Abstract
The MAPKKK Ste11p functions in three Saccharomyces cerevisiae MAPK cascades [the high osmolarity glycerol (HOG), pheromone response, and pseudohyphal/invasive growth pathways], but its activation in response to high osmolarity stimulates only the HOG pathway. To determine what restricts cross-activation of MAPK cascades (cross talk), we have studied mutants in which the pheromone response pathway is activated by high osmolarity (1 m sorbitol). We found that mutations in the HOG1 gene, encoding the p38-type MAPK of the HOG pathway, and in the PBS2 gene, encoding the activating kinase for Hog1p, allowed osmolarity-induced activation of the pheromone response pathway. This cross talk required the osmosensor Sho1p, as well as Ste20p, Ste50p, the pheromone response MAPK cascade (Ste11p, Ste7p, and Fus3p or Kss1p), and Ste12p but not Ste4p or the MAPK scaffold protein, Ste5p. The cross talk in hog1 mutants induced multiple responses of the pheromone response pathway: induction of a FUS1::lacZ reporter, morphological changes, and mating in ste4 and ste5 mutants. We suggest that Hog1p may prevent osmolarity-induced cross talk by inhibiting Sho1p, perhaps as part of a feedback control on the HOG pathway. We have also shown that Ste20p and Ste50p function in the Sho1p branch of the HOG pathway and that a second osmosensor in addition to Sho1p may activate Ste11p. Finally, we have found that pseudohyphal growth exhibited by wild-type (HOG1) strains depends on SHO1, suggesting that Sho1p may be a receptor that feeds into the pseudohyphal growth pathway.
Keywords: Hog1, MAP kinase, yeast pheromone response, yeast high osmolarity response, HOG pathway, signal transduction
Multiple mitogen-activated protein kinase (MAPK) cascades coexist in eukaryotic cells and mediate appropriate cellular responses to distinct environmental inputs (Robinson and Cobb 1997; Banuett 1998). The MAPK cascade consists of a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK). In Saccharomyces cerevisiae, four complete MAPK cascade modules have been identified (Herskowitz 1995; Levin and Errede 1995). The pheromone response pathway is activated by peptide pheromones and prepares cells for mating (Leberer et al. 1997). The pseudohyphal development/invasive growth pathway responds to environmental conditions to allow formation of pseudohyphal cells in a/α diploids and invasive growth in haploids (Liu et al. 1993; Roberts and Fink 1994). The PKC-regulated MAPK pathway responds to heat stress and hypotonic shock (Davenport et al. 1995; Kamada et al. 1995), and the high osmolarity glycerol (HOG) pathway responds to hypertonic stress (Brewster et al. 1993).
The MAPKKK Ste11p plays an essential role in three of the yeast MAPK pathways: the pheromone response pathway, the pseudohyphal/invasive growth pathway, and the HOG pathway (Fig. 1). In response to pheromones, a heterotrimeric G protein encoded by GPA1 (Gα), STE4 (Gβ), and STE18 (Gγ) is activated, liberating free Gβγ to stimulate downstream components (Dietzel and Kurjan 1987; Miyajima et al. 1987). Free Gβγ binds to and is thought to activate Ste20p, a PAK-related protein kinase (Leeuw et al. 1998), and Ste5p, a scaffold protein that binds each component of the MAPK cascade (Whiteway et al. 1995; Inouye et al. 1997; Feng et al. 1998). The MAPKKK Ste11p is activated by a mechanism, probably involving Ste20p-dependent phosphorylation (Wu et al. 1995), which allows Ste11p to phosphorylate Ste7p, the MAPKK (Neiman and Herskowitz 1994). Ste7p then phosphorylates and activates the MAPKs Fus3p or Kss1p (Gartner et al. 1992; Errede et al. 1993), which activate the transcription factor Ste12p (Cook et al. 1996; Tedford et al. 1997), leading to expression of genes required for mating.
Figure 1.
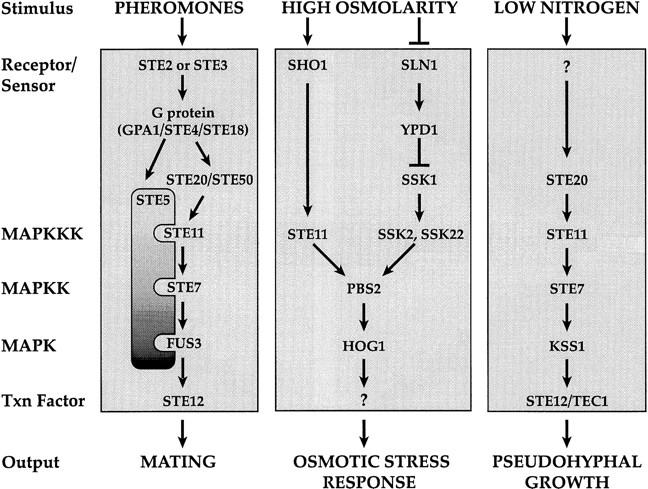
The pheromone response, high osmolarity glycerol (HOG), and pseudohyphal growth pathways.
a/α cells of the Σ1278b strain background are capable of forming pseudohyphae in response to low nitrogen conditions (Gimeno et al. 1992). Haploid cells of this background exhibit agar invasion on rich medium (Roberts and Fink 1994). Components of the pheromone response pathway are required for pseudohyphal development and invasive growth (Liu et al. 1993; Roberts and Fink 1994): Ste20p, Ste11p, Ste7p, and Ste12p are required for both pathways. Kss1p appears to be the MAPK for the pseudohyphal development and invasive growth pathways (Cook et al. 1997; Madhani et al. 1997). The upstream activators of these pathways remain to be identified: The pheromone receptors and associated heterotrimeric G protein are not required for pseudohyphal or invasive growth (Liu et al. 1993; Roberts and Fink 1994). Although the pseudohyphal and invasive growth pathways use a MAPK cascade containing Ste11p, Ste7p, and Kss1p, they do not require Ste5p (G. Fink, pers. comm.).
During elevated osmotic conditions, yeast cells accumulate high concentrations of glycerol to counteract the osmotic differential between the inside of the cell and the external environment (for review, see Blomberg and Adler 1992). High osmolarity causes activation of the MAPK, Hog1p (Brewster et al. 1993), which triggers increased synthesis of glycerol by activating transcription of genes required for glycerol synthesis such as GPD1 (Larsson et al. 1993; Albertyn et al. 1994). The HOG pathway has two known input branches. One involves a phosphorelay system related to the histidyl–aspartyl phosphorelay systems of bacteria and includes the integral membrane protein Sln1p and the response regulator Ssk1p (Maeda et al. 1994). Ssk1p activates two redundant MAPKKKs, Ssk2p and Ssk22p, which subsequently activate Pbs2p, the MAPKK for Hog1p (Maeda et al. 1995; Posas and Saito 1998). The other input is through the osmosensor Sho1p, which is a putative membrane-spanning protein with a carboxy-terminal SH3 domain (Maeda et al. 1995). This domain binds to a proline-rich region in Pbs2p. Activation of Pbs2p by Sho1p requires Ste11p, which phosphorylates Pbs2p. Pbs2p has been termed a scaffold protein because it binds multiple components of the HOG MAPK pathway: Sho1p, Ste11p, and Hog1p (Posas and Saito 1997). Because there are two inputs for activating Hog1p, mutants defective in either the Sho1p branch or the Sln1p branch are not osmosensitive. In contrast, mutants defective in both branches, for example, ssk2 ssk22 sho1 or ssk2 ssk22 ste11 strains are osmosensitive (Maeda et al. 1995; Posas and Saito 1997).
Although Ste11p functions in both the pheromone response and HOG pathways, activation of Ste11p by high osmolarity does not elicit a mating response (Posas and Saito 1997). Similarly, activation of Ste11p by α-factor does not activate the HOG pathway (Posas and Saito 1997). The mechanism by which signal specificity is maintained during high osmolarity stress or during exposure to pheromone remains to be determined. The presence of Ste11p in two different complexes (with Ste5, Ste7p, and Fus3p or Kss1p or with Sho1p, Pbs2p, and Hog1p) has been proposed to be responsible for preventing inappropriate activation of heterologous pathways (Posas and Saito 1997). Here we describe conditions under which the pheromone response pathway is efficiently activated by high osmolarity stress. These studies lead us to propose that Hog1p itself is responsible for limiting cross talk during high osmolarity stress, perhaps by inhibiting the Sho1p branch of the HOG pathway. We define the components of the pheromone response and HOG pathways required for cross talk. Our findings also suggest the existence of a third branch for activating the HOG pathway and raise the possibility that Sho1p is an upstream component of the pseudohyphal development pathway.
