Abstract
The locus control region of the β-globin cluster contains five DNase I hypersensitive sites (5′HS1–5) required for locus activation. 5′HS3 contains six G-rich motifs that are essential for its activity. Members of a protein family, characterized by three zinc fingers highly homologous to those found in transcription factor Sp1, interact with these motifs. Because point mutagenesis cannot distinguish between family members, it is not known which protein activates 5′HS3. We show that the function of such closely related proteins can be distinguished in vivo by matching point mutations in 5′HS3 with amino acid changes in the zinc fingers of Sp1 and EKLF. Testing their activity in transgenic mice shows that EKLF is a direct activator of 5′HS3.
Keywords: Zinc fingers, binding site specificity, locus control region, Sp1, EKLF, transcriptional activation, globin
The human β-globin cluster contains five structural genes in the order 5′-ε (embryonic)–Gγ–Aγ (fetal)–δ–β (adult)-3′. Expression of these genes is completely dependent on the presence of the locus control region (LCR), a 20-kb sequence located upstream of the ε-globin gene (for review, see Grosveld et al. 1993). The LCR is characterized by the presence of five erythroid-specific DNase I hypersensitive sites, termed 5′HS1–5. The activity of the LCR resides in the hypersensitive areas (Grosveld et al. 1993). Our previous work has focused on the functional dissection of 5′HS3 of the LCR. It is the strongest activator when single hypersensitive sites are linked to the β-globin gene (Fraser et al. 1990), and it appears to have a dominant chromatin-opening activity (Ellis et al. 1996). The 225-bp core fragment coinciding with the hypersensitive site retains the essential properties of 5′HS3 (Philipsen et al. 1990). This core contains three footprinted binding sites for the erythroid-specific transcription factor GATA-1, alternated with three footprinted G-rich sequences or G boxes (Philipsen et al. 1990; Strauss and Orkin 1992). The smallest fragment capable of activating a β-globin gene in transgenic mice consists of footprints 1–3 (fp 1–3) of the core fragment (Philipsen et al. 1993). Fp 1 and 3 are GATA-1 binding sites, and fp 2 contains G boxes that are bound by multiple factors. Point mutations in fp 1 and 3 have shown that intactness of these sequences is absolutely essential for activity. From this, we concluded that GATA-1 binds to fp 1 and 3 in vivo. In contrast, point mutations abolishing binding of single factors to the G boxes of fp 2 could not be designed (Philipsen et al. 1993). The G boxes are bound by members of a transcription factor family characterized by the presence of three zinc fingers highly homologous to those found in Sp1. The Sp1 family includes ubiquitously expressed proteins like Sp1 (Kadonaga et al. 1987), Sp3 (Hagen et al. 1992; Kingsley and Winoto 1992), basic transcription element binding protein (BTEBs; Imataka et al. 1992) and basic Krüppel-like factor (BKLF; Crossley et al. 1996), and tissue-restricted factors like gut-enriched Krüppel-like factor (GKLF; Shields et al. 1996), lung Krüppel-like factor (LKLF; Anderson et al. 1995), and erythroid Krüppel-like factor (EKLF; Miller and Bieker 1993). Moreover, a large number of Sp1-like zinc finger domains can be found in the expressed sequence tag (EST) databases. Fp 2 is bound in vitro by Sp1, Sp3, BKLF, EKLF, and other as yet uncharacterized family members, present in erythroid nuclear extracts (Philipsen et al. 1993; N. Gillemans, unpubl.). This plethora of transcription factors precludes a straightforward interpretation of the relationship between binding sites and activators, and thus it is not known which protein binds to fp 2 in vivo.
The apparent redundancy of transcription factors presents a general problem in the interpretation of gene expression data. The analysis of knockout mice is a first step toward the solution of this problem (e.g., Nuez et al. 1995; Perkins et al. 1995; Marin et al. 1997; Tewari et al. 1998), but this approach can only provide indirect answers. Pioneering work in Drosophila has established that a DNA-binding specificity mutant can be used as a tool to demonstrate direct interaction of a transcription factor and a DNA target site in a multicellular organism (Schier and Gehring 1992). In this paper we have applied this approach to mice, utilizing the minimal fragment of 5′HS3 to test the role of putative activator proteins (Sp1 and EKLF) directly. First, we mutated the G boxes in fp 2 so they no longer bind Sp1 family members in vitro. Second, we designed zinc fingers recognizing the mutated sequence in fp 2. We then tested the capacity of the engineered factors (mSp1 and mEKLF) to activate the mutant fp 2 construct in transgenic mice. The results show that EKLF but not Sp1 activates fp 2 of 5′HS3, and we conclude that EKLF plays a very specific role in the activity of 5′HS3 and in the activation of the β-globin locus.
