Abstract
In Euplotes crassus, telomerase is responsible for telomere maintenance during vegetative growth and de novo telomere synthesis during macronuclear development. Here we show that telomerase in the vegetative stage of the life cycle exists as a 280-kD complex that can add telomeric repeats only onto telomeric DNA primers. Following the initiation of macronuclear development, telomerase assembles into larger complexes of 550 kD, 1600 kD, and 5 MD. In the 1600-kDa and 5-MDa complexes, telomerase is more processive than in the two smaller complexes and can add telomeres de novo onto nontelomeric 3′ ends. Assembly of higher order telomerase complexes is accompanied by an extended region of RNase V1 and RNase T1 protection in the telomerase RNA subunit that is not observed with telomerase from vegetatively growing cells. The protected residues encompass a highly conserved region previously proposed to serve as a platform for formation of higher order structures. These findings provide the first direct demonstration of developmentally regulated higher order telomerase complexes with unique biochemical and structural properties.
Keywords: Telomerase, chromosome healing, Euplotes crassus, RNA footprinting, holoenzyme
The integrity of the telomere complex is essential for genome stability in eukaryotes. The primary mechanism for generating and sustaining chromosome ends is through the action of telomerase, a ribonucleoprotein (RNP) that catalyzes the addition of telomeric DNA repeats (Greider 1995; Melek and Shippen 1996). Given the strong correlation between telomerase expression and cellular proliferation capacity in higher eukaryotes and tumorigenesis in humans (Greider 1998; Shippen and McKnight 1998), understanding the functions and interactions of telomerase RNP constituents is of considerable importance.
Telomerase directs telomere repeat addition via reverse transcription of a short templating domain in its RNA subunit (Greider 1995). The most well-studied telomerase RNAs, found in ciliated protozoa, have a conserved secondary structure (Romero and Blackburn 1991; Bhattacharyya and Blackburn 1994; Lingner et al. 1994; Zaug and Cech 1995). Telomerase RNAs identified in yeast and mammals are significantly larger than their ciliate counterparts (Singer and Gottschling 1994; Blasco et al. 1995; Feng et al. 1995), and currently, no phylogenetic or structural data are available for these molecules.
The protein subunits of the telomerase complex are still being cataloged. The catalytic reverse transcriptase subunit (TERT), which was first purified from the hypotrichous ciliate Euplotes aediculatus (Lingner et al. 1997), has subsequently been identified in Tetrahymena, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mammals (Harrington et al. 1997a; Lingner et al. 1997; Nakamura et al. 1997; Meyerson, et al. 1997; Collins and Gandhi, 1998). TERT proteins have several characteristic reverse transcriptase (RT) domains as well as a telomerase-specific motif. Point mutations within conserved residues of the RT domains inactivate telomerase in vitro and reduce telomere length in vivo (Harrington et al. 1997a; Lingner et al. 1997).
Although coexpression of the telomerase RNA and TERT subunits is sufficient to reconstitute telomerase activity (Beattie et al. 1998; Collins and Gandhi 1998; Weinrich et al. 1998), two additional proteins, p80 and p95, have been reported to associate with the telomerase RNP (Collins et al. 1995; Harrington et al. 1997b; Nakayama et al. 1997). The Tetrahymena p80 and p95 bind to single-stranded telomeric DNA and the telomerase RNA in vitro, respectively (Gandhi and Collins 1998). Mammalian homologs of p80 copurify and/or coimmunoprecipitate with telomerase activity suggesting a tight association between p80, TERT, and the telomerase RNA (Harrington et al. 1997b; Nakayama et al. 1997).
In addition to maintaining pre-existing tracts of telomeres, telomerase can also add telomeric repeats directly onto nontelomeric DNA. Because this reaction stabilizes broken chromosomes, it is sometimes referred to as chromosome healing (Harrington and Greider 1991; Melek and Shippen 1996). De novo telomere formation can occur spontaneously following accidental or artificially induced chromosome breakage, or it may be part of a developmental program of coupled chromosome fragmentation and new telomere formation. Developmentally controlled de novo telomere formation is a process integral to the sexual cycle of ciliated protozoa (Prescott 1994).
The hypotrichous ciliate Euplotes crassus provides a useful system for studying both developmentally programmed telomere formation and telomere maintenance. As with other ciliates, E. crassus contains two nuclei, a transcriptionally active macronucleus and a transcriptionally silent germ-line micronucleus. During the sexual phase, the macronucleus is destroyed and a new macronucleus is generated from a copy of the micronucleus. This process involves extensive site-specific fragmentation of the micronuclear genome and de novo addition of telomeres by telomerase onto thousands of linear DNA molecules. In vitro experiments with E. crassus and Tetrahymena enzymes reveal that telomerase forms telomeres on nontelomeric DNA by bypassing the requirement for Watson–Crick base pairing between DNA and the telomerase RNA to initiate synthesis at a specific residue in the RNA template (Melek et al. 1996; Wang and Blackburn, 1997; Wang et al. 1998). During subsequent vegetative growth, telomerase maintains telomeres on the ends of all the linear DNA molecules, replenishing terminal nucleotides lost as a consequence of semiconservative DNA replication (Prescott 1994).
The behavior of the E. crassus telomerase in vitro mimics its distinct functions in vivo. Telomerase in isolated developing macronuclei efficiently extends both telomeric and nontelomeric DNA 3′ termini, consistent with its role in de novo telomere formation. In contrast, telomerase from vegetative macronuclei, although able to extend telomeric DNA substrates, is unable to extend nontelomeric 3′ termini (Melek et al. 1996; Bednenko et al. 1997). Thus, the E. crassus telomerase undergoes a developmentally programmed switch in its specificity for DNA that precisely matches its substrates in vivo. The switch appears to be mediated in part by a soluble chromosome healing factor (CHF), which can be reconstituted with purified telomerase to facilitate telomere repeat addition onto nontelomeric 3′ DNA ends (Bednenko et al. 1997). This observation implies that the Euplotes telomerase is a holoenzyme in the developing macronucleus, consisting of a core RNP particle that associates with additional subunits that modify enzyme activity.
Here, we show that telomerase in the vegetative macronucleus exists as a 280-kD complex that is unable to extend nontelomeric DNA 3′ termini. During development of a new macronucleus, three new larger telomerase particles appear, a 550-kD, 1600-kD, and a ⩾5-MD complex. The larger complexes have structural and biochemical properties distinct from the 280-kD complex. Moreover, in contrast to the 280-kD complex, the 1600-kD, and the 5-MD particles processively elongate both telomeric and nontelomeric DNA 3′ termini. These findings provide the first direct evidence for programmed assembly of telomerase into higher order complexes with distinct biochemical and structural properties.
