Abstract
Synthetic genetic devices that interface with native cellular pathways can be used to change natural networks to implement new forms of control and behavior. The engineering of gene networks has been limited by an inability to interface with native components. We describe a class of RNA control devices that overcome these limitations by coupling increased abundance of particular proteins to targeted gene expression events through the regulation of alternative RNA splicing. We engineered RNA devices that detect signaling through the NF-κB and Wnt signaling pathways in human cells and rewire these pathways to produce new behaviors, thereby linking disease markers to noninvasive sensing and reprogrammed cellular fates. Our work provides a genetic platform that can build programmable sensing-actuation devices enabling autonomous control over cellular behavior.
Cellular decisions, such as differentiation, response to stress, disease progression, and apoptosis, depend on regulatory networks that control enzymatic activities, protein translocation, and genetic responses. Central to the genetic programming of biological systems is the ability to process information within cellular networks and link this information to new cellular behaviors, in essence rewiring network topologies. Altered network topologies have been achieved through engineered transcriptional networks (1, 2) and signal transduction cascades (3). However, these systems are limited to processing transcription factor inputs, which represent a small fraction of the human proteome (4, 5), or require replacing endogenous cellular components. Alternative platforms for constructing sensing-actuation devices based on the detection of broad classes of proteins will have widespread applications in basic research, biotechnology, and medicine.
RNA is a promising substrate for platforms to interface with cellular networks because of the versatile sensing and actuation functions that RNA can exhibit and the ease with which RNA structures can be designed (6, 7). RNA-based sensing-actuation devices have been engineered that respond predominantly to externally applied small molecule (6, 8, 9) and nucleic acid (10–12) inputs and control gene expression through diverse mechanisms. Pre-mRNA splicing is one such mechanism, in which devices responsive to exogenous small molecule and protein inputs can regulate splicing events (8, 13). However, protein-responsive gene regulatory platforms based on programmed alternative splicing must support modular and extensible input/output functionalities, provide regulatory properties that translate to control over cell behaviors, and be sensitive to changes in endogenous protein concentrations or localization. Although RNA aptamers that bind to proteins have been generated through in vitro selection methods (14, 15), such protein-sensing components have not been routinely integrated into RNA-based regulatory devices, leaving a large number of biological signals currently inaccessible.
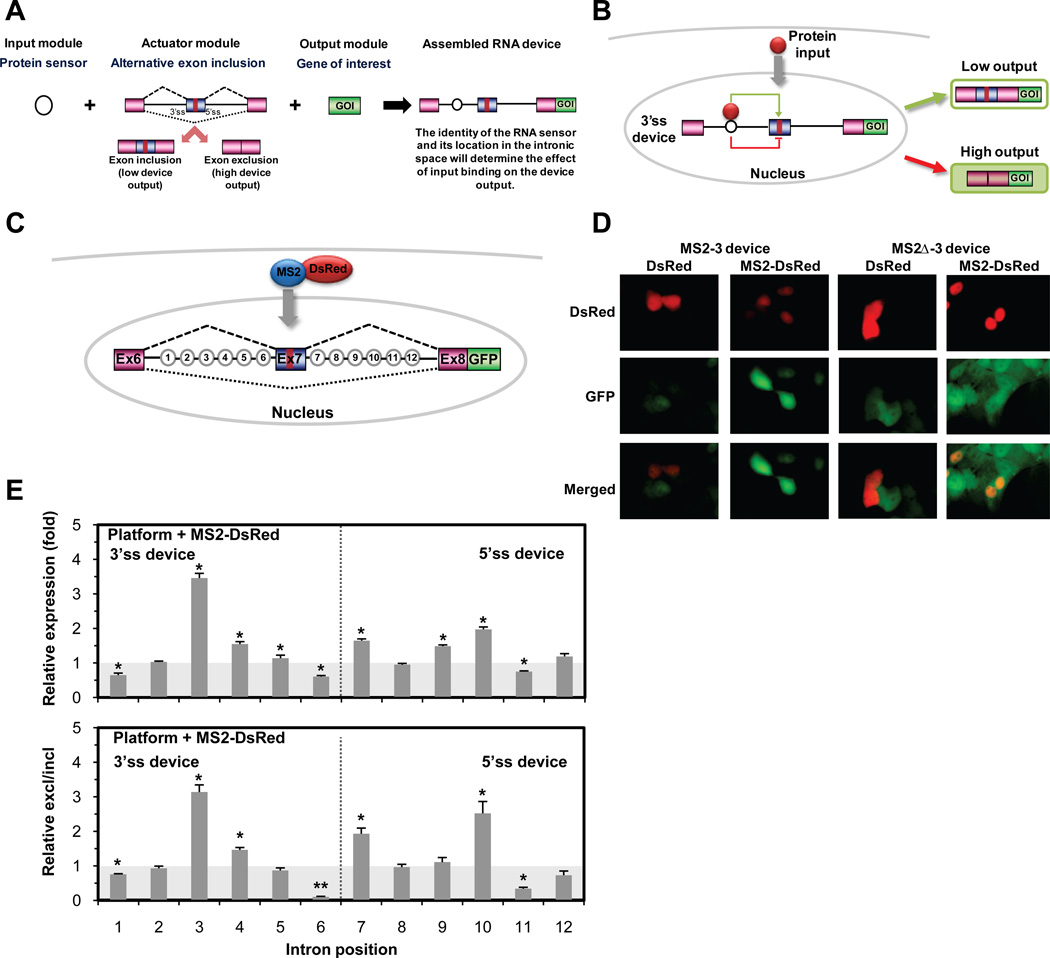