Results
Osmotic stress induces expression of FUS1::lacZ in Hog1p-deficient strains
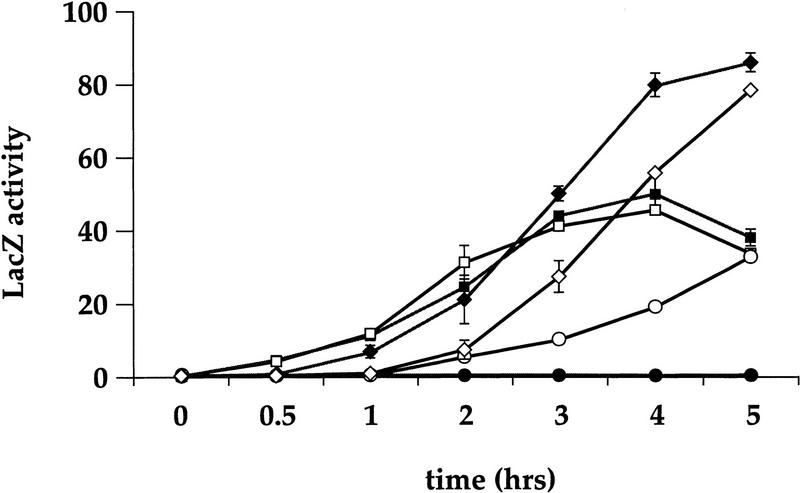
In the course of other studies of hog1 mutants, which revealed functional relationships between the pheromone response pathway and the HOG pathway (S. O’Rourke, unpubl.), we tested whether 1 m sorbitol can activate the pheromone response pathway. In our initial experiments, we assayed activation of the pheromone response pathway by induction of the FUS1::lacZ reporter gene, which is activated by Ste12p. Wild-type cells exposed to 1 m sorbitol did not exhibit any expression of the reporter even after 5 hr of incubation (Fig. 2). In contrast, an isogenic hog1 deletion mutant produced a clearly measurable level of β-galactosidase by 2 hr, ∼20% the level of these strains exposed to α-factor, which increased steadily over 5 hr to reach a level comparable to that observed in the α-factor-treated cells (Fig. 2). Wild-type and hog1 mutants exhibited similar induction in response to α-factor. The presence of both α-factor and sorbitol led to increased expression of the reporter by 5 hr in both wild-type and hog1 strains. Additional experiments showed that 0.5 m NaCl and 0.4 m sorbitol also stimulated FUS1::lacZ induction in hog1 but not wild-type strains (data not shown). These observations demonstrate that high osmolarity induces the pheromone response pathway in strains lacking Hog1p.
Figure 2.
A hog1 strain, but not wild type, induces FUS1::lacZ in response to 1 m sorbitol. a wild-type FUS1::lacZ (SO329) (solid symbols) and a hog1 FUS1::lacZ (SO330) (open symbols) strains were grown to log phase in YEPD medium, and t = 0 samples were taken for β-galactosidase assays. The cells were resuspended in YEPD + α-factor medium: (█ wild type; □ hog1; YEPD + 1 m sorbitol medium: (•, wild type; ○, hog1), and YEPD + 1 m sorbitol + α-factor medium: (♦, wild type; ⋄, hog1). At the indicated times, 1-ml samples were harvested for β-galactosidase assays as described in Materials and Methods. Assays were done in duplicate and the average ± s.d. of three experiments is shown. LacZ activity is shown in Miller units.
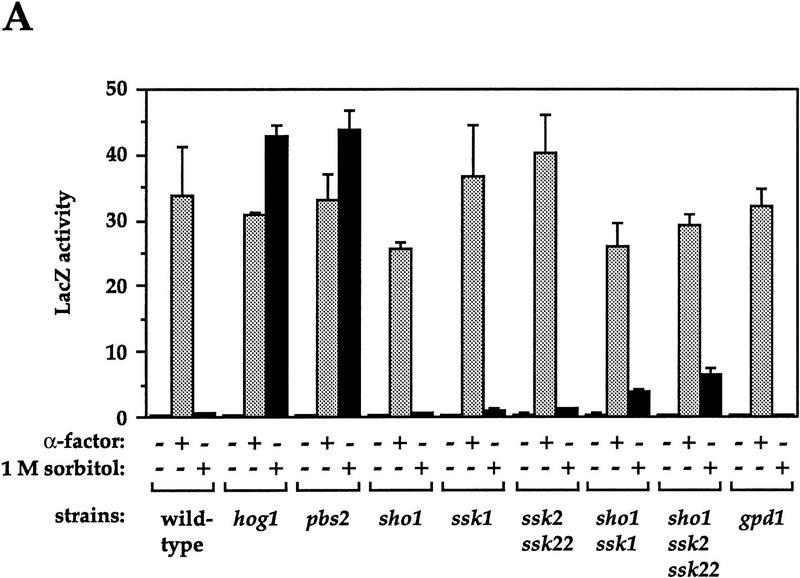
Next, we analyzed a series of strains defective in the HOG pathway to determine whether they also exhibit osmolarity-induced activation of FUS1::lacZ (cross talk) (Fig. 3A). Mutants gave three different responses: (1) The strain lacking Pbs2p behaved like the hog1 mutant, exhibiting high-level induction; (2) mutants defective in either of the known input branches to the HOG pathway, in SHO1 (affecting the Sho1p branch) or in SSK1 or in SSK2 and SSK22 (affecting the Sln1p branch), exhibited negligible cross talk; and (3) mutants defective in both of these branches, because of mutation in both SHO1 and SSK1 or in SHO1, SSK2, and SSK22, exhibited low levels of induction (9%–15% of the level observed in hog1 and pbs2 mutants). All of these strains exhibited similar induction of FUS1::lacZ in response to α-factor. These results indicate that lack of Hog1p activity is required for osmolarity-induced cross talk: Both hog1 and pbs2 mutants are completely lacking in Hog1p activity. Because hog1 and pbs2 mutants are unable to increase intracellular glycerol in response to osmotic stress, we entertained the possibility that reduction of glycerol due to mutation of GPD1 would also allow cross talk. However, we found the strain deleted for GPD1 did not exhibit cross talk (Fig. 3A).
Figure 3.
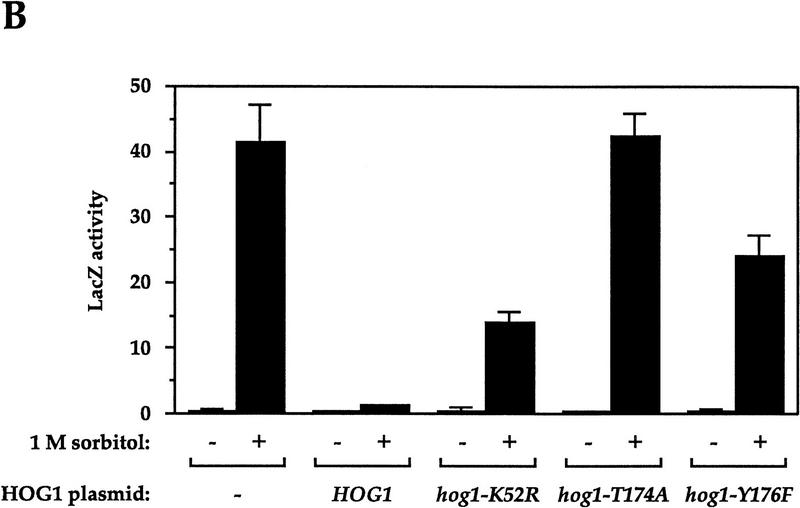
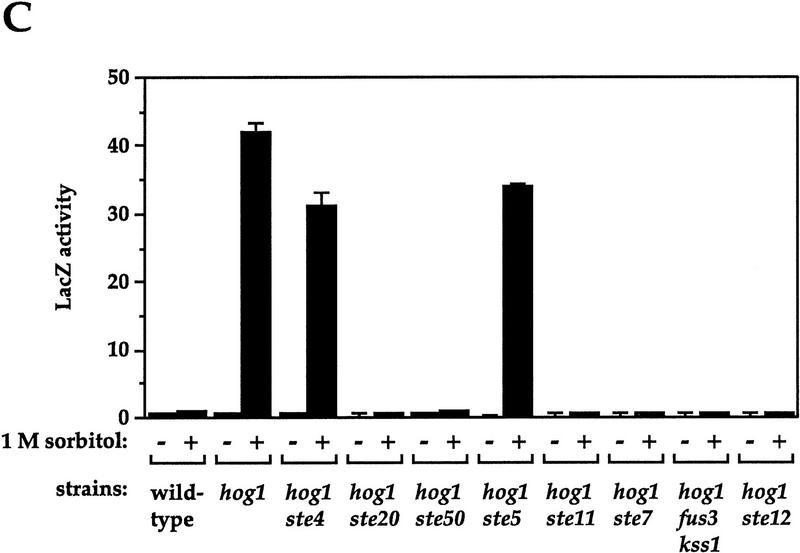
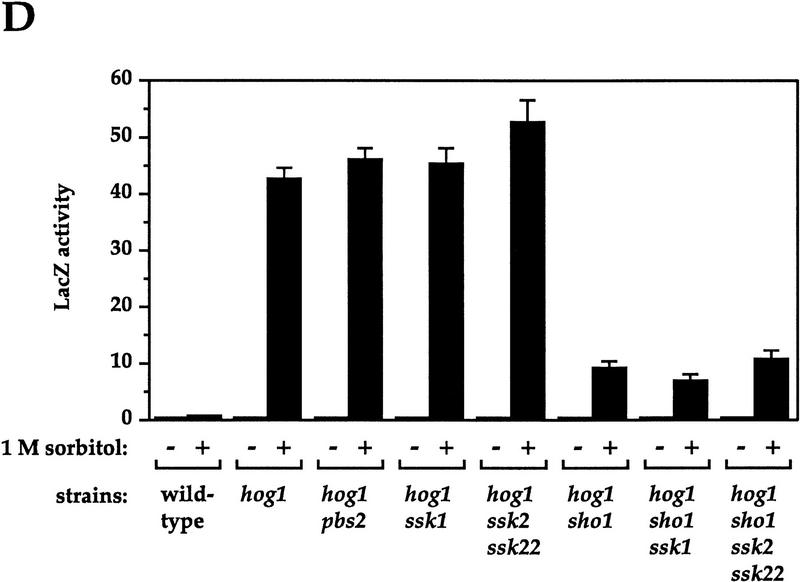
Genetic control of osmolarity-induced cross talk using FUS1::lacZ as a reporter. Cultures of log phase yeast strains were shifted to the indicated media for 5 hr (6 hr for B) and harvested. β-Galactosidase assays were done in triplicate, and the average ± s.d. of three experiments is shown. LacZ activity is plotted in Miller units. (A) Dependence of osmolarity-induced cross talk on inactivation of the HOG pathway. Strains SO329 (wild type), SO330 (hog1), SO382 (pbs2), SO351 (sho1), SO352 (ssk1), SO354 (ssk2 ssk22), SO353 (sho1 ssk1), SO355 (sho1 ssk2 ssk22), and SO385 (gpd1) were tested. (B) Hog1p kinase activity and phosphoacceptor sites are required to prevent cross-activation of FUS1::lacZ in response to 1 m sorbitol. Strain SO330 (hog1 FUS1::lacZ) was transformed with an empty vector (pRS316) or low-copy HOG1-containing plasmids: HOG1 (pJB15), hog1–K52R (pJBM3), hog1–T174A (pJBM4), or hog1–Y176F (pJBM2). Two independent transformants were assayed for each plasmid. (C) Dependence on the pheromone response pathway for osmolarity-induced cross talk in hog1 mutants. Strains SO329 (wild type), SO330 (hog1), SO331 (hog1 ste4), SO391 (hog1 ste20), SO373 (hog1 ste50), SO332 (hog1 ste5), SO333 (hog1 ste11), SO334 (hog1 ste7), SO335 (hog1 fus3 kss1), and SO143 (hog1 ste12) were analyzed. (D) Dependence on the Sho1p osmosensor for osmolarity-induced cross talk in hog1 mutants. The strains tested were SO329 (wild type), SO330 (hog1), SO383 (hog1 pbs2), SO384 (hog1 ssk1), SO358 (hog1 ssk2 ssk22), SO356 (hog1 sho1), SO357 (hog1 sho1 ssk1), SO359 (hog1 sho1 ssk2 ssk22).