Results
Developmentally programmed assembly of higher order telomerase complexes
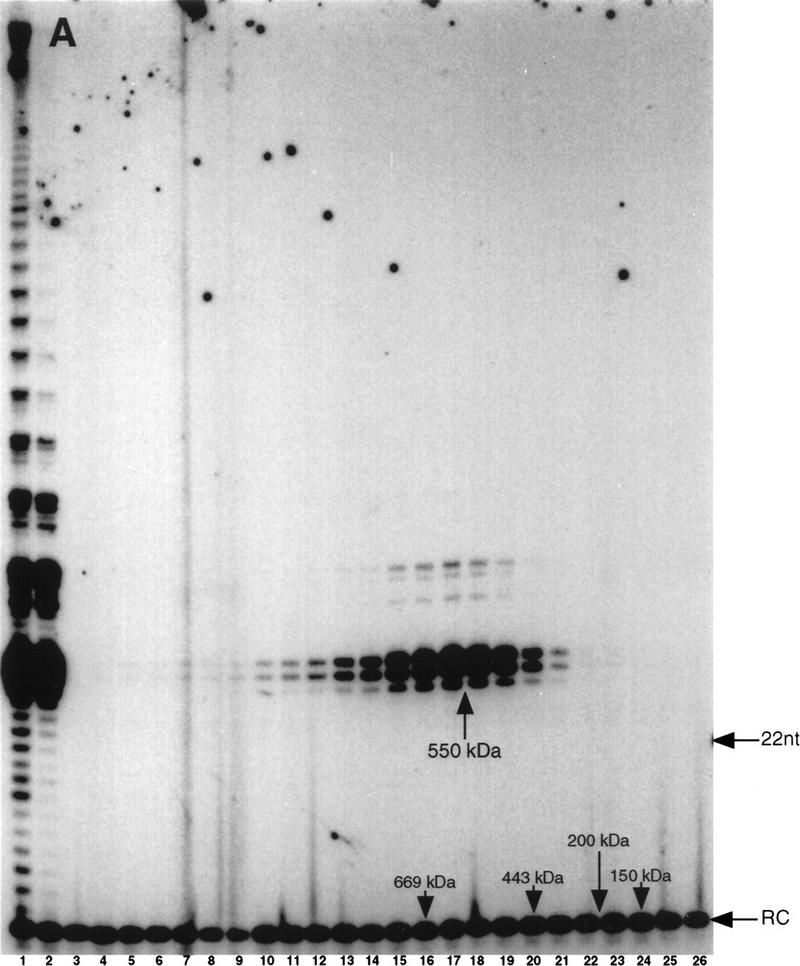
To characterize telomerase interactions with CHF and other cellular components, we attempted to employ gentle purification techniques to copurify factor(s) that might directly bind the telomerase RNA or catalytic subunit by use of gel filtration chromatography. S100 macronuclear lysates were prepared from E. crassus and telomerase activity was fractionated on a superose 6-gel filtration column. We made three changes relative to our previous purification protocol (Bednenko et al. 1997; Greene et al. 1998) that allowed us to detect higher molecular weight complexes. First, we used significantly more material per purification; second, we added NP-40 to a final concentration of 0.1% during chromatography; and third, we added 0.1mg/ml acetylated BSA to the fractions following collection. We suspect that the increased protein concentration of our samples stabilized the higher molecular weight complexes during chromatography (see below). Fractions were assayed with the telomeric DNA primer (G4T4)3. Telomerase activity from vegetatively growing Euplotes eluted predominately as a single, symmetrical peak with an apparent molecular mass of 280-kD (Fig. 1). In addition, a less abundant higher molecular weight complex was observed in the void volume (Fig. 1, lanes 4–6).
Figure 1.

Telomerase from vegetative macronuclei elutes as a 280-kD complex and a high molecular weight complex in gel filtration. S100 macronuclear lysates from vegetatively growing E. crassus were fractionated by gel filtration chromatography on a superose 6 column. Fractions were assayed with (G4T4)3. The positions of a 5′ 32P-labeled 12-mer recovery control (RC) and a terminal transferase 3′32P-labeled 22 nucleotide marker are indicated. (Lane 1) S100; (lanes 2–24) even numbered fractions of the column elution profile. Elution positions of molecular weight standards are indicated. The high molecular mass products in lane 1 are the result of extension of contaminating DNA in the lysate and are not the result of processive extension of the primer (data not shown).
Telomerase fractionated from developing macronuclear lysates showed a radical redistribution in the elution profile relative to telomerase isolated from vegetatively growing Euplotes. Telomerase from developing macronuclei eluted as a broad band of activity ranging from ∼500 kD up into the column void volume (Fig. 2). PhosphorImager quantitation of the first telomeric repeat (G3) revealed a peak of telomerase activity with an apparent molecular mass of 550-kD (Fig. 2, lane 17; data not shown). Two additional peaks were detected, one at 1600 kD and one in the void, based on quantitation of products resulting from addition of the third telomeric repeat (G19) (Fig. 2, lanes 5,13; data not shown).
Figure 2.
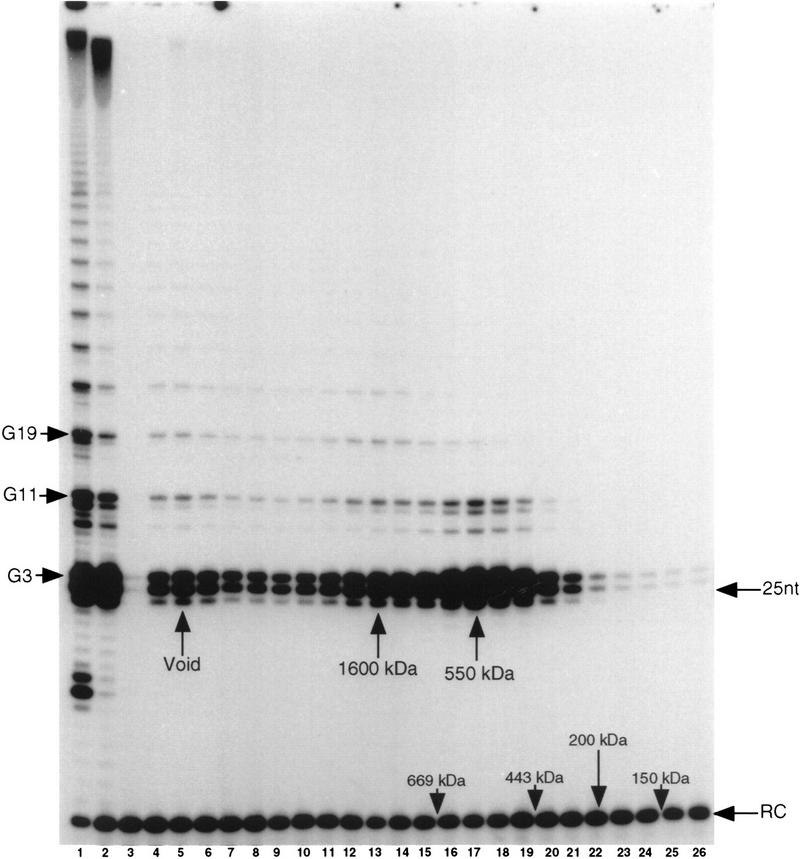
Telomerase from developing macronuclei assembles into higher order complexes. S100 macronuclear lysates from E. crassus undergoing macronuclear development were fractionated by gel filtration chromatography as described in Fig. 1. Assays and molecular weight markers are as in Fig. 1. (Lane (1) Macronuclei; (lane 2) S100; (lanes 3–26) even-numbered fractions from the column elution profile. G3, 11, and 19 correspond to synthesis of the third, eleventh, and nineteenth dG, respectively, in the G4T4 repeated ladder. The peaks corresponding to distinctive telomerase complexes are indicated.