We developed a protein-responsive RNA-based regulatory device by integrating RNA aptamers that bind to protein ligands into key intronic locations of an alternatively spliced transcript, thus linking intracellular protein concentrations to gene expression events. Our regulatory platform consists of an output module, a gene of interest (GOI), placed downstream of the sensing-actuation device, a three-exon, two-intron mini-gene in which the middle exon is alternatively spliced, or excluded (Fig. 1A). The middle exon contains a stop codon, such that expression of the GOI is high when the exon is excluded. Control is exerted by the input module, composed of an RNA aptamer that senses changes in nuclear protein concentrations whereby ligand binding to the aptamer alters the splicing pattern, likely through steric hindrance or recruitment of components involved in spliceosome site (ss) recognition (Fig. 1A, B) (16).
Fig. 1.
An alternative splicing-based RNA control device translates protein inputs to targeted gene expression outputs. (A) Platform composition of an RNA control device based on alternative splicing. The input module, consisting of an RNA-based protein sensor or aptamer, detects changes in nuclear protein concentrations. The sensor transmits information on binding events to the actuator module, consisting of a three-exon, two-intron mini-gene where the alternatively spliced exon contains a stop codon. The actuator controls the expression of the output module, consisting of a gene of interest (GOI). The three modules are physically linked in a transcript to form the assembled RNA control device. (B) Mechanism of RNA device function. A 3’ ss device is shown, where the input module is located in the intron upstream of the alternative exon and binding of the protein input to the sensor alters the splicing pattern by either enhancing (green) or suppressing (red) alternative exon inclusion. Exclusion of the alternative exon results in removal of the stop codon upstream of the GOI, thereby increasing the gene expression output from the device. (C) Determination of optimal input module location within intronic sequence space of the regulatory device. The MS2 aptamer was inserted at 12 intronic positions spaced by 15-nts flanking the alternatively spliced exon. (D) Fluorescence images of the MS2-responsive devices. The increased fluorescence output from an MS2-responsive device is specific to the MS2-DsRed protein input and the wild-type MS2 aptamer in position 3 (MS2-3). (E) The response of the MS2-responsive device to the MS2-DsRed protein is affected by the location of the input module. For all activities reported as relative expression (fold), the ratio of the mean GFP levels of the wild-type RNA device in the presence of ligand (MS2-DsRed) to the absence of ligand (DsRed) is normalized to the same ratio for the mutant device. Transcript isoform analysis of the MS2-responsive devices with qRT-PCR supported the gene expression data (bottom panel). For all qRT-PCR data reported as relative excl/incl (fold), the ratio of the mean expression levels of the exon 7 excluded isoform to the exon 7 included isoform for the wild-type device in the presence of ligand to the absence of ligand is normalized to the same ratio for the mutant device. For all reported activities, mean expression levels from two independent experiments are shown. Error bars represent +/− s.d. from mean values. P-values derived from the Student’s t-test are as follows: *P < 0.05 and **P < 0.01. Unnormalized expression levels for all devices are provided in Tables S1 to S6.
