feder et al.’s (2005) RND for whole XR-chromosome sequences between D. persimilis (per) and both SR D. persimilis (SRper, comparison I: Intraspecies), and D. pseudoobscura (ps, comparison II: Interspecies) (fig. 1A). Greater RND values indicate more differentiation has occurred between the focal species after correcting for divergence to the outgroup (D. miranda), whereas smaller RND values indicate less divergence between the focal species. Each 200-kb window was placed into one of four groups according to whether it was more (A) or less (B) than 2.5 Mb outside the XR inversion and more (D) or less (C) than 2.5 Mb inside the inversion (fig. 1B). Overall, calculations of mean RND values for each region of comparison I resulted in the following rank order: A ≤ B ≤ D < C (fig. 1C). Hence, the regions just inside the inversion breakpoints had significantly higher relative divergence than all other regions. In contrast, comparison II resulted in the same rank order but with different regions being significantly different from one another: A < B ≤ D ≤ C (fig. 1C). Hence, the regions far outside the inversion breakpoints had significantly lower relative divergence than all other regions. As predicted, the RND between D. persimilis—SR and D. pseudoobscura (no inversion difference) was not significantly different for any comparison.
FIg. 1.—
Intraspecific versus interspecific differentiation along XR. (A) Arrows represent the orientation along XR of each of the three genomes compared. (B) Ideogram of the XR-chromosome arm (modified from Schaeffer et al. [2008] fig. 8D and reproduced with permission from Genetics Society of America) with the inverted segment highlighted by the green circle, and the segments compared labeled above the chromosome. (C) Summary of all pairwise comparisons of mean RND within each chromosome region (A, B, C, and D) of intraspecific (I) and interspecific (II) RND values. Each cell gives the mean RND for each region, total number of data points, and P value using a two-sample Wilcoxon test.
Analysis of Recombination Rate Reduction in Single-Generation Hybrids at Inversion Boundaries
Genotypes of markers at inversion boundaries and the markers within each inversion were compared for each individual to determine how far outside the inversion breakpoints strong suppression of recombination extends (see detailed results in supplementary table 2, Supplementary Material online). At the C2 centromeric boundary, no recombinants were recovered among 471 progeny surveyed as far as 2.4 Mb and as far as 2.56 Mb on the telomeric boundary. At the XR centromeric boundary, no recombinants were recovered as far as 2.44 Mb, and on the telomeric boundary, no recombinants were recovered as far as 2.79 Mb. At the XL boundaries, there was an absence of observed recombination as far as 3 Mb on the centromeric side of the inversion, and no recombinants were recovered on the telomeric side of the inversion as far as 2.73 Mb.
The range between minimum recombination suppression and the maximums presented above is summarized in table 1 and compared with previous results from Kulathinal et al. (2009). The results from the current study used more markers to fine map both the lower and upper limit of this range. The third column represents the narrowest range as summarized from both studies.
Table 1.
Summary of Recombination Suppression at Inversion Boundaries as Compared with Kulathinal et al. (2009)
| Kulathinal et al. (2009) (Mb) | Current Study | Narrowest Range (Mb) | |
| Centromeric side XL | 0–2.84 | 2.75–3 Mb | 2.75–2.84 |
| Telomeric side XL | 0.4–2.8 | 2.73 Mb+ | 2.73–2.8 |
| Centromeric side XR | 0–3.35 | 2.18–2.44 Mb | 2.18–2.44 |
| Telomeric side XR | 2.1–2.8 | 2.79 Mb+ | 2.79–2.8 |
| Centromeric side C2 | 0–4.55 | 2.32–2.56 Mb | 2.32–2.56 |
| Telomeric side C2 | 0–2.77 | 2.29–2.4 Mb | 2.29–2.4 |
Note.—The refined range combines the results of the two studies. See supplementary table S2, Supplementary Material online for detailed results.