Results
Experimental strategy
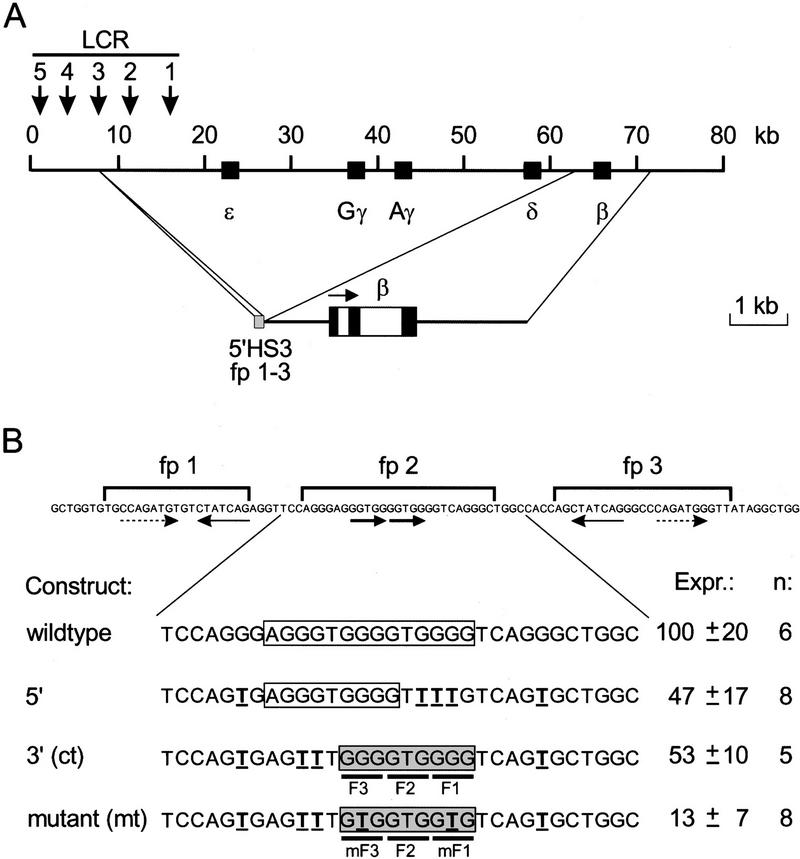
We have used human β-globin gene activation by the minimal fragment of 5′HS3 as the experimental system (Fig. 1A). The functional importance of the GATA-1 sites in fp 1 and 3 and the G boxes in fp 2 (Fig. 1B) has been analyzed extensively in transgenic mice (Philipsen et al. 1993). The results of these experiments show that all three footprinted sequences are required for activation of a linked β-globin gene. Sp1 family members recognize a 9-bp sequence GGG GT/CG GGG (Letovsky and Dynan 1989) and thus it could be predicted that fp 2 contains two overlapping binding sites, in addition to potential weaker binding sites (Fig. 1B, wild type). To minimize the complexity of fp 2, we first determined whether one binding site would suffice for activation. Two mutants with only one binding site (Fig. 1B, 5′ and 3′) were made by disrupting every other potential binding site by G → T transversions (Philipsen et al. 1993). These mutants were placed in their natural context between fp 1 and 3 and the resulting fragments were tested for their capacity to activate the β-globin gene in transgenic mice. Founder fetuses were dissected at 13.5 days of gestation (E13.5), and DNA was prepared from the head, yolk sac, and placenta to determine the transgene copy number and to check for mosaicism (Philipsen et al. 1993). RNA was isolated from the livers of nonmosaic transgenic fetuses, and the level of human β-globin mRNA was quantitated with S1 nuclease protection analysis using the level of the endogenous β-major mRNA as an internal control (Philipsen et al. 1993). The expression level per transgene copy was set at 100 for the wild-type fp 1–3 construct (Fig. 1B). The constructs with only one G box in fp 2 are expressed at approximately twofold lower levels than those observed with the wild-type fp 1–3 construct. Thus, we conclude that each G box contributes to the activity of fp 1–3. Because the 3′ sequence has the best match with the Sp1 consensus binding site (Letovsky and Dynan 1989), we concentrated on this motif. For reasons of clarity, we will refer to this permutation of fp 2 as control (ct in Fig. 1B), hereafter.
Figure 1.
Activity of the fp 1–3 fragment 5′HS3 of the human β-globinLCR in transgenic mice. (A) Schematic drawing of the human β-globin locus. The hypersensitive sites of the LCR are indicated by downward arrows, the genes by solid boxes (top). The activity of fp 1–3 of 5′HS3 was analyzed in transgenic mice using the HpaI–EcoRV fragment of the β-globin gene as the reporter gene (bottom). (B) Quantitation of the expression of fp 1–3 constructs in transgenic mice. The sequence of the fp 1–3 area is shown (top). Strong GATA-1 binding sites are underlined with thin arrows; weak GATA-1 binding sites with broken arrows (Philipsen et al. 1993), the two G-rich motifs in fp 2 are underlined with bold arrows. Permutations of fp 2 analyzed in transgenic founder fetuses are shown below. The G-rich motifs are boxed. Point mutations introduced are in boldface and underlined. A shaded box highlights the mutant binding site. Nucleotides interacting with the zinc fingers of Sp1 family members are underscored with solid lines (F1, F2, F3), as are nucleotides in the mutant binding site interacting with the mutant zinc fingers (mF1, mF3). Expression levels (Expr.) are given with standard deviations, and the number of independent transgenics analyzed is indicated (n).