We systematically analyzed the device architecture to determine the intron positions that enabled aptamer-mediated protein-responsive regulation of alternative splicing. We inserted the aptamer for the bacteriophage coat protein MS2 (17) at 6 positions in each intron (1–12) of the SMN1 mini-gene (18) (Fig. 1C and Fig. S1A), and linked the device to the gene encoding green fluorescent protein (GFP). The SMN1 mini-gene was selected because key regulatory sequences are located in exon regions (19), such that insertion of synthetic sequences into intronic regions is not likely to strongly affect splicing patterns. Human embryonic kidney (HEK)-293 cell lines that stably expressed these devices or the corresponding negative controls expressing mutant aptamers (SOM text S1) displayed differences in fluorescence compared to cells expressing a device containing no aptamer sequence, with insertion at most positions causing increased exon exclusion (Fig. S1B and C), indicating that secondary structure can modulate splicing patterns.
To examine protein-specific effects on splicing, cell lines were transfected with a plasmid encoding the MS2 coat protein fused to fluorescent protein DsRed and a Simian virus 40 (SV40) nuclear localization signal (MS2-DsRed) (Fig. 1C and Fig. S1D). Integration of aptamers into 6 positions resulted in increases in fluorescence (P < 0.05, Student’s t-test) and 3 positions resulted in decreases in fluorescence (P < 0.05) relative to that of cell lines expressing DsRed. Control experiments showed that these effects were specific to the wild-type aptamer (Fig. 1,D and E, Fig. S1E, and Table S1). Transcript isoform analysis by quantitative real-time polymerase chain reaction (qRT-PCR) confirmed the effect on splicing (Fig. 1E and Fig. S1F), and there was a significant correlation between splicing patterns and fluorescence (P ≪ 0.01, Anova). Although the regulatory effects of the devices were modest (~2- to 4-fold), these effects are comparable to those in other splicing (20) and RNA regulatory (21) systems that have key roles in controlling biological processes. We selected positions 3, 6, and 10 as points of input module integration for device tailoring on the basis of their relative amount of protein-mediated splicing regulation and location relative to splicing motifs (3’ ss, 5’ ss, branch point, and polypyrimidine tract).
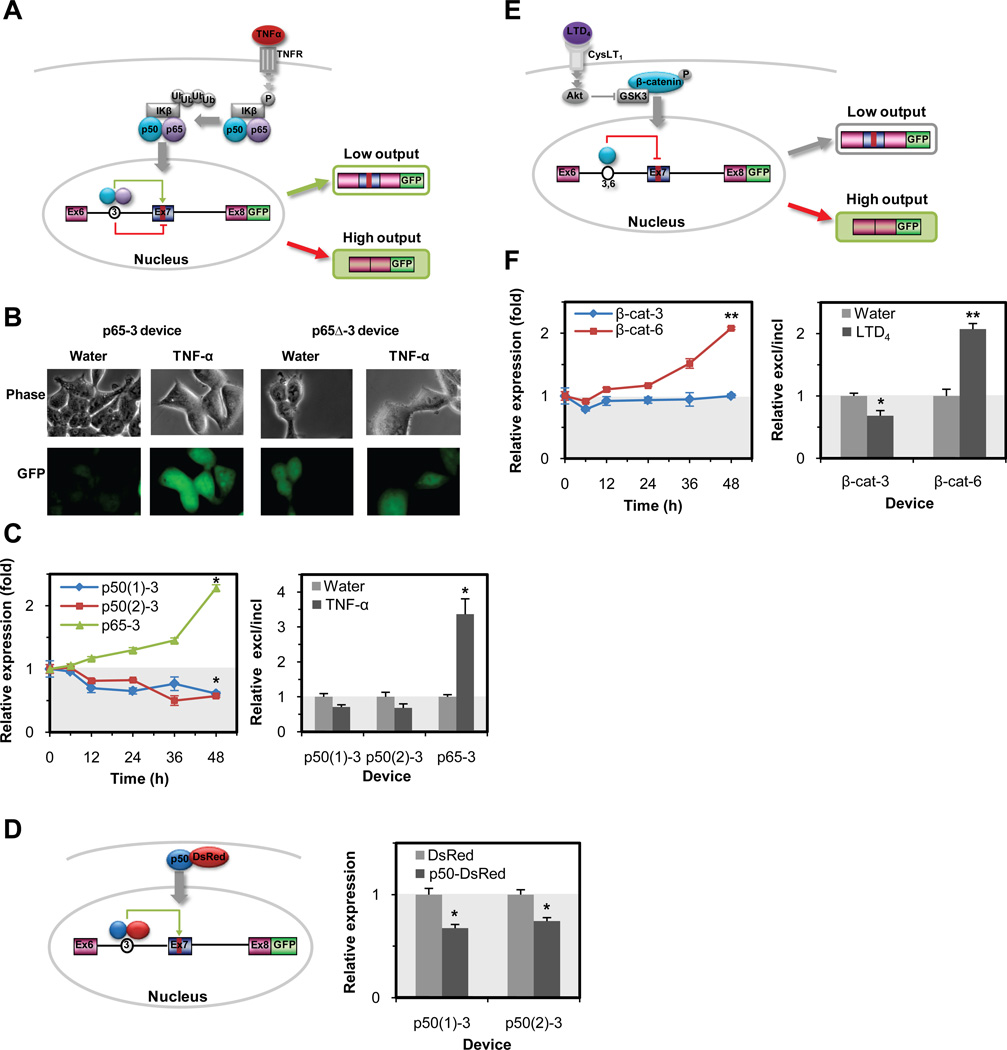
To investigate the modularity of the input processing function of our RNA devices and the ability to detect nuclear localized proteins resulting from activated signaling pathways, we built devices with aptamers that bind the subunits p50 (22, 23) and p65 (24) of the transcription factor NF-κB inserted into position 3 (Fig. 2A). NF-κB p50 and p65 dimers have an important role in disease by binding to κB sites in promoters or enhancers of genes participating in immune and inflammatory responses, cell adhesion, proliferation, and apoptosis (24). NF-κB signaling and subsequent translocation of p50 and p65 to the nucleus was induced in cell lines stably