Because the exclusion size for Superose 6 is ⩾5 MD, this value was used to estimate the minimal molecular mass of telomerase complexes in the void volume. The high molecular weight complex from vegetatively growing cells has the same biochemical properties as the 280-kD complex (see below). However, because the largest and the smallest telomerase complex from developing cells exhibit dramatically different biochemical properties, the larger complex is described in detail in this report. Hereafter, we will refer to this complex from developing cells as the 5-MD complex.
To ensure that the overlapping peaks of telomerase activity in developing macronuclei represented distinct complexes, the 550- and 1600-kD peaks were isolated, concentrated, and rechromatographed (Fig. 3A,B). Each complex eluted as a single symmetrical peak that corresponded to the same molecular mass observed with the S100 lysate. In addition, the 5-MD telomerase complex eluted in the void volume when rechromatographed (data not shown). We estimate a 25% recovery from the second round of chromatography for all the complexes on the basis of the amount of material required to obtain an equivalent signal in the primer elongation assay.
Figure 3.
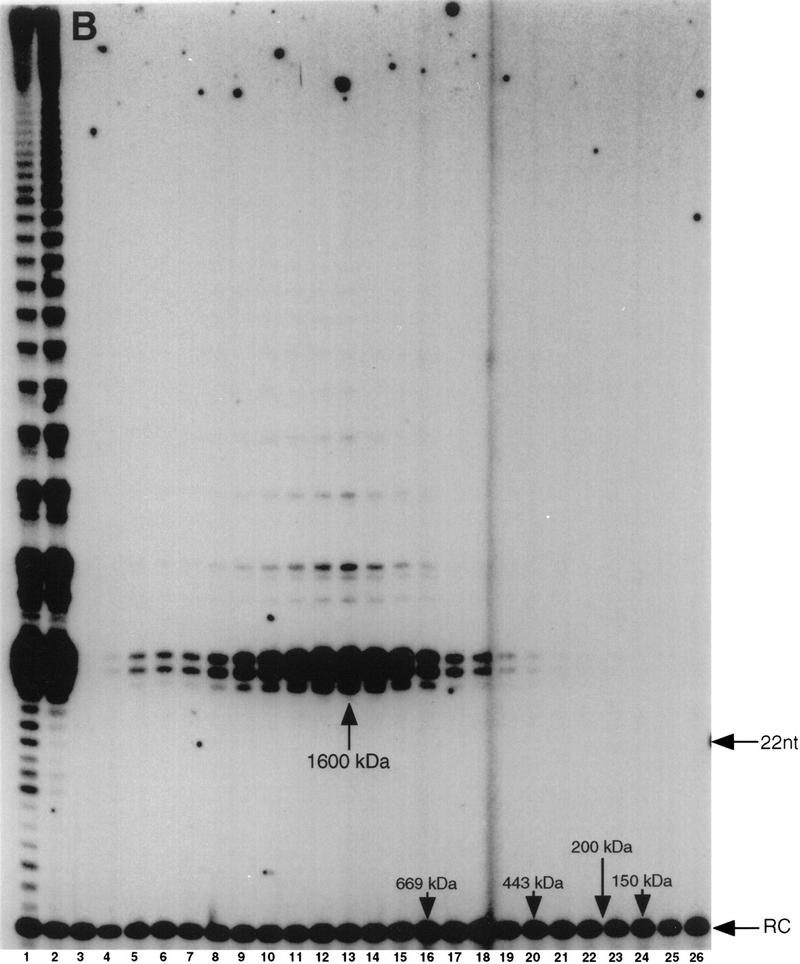
Distinct forms of telomerase are present in developing macronuclei. Individual telomerase complexes fractionated from developing macronuclear lysates were concentrated by dialysis into 40% PEG, rechromatographed by gel filtration and assayed for telomerase activity. Rechromatographed 550-kD (Fig. 2, fractions 16–18) (A) and 1600-kD (Fig. 2, fractions 12–14) (B) complexes are shown. (Lanes 1) Reactions with a macronuclear lysate; (lane 2) reactions with the isolated, concentrated telomerase peaks (see Materials and Methods) prior to rechromatography; (lanes 3–26) superose 6 column elution profile. Assays and molecular mass markers are as in Fig. 1.
The number of telomeric repeats detected with both the 550- and 1600-kD complexes increased following enzyme concentration with polyethylene glycol (cf. Figs. 2, lane 13, with 3B, lane 2, and 2, lane 17, with 3A, lane 2). However, the 1600-kD complex still generated significantly more repeats than the 550-kD complex (Fig. 3A,B, lanes 2). Following the second round of chromatography, the processivity of the individual complexes remained unchanged (see Figs. 2, lane 13, and 3B, lane 13 and 2, lanes 17–18, and 3A, lanes 17–18). These results indicate that the higher order telomerase complexes retain both molecular mass and catalytic properties following the second round of chromatography.
All three peaks of activity contained telomerase RNA, were inhibited by RNase A, and the RNase A inhibition could be reversed by inclusion of the ribonuclease inhibitor RNAsin (data not shown). Additionally, all three peaks of telomerase activity were resistant to treatment with DNase I prior to fractionation and were stable during gel filtration chromatography in the presence of 0.1% NP40 (see Materials and Methods) or 1 mm potassium glutamate (data not shown). Taken together, these data argue that the large telomerase complexes are not constrained by DNA, nor are they nonspecific complexes held together by either electrostatic or hydrophobic interactions.
Previously, we noted a decrease in telomerase processivity following partial purification (Greene et al. 1998). To determine if the decline in processivity could be attributed to the loss of a telomerase-associated factor, the size of telomerase particles purified by ion-exchange chromatography as described previously (Greene et al. 1998) was determined. The partially purified telomerase from developing cells eluted as a single peak of 550 kD, which appeared to be identical to the 550-kD complex obtained directly from S100 lysates following gel filtration in terms of size and biochemical properties (data not shown and see below).
Primer specificities of the individual telomerase complexes
The developmental differences in primer specificity exhibited by the E. crassus telomerase reflect the changing DNA substrates available to telomerase in vivo (Melek et al. 1996; Bednenko et al. 1997). To test whether the different telomerase complexes display biochemical properties consistent with the appropriate stage of the ciliate life cycle, complexes were reacted with primers that mimic the in vivo substrates of telomerase. The telomere maintenance function was examined by reacting complexes with a fully telomeric primer, (G4T4)3. The capacity for new telomere formation was measured with GT-13, a primer that contains the telomeric sequence GGGGTTTT at its 5′ terminus and a stretch of 13 nontelomeric nucleotides at its 3′ terminus. This chimeric primer is efficiently extended by telomerase in developing macronuclei, even though its 3′ terminus cannot form Watson–Crick base pairs with the RNA template. In contrast, telomerase in isolated vegetative macronuclei will not extend GT-13 with telomeric repeats (Melek et al. 1996; Bednenko et al. 1997) ,
The 280-kD telomerase complex from vegetative macronuclei catalyzed the addition of telomeric repeats onto the telomeric primer (Fig. 4, lane 1). The products of this reaction displayed an elongation profile characteristic of telomerase from vegetative E. crassus, wherein the strongest bands in the profile corresponded to an enzyme pause or dissociation after incorporation of the third G and fourth T in each GGGGTTTT repeat (Fig. 4, lane 1; Bednenko et al. 1997). As with intact vegetative macronuclei, the 280-kD complex was unable to extend the GT-13 primer beyond the addition of a single G residue (Fig. 4, lane 2; Bednenko et al. 1997). An additional product (marked with an asterisk) was obtained in these reactions, but it was not primer dependent, nor was it detected on further purification of the 280-kD telomerase complex (data not shown). The high molecular weight complex from vegetative cells displayed the same properties as the 280-kD complex in terms of primer specificity, processivity and profile of elongation products (data not shown). We note that the inability of the 280-kD and high molecular weight complexes to extend nontelomeric DNA directly reflects the properties of telomerase in the intact vegetative macronucleus.