To determine whether mutants defective in genes other than HOG1 and PBS2 would exhibit osmolarity-induced cross talk, we carried out a screen for mutants that express FUS1::lacZ in the presence of 1 m sorbitol (see Materials and Methods). Of 19 mutants that exhibited β-galactosidase activity, 3 were constitutive and were not studied further. Of the 16 remaining mutants, all of which were osmosensitive, 10 were defective in HOG1 and 6 in PBS2 (see Materials and Methods). The mutant hunt therefore did not identify any genes other than HOG1 and PBS2 that are necessary to restrict cross talk.
Hog1p requires catalytic activity and phosphoacceptor sites to prevent osmolarity-induced cross talk
The studies above showed that strains deleted for HOG1 exhibit osmolarity-induced cross talk. Next, we determined whether mutations that affect specific aspects of Hog1p function also exhibit cross talk. The hog1–K52R mutation is predicted to abolish the catalytic activity of Hog1p; the hog1–T174A and hog1–Y176F mutations block the sites at which Pbs2p phosphorylates Hog1p (Schüller et al. 1994). hog1 deletion strains carrying plasmids with different HOG1 alleles were assayed for FUS1::lacZ expression in the presence of 1 m sorbitol (Fig. 3B). The strain carrying the wild-type HOG1 plasmid exhibited ∼3% of the full level of cross talk (probably because of plasmid loss). In contrast, the strain producing catalytically inactive Hog1p (hog1–K52R) produced 34% of the full level of cross talk. The hog1–Y176F and hog1–T174A mutants exhibited even higher levels (58% and 102%, respectively). The two mutants that exhibited lower levels of cross talk (hog1–K52R and hog1–Y176F) appear to retain some Hog1p activity, as they can support growth in the presence of intermediate osmolarity (Schüller et al. 1994). These observations indicate that catalytic activity of Hog1p is required to inhibit cross talk.
Osmolarity-induced cross talk requires components of the pheromone response pathway downstream of Ste4p, but not Ste5p
Activation of the FUS1::lacZ reporter by high osmolarity in Hog1p-deficient strains could in principle occur by activation of the pheromone response pathway at any level, from the receptor, Ste2p, to the transcription factor, Ste12p. To determine at what level the signal from high osmolarity feeds into the pheromone response pathway, we analyzed cross talk in hog1 mutants defective in different components of the pheromone response pathway (Fig. 3C). These hog1 strains exhibited two types of behavior: Those defective also in STE4 or STE5 behaved like a hog1 single mutant and exhibited an essentially full level of cross talk (74% and 81%, respectively). In contrast, hog1 strains also defective in STE20, STE50, STE11, STE7, STE12, or in both FUS3 and KSS1 exhibited no cross talk (<1%). hog1 mutants defective in either FUS3 or KSS1 exhibited osmolarity-induced cross talk like the hog1 single mutant (data not shown), indicating that Fus3p and Kss1p are functionally redundant for this pathway (see Cook et al. 1997; Madhani et al. 1997). These observations show that the pheromone response MAPK cascade and downstream transcription factor Ste12p are necessary for osmolarity-induced cross talk, as are Ste20p and Ste50p, which play an imperfectly defined role in activating Ste11p (Ramer and Davis 1993; Wu et al. 1995; Xu et al. 1996). In contrast, Ste4p, the Gβ component of the heterotrimeric G protein, and Ste5p, the scaffold for the MAPK module, are not required. a/α hog1/hog1 diploids, which do not express STE4 or STE5 (see Bardwell et al. 1994), also exhibit cross talk (data not shown). These observations indicate that the signal for activating transcription of FUS1::lacZ in hog1 mutants enters the pheromone response pathway after Ste4p and Ste5p and prior to Ste20p and Ste50p.
Only one of the known osmosensors, Sho1p, is required for osmolarity-induced cross talk
Because activation of the pheromone response pathway in hog1 and pbs2 mutants occurs in response to osmotic stress rather than being constitutive, we anticipated that components of the HOG pathway involved in sensing high osmolarity stress would be required for induction. To identify these components, we tested strains defective in both HOG1 and in additional genes of the HOG pathway. Strains defective in both HOG1 and PBS2, as expected, behaved the same as singly defective strains (Fig. 3D). Inactivating the Sln1p branch of the pathway, as in hog1 ssk1 and hog1 ssk2 ssk22 strains, similarly did not affect the level of cross talk. In contrast, hog1 strains defective in SHO1 exhibited a 78% reduction in cross talk. Cross talk exhibited by pbs2 mutants was similarly reduced by a mutation in SHO1 (data not shown). Inactivation of the Sln1p branch in the hog1 sho1 strain by deleting SSK1 or SSK2 and SSK22 did not reduce cross talk further. These results indicate that the major source of the high osmolarity signal that triggers the pheromone response pathway in hog1 mutants is the Sho1p branch, which shares components with the pheromone response pathway (see below). The observation that osmolarity-induced cross talk in hog1 sho1 ssk1 mutants remains at 16% of the full level, much higher than observed in hog1 ste11 or hog1 ste50 mutants (Fig. 3C), leads us to propose the existence of a third osmosensing branch in addition to Sho1p and Sln1p (see Discussion).
High osmolarity induces shmoo formation and mating in hog1 mutants
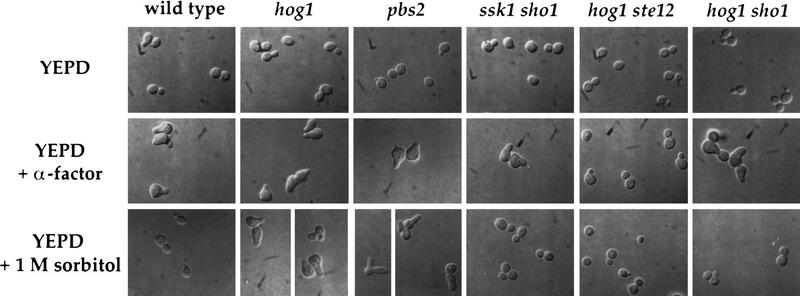
The pheromone response pathway governs not only induction of FUS1::lacZ but also mediates cellular morphology changes and mating. To determine whether osmolarity-induced activation of the pheromone response pathway exhibited by hog1 mutants extends to these processes, we first examined the ability of high osmolarity to induce morphological alterations (formation of pear-shaped cells, shmoos) characteristic of pheromone-treated cells. Shmoos formed by wild-type yeast in response to α-factor are shown in Figure 4 (middle row), in contrast to the budded morphology of cells grown in the absence of pheromone (Fig. 4, top row). Exposure of wild-type yeast cells to 1 m sorbitol had no effect on their morphology (Fig. 4, bottom row). In contrast, 1 m sorbitol induced hog1 and pbs2 mutants to form shmoo-like cells, a response that required functional SHO1 and STE12 genes (Fig. 4, bottom row). In other studies, we have observed that STE20, STE11, STE7, and FUS3 or KSS1 are required for this morphological response in hog1 strains, whereas STE4, STE5, PBS2, SSK1, SSK2, and SSK22 are not (S. O’Rourke, unpubl.). A strain defective in SSK1 and SHO1 did not exhibit a morphological change in response to 1 m sorbitol (Fig. 4, bottom row), just as this strain did not exhibit a high level of FUS1::lacZ induction when stimulated with 1 m sorbitol.
Figure 4.
High osmolarity induces a shmoolike morphology in hog1 and pbs2 mutants. (Top row) Cells incubated in YEPD liquid medium; (middle row) cells treated with α-factor in YEPD for 2 hr; (bottom row) cells treated with 1 m sorbitol in YEPD for 6 hr. Strains SO329 (wild type), SO330 (hog1), SO382 (pbs2), SO353 (ssk1 sho1), SO143 (hog1 ste12), and SO356 (hog1 sho1) were grown to log phase and shifted into fresh liquid media as indicated.