expressing the NF-κB devices with tumor necrosis factor alpha (TNF-α) (25). The p65-3 device displayed increased gene expression (P < 0.01), corresponding to an increase in exon exclusion as a result of p65 binding to the sensor, whereas the p50-3 devices exhibited decreased gene expression (P < 0.05) and exon exclusion as a result of p50 binding to the sensor (Fig. 2,B and C, Fig. S2A, and Table S2). Controls with mutant aptamer devices showed that responses were specific to the wild-type aptamer sequences. Cell lines expressing the p50-responsive devices and a p50-DsRed fusion exhibited decreases in fluorescence when compared to cells expressing DsRed (Fig. 2D, Fig. S2B, and Table S3), supporting that the response observed under TNF-α stimulation is directly mediated by p50 binding. The differing output signals from the p65- and p50-responsive devices may be due to differences in aptamer binding (24), aptamer structure, or interactions of p65 and p50 with spliceosomal components (SOM text S2).
Fig. 2.
RNA control devices detect endogenous protein inputs and signaling through native pathways. (A) Mechanism of the NF-κB-responsive device based on TNF-α stimulation (20 ng/ml) of the NF-κB pathway. Ligand binding to the TNF-α receptor leads to activated signaling and translocation of p50 and p65 into the nucleus. The NF-κB-responsive devices contain NF-κB p65 (p65-3) or p50 (p50(1)-3 and p50(2)-3) aptamers inserted into position 3. (B) Phase (top) and fluorescence (bottom) images of the NF-κB p65-responsive devices. The increased fluorescence output from a NF-κB-responsive device is specific to the pathway stimulation and the wild-type p65 aptamer in position 3 (p65-3). (C) NF-κB-responsive devices exhibited responses to TNF-α stimulation at the level of gene expression (left panel) and splicing pattern (right panel). For all data, relative expression (fold) was determined as described in Fig. 1F. qRT-PCR data is reported as relative excl/incl, the ratio of the mean expression levels of the exon 7 excluded isoform to the exon 7 included isoform for the wild-type device relative to the same ratio for the mutant device under the indicated ligand condition. (D) Mechanism of the NF-κB p50-responsive device based on a p50-DsRed protein input and corresponding device response. The NF-κB p50-responsive devices exhibited responses to a heterologous p50-DsRed protein similar to that observed with TNF-α stimulation. Activities were reported as relative expression by taking the ratio of the mean GFP levels of the wild-type RNA device to that from the mutant device under the indicated ligand condition. (E) Mechanism of the β-catenin-responsive device based on LTD4 stimulation (80 nM) of the Wnt pathway. LTD4 stimulation leads to stabilization of β-catenin and accumulation in the nucleus. The β-catenin-responsive devices contain the β-catenin aptamer in positions 3 (β-cat-3) and 6 (β-cat-6). (F) β-catenin-responsive devices exhibited responses to LTD4 stimulation at the level of gene expression (left panel) and splicing pattern (right panel).