Analysis of ICE Using High-Throughput Genotyping
The recombination rate of hybrids along C2 ranged from 0 (within inverted regions) to 18.92 cM/Mb with an average interval size of 500 kb. The comparable recombination rate in the same intervals for pure species ranged from 0.15 to 20.54 cM/Mb. Although the range of recombination rate values was similar, the distribution of these events was significantly different in the hybrids in 31 of 47 intervals. The ratio of cM/Mb in hybrids to cM/Mb in pure species (D. persimilis) yielded a calculated fold change, which ranged from 0 to 9.14. The fold change is highest (>2-fold) in the first 4 Mb and the last 5.5 Mb of the chromosome. Figure 2 shows a plot comparing recombination rates in D. persimilis and hybrids as well as a plot of the log (base 10)-normalized fold-change of recombination rate in hybrids relative to pure species. For this study, differences in recombination rates of hybrids along chromosome 2 (bearing an inversion) can be attributed to inversion heterozygosity of C2 (intrachromosomal effect) and inversion heterozygosity of XL and XR (interchromosomal effect).
Because previous studies on ICE have also observed large effects in distal and proximal regions (which normally have low recombination in Drosophila), we tested for an association between recombination rate with D. persimilis and the fold change observed in hybrids. We found that regions with low recombination rate in D. persimilis were among those with the highest log fold change in hybrids (P < 0.0001; r2 = 0.373; N = 36), suggesting these regions are most susceptible to changes in recombination. We excluded regions with no observed recombination in hybrids (inside the inversion and within 3 Mb of the inversion breakpoints).
Next, we tested for an association of nucleotide divergence between species and hybrid recombination rate. We calculated divergence between D. pseudoobscura and D. persimilis at 4-fold degenerate third position codons (C4). Further, we calculated a corresponding diversity measure along the same regions between two sequences of D. pseudoobscura at C4 sites, to correct for variation in levels of diversity contributing to variation in divergence. In a multiple regression analysis with log fold change as the response variable and the diversity and divergence calculations as predictor variables, we found log fold change explains a significant proportion of the variation in divergence between species (P = 0.0088; N = 31) but does not account for the variation in diversity within species (P = 0.0831), suggesting that the correlation observed was not driven by lower segregating ancestral diversity in chromosomal ends (Noor and Bennett 2009). In the above analysis, regions with no recombination in hybrids were excluded, along with one interval with no C4 bases for the diversity measure, and the last 4 intervals of the chromosome which are misassembled in the D. pseudoobscura genome making it difficult to obtain reliable divergence estimates. When we further reduced the analysis to exclude chromosome ends, the results remained the same with a significant result for divergence (P = 0.0049; N = 18), but not for diversity (P = 0.0569), despite the reduced variation in the log fold change variation because ICE affects the ends most strongly.
Discussion
Between-Species Exchange across XR Inversion Detected in One Generation; However, Prolonged Evidence of Exchange Not Observed
We found that the observed rate of exchange via double crossovers along the largest inversion (XR inversion, 12.5 Mb) differentiating D. pseudoobscura and D. persimilis was similar to published rates of exchange across inversions segregating within species (Levine 1956; Ishii and Charlesworth 1977) but much lower than recombination in collinear regions greater than ∼2.5 Mb outside of the inversion or within the same region in homokaryotypes (Kulathinal et al. 2008; Stevison and Noor 2010). This observed rate of double-crossover in inversion heterokaryotypes approaches the expected genomic rate of gene conversion (∼10−5–10−6), estimated using the rosy locus in Drosophila (Smith et al. 1970; Chovnick 1973). Our estimate of 10−4 is also much higher than estimates previously used in models examining the role of inversions in persistence of species with gene flow (Navarro et al. 1997; Kirkpatrick and Barton 2006; Feder and Nosil 2009).
Higher Rates of Long-term Exchange within XR Inversion in Intraspecies versus Interspecies Genomic Comparisons
One way to test if genetic background plays a role in differentiation in heterokaryotypes is to compare differentiation along segments of an inversion segregating within species with differentiation along the same segments of the same inversion between species. We did this by comparing the XR-chromosome arm of D. persimilis with D. pseudoobscura (heterokaryotype between species) and D. persimilis with D. persimilis—SR (heterokaryotype within species) and examining how the pattern of differentiation varied between these comparisons. As expected, divergence relative to an outgroup (RND) is higher in all regions in the interspecies comparison between D. persimilis and D. pseudoobscura versus the intraspecies comparison between D. persimilis and D. persimilis—SR.