Having found that high osmolarity can induce expression of FUS1::lacZ and a shmoo-like morphology in hog1 mutants, we tested whether high osmolarity could also restore mating to hog1 strains also defective in various genes required for mating. Wild-type and hog1 strains mated efficiently in both the presence and absence of 1 m sorbitol (Table 1). As expected, HOG1 strains defective in STE4, STE5, STE11, STE7, STE12, or both FUS3 and KSS1 were defective in mating in the presence or absence of 1 m sorbitol (Table 1; data not shown); hog1 strains defective in different STE genes exhibited three different responses: (1) hog1 strains defective in STE11, STE7, STE12, or FUS3 and KSS1 were defective in mating both in the presence and absence of 1 m sorbitol (Table 1; data not shown); (2) the substantial residual mating exhibited by hog1 ste20 strains (∼10% that of hog1 STE20 strains) was reduced >70-fold by 1 m sorbitol (1 m sorbitol also reduced mating of the HOG1 ste20 strain 4-fold); and (3) mating by hog1 ste4 was increased 670-fold in the presence of 1 m sorbitol, from 2.7 × 10−7 to 1.8 × 10−4. An even more striking improvement in mating was displayed by the hog1 ste5 strain, which exhibited a 16,000-fold increase in mating in the presence of 1 m sorbitol, from 1.6 × 10−7 to 2.6 × 10−3. The improved mating due to 1 m sorbitol resulted from osmolarity-induced cross talk, as it was dependent on SHO1: The hog1 sho1 ste4 strain mated equally poorly in the absence or presence of 1 m sorbitol, and the hog1 sho1 ste5 strain exhibited only a threefold improvement in mating in the presence of 1 m sorbitol. Interestingly, 1 m sorbitol slightly improved mating by a HOG1 ste4 strain (a 4.5-fold incease), suggesting that a low level of osmolarity-induced cross talk might also occur in HOG1 cells. Thus, the mating assays parallel our results obtained with the FUS1::lacZ and morphological assays of cross talk: hog1 strains defective in STE4 or STE5 exhibit osmolarity-induced cross talk, but hog1 strains defective in SHO1, STE20, or downstream pheromone response pathway genes do not exhibit cross talk.
Table 1.
A hog1 mutation suppresses the mating defects of ste4 and ste5 strains in the presence of 1 m sorbitol
|
a strainsa
|
YEPD mating (%)b
|
YEPD + 1 m sorbitol mating (%)b
|
Effect of 1 m sorbitolc
|
|---|---|---|---|
| Wild type | 58 ± 5 | 74 ± 12 | 1.3 |
| ste4 | 1.4 ± 0.4 × 10−5 | 6.8 ± 2 × 10−5 | 4.9 |
| ste5 | 4.7 ± 2 × 10−6 | 6.6 ± 2 × 10−6 | 1.4 |
| ste20 | 2.9 ± 0.6 | 0.79 ± 0.3 | 0.27 |
| ste11 | <3.3 ± 2 × 10−6 | 5.3 ± 2 × 10−6 | — |
| hog1 | 68 ± 7 | 63 ± 9 | 0.93 |
| hog1 ste4 | 2.7 ± 1 × 10−5 | 1.8 ± 0.3 × 10−2 | 670 |
| hog1 ste5 | 1.6 ± 0.6 × 10−5 | 0.26 ± 0.08 | 16,000 |
| hog1 ste20 | 6.6 ± 2 | 9.4 ± 3 × 10−2 | 0.014 |
| hog1 ste11 | <3.8 ± 1 × 10−6 | 1.2 ± 0.7 × 10−5 | — |
| hog1 sho1 | 72 ± 8 | 61 ± 9 | 0.85 |
| hog1 sho1 ste4 | 3.5 ± 2 × 10−5 | 3.1 ± 0.8 × 10−5 | 0.89 |
| hog1 sho1 ste5 | 2.0 ± 2 × 10−5 | 6.1 ± 4 × 10−5 | 3.1 |
Strains tested were SO329 (wild type), SO341 (ste4), IH2731 (ste5), IH2735 (ste20), SO336 (ste11), SO330 (hog1), SO331 (hog1 ste4), SO332 (hog1 ste5), SO391 (hog1 ste20), SO333 (hog1 ste11), SO356 (hog1 sho1), SO344 (hog1 sho1 ste4), and SO398 (hog1 sho1 ste5).
Mating efficiency is the number of diploids formed divided by the number of total cells at the end of the experiment. Strains were mated to wild-type α strain IH1793.
The effect of 1 m sorbitol was calculated by dividing the mating efficiency on YEPD + 1 m sorbitol by the mating efficiency on YEPD.
Ste50p and Ste20p are components of the Sho1p-dependent branch of the HOG pathway
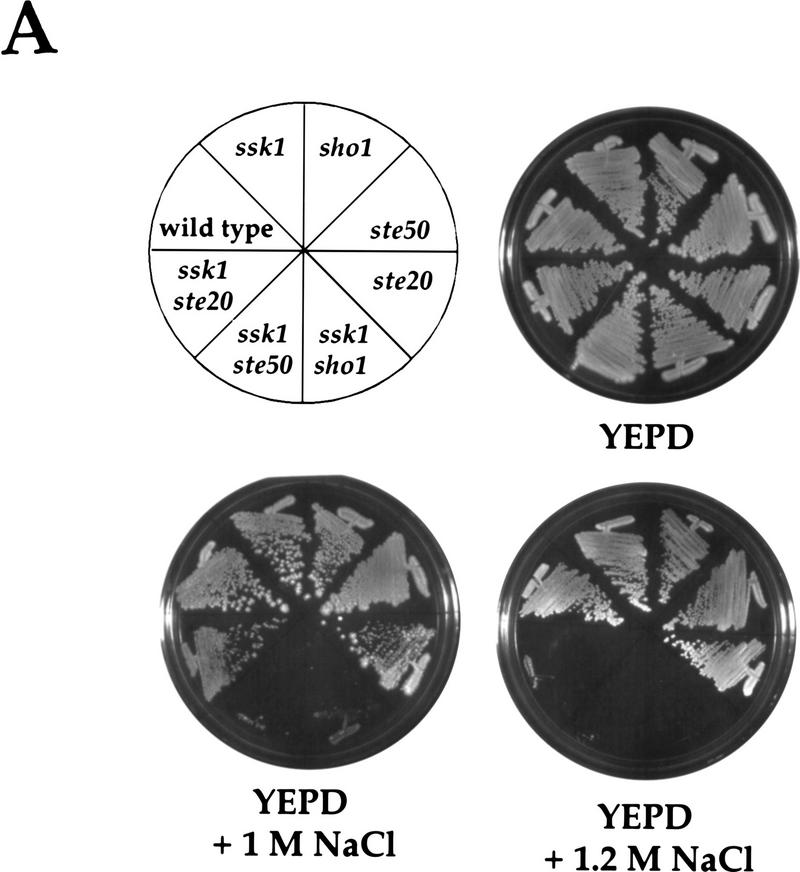
Because STE50 and STE20 are required for osmolarity-induced cross talk (Fig. 3C) and function upstream of Ste11p, we reasoned that these genes may normally function in the HOG pathway. The role of STE11 in the HOG pathway is revealed in mutants that are defective also in the Sln1p branch: ste11 ssk2 ssk22 and ste11 ssk1 mutants are osmosensitive, as is a sho1 ssk1 strain (Posas and Saito 1997; see below). As shown in Figure 5A, wild-type, ssk1, sho1, ste50, and ste20 strains grew equally well on YEPD and YEPD + NaCl plates. In contrast, the ste50 ssk1 double mutant was osmosensitive on YEPD + 1 m NaCl plates, as was the sho1 ssk1 double mutant. The ste50 sho1 double mutant was as osmoresistant as the ste50 and sho1 single mutants (data not shown), suggesting that Ste50p functions specifically in the Sho1p-dependent branch of the HOG pathway. As described previously, a ste20 mutation did not increase osmosensitivity when present in a strain defective for the Sln1p branch (Posas and Saito 1997). We verified this result in our strain background for growth on YEPD + 1 m NaCl medium. However, we observed that the ssk1 ste20 strain was osmosensitive on YEPD + 1.2 m NaCl medium (Fig. 5A). ste50 and ste20 mutations also caused osmosensitivity in strains lacking SSK2 and SSK22 (data not shown).
Figure 5.
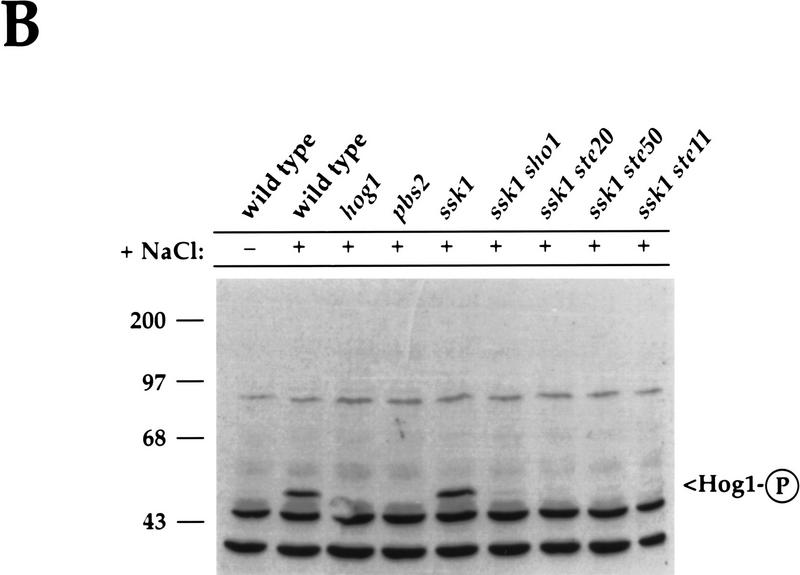
Ste50p and Ste20p function in the Sho1p branch of the HOG pathway. (A) Osmosensitivity of ste50 and ste20 mutants. Yeast strains were streaked on YEPD, YEPD + 1 m NaCl, and YEPD + 1.2 m NaCl plates, as indicated, and grown for 3 days (YEPD), 6 days (YEPD + 1 m NaCl) or 8 days (YEPD + 1.2 m NaCl), at 30°C to assay growth. SO329 (wild type), SO352 (ssk1), SO351 (sho1), SO360 (ste50), IH2735 (ste20), SO353 (ssk1 sho1), SO361 (ssk1 ste50), and SO387 (ssk1 ste20) were examined. The osmosensitivities of the ste50 ssk1 and ste20 ssk1 strains were complemented by low-copy STE50 and STE20 plasmids, respectively (data not shown), indicating that the phenotypes are attributable to the ste50 and ste20 mutations. (B) Ste50p- and Ste20p-dependent activation of Hog1p in an SSK1-deficient strain. Yeast cultures were grown in YEPD and treated with 0.7 m NaCl for 5 min, as indicated, prior to preparing total protein. The presence of activated Hog1p was visualized by probing a blot with an anti-phospho-specific p38 antibody as described in Materials and Methods. Molecular size markers are shown in kD. Strains analyzed were SO329 (wild type), SO330 (hog1), SO382 (pbs2), SO352 (ssk1), SO353 (ssk1 sho1), SO387 (ssk1 ste20), SO361 (ssk1 ste50), and SO399 (ssk1 ste11).