Figure 4.
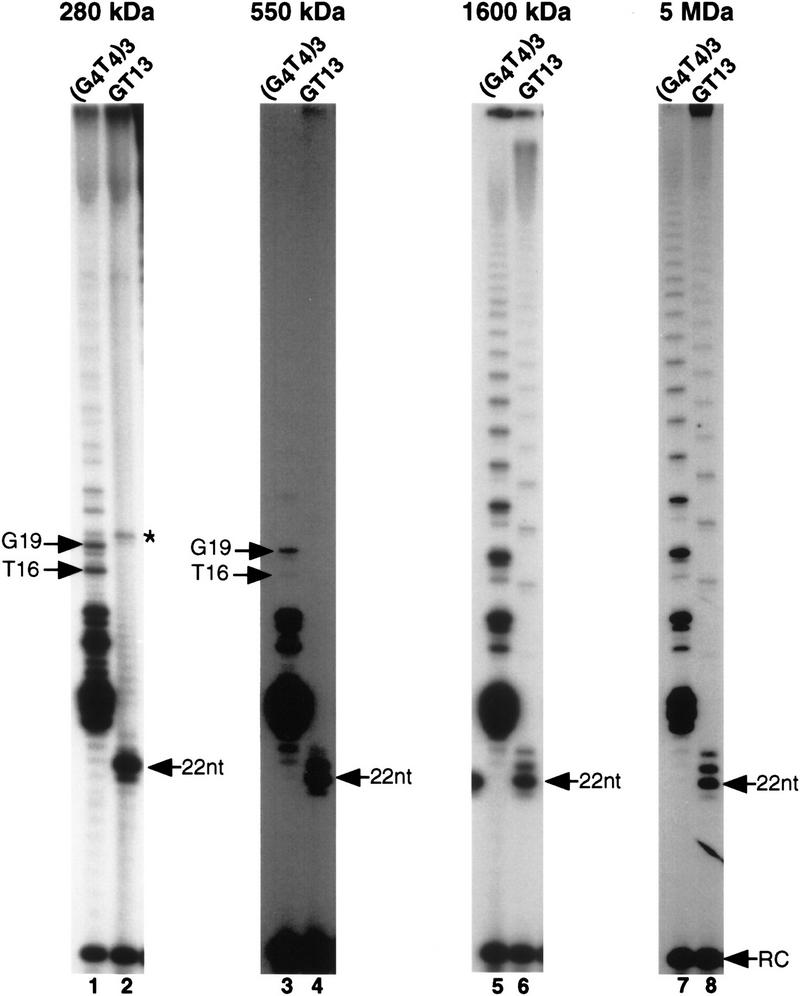
Individual telomerase complexes show unique biochemical properties with respect to primer utilization and processivity. Telomerase complexes fractionated by gel filtration were assayed with (G4T4)3 or the chimeric primer GT-13 (GGGGTTTTACTACGCGATCAT). (Lanes 1,2) Reactions with 280-kD vegetative complex; (lanes 3,4) 550-kD developmental complex; (lanes 5,6) 1600-kD developmental complex; (lanes 7,8) the developmental 5-MD complex. The migration position of the fourth T and/or third G residue in the TTTTGGGG elongation profile are indicated (T16 and G19). (*)The band in the 280-kD preparation is not primer dependent and does not copurify further with telomerase activity. An overexposure of 550-kD complex assays is shown to emphasize the lack of processive products.
Like the 280-kD complex, the 550-kD complex isolated from developing macronuclei was not able to extend GT-13 beyond one to two dG residues (Fig. 4, lane 4). In addition, the 550-kD complex was less processive in extending a telomeric primer than the S100 lysate and only catalyzed the addition of approximately one to three telomeric repeats (cf. Figs. 2, lane 2, and 4, lane 3). The 550-kD complex, like the other development-specific complexes, generated elongation products in which pauses occurred predominantly after the addition of the third G in the TTTTGGGG repeat (Fig. 4, cf. lanes 1 and 3). This elongation profile is characteristic for telomerase from developing macronuclei (Bednenko et al. 1997).
The two larger telomerase complexes of 1600 kD and 5 MD displayed strikingly different biochemical properties from the smaller telomerase complexes. Both of the larger complexes added multiple telomeric repeats onto the telomeric primer (G4T4)3 (Fig. 4, lane 5,7), exhibiting significantly greater processivity than was observed for either the 280- or the 550-kD particles (Fig. 4, lanes 1,3). All of our telomerase reactions were carried out under the same conditions. Nevertheless, such dramatic differences in enzyme processivity for the E. crassus telomerase have not been observed with subtle changes in primer or salt concentration (E. Greene, J. Bednenko, and D. Shippen, unpubl.). Furthermore, in contrast to findings reported for the E. aediculatus telomerase (Hammond and Cech 1997), we failed to detect any change in E. crassus telomerase processivity when dGTP concentrations were varied over a 400-fold range (J. Bednenko and D. Shippen, unpubl.). The only significant differences in enzyme processivity that we have observed occurred during purification of E. crassus telomerase (Greene et al. 1998).
The most remarkable distinction between the two smaller complexes and the 1600-kD and 5-MD complexes was in primer specificity. The 1600-kD and 5-MD complexes efficiently and processively elongated GT-13 (Fig. 4, lane 6,8). The ability of these complexes to directly elongate nontelomeric 3′ ends indicates that CHF is associated with higher order telomerase assemblies isolated from developing macronuclei.
Each telomerase complex was tested for the ability to utilize fully nontelomeric substrates. In the absence of a G-rich telomeric cassette at the primer 5′ terminus, none of the complexes efficiently extended a nontelomeric 3′ end (data not shown). In addition to its RT activity, the E. crassus telomerase is associated with an endonuclease activity that preferentially cleaves at the junctions of telomeric and nontelomeric DNA (Melek et al. 1996; Greene et al. 1998). This activity has been postulated to serve a proofreading function for telomerase and/or to participate in chromosome fragmentation. All of the telomerase complexes catalyzed endonucleolytic cleavage of DNA oligonucleotides (data not shown), supporting the conclusion that the nuclease activity is intrinsic to the core telomerase RNP (Melek et al. 1996; Bednenko et al. 1997; Greene et al. 1998).
Structurally distinct telomerase RNP complexes in developing and vegetative macronuclei
The ciliate telomerase RNAs share a phylogenetically conserved secondary structure and contain several conserved sequences predicted to be important for telomerase function (Romero and Blackburn 1991; Lingner et al. 1994). In addition, the Tetrahymena thermophila telomerase RNA has been examined in vivo and in vitro and footprinting studies support the phylogenetically predicted structure (Bhattacharyya and Blackburn 1994; Zaug and Cech 1995). Because our results indicated that the Euplotes telomerase exhibits different biochemical properties as a function of development, we reasoned that additional levels of modification might be detected in telomerase RNP architecture. Therefore, we examined the accessibility of the telomerase RNA to chemical modification and ribonuclease cleavage in the context of the RNP particle.