The expected effect of gene flow through double crossing-over in inversion heterozygotes should reduce divergence at the midsegment of the inverted region relative to regions near the breakpoints (regions C vs. D in fig. 1), where double crossing-over is significantly reduced (Navarro et al. 1997). This prediction is consistent with our observed differences in RND between regions just inside the breakpoint and near the midsegment of the inversion, showing that differentiation among regions within the inverted region (C and D vs. A and B) persists both within- and between-species. The observation that the RND at region C is significantly higher than at region D for the intraspecies comparison (P = 0.0001) but not for the interspecies comparison (P = 0.111), suggests that higher rates of exchange via double crossing-over have occurred within species than between species. These differences are likely due to the higher frequency of inversion heterozygotes forming within species, the higher rates of exchange in the more closely related homologous chromosomes, and the putative lack of selective consequences of exchange products within species. These results are also consistent with a recent population genetics analysis, which failed to find evidence for historical gene flow between D. persimilis and D. pseudoobscura within the same inverted segment (Stevison 2011).
The mean difference between regions in collinear parts of the chromosome and regions within 2.5 Mb outside the inversion (A vs. B) is significantly higher in the interspecies comparison (P = 0.025). However, in the intraspecies comparison, these regions are not significantly different (P = 0.913), suggesting that the impact of recombination suppression just outside the inversion boundaries may also be dependent on the frequency of inversion heterozygotes. Furthermore, in comparison I, only group C showed a mean RND significantly different from the collinear region A (P < 0.0001). In comparison II, however, all other groups showed significantly different mean RND values from group A (A vs. B, P = 0.025; A vs. C, P = 0.0008; and A vs. D, P = 0.0151). These combined observations suggest that the overall strength of heightened divergence for between species inversions is dependent on the frequency of inversion heterozygotes. Finally, both the pattern of increased exchange at the midsegment relative to the breakpoint and significantly increased divergence at the breakpoints relative to collinear regions are consistent with previous studies modeling gene flow across inversions (i.e., Navarro et al. 1997) and experimental results (Schaeffer and Anderson 2005; Machado et al. 2007).
Recombination Suppression Extends 2.5–3 Mb beyond Between-Species Inversion Boundaries
One of the more puzzling phenomena associated with inversion heterozygosity is the extension of recombination suppression beyond inversions, outside the breakpoints. Here, we refined the known ranges of recombination suppression at inversion boundaries for the three major inversions differentiating D. pseudoobscura and D. persimilis (table 1). The results in table 1 clearly show that the level of recombination suppression due to inversion heterozygosity cumulatively extends 2.5–3 Mb beyond the breakpoints of each inversion, adding 5–6 Mb to the total expected size of the inversion itself. Because recombination is not suppressed in these boundary regions in the absence of an inversion (Stevison and Noor 2010), it is not immediately obvious how recombination suppression would extend this far outside of the inversion loop. Previous studies have accredited this result to the difficulty of the synaptonemal complex from forming at inversion boundaries (Roberts 1976). Another possibility is that because inversion boundaries tend to accumulate in unstable/repetitive regions of chromosomes (Andolfatto et al. 1999; Caceres et al. 1999; Ranz et al. 2007), inversion boundaries could serve to recruit recombination suppression over the length of the inversion, triggered perhaps by heterozygosity immediately outside the inversion boundary. If inversion boundaries were indeed the molecular trigger for reduced recombination inside inverted regions, it would follow that these regions are also susceptible to recombination suppression.
Interchromosomal Effect Is Highest in Regions of Low Recombination and Corresponds to Variation in Between-Species Divergence
Our study investigates both intra- and interchromosomal effects (ICE) using ∼50 markers along chromosome 2. Because we analyze hybrids between D. pseudoobscura and D. persimilis, the ICE we observe is due to heterozygosity at three inversions—two on the X and one on the second (focal chromosome). Previous studies have observed ICE yielding differences in recombination rates as high as 250% higher than standard map distance (Schultz and Redfield 1951), whereas we observed greater than 800% higher recombination rates, with an average of 224%.
The higher proportional increase in recombination rate in this study relative to earlier studies could result from 1) heterozygosity for more inversions (other studies observe ICE with only 1–2 heterozygous inversions), 2) the size of the intervals at which we analyzed recombination differences in hybrids, and/or 3) higher differentiation in collinear regions due to our use of inversions differentiating species rather than populations of the same species. Previous studies averaged ICE over larger intervals and therefore may have masked the effects of smaller regions with very strong ICE. In our study, the average percent change in the first and last 5 Mb is ∼250% higher recombination in inversion heterozygotes (more closely matching differences within species for single inversions); therefore, the size of intervals assayed in the current study is most likely responsible for the higher observed change in recombination rates.