The zinc fingers of Sp1 family members are structurally closely related to those of the Zif268 protein (Narayan et al. 1997; Sjottem et al. 1997). From the crystal structure of the DNA-binding domain of Zif268 it is known that one zinc finger occupies 3 bp in the recognition sequence (Pavletich and Pabo 1991). We therefore assumed that fingers 1 and 3 recognize the triplet GGG, and finger 2 the triplet GTG in fp 2 (Fig. 1B). Hence, mutation of GGG to GTG triplets should interfere with recognition by fingers 1 and 3 of Sp1 family members. We tested the activity of this fp 2 mutant in transgenic mice. The analysis of eight founder fetuses shows that the fp 2 mutant results in a further fourfold reduction of β-globin reporter expression as compared with control fp 2 (Fig. 1B). These data are consistent with earlier observations in transgenic mice containing more drastic mutations in fp 2 (Philipsen et al. 1993). Thus, we conclude that the lower expression level is due to decreased binding of an activator protein to the mutant fp 2 sequence and that the fp 2 mutant could be used to test the transcriptional effect of modified activator proteins engineered to bind specifically to this sequence.
Construction of transcription factors with altered DNA-binding site specificity
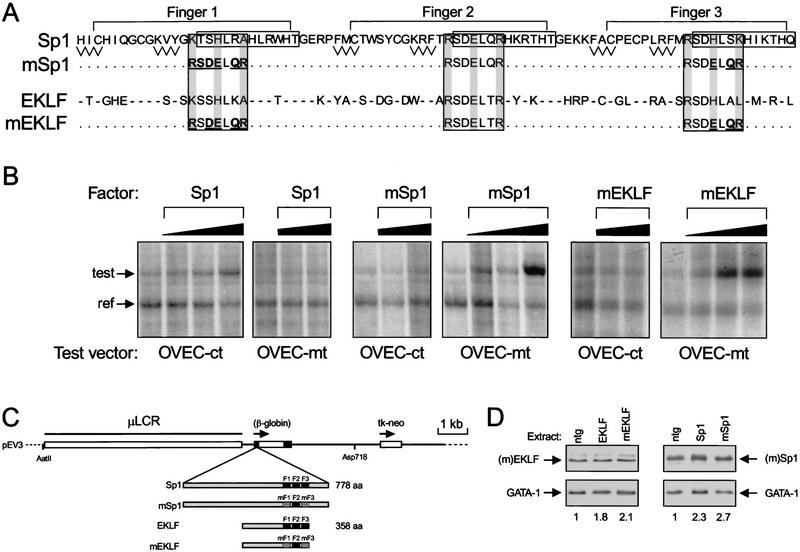
We set out to change the DNA-binding site specificity of two candidate activators of 5′HS3: Sp1, the ubiquitously expressed archetypal transcription factor, and EKLF, the only erythroid-specific Sp1 family member described to date. The fp 2 mutant contains a 9-bp sequence consisting of three GTG triplets (Fig. 1B). In wild-type Sp1 family members, zinc finger 2 recognizes this sequence (Li et al. 1991; Feng et al. 1994; Narayan et al. 1997; Sjottem et al. 1997). Thus, if we would change the specificity of fingers 1 and 3 of EKLF and Sp1 to that of finger 2, we should obtain proteins that interact specifically with the fp 2 mutant. However, such changes may have unexpected consequences. A simple triplication of finger 2 of Sp1 binds to such a sequence albeit with reduced affinity (Desjarlais and Berg 1993), which might be due to structural determinants provided by fingers 1 and 3, and/or the spacer amino acids between the fingers (Jacobs 1992; Choo and Klug 1993; Isalan et al. 1997). In addition, zinc fingers are involved in protein–protein interactions (Perkins et al. 1994; Merika and Orkin 1995) and notably, the interactions with GATA-1 are of direct relevance to this work (Merika and Orkin 1995). Therefore, we substituted the minimum number of amino acids in the recognition helices of fingers 1 and 3 to make their DNA-binding specificity identical to that of finger 2 of Sp1 (Pavletich and Pabo 1991; Narayan et al. 1997; Sjottem et al. 1997). No other changes to the zinc finger domains of Sp1 and EKLF were made to preserve presumptive functions other than direct DNA recognition (Fig. 2A; mSp1 and mEKLF).
Figure 2.