Because osmosensitivity can arise from a variety of physiological defects and not necessarily because of inability to activate Hog1p, we tested the role of Ste20p and Ste50p in the HOG pathway by assessing phosphorylation of Hog1p directly. As described previously, incubation of yeast cells in high osmolarity medium results in tyrosine phosphorylation of Hog1p (Brewster et al. 1993). We utilized an anti-phospho-specific p38 antibody that recognizes the phosphorylated form of p38 to detect Hog1p activation. In Figure 5B, we show that the antibody recognized phosphorylated Hog1p in extracts from wild-type yeast treated with 0.7 m NaCl for 5 min, but not in untreated cells or in extracts prepared from NaCl-treated hog1 or pbs2 mutants. As shown previously for ssk2 ssk22 strains (Maeda et al. 1995) and in Figure 5B for ssk1 strains, Hog1p is phosphorylated efficiently in mutants defective in the Sln1p branch, presumably by the Sho1p pathway. However, when SHO1, STE20, STE50, or STE11 is deleted in an ssk1 mutant, Hog1p was not phosphorylated during exposure to 0.7 m NaCl (Fig. 5B). The inability to phosphorylate Hog1p and the osmosensitivity data indicate that Sho1p, Ste20p, Ste50p, and Ste11p all function to transduce the osmotic stress signal to Hog1p during high extracellular osmolarity. In contrast, we found no role for Ste7p in the HOG pathway: A ste7 ssk1 double mutant phosphorylated Hog1p in response to 0.7 m NaCl and was osmoresistant on medium containing 1.2 m NaCl (data not shown).
SHO1 is required for pseudohyphal growth in wild-type and Hog1p-deficient strains
a/α strains of the Σ1278b background exhibit pseudohyphal growth when starved for nitrogen (on SLAHD medium; Gimeno et al. 1992). Pseudohyphal development requires STE20, STE11, STE7, KSS1, and STE12 but not STE4, STE5, or STE18 (Liu et al. 1993; Cook et al. 1997; Madhani et al. 1997; G. Fink, pers. comm.). a/α hog1/hog1 strains were reported recently to exhibit a hyper-pseudohyphal phenotype on SLAHD medium, which is at low osmolarity (Madhani et al. 1997; Fig. 6A, B). We reasoned that Sho1p-dependent cross talk may be responsible for this hyperpseudohyphal growth. The a/α hog1/hog1 sho1/sho1 strain exhibited greatly reduced pseudohyphal development (Fig. 6D): Only 0.5% of the colonies had extensive pseudohyphae, compared to 84.5% of colonies produced by the isogenic SHO1/SHO1 strain (Table 2). The hyperpseudohyphal growth of hog1/hog1 strains thus requires function of Sho1p under conditions of low osmolarity. We observed furthermore that HOG1 strains also require SHO1 for pseudohyphal development (Fig. 6C): Only 2% of the colonies formed by the HOG1/HOG1 sho1/sho1 strain exhibited robust pseudohyphal development, and 92.5% lacked pseudohyphae altogether (Table 2). In contrast, 43% of wild-type colonies exhibited pseudohyphae like those in Figure 6A, and 31.5% lacked pseudohyphae. These observations demonstrate that SHO1 is necessary both for the hyperpseudohyphal response of hog1/hog1 strains as well as for normal pseudohyphal development in wild-type strains.
Figure 6.
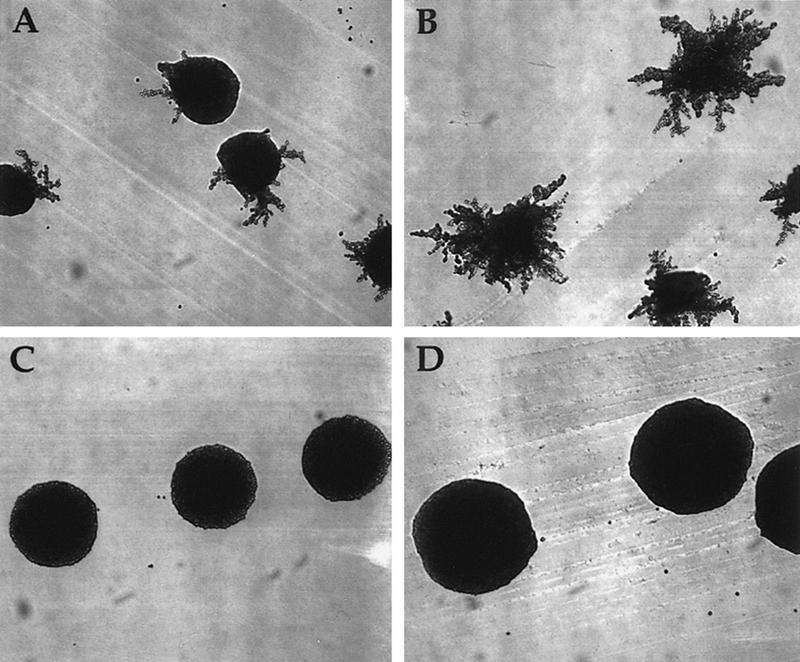
Hyperfilamentous growth of hog1/hog1 a/α Σ1278b strains is dependent on SHO1. (A) wild type (SO392) + YCplac22, (B) hog1/hog1 (SO393) + YCplac22, (C) sho1/sho1 (SO394), and (D) hog1/hog1 sho1/sho1 (SO395) strains in the Σ1278b genetic background were streaked on a SLAHD plate and grown for 5 days.
Table 2.
SHO1 is required for pseudohyphal growth
| Strainsa
|
Filamentation (% of total colonies)
|
||
|---|---|---|---|
| robust
|
weak
|
none
|
|
| Wild type | 43 | 25.5 | 31.5 |
| hog1/hog1 | 84.5 | 15 | 0.5 |
| sho1/sho1 | 2 | 6 | 92.5 |
| hog1/hog1 sho1/sho1 | 0.5 | 15 | 84.5 |
a/α Σ1278b strains assayed were wild type (SO392), hog1/hog1 (SO393), sho1/sho1 (SO394), and hog1/hog1 sho1/sho1 (SO395). Two hundred colonies were counted for each strain. The SO392 and SO393 strains harbored plasmid YCplac22 to complement the trp1 auxotrophy in these strains. Robust, as in Fig. 6, A and B; weak, one filament per colony.
Discussion
The presence of Ste11p in three different MAPK pathways (the pheromone response pathway, the pseudohyphal development pathway, and the HOG pathway; see Fig. 7) poses the problem of cross talk acutely: What is the mechanism that restricts Ste11p to activate only a subset of potential targets? We have found that Hog1p, the MAPK of the HOG pathway, limits activation of the pheromone response pathway by high osmolarity (cross talk). In particular, hog1 mutants exhibit inappropriate signaling (diagrammed as a dashed line in Fig. 7), which is manifested in several different ways: activation of FUS1::lacZ, induction of shmoo morphology, suppression of mating defects in ste4 and ste5 mutants, and hyperpseudohyphal growth. Our studies have focused on three aspects of this phenomenon. First of all, we have identified the components of the HOG pathway and the pheromone response pathway that are required for osmolarity-induced cross talk in hog1 mutants. Second, we have exploited features of this cross talk phenomenon to reveal a role for Ste50p and Ste20p in the HOG pathway and to suggest the existence of a third input to the HOG pathway. Finally, our experiments with Σ1278b strains suggest that Sho1p provides input not only for the HOG pathway but for the pseudohyphal pathway as well.
Figure 7.
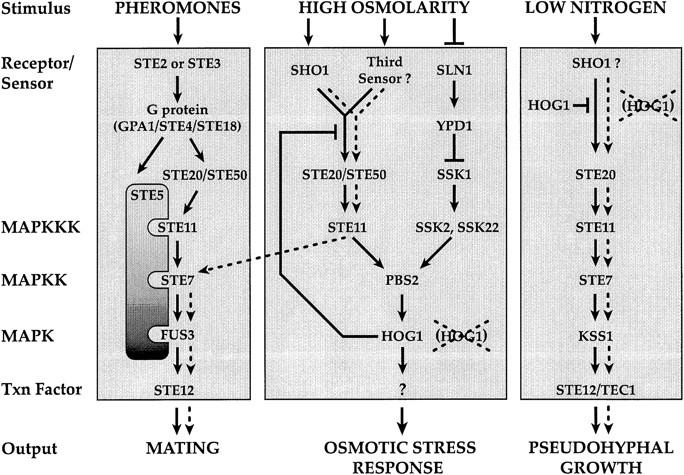
A revised model of the pheromone response, HOG, and pseudohyphal growth pathways. The diagram shows feedback regulation of the Sho1p branch of the HOG pathway by Hog1p. Ste50p and Ste20p are shown as components of both the pheromone response and HOG pathways. Sho1p is shown as a sensor for both the pseudohyphal growth and HOG pathways. Processes that occur inappropriately in hog1 mutants (osmolarity-induced cross talk to the pheromone response pathway and hyperpseudohyphal growth) are shown as broken lines.