Several studies on inversions segregating within Drosophila melanogaster have shown that ICE has a stronger impact on proximal (centromeric) and distal (telomeric) portions of chromosomes (Schultz and Redfield 1951; Lucchesi and Suzuki 1968; Portin and Rantanen 1990; Krimbas and Powell 1992) and that these regions tend to have more restricted recombination within homokaryotypes. Similar to previously published results, we found that ICE was strongest in regions distal and proximal on chromosome 2. Because these regions are known to have lower recombination rates overall, we showed that, irrespective of chromosome position, regions of low recombination were most susceptible to ICE, supporting that these regions may be less resilient to disruptions in recombination rate. The observation of heightened effect on chromosome ends/centromeric regions has also been observed in maize and grasshoppers (Lucchesi and Suzuki 1968); however, restricted recombination in these regions is not universal. For example, although in Drosophila, both centromeric and telomeric regions tend to have reduced recombination (Begun et al. 2007), in mammals, telomeric regions tend to have higher than average recombination (Kong et al. 2002). Therefore, future research could examine if ICE impacts telomeric regions in mammals.
Finally, we examined how changes in recombination rates in hybrids correspond to patterns of gene flow and differentiation, as an extension of how inverted regions (which have low recombination in hybrids) often bear high divergence between species. To test for this, we calculated divergence between D. pseudoobscura and D. persimilis and found a strong correlation with log fold change of recombination in hybrids relative to pure species. When we corrected for regions of low within species diversity (e.g., chromosome ends), we still observed a significant association, showing that low diversity at chromosomal ends (where ICE is strongest) does not influence the relationship between divergence and recombination rate changes in hybrids. Hence, it appears that the interchromosomal effect may actually “increase” interspecies gene flow outside of the inverted regions—a factor not considered previously in the effects of inversions on species persistence.
Conclusions
Our results suggest that models determining the role of inversions in maintaining species should place less emphasis on single-generation recombination rates and more emphasis on both the expected frequency of heterokaryotes and potential fitness consequences that prevent the success of progeny that carry exchange products from propagating in the next generation (see also Faria and Navarro 2010; Jackson 2011). Although exchange products are rarely observed in inversion heterozygotes, the frequency of heterozygotes forming dictates the frequency of exchange products in the population. This frequency is a function of both the expected level of migration between species and any premating barriers to exchange. In D. pseudoobscura and D. persimilis, we know that the rate of formation of hybrids in sympatry is 10−4 (Powell 1983); however, the overall species hybridization rate would need to account for the proportion of D. pseudoobscura found in sympatry. Therefore, for between-species inversions, exchange is the product of two rare events (rare formation of inversion heterozygotes and rare double-crossover events). Whereas, for within-species inversions, it is the product of one rare and one common event, with inversion heterozygote formation expected to be higher within-species.
Furthermore, because the accumulation of genetic incompatibilities occurs gradually over time, selection does not need to be very strong in early generations when migration is nearly zero. However, upon secondary contact, higher rates of migration, and thus selection are likely, indicating that these parameters should be considered nonindependent. These three factors—the frequency of heterokaryotypes/migrants, the fitness consequences of exchange in heterokaryotypes, and the rate at which genetic incompatibilities accumulate—are likely the most important factors contributing to the absence of long-term exchange detected between D. pseudoobscura and D. persimilis along the inverted region on the XR-chromosome arm.
In the past 10 years, interest in the role of chromosomal inversions in speciation has been rekindled based on the inherent properties of inversions to restrict recombination in heterokaryotypes. This reduced recombination protects existing adaptive complexes and genetic incompatibilities and allows for the accumulation of additional incompatibilities between species. The research presented here confirms that some features found in inversions segregating within species apply to interspecies inversion differences but also identifies several potential differences and hypothesizes their causes. Further research should explore some of the patterns suggested here, particularly considering 1) which factors may maintain divergence in inverted regions between species despite detectable double crossover events, 2) the relationship between size of recombination intervals on the intensity of inter- and intrachromosomal effects (ICE) observed, and 3) how much ICE increases interspecies gene exchange outside inverted regions.