Sp1 family members with novel binding site specificity. (A) The amino acid sequence of the three zinc fingers of Sp1 is shown (top). β-Sheets are shown as zigzag lines and α-helices are boxed (adapted from Pavletich and Pabo 1991). Large rectangles include all of the amino acids that are known to make base contacts; the shading indicates residues that are thought to be particularly important for binding site recognition. Amino acid substitutions in fingers 1 and 3 of Sp1 are underlined and bold (mSp1). The amino acid sequence of the EKLF zinc fingers is shown below. Residues identical to those found in Sp1 are indicated by dashes. Amino acid substitutions in fingers 1 and 3 of EKLF are underlined and bold (mEKLF). Unchanged residues are indicated by dots. (B) mSp1 and mEKLF are functional transcriptional activators. CHO cells were transfected with 3 μg of OVEC test vector containing either the control (OVEC–ct) or the mutant (OVEC–mt) binding site, 0.15 μg of OVEC reference vector and 0.1, 0.5, or 2 μg of the indicated pRc/CMV expression vector (factor). After 72 hr, the cells were harvested and RNA synthesized from the OVEC vectors was analyzed by the RNase protection assay (Westin et al. 1987). The RNA probe detects the signals of both the reference and test vector; these are indicated by arrows (ref and test, respectively). (C) Schematic drawing of the pEV3 vector used to express transcription factors in erythroid cells. The cDNAs encoding Sp1, mSp1, EKLF, and mEKLF were inserted in the first exon of a modified β-globin gene. Erythroid-specific expression is driven by a 6.5-kb version of the LCR (μLCR; Talbot et al. 1989). The tk-neo gene is used for selection of stably transfected MEL cells. The AatII and Asp718 restriction sites were used to isolate the microinjection fragment for transgenesis. (D) Western blot analysis of transcription factor expression in transgenic mice. Nuclear protein extracts were made of E13.5 livers with the indicated pEV transgene; (ntg) Nontransgenic fetal liver. Twenty micrograms of protein was used for Western blotting. The blots were probed with anti-EKLF (top left) or anti-Sp1 rabbit polyclonal antibodies (top right). As a loading control, they were reprobed with an anti-GATA-1 rat monoclonal antibody (bottom). (Bottom) Relative levels of EKLF and Sp1 as determined by densitometric scanning of the autoradiographs and normalized to the GATA-1 loading control (ntg = 1).
The mutant transcription factors are transcriptional activators
First, we determined whether mEKLF and mSp1 would be active as transcriptional activators in a binding site-dependent manner. We inserted the control and mutant binding site 20 bp upstream of the TATA box in the promoter of the OVEC test vector (Westin et al. 1987) to assess the capacity of the factors to act as classical transcriptional transactivators. The cDNAs encoding the factors were cloned in the expression vector pRc/CMV. These constructs were cotransfected in CHO cells with the OVEC test vectors and the OVEC–REF vector, which serves as an internal control for transfection efficiency. RNA was isolated 72 hr after transfection, and the RNA synthesized from the OVEC test- and OVEC–REF vectors was quantitated by RNase protection analysis (Westin et al. 1987). This shows that increasing the amount of mSp1 stimulates transcription of the OVEC vector with the mutant binding site (OVEC-mt; Fig. 2B) but not of the OVEC vector with the control binding site (OVEC-ct; Fig. 2B). On the contrary, Sp1 has no effect on transcription of the OVEC-mt construct but stimulates transcription of the OVEC-ct construct (Fig. 2B). Transactivation by Sp1 appears to be less effective than that observed with mSp1 (Fig. 2B), most likely because of competition with transcriptional repressors like Sp3 for binding to the control sequence but not the mutant sequence (Hagen et al. 1994). A very similar result is obtained with mEKLF transactivation (Fig. 2B; data not shown).
We conclude that the altered DNA-binding specificity mutants of EKLF and Sp1 are functional transcriptional activators that can stimulate transcription through the mutant fp 2 sequence in vivo.
Erythroid-specific expression and DNA-binding properties of the transcription factors
To express the transcription factors in red cells, we cloned cDNAs encoding the wild-type and mutant proteins in the erythroid-specific expression vector pEV3 (Needham et al. 1992), resulting in Sp1–pEV, mSp1–pEV, EKLF–pEV, and mEKLF–pEV (Fig. 2C). Murine erythroleukaemia (MEL) cells were stably transfected with these constructs to check for the expression and DNA-binding properties of the transgene-derived transcription factors (Fig. 3A; data not shown); they were then used to generate transgenic mice. For each construct, one transgenic line was analyzed further; these lines will be referred to as Sp1–pEV, mSp1–pEV, EKLF–pEV, and mEKLF–pEV.
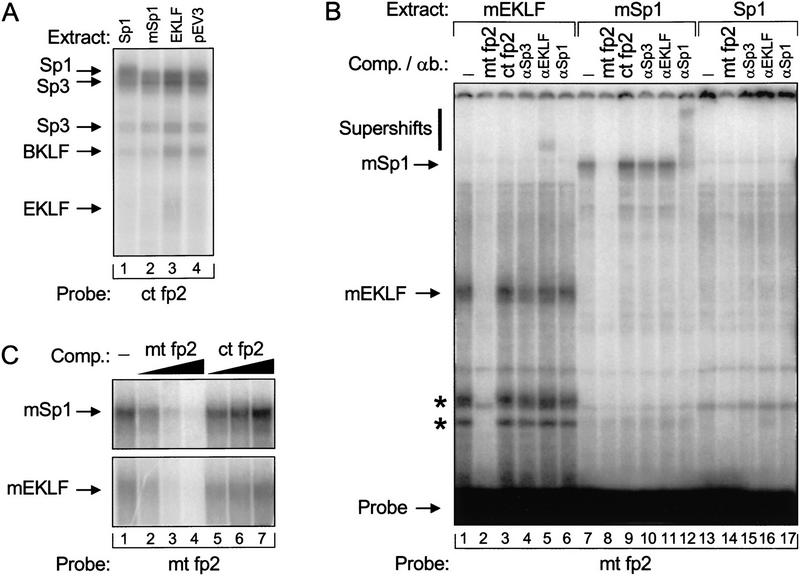
Figure 3.