First, chemical probing was undertaken to identify single-stranded, accessible regions of the RNA in intact macronuclei. Intact macronuclei were treated with dimethylsulfoxide (DMS), RNA was isolated, and modifications were detected by primer extension (Zaug and Cech 1995). The resulting pattern of modification was in good agreement with the proposed secondary structure for the Euplotes telomerase RNA (Lingner et al. 1994). No significant differences between telomerase from developing and vegetative macronuclei were observed (data not shown).
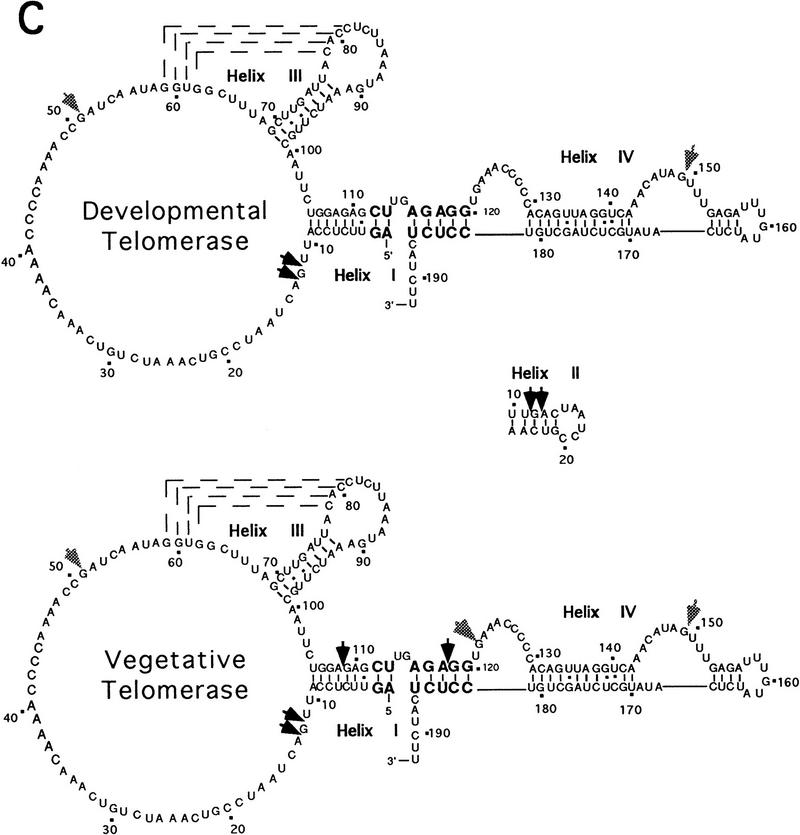
Additional structure probing was performed with endoribonucleases. In this case, macronuclei were incubated with limiting amounts of either RNase V1 (cleaves either double-stranded RNA or single-stranded, structured RNA) or RNase T1 (cleaves single-stranded guanines). Only limited regions of the RNA were accessible to nuclease digestion; the vast majority of RNA was completely protected. RNase V1 treatment revealed an accessible region near the 5′ end of the RNA (nucleotides 11 and 12), which was cleaved in both vegetative and developing macronuclei (Fig. 5A,C). These cleavage sites localized to a region predicted to be single stranded on the basis of the phylogenetic model. However, using computer modeling for lowest free-energy structures (Zucker 1989), we noted the potential for a thermodynamically stable helix in this region (Fig. 5C).
Figure 5.


Accessibility of the telomerase RNA in intact macronuclei is developmentally regulated. (A) Footprinting reactions with RNase V1. (Lanes 1,6) Extension reactions without prior ribonuclease treatment; (lanes 5,9) (labeled stop) reaction termination controls (see Materials and Methods); (lanes 2–4) 5-, 10-, and 15-min RNase V1 footprint reactions, respectively, with developing macronuclei; (lanes 7,8) 5- and 10-min RNase V1 footprint reactions with vegetative macronuclei. Cleavage sites are indicated with arrowheads and sequencing ladders used to map the sites of ribonuclease accessibility are denoted. (B) Footprinting reactions with RNase T1. Control lanes and sequencing markers are labeled as in A. (Lanes 2–4,6–8) 5-, 10-, and 15-min reactions with RNase T1; (lane 9) stop reaction. Predominant cleavage sites are indicated with arrowheads. The RNase T1 cleavage site at position 122 in the vegetative macronuclei is near a nonspecific band at nucleotide 125. The background level of damage at this site varied from experiment to experiment and PhosphorImager analysis confirmed the increased intensity of this band in the presence of RNase T1 (data not shown), ruling out the possibility that it is solely the result of nonspecific RNA damage. (C) Ribonuclease cleavage sites superimposed on the phylogentically predicted structure of the E. crassus telomerase RNA (Shippen-Lentz and Blackburn 1990; Lingner et al. 1994). Conserved nucleotides 112–120 are shown in larger, boldface type and a potential thermodynamically stable helix corresponding to nucleotides 10–25 is depicted. Nucleotides 36–50 comprise the templating domain. RNase V1 cleavage sites, black arrowheads, and RNase T1 sensitive sites, gray arrowheads, are indicated.
Reactions with RNase V1 uncovered a dramatic difference in telomerase RNP particles present in intact developing and vegetative macronuclei. Nucleotides 109 in helix I and 119 in the adjacent portion of helix IV were accessible to cleavage by RNase V1 in intact macronuclei from vegetative cells, but were not accessible in intact developing macronuclei (Fig. 5A,C). These nucleotides encompass a conserved region (Fig. 5C, larger, bold type) predicted previously to serve as a site for the binding of telomerase-associated factors (Lingner et al. 1994).
Another developmentally regulated structural distinction was observed in reactions with RNase T1. Prominent cleavage sites were detected at nucleotides 51 and 149 in both intact developing and vegetative macronuclei (Fig. 5B,C). However, a cleavage site at nucleotide 122 (Fig. 5B, cf. lanes 6–8 and lane 9), immediately adjacent to the RNase V1 accessible region, was observed only in vegetative macronuclei (Fig. 5B,C). We conclude from these data that an extended region of nuclease protection encompasses a conserved region in the telomerase RNA during macronuclear development.
The appearance of the protected region of helices I and IV of the telomerase RNA in intact macronuclei was temporally coupled to the disappearance of the 280-kD telomerase complex during macronuclear development (Fig. 1,2,5; data not shown). To determine if the extended footprint was directly related to the increased apparent molecular mass of telomerase during development, RNase V1 footprints were performed on the individual telomerase complexes, isolated by gel filtration. RNase V1 treatment revealed two structural distinctions in telomerase present in intact macronuclei relative to isolated telomerase complexes (Fig. 6). First, all of the isolated telomerase complexes showed decreased sensitivity to RNase V1 at nucleotides 11 and 12 compared with telomerase present in intact macronuclei (cf. Figs. 5A and 6A–C). Second, a new region of RNase V1 sensitivity at nucleotide 53 was detected in the isolated 550- and 1600-kD complexes that was not observed with intact developing macronuclei (cf. Figs. 5A and 6B and C). This site shows significant background cleavage in both the vegetative macronuclei and the isolated 280-kD complex (Figs. 5A and 6A).