Supplementary Material
Supplementary file and table S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank S. McGaugh for preparation of samples for genomic sequencing and alignment of sequences to reference genome. We thank S. Bennett and A. Boehling for technical support. We also thank C. Smukowski, S. McGaugh, and T. Mitchell-Olds for comments on the manuscript. Funding for this work was provided by grants from the National Institutes of Health (GM076051, GM086445, and GM092501) and from the National Science Foundation (DEB0715484 to M.A.F.N.).
References
- Andolfatto P, Depaulis F, Navarro A. Inversion polymorphisms and nucleotide variability in Drosophila. Genet Res. 2001;77:1–8. doi: 10.1017/s0016672301004955. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Wall JD. Linkage disequilibrium patterns across a recombination gradient in African Drosophila melanogaster. Genetics. 2003;165:1289–1305. doi: 10.1093/genetics/165.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P, Wall JD, Kreitman M. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics. 1999;153:1297–1311. doi: 10.1093/genetics/153.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquadro CF, Weaver AL, Schaeffer SW, Anderson WW. Molecular evolution of inversions in Drosophila pseudoobscura—the amylase gene region. Proc Natl Acad Sci U S A. 1991;88:305–309. doi: 10.1073/pnas.88.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:2534–2559. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Rozas J, Navarro A, Barbadilla A. The estimation of the number and the length distribution of gene conversion tracts from population DNA sequence data. Genetics. 1997;146:89–99. doi: 10.1093/genetics/146.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A, et al. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, O'Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Caceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- Carneiro M, Blanco-Aguiar JA, Villafuerte R, Ferrand N, Nachman MW. Speciation in the European rabbit (Oryctolagus cuniculus): islands of differentiation on the X chromosome and autosomes. Evolution. 2010;64:3443–3460. doi: 10.1111/j.1558-5646.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson HL. The selective elimination of inversion dicentric chromatids during meiosis in the eggs of Sciara impatiens. Genetics. 1946;31:95–113. doi: 10.1093/genetics/31.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick A. Gene conversion and transfer of genetic information within inverted region of inversion heterozygotes. Genetics. 1973;75:123–131. doi: 10.1093/genetics/75.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Kliman RM, Noor MAF. Fine-scale crossover rate heterogeneity in Drosophila pseudoobscura. J Mol Evol. 2007;64:129–135. doi: 10.1007/s00239-006-0142-7. [DOI] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Meyers W, Crittenden AP, Sniegowski P. The fertility effects of pericentric inversions in Drosophila melanogaster. Genetics. 1993;134:487–496. doi: 10.1093/genetics/134.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Procé SM, Halligan DL, Keightley PD, Charlesworth B. Patterns of DNA-sequence divergence between Drosophila miranda and D. pseudoobscura. J Mol Evol. 2009;69:601–611. doi: 10.1007/s00239-009-9298-2. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Epling C. The suppression of crossing over in inversion heterozygotes of Drosophila pseudoobscura. Proc Natl Acad Sci U S A. 1948;34:137–141. doi: 10.1073/pnas.34.4.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T, Sturtevant AH. Inversions in the chromosomes of Drosophila pseudoobscura. Genetics. 1938;23:28–64. doi: 10.1093/genetics/23.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky TG. Genetics and the origin of species. New York: Columbia University Press; 1937. [Google Scholar]
- Dobzhansky TG, Epling C. Contributions to the genetics, taxonomy, and ecology of Drosophila pseudoobscura and its relatives. Washington (DC): Carnegie Institute: 1944. [Google Scholar]
- Fan JB, et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Faria R, Navarro A. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25:660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Feder JL, Nosil P. Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution. 2009;63:3061–3075. doi: 10.1111/j.1558-5646.2009.00786.x. [DOI] [PubMed] [Google Scholar]
- Feder JL, et al. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci U S A. 2005;102:6573–6580. doi: 10.1073/pnas.0502099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick CL, Stevison LS, Noor MAF. Fine-scale crossover rate and interference along the XR-chromosome arm of Drosophila pseudoobscura. Drosoph Inf Serv. 2009;92:27–29. [Google Scholar]