Gel retardation analysis of Sp1 family members binding to the mutant fp 2 binding site. (A) Extracts were made from MEL cells transfected with pEV3, EKLF–, Sp1–, and mSp1–pEV. In lanes 1 and 2, 2 μg of extract was incubated with the control fp 2 probe (ct; see Fig. 1). In lanes 3 and 4, 4 μg of extract was used. These reactions contained a fivefold molar excess of Sp1 competitor oligonucleotide to reduce the background caused by Sp1 family members that bind more avidly to GC than to GT boxes. The binding reactions were separated on a 4% PAA/0.5× TBE gel. The positions of bands corresponding to Sp1, Sp3, BKLF, and EKLF are indicated (see Crossley et al. 1996; Marin et al. 1997). (B) Extracts were made from fetal livers transgenic for Sp1–, mSp1–, or mEKLF–pEV. In lanes 1–6, 10 μg of extract was incubated with the mutant fp 2 probe (mt; see Fig. 1). In lanes 7–12, 2 μg of extract was used, and in lanes 13–17, 5 μg of extract was added to the reactions. In addition, the reactions contained a 50-fold molar excess of cold mutant fp 2 double-stranded competitor oligonucleotide (mt fp2; lanes 2,8,14) or control fp2 control competitor (ct fp2; lanes 3,9), or antibodies directed against Sp3 (αSp3; lanes 4,10,15), EKLF (αEKLF; lanes 5,11,16) and Sp1 (αSp1; lanes 6,12,17). The positions of mEKLF, mSp1, and the free probe are indicated by arrows; a vertical bar denotes the positions of supershifts after the addition of antibody. Apparent degradation products of mEKLF are marked with an asterisk. (C) Binding specificity of mSp1 and mEKLF to the mutant fp 2 sequence. Two micrograms of mSp1–pEV or 4 μg of mEKLF–pEV fetal liver extract was incubated with the mutant fp 2 probe in the presence of increasing amounts of cold competitor oligonucleotides. (Lane 1) No competitor added; (lanes 2–4) 2-, 10-, and 50-fold molar excess of mutant fp 2 competitor; (lanes 5–7) 2-, 10-, and 50-fold molar excess of control fp 2 competitor, respectively.
The expression levels of transgene-derived transcription factors were assessed by Western blot analysis of E13.5 fetal liver extracts. The blots were probed with rabbit polyclonal antisera directed against EKLF or Sp1. The epitopes recognized by these antibodies are amino-terminal to the zinc finger domains and they therefore detect both the wild-type and mutant transcription factors. As a loading control, the blots were reprobed with a rat monoclonal antibody specific for the erythroid-specific transcription factor GATA-1. The results show that the expression levels of the transgene-derived transcription factors are comparable to those of the endogenous factors, as we observe a moderate (approximately twofold) increase of the signals in the transgenic samples, relative to the GATA-1 control (Fig. 2D). We conclude that the transgene-derived transcription factors are expressed at physiological levels, thus allowing comparison of the biological effects of these factors.
Protein extracts derived from transfected MEL cells and E13.5 transgenic livers were used for gel retardation analysis with oligonucleotide probes containing the control or the mutant fp 2 sequence. Transfected MEL cell extracts show that transgene-derived Sp1 and EKLF proteins bind to the control fp 2 sequence (Fig. 3A); this could not be demonstrated in fetal liver extracts because of the large background caused by other proteins present in these extracts (data not shown).
Very little, if any, specific binding to the mutant fp 2 oligonucleotide is observed with extracts from Sp1–pEV fetal livers (Fig. 3B), or with extracts from EKLF–pEV and nontransgenic livers (data not shown). Thus, Sp1 family members do not appear to interact efficiently with the mutant sequence, in agreement with the functional data obtained in transgenic mice (Fig. 1B) and CHO cells (Fig. 2B). In contrast, transgene-derived mSp1 and mEKLF proteins do interact with the mutant fp 2 sequence. The specificity of these interactions is demonstrated by the addition of competitor oligonucleotides and antibodies to the reactions (Fig. 3B,C). Thus, we conclude that mEKLF and mSp1 bind specifically to the mutant fp 2 sequence in vitro and not to the control fp 2 sequence, whereas the opposite is true for wild-type EKLF and Sp1.