Osmolarity-induced cross talk uses components of the HOG pathway, including some newly identified components
Osmolarity-induced activation of the pheromone response pathway exhibited by hog1 mutants begins with an osmosensing component of the HOG pathway (Sho1p) and culminates with the pheromone response MAPK cascade. The importance of Sho1p is demonstrated by the observation that mutation of SHO1 causes a 78% reduction in the level of cross talk in hog1 strains. The Sln1p branch of the HOG pathway apparently plays no role in osmolarity-induced cross talk: hog1 mutants defective in SSK1 or in SSK2 and SSK22 still exhibit high levels of cross talk.
The finding that Ste50p and Ste20p are required for osmolarity-induced cross talk in hog1 mutants led us to test their role in the HOG pathway of wild-type cells. Such a role could be seen in strains lacking the Sln1p branch of the HOG pathway as measured by osmosensitivity and phosphorylation of Hog1p (Fig. 5). In particular, a STE50 mutation caused osmosensitivity in a strain lacking SSK1. Although prior studies (Posas and Saito 1997) had failed to find a requirement of STE20 in osmotolerance, we observed that ste20 ssk1 strains are much more sensitive than STE20 ssk1 strains to 1.2 m NaCl. In addition to being osmosensitive, the ste20 ssk1 and ste50 ssk1 strains similarly failed to activate Hog1p during high osmolarity stress, as was found previously for strains defective for both the Sln1p branch (ssk2 ssk22) and SHO1 or STE11 (Posas and Saito 1997). These observations indicate that Ste20p and Ste50p are involved in activation of Ste11p in both the HOG and pheromone response pathways.
The HOG pathway may have a third input branch
Three different observations lead us to propose the existence of a third input branch to the HOG pathway. First, in a hog1 background, eliminating the two known input branches to the pathway, by mutation in SHO1 and SSK1 or in SHO1, SSK2, and SSK22, reduced but did not eliminate osmolarity-induced cross talk. Second, we observed that sho1 ssk1 and sho1 ssk2 ssk22 strains exhibited a significant level of cross talk. Because the cross talk observed in these strains is osmolarity-induced, it is clear that an osmosensing function remains active even in the absence of the two known inputs to the HOG pathway. Finally, we observed that ste11 ssk1 and ste50 ssk1 strains display a stronger osmosensitive phenotype than sho1 ssk1 strains (S. O’Rourke, unpubl.), suggesting that the hypothetical third input to the HOG pathway feeds into Ste11p and Ste50p.
Osmolarity-induced cross talk uses some elements of the pheromone response pathway, but not Ste5p
As expected, osmolarity-induced cross talk utilizes components of the pheromone response pathway in addition to those shared with the HOG pathway: The MAPK module and the transcription factor Ste12p are also required. Given that Sho1p is thought to normally activate Ste11p for the osmotic stress response, it is not surprising that a more upstream component of the pheromone response pathway, Gβ (Ste4p), is not required for cross talk. It is striking that Ste5p is not required for osmolarity-induced activation of the pheromone response pathway reporter FUS1::lacZ, given that it is required when the pheromone response pathway MAPK cascade, composed of Ste11p, Ste7p, and the MAPK Fus3p, is activated by pheromone (McCaffrey et al. 1987). This observation parallels the finding that the MAPK module of the pseudohyphal development pathway, containing Ste11p and Ste7p along with the MAPK Kss1p, also does not require Ste5p (G. Fink, pers. comm.). These observations lead us to propose that Sho1p may perform some of the same functions as Ste5p, in particular, for linking the activation of Ste11p to the external environment and for activation of Ste7p. As discussed further below, we propose that Sho1p performs these functions not only during osmolarity-induced cross talk in hog1 mutants, but also during pseudohyphal growth in wild-type (HOG1) cells.
hog1 mutants induce multiple STE12-dependent processes
High osmolarity induces all of the same processes in hog1 mutants as are induced by mating pheromones in wild-type cells. The classical reporter for transcriptional activation, FUS1::lacZ, is induced more slowly by 1 m sorbitol than by α-factor, but the levels of induction are comparable after a few hours (Fig. 2). Likewise, we observed that 1 m sorbitol induces hog1 and pbs2 mutants to form projections similar to those induced by α-factor. Perhaps most strikingly, high osmolarity improved mating by ste4 hog1 and ste5 hog1 mutants 670- and 16,000-fold, respectively (Table 1). The greater increase seen in the ste5 mutant than in the ste4 mutant may be due to roles of Ste4p during mating in addition to stimulating the MAPK module (Schrick et al. 1997). The ability to activate the pheromone response MAPK module in the absence of Ste4p and Ste5p using sorbitol as a mating pheromone offers some novel opportunities to dissect early steps in the mating pathway.
In contrast to the ability of high osmolarity to improve mating in ste4 hog1 and ste5 hog1 mutants, 1 m sorbitol did not suppress the mating defect of ste11 hog1, ste7 hog1, fus3 kss1 hog1, or ste12 hog1 strains. This lack of suppression is consistent with the idea that the cross talk signal enters the pheromone response pathway upstream of Ste11p.
We observed that the hyperpseudohyphal growth of hog1/hog1 strains (Madhani et al. 1997) is dependent on SHO1 and thus may represent osmolarity-independent activation of Ste11p by Sho1p. To our surprise, we observed that SHO1 is also required for pseudohyphal development in wild-type (HOG1) strains. These results indicate that Sho1p provides input to the pseudohyphal development pathway not only under the unnatural conditions of a hog1p-mutant strain but in wild-type strains as well. According to this view, the hyperpseudohyphal growth of hog1/hog1 strains would not represent cross talk but, rather, constitutive activation of the pathway by loss of a negative regulator (HOG1). It will be of interest to determine whether Sho1p and Hog1p respond not only to osmotic stress but to other stimuli as well. In Schizosaccharomyces pombe, the pathway that senses high osmolarity responds to multiple stimuli, including nutrient limitation (for review, see Banuett 1998).
Hog1p may down-regulate the Sho1p branch of the HOG pathway to limit cross talk
It has been proposed that the presence of Ste11p in different protein complexes (with Ste7p, Fus3p, and Ste5p and with Pbs2p, Hog1p, and Sho1p) restricts cross activation (Posas and Saito 1997). Activated Ste11p might phosphorylate only the MAPKK (Ste7p or Pbs2p) present within its complex. Evidence that Ste5p can sequester Ste7p comes from the observation that the ability of certain forms of Ste7p to partially bypass a mutation in the PKC pathway (bck1) is greater in ste5 mutants than in STE5 strains (Yashar et al. 1995). We have shown that high levels of cross talk are observed in the complete absence of Hog1p or with mutant proteins that are defective in its catalytic activity or in its activation. Because the hog1–K52R and hog1–T174A proteins are tyrosine phosphorylated during osmotic stress (Schüller et al. 1994), they presumably form a complex with Pbs2p, Sho1p, and Ste11p. If they do form a complex, then scaffolding per se appears not be sufficient to maintain signal specificity. Instead, we propose that phosphorylation of a target protein by Hog1p is responsible for limiting cross talk in wild-type strains.
In principle, the target of Hog1p responsible for limiting cross talk might be any component of the osmolarity-induced cross talk pathway (from Sho1p to Ste12p). If Hog1p inhibited a component of the pheromone response MAPK cascade or other proteins necessary for mating, we might expect 1 m sorbitol to inhibit mating or induction of FUS1::lacZ by α-factor. Such inhibition has not been observed (Fig. 2; Table 1; S. O’Rourke, unpubl.). Based on these observations, we suggest that Hog1p may down-regulate components of the HOG pathway, specifically the Sho1p branch, after stimulation by high osmolarity. Although we cannot rigorously exclude Ste50p, Ste20p, and Ste11p as targets, a particularly appealing possibility is that Sho1p is phosphorylated by Hog1p.
Feedback regulation of receptor activity by protein phosphorylation has been well documented for G-protein-coupled receptors (for review, see Ferguson et al. 1996). We propose that inhibition of the Sho1p branch of the HOG pathway ensures that activation of the pathway is transient. Sustained activation of the HOG pathway results in lethality (Maeda et al. 1994; Wurgler-Murphy et al. 1997). Thus, mechanisms that down-regulate the pathway after the appropriate responses have occurred are essential. The protein phosphatases Ptp2p and Ptp3p inhibit Hog1p activity (Jacoby et al. 1997; Wurgler-Murphy et al. 1997), which is sufficient for inhibiting the Ssk1p-dependent branch, as Hog1p is the only known target of this branch. However, if Hog1p were the only point of down-regulation, high osmolarity would cause chronic stimulation of the Sho1p branch (including Ste20p, Ste50p, and Ste11p), which could lead to inappropriate activation of the pheromone response and pseudohyphal growth pathways in wild-type cells. We thus propose that Hog1p plays a critical role in adaptation of yeast to high osmolarity conditions by inhibiting the Sho1p branch of the HOG pathway.