We also inserted aptamers that recognize the signaling protein β-catenin (26) into sites 3 and 6 to build β-catenin-responsive devices (Fig. 2E). β-catenin is a central component of the Wnt signaling pathway and is localized to the nucleus upon pathway activation to aid in the transcription of genes that regulate cell growth, differentiation, and tumorigenesis (26). We examined the effect of stimulating the β-catenin pathway with leukotriene D4 (LTD4) on the response of our engineered β-catenin-responsive devices. The β-cat-6 device exhibited increased gene expression (P < 0.05), corresponding to an increase in exon exclusion (Fig. 2F, Fig. S2C, and Table S4), whereas the β-cat-3 device did not respond to LTD4 stimulation. Control experiments demonstrated that the β-cat-6 device response was specific to the wild-type aptamer sequences. Results from the MS2, NF-κB, and β-catenin studies demonstrate that particular protein ligands can have distinct positional and functional effects on splicing, such that aptamer position and the target protein provide device tuning capability. These studies verify the flexibility of our synthetic devices to be interfaced with cellular signaling pathways and their ability to detect disease biomarkers and link this detection to regulated gene expression events.
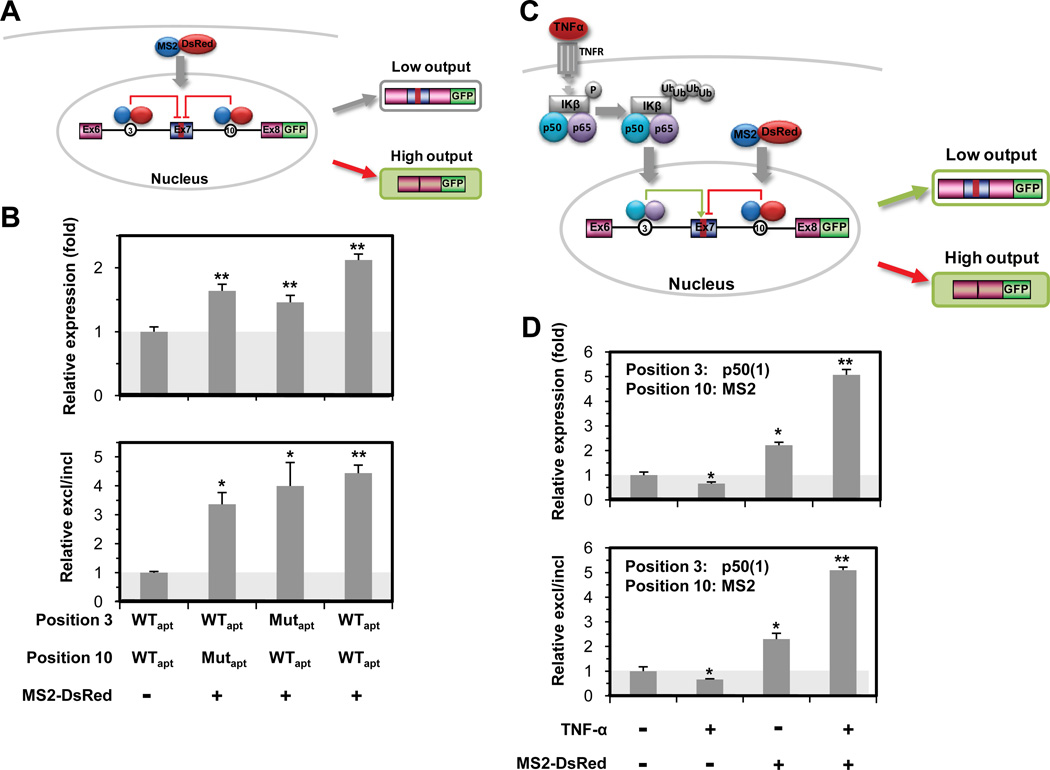
To examine the extension of our device platform to multi-input processing, we constructed devices containing combinations of the wild-type and mutant MS2 aptamers in positions 3 and 10 (Fig. 3A). Devices containing the wild-type aptamer in either position displayed significant increases in gene expression (P < 0.01) and exon exclusion in the presence of MS2-DsRed compared to that in the absence of ligand (Fig. 3B, Fig. S3A, and Table S5). A device with aptamers in both positions showed a ~30–45% increase in gene expression and exon exclusion compared to that of the single-aptamer devices. We also built multi-input devices to detect heterologous MS2-DsRed and endogenous NF-κB p50 (Fig. 3C). We inserted the wild-type and mutant p50(1) and MS2 aptamers into sites 3 and 10, respectively. TNF-α stimulation led to a decrease in gene expression and exon exclusion, whereas expression of MS2-DsRed led to a significant increase in gene expression (P < 0.05) and exon exclusion from this device (Fig. 3D, Fig. S3B and C, and Table S6). The device response in the presence of both ligands was greater than the sum of the individual ligand output signals, suggesting that the combined inputs have a synergistic effect on the output signal. These studies indicate that our device platform can support combinatorial regulation of gene expression in response to multiple protein inputs.