Functional analysis in transgenic mice
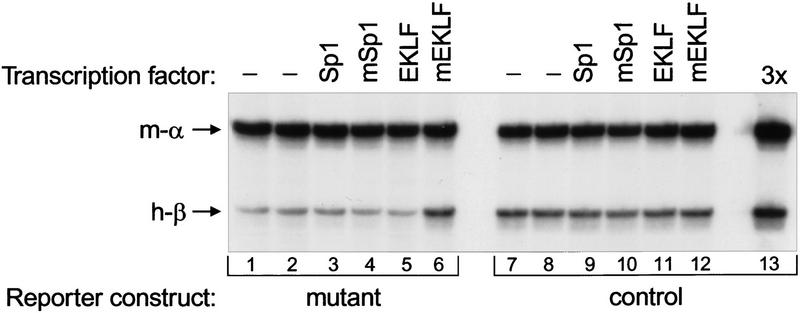
To analyze the effect of EKLF and Sp1 on the activity of fp 1–3 constructs in transgenic mice, we generated reporter mice with the control or the mutant fp 1–3 fragment driving expression of the human β-globin gene (see Fig. 1). For each construct, two independent lines were established. These lines were crossed with the Sp1–pEV, mSp1–pEV, EKLF–pEV, and mEKLF–pEV lines, and the effect of transgene-derived transcription factors on human β-globin gene expression was determined by quantitative S1 nuclease protection analysis (Fig. 4; Table 1). The level of mouse α-globin mRNA was used as an internal control. Expression of the control reporter gene is not altered by the presence of Sp1–pEV, mSp1–pEV, EKLF–pEV, or mEKLF–pEV transgenes. Thus, the transgene-derived transcription factors have no influence on the control reporter gene, excluding the possibility of a dominant effect on reporter gene expression. In contrast, the expression levels of the mutant reporter gene are affected by mEKLF, but not by Sp1, mSp1, and EKLF. This demonstrates that the observed differences are dependent on the altered zinc finger domains of mEKLF and that the effect is specific to EKLF, as mSp1 has no influence on the mutant reporter gene. Expression of the mutant reporter gene is increased threefold in the presence of mEKLF, approaching the level observed with the control construct (Fig. 4; Table 1). From this we conclude that EKLF directly activates 5′HS3 by binding to fp 2 in vivo. Interestingly, the mSp1 protein has no effect on β-globin gene expression through fp 2 of 5′HS3, even though it is a functional activator when the mutant binding site is placed in the context of the promoter of the OVEC vector (Fig. 2B). Thus, it appears that the specific combination of GATA-1 bound at fp 1 and 3 and EKLF bound at fp 2 is required for the activity of 5′HS3 in erythroid cells.
Figure 4.
Quantitative S1 nuclease protection assay of human β-globin transgene expression. RNA was isolated from E13.5 fetal livers containing the mutant or the control β-globin reporter construct and the Sp1–, mSp1–, EKLF–, or mEKLF–pEV transgenes as indicated. One microgram of RNA was used for the S1 nuclease protection assay with probes detecting endogenous mouse α-globin mRNA (m-α) and human β-globin mRNA (h-β). An increased level of human β-globin mRNA is only observed for the mutant reporter in the presence of mEKLF (lane 6); no changes are found in any of the other combinations. Lane 13 contains 3 μg of the RNA sample used in lane 7 to demonstrate probe excess.
Table 1.
Expression levels of human β-globin mRNA in E13.5 fetal liver cells of reporter/transcription factor transgenic lines
| pEV transgene
|
Reporter transgene (copy no.)
|
|||
|---|---|---|---|---|
| control line 1 (5)
|
control line 2 (17)
|
mutant line 1 (7)
|
mutant line 2 (4)
|
|
| None | 56 | 40 | 15 | 11 |
| Sp1 | 55 | 42 | 14 | 10 |
| mSp1 | 55 | 42 | 16 | 11 |
| EKLF | 55 | 40 | 15 | 11 |
| mEKLF | 54 | 41 | 43 | 32 |
Expression of the reporter genes was calculated as the ratio of human β-globin to mouse α-globin signals, corrected for transgene copy number, and normalized to the expression level of the wild-type fp 1–3 construct (=100; see Fig. 1B).
Analysis of reporter gene expression at the single cell level
The S1 nuclease protection assay described above provides a quantitative measurement of reporter gene expression in populations of cells. Therefore, the reporter gene might be expressed at a high level in a subset of the cells and not in the remaining cells. In this scenario, mEKLF would act by increasing the fraction of cells expressing the mutant reporter gene. Alternatively, the reporter gene might be expressed in the large majority of erythroid cells, in which case mEKLF could exert its effect by increasing the rate and/or the duration of active transcription of the mutant reporter gene (Milot et al. 1996b). To distinguish between these possibilities, we analyzed reporter gene expression at the single cell level by primary transcript in situ hybridization (Wijgerde et al. 1995). Oligonucleotide probes detecting nuclear transcription spots of the endogenous α-globin genes were used to determine the number of erythroid cells (Tewari et al. 1996). Human β-globin nuclear transcription signals were visualized with four oligonucleotides derived from intron 1; as two of these probes overlap partially with exon sequences, cytoplasmic β-globin mRNA can also be detected (Dillon et al. 1997). The presence of cytoplasmic mRNA shows that a cell has expressed the transgene at some point in time, whereas the presence of nuclear precursor RNA at the site of transcription is indicative of active transcription because precursor RNAs are rapidly processed (Wijgerde et al. 1995). The results of this analysis are summarized in Figure 5 and Table 2.
Figure 5.