Hall et al. (1996) reported recently that high osmolarity (0.5 m NaCl) can induce the pheromone response pathway. These workers observed a twofold increase in phosphorylation of Fus3p on tyrosine-182, which is phosphorylated by Ste7p (Gartner et al. 1992). Strikingly, phosphorylation was increased 13-fold after osmotic shock of a hog1Δ strain. Hall et al. (1996) interpreted these observations to indicate that high osmolarity induced cross talk and that this cross talk was inhibited by the HOG pathway. Other analyses led them to suggest that this inhibition might be exerted both on Fus3p and on an additional component. We suggest that this inhibition is mediated by Hog1p acting early in the pathway, perhaps on Sho1p itself. Although there are aspects of the observations by Hall et al. (1996) that we do not understand (in particular, why they did not observe induction of FUS1::lacZ by high osmolarity in their hog1 strain), we believe that their findings and ours on cross talk in hog1 strains concern the same phenomenon. In particular, we propose that the transient activation of Fus3p observed by Hall et al. (1996) with 0.5 m NaCl treatment of wild-type strains represents a normal level of cross activation of the pheromone response pathway that is subsequently damped by Hog1p.
The observation that mutations in HOG1 and PBS2 allow high levels of cross talk explains prior observations, particularly on enhanced pseudohyphal development exhibited by hog1 mutants (Madhani et al. 1997) and on increased basal expression of the pheromone response pathway seen in mutants defective in PBS2 (Stevenson et al. 1995). Our discovery that osmolarity-induced cross talk works via the Sho1p branch of the HOG pathway made it possible to uncover the role of Ste20p and Ste50p in this branch of the HOG pathway. We anticipate that Sho1p may communicate similarly with Ste20p and Ste50p to activate the pseudohyphal development pathway in wild-type cells.
Materials and methods
Strains, media, and genetic techniques
Yeast strains were grown in YEPD medium (1% yeast extract, 2% Bacto-Peptone, 2% glucose) at 30°C. Synthetic complete medium (Rose et al. 1990) was used for maintaining plasmids and selecting gene replacements. d-sorbitol and NaCl (Sigma) were used at final concentrations of 1 or 1.2 m as indicated. For α-factor treatments, cells were grown in liquid YEPD medium, and 0.5 mg/ml α-factor in 0.01 m HCl was added to a final concentration of 0.005 mg/ml. Yeast transformations were done by the lithium acetate procedure (Schiestl and Gietz 1989). Yeast strains (Table 3) were derived from the EG123 strain background (trp1-1 leu2-3, 112 ura3-52 his4 can1; Siliciano and Tatchell 1984), except for the Σ1278b-derived strains SO392, SO393, SO394, and SO395, and the MATα mating tester (IH1793). The four Σ1278b strains were derivatives of RRY1045 (MATa/MATα ura3-52/ura3-52 his3::hisG/his3::hisG leu2::hisG/LEU2trp1::hisG/TRP1), which was obtained from G. Fink (Whitehead Institute, MIT, Cambridge, MA). Gene disruptions were confirmed by phenotypic analysis and/or PCR reactions with gene-specific primers.
Table 3.
Strains used
| Strain
|
Relevant genotype
|
Source
|
|---|---|---|
| IH1793 | MATα lys1 | Ira Herskowitz (IH) collection |
| IH2731 | MATa ste5::LEU2 | IH collection |
| IH2735 | MATa ste20::TRP1 | IH collection |
| SO143 | MATa hog1::hisG ste12::TRP1 FUS1::lacZ::LEU2 | this study |
| SO329 | MATa FUS1::lacZ::LEU2 | this study |
| SO330 | MATa hog1::hisG FUS1::lacZ::leu2 | this study |
| SO331 | MATa hog1::hisG ste4::LEU2 FUS1::lacZ::leu2 | this study |
| SO332 | MATa hog1::hisG ste5::LEU2 FUS1::lacZ::LEU2 | this study |
| SO333 | MATa hog1::hisG ste11::URA3 FUS1::lacZ::LEU2 | this study |
| SO334 | MATa hog1::hisG ste7::LEU2 FUS1::lacZ::leu2 | this study |
| SO335 | MATa hog1::hisG fus3::URA3 kss1::URA3 FUS1::lacZ::leu2 | this study |
| SO336 | MATa ste11::URA3 FUS1::lacZ::leu2 | this study |
| SO341 | MATa ste4::LEU2 FUS1::lacZ::leu2 | this study |
| SO344 | MATa hog1::hisG sho1::TRP1 ste4::LEU2 FUS1::lacZ::leu2 | this study |
| SO351 | MATa sho1::TRP1 FUS1::lacZ::leu2 | this study |
| SO352 | MATa ssk1::LEU2 FUS1::lacZ::leu2 | this study |
| SO353 | MATa ssk1::LEU2 sho1::TRP1 FUS1::lacZ::leu2 | this study |
| SO354 | MATa ssk2::LEU2 ssk22::LEU2 FUS1::lacZ::leu2 | this study |
| SO355 | MATa ssk2::LEU2 sk22::LEU2 sho1::TRP1 FUS1::lacZ::leu2 | this study |
| SO356 | MATa hog1::hisG::URA3::hisG sho1::TRP1 FUS1::lacZ::leu2 | this study |
| SO357 | MATa hog1::hisG::URA3::hisG ssk1::LEU2 sho1::TRP1 FUS1::lacZ::leu2 | this study |
| SO358 | MATa hog1::hisG::URA3::hisG ssk2::LEU2 ssk22::LEU2 FUS1::lacZ::leu2 | this study |
| SO359 | MATa hog1::hisG::URA3::hisG ssk2::LEU2 ssk22::LEU2 sho1::TRP1 FUS1::lacZ::leu2 | this study |
| SO360 | MATa ste50::hisG::URA3::hisG FUS1::lacZ::leu2 | this study |
| SO361 | MATa ste50::hisG::URA3::hisG ssk1::LEU2 FUS1::lacZ::leu2 | this study |
| SO373 | MATa hog1::hisG ste50::hisG::URA3::hisG FUS1::lacZ::leu2 | this study |
| SO382 | MATa pbs2::LEU2 FUS1::lacZ::leu2 | this study |
| SO383 | MATa hog1::hisG::URA3::hisG pbs2::LEU2 FUS1::lacZ::leu2 | this study |
| SO384 | MATa hog1::hisG::URA3::hisG ssk1::LEU2 FUS1::lacZ::leu2 | this study |
| SO385 | MATa gpd1::TRP1 FUS1::lacZ::LEU2 | this study |
| SO387 | MATa ssk1::LEU2 ste20::TRP1 FUS1::lacZ::LEU2 | this study |
| SO391 | MATa hog1::hisG ste20::TRP1 FUS1::lacZ::LEU2 | this study |
| SO392 | MATa/MATα leu2/LEU2 | this study |
| SO393 | MATa/MATα leu2/LEU2 hog1::hisG::URA3::hisG/hog1::hisG::URA3::hisG | this study |
| SO394 | MATa/MATα leu2/LEU2 sho1::TRP1/sho1::TRP1 | this study |
| SO395 | MATa/MATα leu2/LEU2 sho1::TRP1/sho1::TRP1 hog1::hisG::URA3::hisG/hog1::hisG::URA3::hisG | this study |
| SO398 | MATa hog1::hisG::URA3::hisG ste5::LEU2 sho1::TRP1 | this study |
| SO399 | MATa ste11::ura3::TRP1 ssk1::LEU2 FUS1::lacZ::leu2 | this study |
Strains SO392–SO395 are in the Σ1278b strain background, whose full genotype is trp1 leu2 ura3 his3. All other strains (except IH1793) are in the EG123 strain background, whose full genotype is trp1 leu2 ura3 his4 can1.
Escherichia coli strain DH5α was used for propagation of plasmids (Table 4). To construct the sho1::TRP1 disruption plasmid, a 1.7-kb fragment encompassing the SHO1 locus from −392 bp from the initiating ATG to +229 bp from the stop codon was amplified from genomic DNA prepared from yeast strain SO329 by PCR using Expand polymerase in Expand buffer no. 1. The 5′ primer had the sequence ATAGACCCTTGAACCTACATATCCG, and the 3′ primer had the sequence GTCAAGTCAATGACATGAGAGTGC. The PCR product was ligated into the pCR2.1 vector (Invitrogen) to yield pSO19. pSO19 was cleaved with SnaBI and ClaI to remove the entire SHO1 coding region except for the DNA encoding the 11 carboxy-terminal amino acids. After treating the cleaved pSO19 with the Klenow fragment, a Klenow-treated, 0.85-kb, BamHI TRP1 DNA fragment from YDp-W (Berben et al. 1991) was ligated into the cleaved pSO19 to yield pSO72. The orientation of the TRP1 gene in pSO72 was determined to be opposite that of the SHO1 gene. Treatment of pSO72 with NotI and SpeI liberated the sho1::TRP1 deletion allele that was used for yeast transformations.
Table 4.