- Foss E, Lande R, Stahl FW, Steinberg CM. Chiasma interference as a function of genetic-distance. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg H. A new inversion of the X-chromosome in Drosophila melanogaster. J Genet. 1935;31:163–184. [Google Scholar]
- Hasson E, Eanes WF. Contrasting histories of three gene regions associated with In(3L)Payne of Drosophila melanogaster. Genetics. 1996;144:1565–1575. doi: 10.1093/genetics/144.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AJ, et al. Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics. 1994;137:1019–1024. doi: 10.1093/genetics/137.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Charlesworth B. Associations between allozyme loci and gene arrangements due to hitch-hiking effects of new inversions. Genet Res. 1977;30:93–106. [Google Scholar]
- Jackson BC. Recombination-suppression: how many mechanisms for chromosomal speciation? Genetica. 2011;139:393–402. doi: 10.1007/s10709-011-9558-0. [DOI] [PubMed] [Google Scholar]
- Joyce EF, McKim KS. Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS Genet. 2010;6:e1001059. doi: 10.1371/journal.pgen.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Krimbas CV, Powell JR. Drosophila inversion polymorphism. Boca Raton (FL): CRC Press; 1992. [Google Scholar]
- Kulathinal RJ, Bennett SM, Fitzpatrick CL, Noor MAF. Fine-scale mapping of recombination rate in Drosophila refines its correlation to diversity and divergence. Proc Natl Acad Sci U S A. 2008;105:10051–10056. doi: 10.1073/pnas.0801848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5:e1000550. doi: 10.1371/journal.pgen.1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laayouni H, Hasson E, Santos M, Fontdevila A. The evolutionary history of Drosophila buzzatii. XXXV. Inversion polymorphism and nucleotide variability in different regions of the second chromosome. Mol Biol Evol. 2003;20:931–944. doi: 10.1093/molbev/msg099. [DOI] [PubMed] [Google Scholar]
- Leman SC, Chen Y, Stajich JE, Noor MA, Uyenoyama MK. Likelihoods from summary statistics: recent divergence between species. Genetics. 2005;171:1419–1436. doi: 10.1534/genetics.104.040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP. Crossing over and inversions in coadapted systems. Am Nat. 1956;90:41–45. [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JT, Suzuki DT. The interchromosomal control of recombination. Annu Rev Genet. 1968;2:53–86. [Google Scholar]
- Machado CA, Haselkorn TS, Noor MAF. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D. persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Maynard Smith S. Genetics and cytology of Drosophila subobscura.VIII. Heterozygosity, viability and rate of development. J Genet. 1954;52:152–164. [Google Scholar]
- McGaugh SE, Machado CA, Noor MAF. Genomic impacts of chromosomal inversions in parapatric Drosophila species. Trans R Soc B Biol Sci. forthcoming doi: 10.1098/rstb.2011.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betran E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega C, Khadem M, Aguade M, Segarra C. Genetic exchange versus genetic differentiation in a medium-sized inversion of Drosophila: the A(2)/A(st) arrangements of Drosophila subobscura. Mol Biol Evol. 2008;25:1534–1543. doi: 10.1093/molbev/msn100. [DOI] [PubMed] [Google Scholar]
- Noor MAF, Bennett SM. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity. 2009;103:439–444. doi: 10.1038/hdy.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitski E, Braver G. An analysis of crossing over within a heterozygous inversion in Drosophila melanogaster. Genetics. 1954;39:197–209. doi: 10.1093/genetics/39.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Chang AS, Noor MAF. A recombinational portrait of the Drosophila pseudoobscura genome. Genet Res. 2006;87:23–31. doi: 10.1017/S0016672306007932. [DOI] [PubMed] [Google Scholar]
- Pegueroles C, Ordonez V, Mestres F, Pascual M. Recombination and selection in the maintenance of the adaptive value of inversions. J Evol Biol. 2010;23:2709–2717. doi: 10.1111/j.1420-9101.2010.02136.x. [DOI] [PubMed] [Google Scholar]
- Portin P, Rantanen M. Further-studies on the interchromosomal effect on crossing over in Drosophila melanogaster affecting the preconditions of exchange. Genetica. 1990;82:203–207. doi: 10.1007/BF00056363. [DOI] [PubMed] [Google Scholar]