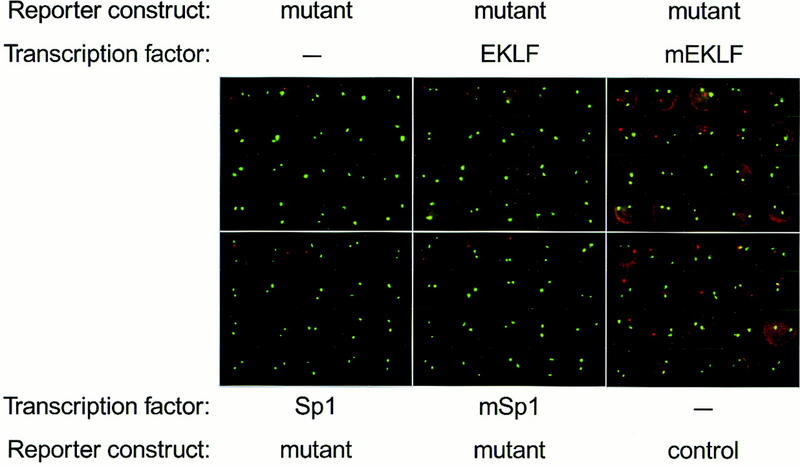
Primary transcript in situ hybridization detecting human β-globin and mouse α-globin transcription. E13.5 fetal liver cells with the indicated transgene makeup (see also Fig. 4) were disrupted in PBS, spotted on polylysine-coated slides, fixed, and subjected to primary transcript in situ hybridization with two sets of oligonucleotide probes detecting mouse α-globin (green) and human β-globin (red) nuclear transcription signals. The probes for human β-globin also detect cytoplasmic mRNA; this is particularly evident in the mutant reporter/mEKLF and the control reporter cells. Representative examples of 20 cells are shown for each transgene combination.
Table 2.
Primary transcript in situ hybridization of E13.5 fetal liver cells from reporter/transcription factor transgenic lines
| pEV transgene
|
Reporter transgene (copy no.)
|
|||
|---|---|---|---|---|
| control line 1 (5)
|
control line 2 (17)
|
mutant line 1 (7)
|
mutant line 2 (4)
|
|
| None | 57 | 86 | 13 | 7 |
| Sp1 | 59 | N.D. | 14 | 8 |
| mSp1 | 55 | N.D. | 12 | 6 |
| EKLF | 56 | N.D. | 14 | 7 |
| mEKLF | 55 | N.D. | 39 | 23 |
The percentage of cells with human β-globin transcription signals was calculated from at least 300 cells with α-globin transcription signals (see Fig. 5) for each data point. The error margin was <10%.
Cells expressing mEKLF show a three- to fourfold increased number of nuclei with transcriptional foci of the mutant reporter gene. Moreover, the cytoplasm of the cells shows a more intense red staining than cells with just the mutant reporter construct, reflecting the increased amounts of human β-globin mRNA in the presence of mEKLF. Human β-globin signals are not affected if mSp1 is expressed in these cells, in agreement with the S1 nuclease analysis. All cells with the control reporter gene contain β-globin mRNA in the cytoplasm, as do cells with the mutant reporter containing mEKLF. Expression of any of the transcription factors has no effect on the control reporter gene, nor does expression of Sp1 or EKLF have any effect on the mutant reporter gene, in agreement with the S1 nuclease data.
Thus, the in situ hybridization analysis shows that the fp 1–3 reporter transgenic lines used in this study do not show classical position effect variegation. Such effects can be observed when LCR mutants in the context of the 70-kb β-globin locus are integrated in heterochromatic regions (Milot et al. 1996b). The fact that we observe primary transcription signals in only part of the cells shows that transcription of the reporter genes is discontinuous. Importantly, mEKLF appears to activate the mutant reporter gene in all cells because the cytoplasmic staining increases in all cells (Fig. 5).
DNase I hypersensitivity analysis
The core fragments of the LCR coincide with strong, erythroid-specific DNase I hypersensitive sites. In E13.5 liver cells of the reporter lines a hypersensitive site is found at the control fp 1–3 sequence, whereas a weaker hypersensitive site is found at the mutant fp 1–3 sequence (data not shown). Thus, we asked whether binding of mEKLF to the mutant fp 1–3 sequence would result in increased DNase I hypersensitivity. To investigate this, nuclei isolated from transgenic E13.5 liver cells were subjected to a DNase I fade-out analysis (see Ellis et al. 1996). Purified DNA of these DNase I series was digested with NcoI and Southern blotted (Fig. 6). In the absence of mEKLF, a weak hypersensitive site is detectable at the mutant fp 1–3 area (Fig. 6B, mutant); hypersensitivity is increased modestly in the compound transgenic (Fig. 6B, mutant + mEKLF). As a control for DNase I digestion, the same blot was stripped and rehybridized with a probe detecting a 0.85-kb fragment of the mouse α1-globin gene, as DNase I sensitivity of this gene should not be affected by mEKLF (Fig. 6B, bottom). Quantitation of the signals in the hypersensitive site area relative to the corresponding mouse α1-globin signals indicates that hypersensitivity is increased two- to threefold in the presence of mEKLF (Fig. 6C). We conclude from these data that direct binding of EKLF is implicated in the chromatin remodeling activity of 5′HS3 of the β-globin LCR (Ellis et al. 1996).
Figure 6.