Plasmids used
| Plasmid
|
Description
|
Source/Reference
|
|---|---|---|
| pRS316 | CEN6 URA3 | Sikorski and Hieter (1989) |
| pJB15 | pRS316 + HOG1 | Brewster et al. (1993) |
| pJBM3 | pRS316 + hog1–K52R | Schüller et al. (1994) |
| pJBM4 | pRS316 + hog1–T174A | Schüller et al. (1994) |
| pJBM2 | pRS316 + hog1–Y176F | Schüller et al. (1994) |
| pJB41 | pRS316 + PBS2 | Brewster et al. (1993) |
| pste4::LEU2 | ste4::LEU2 | Whiteway et al. (1989) |
| pDM4U | ste50::hisG::URA3::hisG | Xu et al. (1996) |
| pNC202 | ste11::URA3 | Beverly Errede (University of North Carolina, Chapel Hill) |
| pDH90 | ste7::LEU2 | D. Higgins and K. Tatchell (Louisiana State University, Baton Rouge) |
| pBC99 | hog1::hisG::URA3::hisG | Bruce Cree/Ira Herskowitz (IH) collection |
| pMA11 | pbs2::LEU2 | Brewster et al. (1993) |
| pSO72 | sho1::TRP1 | this study |
| pDSS14 | ssk1::LEU2 | H. Saito (Dana-Farber Cancer Institute, Boston, MA) |
| pDSS24 | ssk2::LEU2 | H. Saito |
| pDYC734 | ssk22::LEU2 | H. Saito |
| pUCgpd1::TRP1 | gpd1::TRP1 | Albertyn et al. (1994) |
| pFC23 | FUS1::lacZ::LEU2 | Fred Chang/IH collection |
| p307 | leu2Δ::URA3 | Phil Hieter (University of British Columbia, Vancouver, Canada) |
| pUT11 | ura3Δ::TRP1 | Cross (1997) |
| YCplac22 | CEN4 TRP1 | Gietz and Sugino (1988) |
The FUS1::lacZ::LEU2 integrating reporter gene, from plasmid pFC23, was introduced into a wild-type strain (IH1783), which was crossed to other strains to create isogenic strains with respect to the FUS1::lacZ::LEU2 gene. For creating the FUS1::lacZ::leu2Δ reporter, plasmid p307 (LEU2::URA3::leu2Δ) was transformed into yeast containing the FUS1::lacZ::LEU2 reporter and plated on synthetic complete medium lacking uracil. Strains were then patched onto 5-fluoro-orotic acid plates to select for loopouts of the URA3 gene (Boeke et al. 1984). A leu2 isolate was used in subsequent crosses to obtain strains containing the FUS1::lacZ::leu2Δ reporter gene. The activity of the FUS1::lacZ reporter gene was identical when marked with either the LEU2 or leu2Δ allele.
β-Galactosidase assays
LacZ expression was measured as described previously (Stern et al. 1984), except that log-phase cells were treated for 5 or 6 hr as indicated by diluting into fresh medium, medium containing α-factor, or medium containing 1 m sorbitol prior to harvesting.
Yeast mutagenesis and screening for hog1-like mutants
A FUS1::lacZ a strain (SO329) was mutagenized with methanesulfonic acid ethyl ester (Sigma) in four separate pools to 22%–86% killing and plated on YEPD plates at a density of ∼300 colonies per plate (Lawrence 1991). After growth for 3–7 days, colonies were patched onto a YEPD plate at a density of 50/plate. The plates were incubated at 30°C overnight and replica plated to a No. 3 Whatman filter on a YEPD + 1 m sorbitol plate and grown overnight. β-Galactosidase assays were performed on the filters to identify strains expressing FUS1::lacZ. Of 2400 strains examined, 16 were isolated that induced FUS1::lacZ in response to 1 m sorbitol. These were tested for complementation of the FUS1::lacZ induction phenotype by HOG1- and PBS2-containing plasmids (Brewster et al. 1993). The sensitivity of the screen was demonstrated by the isolation of one apparent hog1 mutant that exhibited 22% of the cross talk observed for the hog1 deletion strain.
Quantitative mating analysis
Mating efficiencies were performed as described (Valtz and Herskowitz 1996), except that 6.0 × 106 log-phase cells were used per mating reaction, and mating reactions were performed for 24 hr. Additionally, sterile filters containing the mating mixes were cut in half: One-half was incubated on YEPD, and the other half was incubated on YEPD + 1 m sorbitol.
Microscopy
Yeast cells were photographed with a Zeiss axioskop microscope with a 100× objective lens. Colonies on plates were photographed with a Zeiss Axioskop microscope using a 10× objective lens.
Detection of phosphorylated Hog1p
To assay Hog1p activation, 1.0 OD600 unit of log phase yeast was harvested for each strain and concentrated in 1 ml of YEPD medium. For NaCl treatments, 0.25 ml of 3.5 m NaCl was added to the cells for 5 min at room temperature in microcentrifuge tubes. Protein extracts were prepared by centrifuging the tubes for 30 sec and aspirating the supernatants. Cell pellets were incubated on ice for 5 min prior to resuspension in 150 μl of 1.85 m NaOH, 7.4% β-mercaptoethanol. After incubation for 10 min on ice, 150 μl ice-cold 50% TCA was added. Samples were incubated on ice for 10 min followed by centrifugation at 4°C for 2 min. Pellets were washed with 1 ml of ice-cold acetone, centrifuged at 4°C for 2 min, and resuspended in 100 μl of 1× sample buffer containing 1 mm Na3VO4. After boiling the samples for 5 min and briefly centrifuging, 15 μl was run on a 10% polyacrylamide gel that was transferred to a nitrocellulose membrane. After blocking the nitrocellulose in 5% BSA, 20 mm Tris (pH 7.5), 428 mm NaCl, 0.2% Tween 20 (TBST) for 1 hr at room temperature, the membrane was probed with the anti-phospho-specific p38 antibody (New England Biolabs) in TBST overnight. Immunoreactivity was localized with the ECL system (Amersham) using horseradish peroxidase-conjugated anti-rabbit secondary antibodies.
Acknowledgments
We thank Michael Gustin, Linda Huang, Massoud Ramezani Rad, and Haruo Saito for generously providing plasmids, Bruce Cree, Aaron Neiman, and Cora Styles for strains, and Gerry Fink for personal communication. We also thank members of our laboratory, Shaun Coughlin and Wendell Lim, for valuable discussion, and Flora Banuett, Linda Huang, Kenji Irie, Doug Jeffery, and Mary Maxon, for comments on the manuscript. This work was supported by National Institutes of Health (NIH) grant AI18738 (to I. H.). S.O’R. was supported by an NIH training grant, the Markey Program in Biological Sciences, and the Herbert W. Boyer Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ira@cgl.ucsf.edu; FAX (415) 476-0943.
References
- Albertyn J, Hohmann S, Thevelein JM, Prior BA. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Signalling in the yeasts: An informational cascade with links to the filamentous fungi. Microbiol. Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Inouye CJ, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Blomberg A, Adler L. Physiology of osmotolerance in fungi. Adv Microbiol Physiol. 1992;33:145–212. doi: 10.1016/s0065-2911(08)60217-9. [DOI] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol & Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Kron SJ, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes & Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- Cross FR. “Marker swap” plasmids: Convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Davenport KR, Sohaskey M, Kamada Y, Levin DE, Gustin MC. A second osmosensing signal transduction pathway in yeast. Hypotonic shock activates the PKC1 protein kinase-regulated cell integrity pathway. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- Dietzel C, Kurjan J. The yeast SCG1 gene: α Gα-like protein implicated in the a- and α-factor response pathway. Cell. 1987;50:1001–1010. doi: 10.1016/0092-8674(87)90166-8. [DOI] [PubMed] [Google Scholar]
- Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- Feng Y, Song LY, Kincaid E, Mahanty SK, Elion EA. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, Barak LS, Zhang J, Caron MG. G-protein-coupled receptor regulation: Role of G-protein-coupled receptor kinases and arrestins. Can J Physiol Pharmacol. 1996;74:1095–1110. doi: 10.1139/cjpp-74-10-1095. [DOI] [PubMed] [Google Scholar]
- Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes & Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Hall JP, Cherkasova V, Elion E, Gustin MC, Winter E. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: Isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol Cell Biol. 1996;16:6715–6723. doi: 10.1128/mcb.16.12.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: Role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- Jacoby T, Flanagan H, Faykin A, Seto AG, Mattison C, Ota I. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes & Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- Larsson K, Ansell R, Eriksson P, Adler L. A gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) complements an osmosensitive mutant of Saccharomyces cerevisiae. Mol Microbiol. 1993;10:1101–1111. doi: 10.1111/j.1365-2958.1993.tb00980.x. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Classical mutagenesis techniques. In: Guthrie C, Fink GR, editors. Guide to yeast genetics and molecular biology. San Diego, CA: Academic Press; 1991. pp. 273–281. [Google Scholar]
- Leberer E, Thomas DY, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- Leeuw T, Wu C, Schrag JD, Whiteway M, Thomas DY, Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Levin DE, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:2680–2690. doi: 10.1128/mcb.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima I, Nakafuku M, Nakayama N, Brenner C, Miyajima A, Kaibuchi K, Arai K, Kaziro Y, Matsumoto K. GPA1, a haploid-specific essential gene, encodes a yeast homolog of mammalian G protein which may be involved in mating factor signal transduction. Cell. 1987;50:1011–1019. doi: 10.1016/0092-8674(87)90167-x. [DOI] [PubMed] [Google Scholar]
- Neiman AM, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: Activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- ————— Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 1998;17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramer SW, Davis RW. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes & Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Schrick K, Garvik B, Hartwell LH. Mating in Saccharomyces cerevisiae: The role of the pheromone signal transduction pathway in the chemotropic response to pheromone. Genetics. 1997;147:19–32. doi: 10.1093/genetics/147.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller C, Brewster JL, Alexander MR, Gustin MC, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano PG, Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984;37:969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Stevenson BJ, Ferguson B, De Virgilio C, Bi E, Pringle JR, Ammerer G, Sprague GF., Jr Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes & Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Hougan L, Dignard D, Thomas DY, Bell L, Saari GC, Grant FJ, O’Hara P, MacKay VL. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Wu C, Whiteway M, Thomas DY, Leberer E. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Jansen G, Thomas DY, Hollenberg CP, Ramezani Rad M. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1996;20:773–783. doi: 10.1111/j.1365-2958.1996.tb02516.x. [DOI] [PubMed] [Google Scholar]
- Yashar B, Irie K, Printen JA, Stevenson BJ, Sprague GF, Jr, Matsumoto K, Errede B. Yeast MEK-dependent signal transduction: Response thresholds and parameters affecting fidelity. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]