Figure 6.
Ribonuclease protection in isolated telomerase complexes. Isolated telomerase complexes were footprinted separately as in Fig. 5. RNase V1 footprints of the 280-(A), 550- (B), and 1600-kD (C) complexes are shown. Lane designations are as in Fig. 5. The position of nucleotide 109 is indicated with an arrowhead. For each panel, equivalent amounts of sequencing ladder were loaded and the exposures were adjusted until the signals of full-length RNA were approximately the same.
As with telomerase present in intact vegetative macronuclei, telomerase RNA in the isolated 280-kD complex exhibited sensitivity to RNase V1 in helix I (nucleotide 109) (Figs. 5A and 6A). Slight background at nucleotide 119 obscured any cleavage that may have been the result of RNase V1 treatment in the isolated 280-kD complex (Fig. 6A, lanes 1,4,5). Significantly, nucleotide 109 was completely protected from RNase V1 digestion in isolated 550-kD, 1600-kD, and 5-MDa complexes (Fig. 6B,C; data not shown) as it was in the intact developing macronucleus (Fig. 5A). Thus, the increased apparent molecular mass of telomerase RNP complexes present during macronuclear development is directly related to the nuclease protection in helix I of the telomerase RNA subunit.
Discussion
E. crassus is one of the few model systems in which the two distinct modes of telomere synthesis, telomere maintenance, and de novo telomere formation can be readily observed. Furthermore, because the biochemical properties of telomerase switch in response to the changing in vivo substrates for the enzyme (Melek et al. 1996; Bednenko et al. 1997), E. crassus provides a unique opportunity to investigate the mechanism of DNA recognition and processing by telomerase. In this study, we used protein purification in conjunction with RNA structure probing to identify development-specific telomerase complexes and putative binding sites for potential new telomerase-associated factors that may be intimately involved in the switch in telomerase’s DNA specificity.
Higher order telomerase complexes
During vegetative growth of E. crassus, telomerase predominately exists as a single 280-kD complex capable of extending telomeric primers with additional telomeric DNA. Although we have not yet defined all of the components of this complex, it resembles the telomerase RNP purified to homogeneity from vegetatively growing E. aediculatus, in terms of its apparent molecular mass (280 kD vs. 230 kD) and relatively low processivity with telomeric DNA primers (Lingner and Cech 1996; Hammond et al. 1997). The E. aediculatus complex harbors the telomerase RNA, a 123-kD TERT, and a 43-kD protein subunit (Lingner and Cech 1996; Lingner et al. 1997). In the early steps of purification, a small (∼45 kD) decrease in the size of the E. aediculatus RNP particle was noted that was attributed to the loss of a subunit or proteolytic removal of a nonessential fragment. However, no specific differences in telomerase activity were reported to coincide with the reduction in apparent molecular mass (Lingner and Cech 1996). Given the small change in particle size and the fact that the enzyme was purified from vegetatively growing cells, it is unlikely that the putative subunit/fragment is related to the higher order telomerase complexes that we observed during E. crassus macronuclear development. Currently, it is impossible to obtain synchronized preparations of conjugating E. aediculatus in the laboratory, and hence, a comparative analysis of telomerase complexes from developing cells of the two Euplotes species is not feasible. Nevertheless, the available information for telomerase from vegetative cells argues strongly that the 280-kD complex represents a core telomerase particle, containing a minimum compliment of factors in addition to the telomerase RNA and the catalytic TERT subunit.
During macronuclear development, E. crassus telomerase assembles into a series of distinct higher order complexes with apparent molecular masses of 550 kD, 1600 kD, and 5 MD. Like the 280-kD complex from vegetative cells, the 550-kD complex fails to utilize nontelomeric 3′ ends. In addition to their size differences, the 280-kD and 550-kD complexes can be distinguished mechanistically by the profiles of their products (see Fig. 4, lanes 1,3). This variation in telomerase product profiles was noted previously in assays with intact vegetative and developing macronuclei (Melek et al. 1996; Bednenko et al. 1997). The larger mass of the 550-kD complex is consistent with binding of one or more additional subunits that alter the basic mechanism of telomere synthesis. Another possibility is that the telomerase RNA and/or the catalytic subunit is modified during development in a way that results in subtle changes in how T4G4 repeats are generated.
The 1600-kD complex exhibits structural and biochemical properties clearly distinct from the smaller telomerase RNP particles. Specifically, this complex processively extends both telomeric and nontelomeric 3′ termini. Therefore, we speculate that it represents a telomerase holoenzyme. Although telomerase complexes that elute in the void of a superose 6-gel filtration column can be isolated from the macronuclei of both vegetative and developing cells, the available information does not indicate whether these complexes represent specific telomerase assemblies or aggregates. In all of our assays, the biochemical properties of the higher order vegetative complex were identical to the 280-kD complex. Similarly, the 5-MD complex from developing cells displayed the same properties as the 1600-kD complex.
The S. cerevisiae telomerase RNP has been shown recently to exist as a multimer (Prescott and Blackburn 1997). It is conceivable that the higher order telomerase complexes from Euplotes represent a developmentally regulated multimerization of the RNP. The apparent molecular masses of the complexes (280 kD, 550 kD, and 1600 kD) are not inconsistent with a monomer-to-dimer-to-tetramer assembly. To investigate this idea further, we examined the isolated 550-kD complex under a variety of chromatography conditions (Greene et al. 1998; E. Greene and D. Shippen, unpubl.). In no case did we detect breakdown of the 550-kD complex into a 280-kD particle or aggregation into a 1600-kD or 5-MD complex (data not shown), indicating that the Euplotes telomerase complexes are stable. Although we have no direct evidence for or against RNP multimerization, the developmental stage specificity of the higher order complexes argues that additional cofactors and/or modifications are required for assembly. Recently, we optimized conditions that allow purification of telomerase over seven sequential chromatography steps without significantly altering either the biochemical properties or the sizes of the individual telomerase complexes (E.C. Greene and D.E. Shippen, unpubl.). Analysis of complex constituents is currently underway and should reveal whether the Euplotes telomerase associates with a novel set of proteins during macronuclear development.
Two obvious candidates that could be members of a telomerase higher order complex are p80 and p95, the nucleic acid binding proteins associated with the Tetrahymena and mammalian telomerases (Collins et al. 1995; Harrington et al. 1997b; Nakayama et al. 1997; Gandhi and Collins 1998). Antibodies against the Tetrahymena p80 and p95 fail to cross-react with proteins in E. crassus extracts (E.C. Greene, D.E. Shippen, and K. Collins, unpubl.). In addition, the region of the Euplotes telomerase RNA protected from ribonuclease digestion in developing macronuclei is not conserved in the Tetrahymena telomerase RNA (Lingner et al. 1994). Thus, if a p80 homolog is associated with the E. crassus telomerase, it does not assemble in the same way in Euplotes and Tetrahymena telomerase RNP particles. One important indication of the dramatic divergence of these two RNPs is that the Tetrahymena telomerase, in contrast to the Euplotes enzyme, has an intrinsic ability to add telomeric sequences onto completely nontelomeric DNA substrates in vitro. This activity is apparently not modified during development (Wang and Blackburn 1997).