- Powell JR. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow—evidence from Drosophila. Proc Natl Acad Sci U S A. 1983;80:492–495. doi: 10.1073/pnas.80.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR. Inversion polymorphisms in Drosophila pseudoobscura and Drosophila persimilis. In: Krimbas CV, Powell JR, editors. Drosophila inversion polymorphism. Boca Raton (FL): CRC Press; 1992. pp. 73–126. [Google Scholar]
- Ranz JM, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:1366–1381. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, et al. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Robbins LG. Exchange within heterozygous inversions in Drosophila melanogaster. Genetics. 1974;77:105–114. doi: 10.1093/genetics/77.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PA. The genetics of chromosome aberration. In: Ashburner M, Novitski E, Wright TRF, editors. The genetics and biology of Drosophila. London: Academic Press; 1976. pp. 67–184. [Google Scholar]
- Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 2009;5:e1000419. doi: 10.1371/journal.pgen.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Aguade M. Gene conversion is involved in the transfer of genetic information between naturally-occurring inversions of Drosophila. Proc Natl Acad Sci U S A. 1994;91:11517–11521. doi: 10.1073/pnas.91.24.11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, Anderson WW. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics. 2005;171:1729–1739. doi: 10.1534/genetics.105.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, et al. Polytene chromosomal caps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Redfield H. Interchromosomal effects on crossing over in Drosophila. Cold Spring Harb Symp Quant Biol. 1951;16:175–197. doi: 10.1101/sqb.1951.016.01.015. [DOI] [PubMed] [Google Scholar]
- Smith PD, Finnerty VG, Chovnick A. Gene conversion in Drosophila—non-reciprocal events at maroon-like cistron. Nature. 1970;228:442–444. doi: 10.1038/228442a0. [DOI] [PubMed] [Google Scholar]
- Spurway H, Philip U. Genetics and cytology of Drosophila subobscura. 7. Abnormal gene arrangements in element A. J Genet. 1952;51:198–215. [Google Scholar]
- Stephan W, Langley CH. DNA polymorphism in lycopersicon and crossing-over per physical length. Genetics. 1998;150:1585–1593. doi: 10.1093/genetics/150.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison LS. [Durham (NC)]: Duke University; 2011. Causes and consequences of recombination rate variation in Drosophila [dissertation] [Google Scholar]
- Stevison LS, Noor MAF. Genetic and evolutionary correlates of fine-scale recombination rate variation in Drosophila persimilis. J Mol Evol. 2010;71:332–345. doi: 10.1007/s00239-010-9388-1. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Scotti-Saintagne C, Scotti I, Lai Z, Rieseberg LH. Genomic patterns of adaptive divergence between chromosomally differentiated sunflower species. Mol Biol Evol. 2009;26:1341–1355. doi: 10.1093/molbev/msp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, et al. Genetic exchange in 2La inversion heterokaryotypes of Anopheles gambiae. Insect Mol Biol. 2007;16:703–709. doi: 10.1111/j.1365-2583.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. Inherited linkage variation in the second chromosome. In: Morgan TH, Bridges CB, and Sturtevant AH, editors. Contributions to the genetics of Drosophila melanogaster. III. Washington (DC): Carnegie Institute; 1919. p. 305–341. [Google Scholar]
- Sturtevant AH, Beadle GW. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics. 1936;21:554–604. doi: 10.1093/genetics/21.5.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant AH, Dobzhansky T. Geographical distribution and cytology of “sex ratio” in Drosophila pseudoobscura and related species. Genetics. 1936;21:473–490. doi: 10.1093/genetics/21.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein A. Coincidence of crossing over in Drosophila melanogaster (Ampelophila) Genetics. 1918;3:135–172. doi: 10.1093/genetics/3.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley CS, Eanes WF. Isolation and analysis of the breakpoint sequences of chromosome inversion in(3l)Payne in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1994;91:3132–3136. doi: 10.1073/pnas.91.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, et al. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics. 2009;183:275–288. doi: 10.1534/genetics.109.105817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD, Morley FHW. Effects of pericentric rearrangements on recombination in grasshopper chromosomes. Genetics. 1955;40:604–619. doi: 10.1093/genetics/40.5.604. [DOI] [PMC free article] [PubMed] [Google Scholar]