Fig. 3.
RNA devices implement combinatorial control schemes through multi-input processing. (A) Mechanism of the MS2 multi-input processing regulatory device. Wild-type and mutant MS2 aptamers were inserted into positions 3 and 10. (B) The MS2 multi-input processing device responds to the heterologous MS2-DsRed protein to increase the gene expression output (top panel). Transcript isoform analysis of the MS2 multi-input processing device supports gene expression data (bottom panel). For all data, relative expression (fold) and relative ratios of exon excluded to included transcript isoforms (fold) were determined as described in Fig. 1F. (C) The MS2 / NF-κB p50 multi-input processing regulatory device allows integration of complex input signals and amplification of device response. The NF-κB p50 and MS2 aptamers were inserted into positions 3 and 10, respectively. (D) The MS2 / NF-κB p50 multi-input processing device responds to both inputs to increase the gene expression output (top panel). Transcript isoform analysis of the MS2 / NF-κB p50 multi-input processing device supports gene expression data (bottom panel).
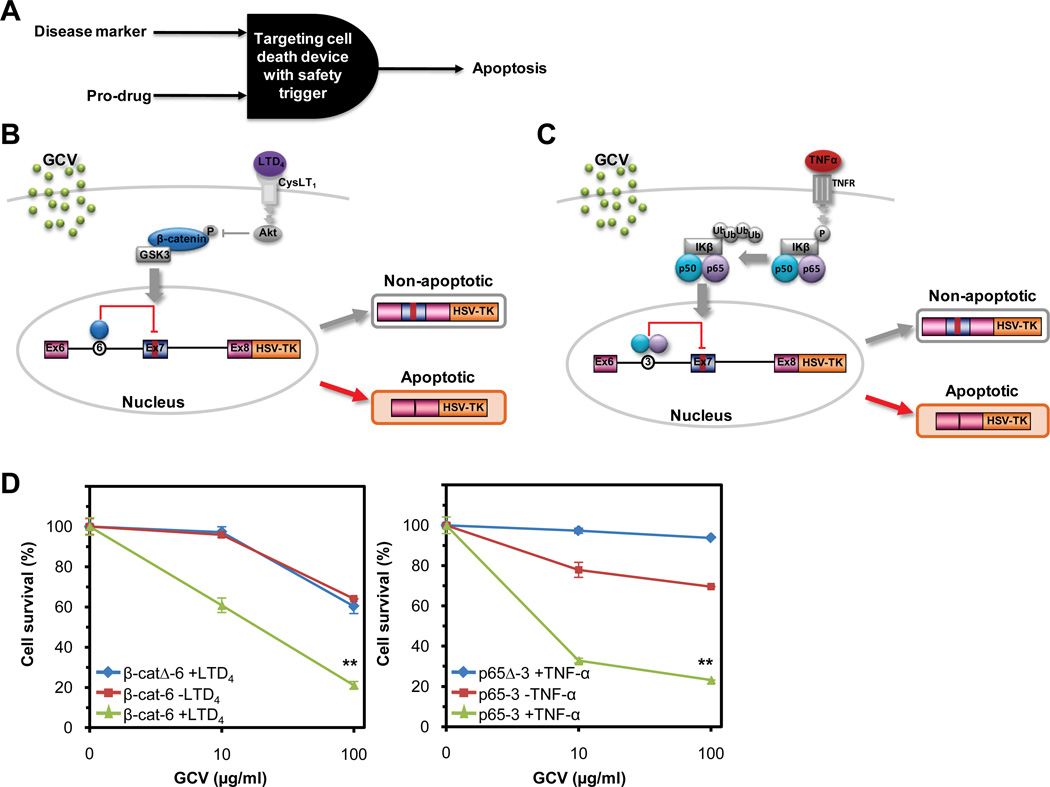
To examine whether our protein-responsive RNA devices can be used to regulate cell-fate decisions, we developed devices that integrated across two therapeutic inputs – increased signaling through a disease-associated pathway and the presence of an exogenously-applied, inactive “pro-drug” - to trigger targeted cell death (Fig. 4A). We constructed β-catenin- and NF-κB-responsive devices that trigger apoptosis by replacing the output module with a gene encoding the herpes simplex thymidine kinase (HSV-TK) (Fig. 4B and C). HSV-TK confers sensitivity to the pro-drug ganciclovir (GCV), which induces apoptosis (27) and has been used in clinical trials to treat tumors (28). Cells stably expressing the β-cat-6 and p65-3 devices exhibited increased sensitivity to GCV under pathway stimulation (Fig. S4A and B) and a cell survival of ~20% at 100 µg/ml GCV (P < 0.01) (Fig. 4D), similar to that of cells overexpressing HSV-TK (Fig. S4C). Cells expressing the devices in the absence of pathway stimulation and mutant devices under pathway stimulation displayed no observable phenotypic effects and survival rates between 60–90% at 100 µg/ml GCV. We demonstrated the modularity of the output function of the device platform by replacing the output module with the proapoptotic gene Puma (Fig. S5).
Fig. 4.