Changes in telomerase RNP architecture during Euplotes development
The ciliate telomerase RNAs share a common secondary structure and several nucleotides that are absolutely conserved (Romero and Blackburn 1991; Lingner et al. 1994). In the hypotrichous ciliate telomerase RNAs, one conserved sequence is located between nucleotides 112–120, encompassing some of the residues in helices I and IV (Fig. 5C). As a consequence of its strict sequence conservation, this domain was postulated to serve as a platform for the formation of a telomerase higher order complex (Lingner et al. 1994; Nugent and Lundblad 1998). Our results support this hypothesis in that ribonuclease accessibility to this region displays a dynamic and developmentally controlled protection profile.
Protection of this region in the telomerase RNA coincides with changes in both apparent molecular mass and the biochemical properties of purified RNP particles. In every case that we examined, ribonuclease accessibility of helices I and IV in intact macronuclei accurately predicted the presence or absence of the 280-kD telomerase complex, suggesting that these two properties may be functionally related. Furthermore, because the higher order telomerase complexes (1600 kD and 5 MD) exhibit a ribonuclease protection profile similar to that found in intact developing macronuclei, the increase in apparent molecular mass is likely to be mediated in part by direct binding of one or more factors to helix I of the telomerase RNA. Although our results do not rule out a drastic alteration of the RNA secondary and/or tertiary structure rather than direct protection from a bound factor, such an alteration would be surprising in the absence of a causative factor. In either case, the data argue strongly for the existence of developmentally regulated factor(s) that directly interact with telomerase.
Possible in vivo roles for telomerase higher order complexes
In the developing macronucleus of ciliates, there are at least two opportunities for telomerase to interact with other cellular machineries. First, studies in Tetrahymena and Euplotes reveal that chromosome fragmentation and de novo telomere formation are tightly coupled. Intermediates lacking telomeres have never been detected, implying an intimate association between the fragmentation machinery and telomerase (Roth and Prescott 1985; Yao et al. 1990; Fan and Yao 1996). Second, aphidicolin treatment of developing Euplotes macronuclei alters telomere synthesis by telomerase, indicating telomeric G-strand synthesis is closely associated with synthesis of the C-rich telomeric strand by Pol-α/primase (Fan and Price 1997).
Given the distinct biochemical properties of Euplotes telomerase complexes, we can make predictions concerning their in vivo roles. The 280-kD core telomerase complex displays properties expected for an enzyme involved in telomere maintenance. This role requires interaction with pre-existing telomeric DNA, but not with nontelomeric 3′ ends. The 550-kD complex, the smallest telomerase particle detected during macronuclear development, nonprocessively extends telomeric substrates, but cannot utilize non-telomeric substrates. This complex could fulfill a function in telomere maturation following initial telomere addition, or it could have a role in telomere maintenance during the final rounds of DNA replication before macronuclear development is completed (Prescott 1994). Alternatively, the 550-kD complex may represent an intermediate on the pathway to assembly of the larger complexes. One piece of evidence supporting this later model is that all of the telomerase complexes from developing macronuclei generate the same pausing profile of elongation products, indicating that the same mechanism of repeat synthesis is employed.
The 1600-kD and the 5-MD telomerase complexes perform processive synthesis on both telomeric and nontelomeric primers 3′ ends. Extension of nontelomeric DNA by the E. crassus telomerase in vitro mimics the de novo telomere formation reaction in vivo. In both cases, telomerase initiates synthesis by adding a specific permutation of the eight nucleotide telomeric repeat, GGGGTTTT (Klobutcher et al. 1981; Vermeesch and Price 1994; Melek et al. 1996). In our assays, a 5′ G-rich sequence is required for extension of nontelomeric 3′ ends in vitro (Melek et al. 1996); none of the isolated telomerase complexes elongate an entirely nontelomeric primer. Because a corresponding G-rich region is absent from chromosome break sites in vivo (Klobutcher et al. 1981), we and others have proposed that an unidentified protein cofactor or another unknown cis-acting DNA sequence element recruits telomerase to the break (Coyne et al. 1996; Melek et al. 1996; Bednenko et al. 1997). On the basis of their biochemical properties, the 1600-kD and the 5-MD telomerase complexes from developing macronuclei contain CHF and perhaps other components of the machinery involved in chromosome fragmentation and telomeric C-strand synthesis. The more processive nature of these complexes compared with the 550-kD particle is also consistent with an enzyme that generates the oversized telomeres initially added to DNA ends during macronuclear development in Euplotes (Roth and Prescott 1985; Vermeesch and Price 1994).
Finally, it should be noted that many telomerases exhibit apparent molecular masses significantly larger than predicted from the size of the telomerase RNA and a TERT (Lue and Peng 1997; Nakayama 1997; Wang and Blackburn 1997; Nakamura and Cech 1998). Thus, telomerase is likely to exist as a holoenzyme in other organisms.
Materials and methods
Macronuclei preparation
E. crassus stains LW13 and LW14 were grown in 40- to 80-liter aliquots at a density of ∼103 cells/ml. Prior to harvesting, cells were starved 5–6 days and then mated as described previously (Greene et al. 1998). For preparation of vegetative macronuclei, E. crassus strain LW13 was grown overnight in 20-liter aliquots of dense algae and cells were harvested before the algae was completely consumed. Vegetative macronuclei were isolated as described previously (Bednenko et al. 1997). Macronuclei were resuspended in TMG buffer [20% glycerol, 30 mm Tris-HCl (pH 7.5), 0.1 mm MgCl2, 1 mm DTT] containing protease and ribonuclease inhibitors (0.1 mm PMSF, 12.5 μg/ml antipain, 1.7 μg/ml aprotinin, 10 μg/ml chymostatin, 10 μg/ml E64, 1 μg/ml leupeptin, 7 μg/ml pepstatin A and 10 mm vanadyl ribonucleoside complex), frozen with liquid nitrogen and stored at −80°C until use. Prior to S100 lysate preparation, macronuclei were adjusted to 1% NP-40 and 0.5 m potassium glutamate (KGlu). Macronuclei were lysed by French Press at 16,000 psi and lysates centrifuged at 40,000 rpm for 1 hr in a TL100 rotor prechilled to 4°C. The resulting lysates were divided into 250-μl aliquots, frozen with liquid nitrogen, and stored at −80°C until use. Protein concentrations were determined by Bradford assay.
Fractionation of telomerase complexes
Telomerase complexes were fractionated by gel filtration at 4°C on a Superose 6 (separation range: 5 × 103–5 × 106 Daltons) HR 10/30 column (Pharmacia) with FPLC. Columns were pre-equilibrated with TMG containing 150 mm NaCl, 0.1 mm PMSF and 0.1% NP-40. S100 lysates (0.2 mg protein) were loaded in 200-μl aliquots and separated at a flow rate of 0.1 ml/min. Fractions were adjusted to 0.1 mg/ml acetylated BSA (U.S. Biochemical) following collection. Column calibration was performed with blue dextran (void volume), thyroglobulin (669 kD), apoferritin (443 kD), β-amylase (200 kD), and ADH (150 kD) as size standards. The elution volumes of the size standards were determined by monitoring absorbance at 280 nm. For rechromatography, individual peaks were collected and concentrated four- to fivefold by dialysis into TMG containing 40% polyethylene glycol 8000 (PEG). The following fractions (see Fig. 2) were combined as peak activities: 550-kD complex (fractions 16–18), 1600-kD complex (fractions 12–14), and 5-MD complex (4–6). The individual complexes were rechromatographed under conditions identical to those for S100 lysates with the exception that acetylated BSA (0.1 mg/ml) was included in the running buffer as a stabilizer. The largest telomerase complex eluted outside the effective fractionation range of the column. Therefore, we estimated the minimal molecular mass of this complex as 5 MD.
Telomerase assays
Telomerase reactions were performed in a total volume of 40 μl and incubated at 30°C for 1 hr. The final concentrations of reaction components were 50 mm Tris-HCl (pH 8.0), 10% glycerol, 5 mm MgC2, 20 mm EGTA, 75 mm NaCl, 0.05% NP-40, 0.1 mm dTTP, 0.25 μm [α-32P]dGTP (800 Ci/mmole), 0.1 mg/ml acetylated BSA (U.S. Biochemical), 1 mm DTT, 1 mm spermidine and 0.4 μm primer. Reactions were stopped by the addition of EDTA to 10 mm and products isolated by phenol/chloroform extraction followed by ethanol precipitation. A 5′32P-labeled 12-mer was added to the EtOH precipitation as a recovery control. Products were resolved on 10% sequencing gels and detected by autoradiography. The DNA primer was in large excess relative to telomerase to ensure that products containing more than a single telomeric repeat result from multiple rounds of processive synthesis (Morin 1989; Lee and Blackburn 1993; Melek et al. 1996).
Footprinting reactions
Macronuclei were adjusted to ∼104 nuclei/ml in TMG. RNase V1 (Pharmacia) or RNase T1 (GIBCO BRL) were added to 3 units or 10 units, respectively, per 100 μl of macronuclei. Footprint reactions with isolated telomerase complexes were performed with 0.01 units of RNase V1 per 150-μl sample (∼10-fold less telomerase than present in the footprints with intact macronuclei, data not shown). Reactions were incubated at room temperature and 100-μl aliquots removed at the indicated intervals. Footprinting reactions were adjusted to 0.3 m NaOAc (pH 5.2) in a total volume 200 μl and RNA isolated by sequential phenol (pH 4.3)/chloroform and chloroform extractions. Reaction termination controls (stop) were performed in which ribonuclease was added to samples and reactions terminated with no incubation to ensure that no RNA damage was incurred during deproteinization and RNA isolation. No RNA damage was detected during the course of the incubations in the absence of exogenously added ribonuclease (data not shown). RNA was precipitated from the aqueous phase by addition of 360 μl of isopropanol, 40 μl 3 m NaOAc (pH 5.2), and 2 μl of glycogen (∼7 μg/μl). After a 1-hr incubation at −20°C, the RNA was precipitated by centrifugation at 14,000 rpm at 4°C for 30 min. Pellets were washed with 70% EtOH and allowed to air-dry. The RNA was then resuspended into 10 μl of DEPC H2O. Annealing reactions were performed in 65 mm Tris-HCl (pH 8.3), 100 mm KCl, 4 mm MgC2 contained 200 fmoles of gel purified 5′32P-labeled primer 1 (AAGATGAGAGGACAGCTAGAGCATATGAG), incubated 1 min at 95°C and chilled on ice. Reactions were then adjusted to 0.5 mm dNTPs and 10 mm DTT. Primer extension was performed by the addition 20 units of Superscript II RNaseH− reverse transcriptase (GIBCO BRL) and incubated at 50°C for 10 min. Products were isolated by phenol extraction and EtOH precipitation and resolved on 6% sequencing gels. Sequencing ladders were generated with 5′32P-labeled primer 1 for PCR cycle sequencing (Epicentre Technologies) of a plasmid containing a cloned copy of the E. crassus telomerase RNA gene. Cleavage sites nearest the 5′ end of the telomerase RNA were definitively mapped with a primer complementary to the telomerase RNA nucleotides 69–100 for reverse transcription (data not shown).
Acknowledgments
We are grateful to Jeff Kapler and members of the Shippen laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health grant GM49157 to D.E.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL dshippen.tamu.edu; FAX (409) 845-9274.
References
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Bednenko J, Melek M, Greene EC, Shippen DE. Developmentally regulated initiation of DNA synthesis by telomerase: Evidence for factor-assisted de novo telomere formation. EMBO J. 1997;16:2507–2518. doi: 10.1093/emboj/16.9.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH. Architecture of telomerase RNA. EMBO J. 1994;13:5721–5730. doi: 10.1002/j.1460-2075.1994.tb06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Funk W, Villeponteau B, Greider CW. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Chalker DL, Yao M-C. Genome downsizing during ciliate development nuclear division of labor through chromosome restructuring. Annu Rev Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- Fan Q, Yao M-C. New telomere formation coupled with site-specific chromosome breakage in Tetrahymena thermophila. Mol Cell Biol. 1996;16:1267–1274. doi: 10.1128/mcb.16.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XQ, Price CM. Coordinate regulation of G- and C strand length during telomere synthesis. Mol Biol Cell. 1997;8:2145–2155. doi: 10.1091/mbc.8.11.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang S-S, Weinrich SL, Avilion AA, Chiu C-P, Adams RR, Chang E, Allsopp RS, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes & Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EC, Bednenko J, Shippen DE. Flexible positioning of the telomerase-associated nuclease leads to preferential elimination of nontelomeric DNA. Mol Cell Biol. 1998;18:1544–1552. doi: 10.1128/mcb.18.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW. Telomerase biochemistry and regulation. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1995. pp. 35–68. [Google Scholar]
- Greider CW. Telomerase activity, cell proliferation, and cancer. Proc Natl Acad Sci. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond PW, Cech TR. dGTP-dependent processivity and possible template switching of Euplotes telomerase. Nucleic Acids Res. 1997;25:3698–3704. doi: 10.1093/nar/25.18.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes & Dev. 1997a;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda L, Robinson MO. A mammalian telomerase-associated protein. Science. 1997b;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Swanton MT, Donini O, Prescott DM. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permutated template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lue NF, Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek M, Shippen DE. Chromosome healing: Spontaneous and programmed de novo telomere formation by telomerase. BioEssays. 1996;18:301–308. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- Melek M, Greene EC, Shippen DE. Processing of nontelomeric 3′ ends by telomerase: Default template alignment and endonucleolytic cleavage. Mol Cell Biol. 1996;16:3437–3445. doi: 10.1128/mcb.16.7.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, et al. hEST2, the putative human telomerase catalytic subunit gene, is upregulated in tumor cells during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SI, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TPL1: A gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: Components and regulation. Genes & Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes & Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Roth M, Prescott DM. DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell. 1985;41:411–417. doi: 10.1016/s0092-8674(85)80014-3. [DOI] [PubMed] [Google Scholar]
- Shippen DE, McKnight TD. Telomeres, telomerase and plant development. Trends Plant Sci. 1998;3:126–130. [Google Scholar]
- Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- Singer M, Gottschling DE. TCL1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Price CM. Telomeric DNA sequence and structure following de novo telomere synthesis in Euplotes crassus. Mol Cell Biol. 1994;14:554–566. doi: 10.1128/mcb.14.1.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Blackburn EH. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gilley D, Blackburn EH. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Yao M-C, Yao C-H, Monks B. The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell. 1990;63:763–772. doi: 10.1016/0092-8674(90)90142-2. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–364. [PMC free article] [PubMed] [Google Scholar]
- Zucker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]