RNA devices detect endogenous markers of disease and trigger targeted cell death. (A) Functional representation of a targeted therapeutic device that integrates across two therapeutic inputs – disease biomarker and an exogenously-applied, inactive pro-drug - to trigger targeted cell death. (B, C) Mechanisms of the β-catenin- (β-cat-6) (B) and NF-κB-responsive (p65-3) (C) devices fused to a suicide gene therapy output module (HSV-TK), which control cell survival in response to detection of disease markers and GCV, a pro-drug trigger. (D) Dose-response curves of cell survival percentages for the β-catenin- and NF-κB-responsive devices fused to HSV-TK indicate a decrease in cell survival as a result of increased signaling through the targeted pathway and the presence of GCV. For all reported data, the mean cell survival levels from two independent experiments are shown.
These results demonstrate that our RNA devices can effectively rewire signaling through disease-associated pathways to trigger apoptosis using clinically-relevant genetic systems. There was a slight reduction in survival in cells expressing our devices in the absence of pathway stimulation, which may reflect effects of high GCV concentrations on cell viability (Fig. S4D) (29) and basal expression of HSV-TK from our devices. Possible therapeutic utility and safety of these targeted cell death devices is supported by effective cell-killing efficacy in the presence of both inputs and minimal background activity in the absence of one or both inputs. Our results demonstrate that synthetic RNA controllers with moderate gene regulatory activities (~2–4-fold) can achieve substantial alterations in downstream functional behaviors through their coupling to potent genetic targets, and effects that are amplified through associated cellular pathways.
Our system demonstrates that heterologous and endogenous proteins not associated with splicing regulation can be directed to alter splicing patterns through synthetic protein-binding sequences. In contrast to protein-based transcriptional control systems, RNA-based systems present advantages in enabling response to proteins other than transcription factors, direct tailoring of input/output processing functions without device redesign, extension to combinatorial processing, and practical implementation in clinical applications (9, 30). The extension of our framework to the processing of multiple protein inputs can be used to engineer RNA-based devices with sophisticated information processing activities and to design and build complex regulatory networks to interrogate and program cellular function.
Supplementary Material
Acknowledgments
We thank K. Hertel for providing the pHis-BIVT-MS2-RSp55 construct, M. Jensen for providing pCD19t-Tk-T2A-IL15op_epHIV7, L. J Maher for providing pGAD24 and B. Stewart for laboratory assistance. This work was supported by the Caltech Joseph Jacobs Institute for Molecular Engineering for Medicine (grant to C.D.S.), the National Institutes of Health (fellowship to K.G.H., grant to C.D.S.), the Department of Defense (grant to C.D.S.), the Alfred P. Sloan Foundation (fellowship to C.D.S.), and the Bill and Melinda Gates Foundation (grant to C.D.S.).
References and Notes
- 1.Weber W, Fussenegger M. Chem Biol. 2009 Mar 27;16:287. doi: 10.1016/j.chembiol.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Khalil AS, Collins JJ. Nat Rev Genet. 2010 May;11:367. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008 Mar 14;319:1539. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 4.Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Curr Opin Struct Biol. 2004 Jun;14:283. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. Nat Rev Genet. 2009 Apr;10:252. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 6.Link KH, Breaker RR. Gene Ther. 2009 Jul 9; doi: 10.1038/gt.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson EA, Ellington AD. Nat Chem Biol. 2007 Jan;3:23. doi: 10.1038/nchembio846. [DOI] [PubMed] [Google Scholar]
- 8.Suess B, Weigand JE. RNA Biol. 2008 Jan–Mar;5:24. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 9.Win MN, Liang JC, Smolke CD. Chem Biol. 2009 Mar 27;16:298. doi: 10.1016/j.chembiol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs FJ, et al. Nat Biotechnol. 2004 Jul;22:841. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 11.Brown BD, et al. Nat Biotechnol. 2007 Dec;25:1457. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 12.Rinaudo K, et al. Nat Biotechnol. 2007 Jul;25:795. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 13.Villemaire J, Dion I, Elela SA, Chabot B. J Biol Chem. 2003 Dec 12;278:50031. doi: 10.1074/jbc.M308897200. [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. Nature. 1990 Aug 30;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Tuerk C, Gold L. Science. 1990 Aug 3;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Matlin AJ, Clark F, Smith CW. Nat Rev Mol Cell Biol. 2005 May;6:386. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 17.Carey J, Lowary PT, Uhlenbeck OC. Biochemistry. 1983 Sep 27;22:4723. doi: 10.1021/bi00289a017. [DOI] [PubMed] [Google Scholar]
- 18.Culler SJ, Hoff KG, Voelker RB, Berglund JA, Smolke CD. Nucleic Acids Res. 2010 Apr 12; doi: 10.1093/nar/gkq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Am J Hum Genet. 2006 Jan;78:63. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DS, Gusti V, Dery KJ, Gaur RK. BMC Mol Biol. 2008;9:23. doi: 10.1186/1471-2199-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arndt GM, et al. BMC Cancer. 2009;9:374. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi J, et al. Nucleic Acids Res. 2006;34:3577. doi: 10.1093/nar/gkl482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan R, et al. Nucleic Acids Res. 2006;34:e36. doi: 10.1093/nar/gnj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurster SE, Maher LJ., 3rd Rna. 2008 Jun;14:1037. doi: 10.1261/rna.878908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellweg CE, Arenz A, Bogner S, Schmitz C, Baumstark-Khan C. Ann N Y Acad Sci. 2006 Dec;1091:191. doi: 10.1196/annals.1378.066. [DOI] [PubMed] [Google Scholar]
- 26.Lee HK, et al. Cancer Res. 2007 Oct 1;67:9315. doi: 10.1158/0008-5472.CAN-07-1128. [DOI] [PubMed] [Google Scholar]
- 27.Beltinger C, et al. Proc Natl Acad Sci U S A. 1999 Jul 20;96:8699. doi: 10.1073/pnas.96.15.8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lifang Y, Min T, Midan A, Ya C. J Exp Clin Cancer Res. 2008;27:42. doi: 10.1186/1756-9966-27-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y, Kong B, Ma D, Qu X, Jiang S. Int J Gynecol Cancer. 2006 Jan–Feb;16:156. doi: 10.1111/j.1525-1438.2006.00470.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen YY, Jensen MC, Smolke CD. Proc Natl Acad Sci U S A. 2010 Apr 26; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.