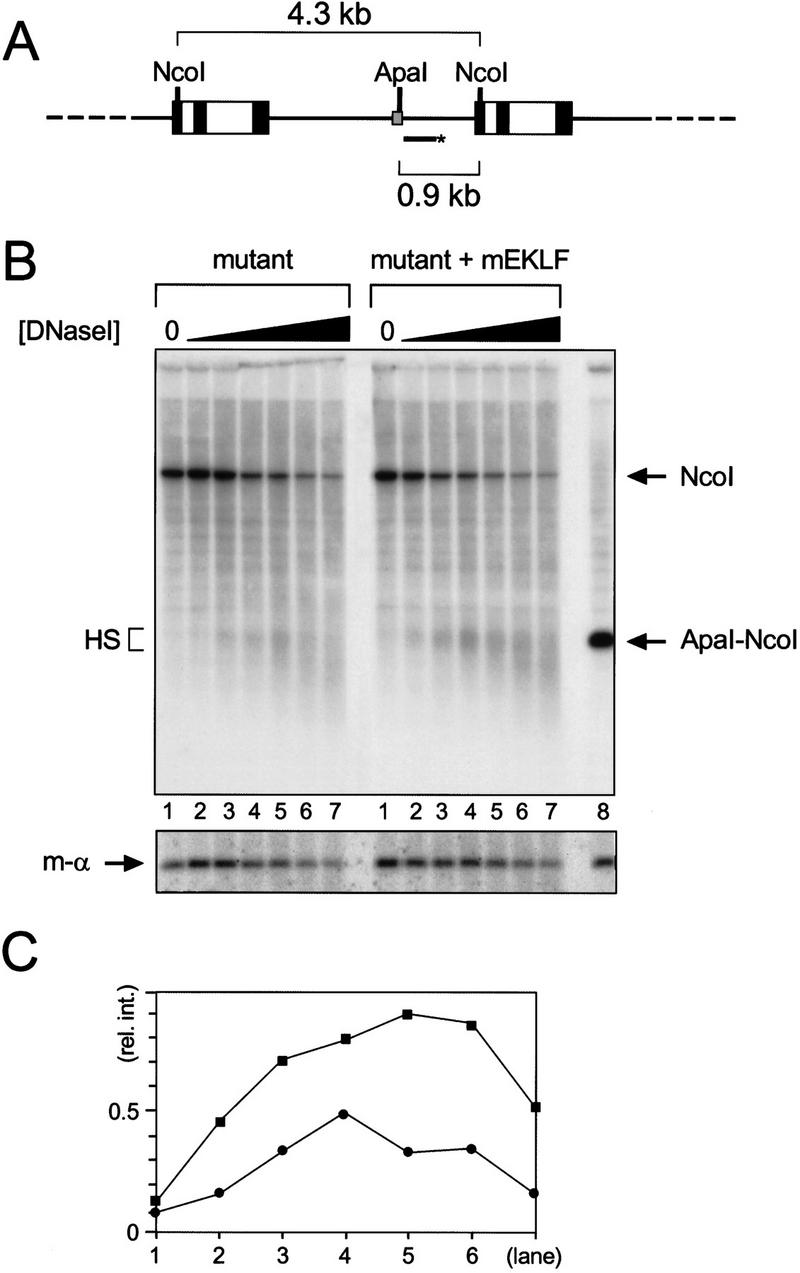
Hypersensitive site formation at the mutant fp 1–3 sequence in the presence of mEKLF. (A) Schematic drawing of the mutant reporter transgene array and the strategy used to detect hypersensitivity at fp 1–3. The HpaI–AccI probe does not detect the mEKLF–pEV transgene (Needham et al. 1992). The probe (bar with asterisk) hybridizes to the 4.3-kb NcoI repeat fragment of the mutant reporter gene. The 0.9-kb ApaI–NcoI fragment marks the position of the hypersensitive site (see B). (Other details are as for Fig. 1.) (B) Southern blot analysis of the hypersensitive site at the mutant fp 1–3 sequence. Nuclei were isolated from E13.5 fetal livers from mutant reporter and mutant reporter/mEKLF–pEV transgenics as indicated. Nuclei were digested with increasing amounts of DNase I, and DNA was purified, cut with NcoI, and analyzed by Southern blotting. The DNA in lane 8 was digested with NcoI and ApaI to mark the position of fp 1–3 (arrow, ApaI–NcoI). The repeat band of the mutant reporter transgene is indicated by an arrow (NcoI); the hypersensitive site at mutant fp 1–3 by a bracket (HS). (Bottom) The blot was stripped and rehybridized with a probe detecting an 0.85-kb NcoI fragment of the mouse α1-globin gene (m-α). (C) Quantitation of the hypersensitive site by PhosphorImager analysis. The signal in the hypersensitive site was divided by the signal in the corresponding mouse α1-globin band [(y-axis) relative intensity of the hypersensitive site] and plotted against the lane number (x-axis). (•) Mutant reporter; (█) mutant reporter plus mEKLF–pEV.
Discussion
Binary transgenic systems
In this paper we have used binary transgenic mice to investigate the apparent redundancy of transcription factors binding to an important genetic element, 5′HS3 of the β-globin LCR. Our approach encompasses the use of engineered zinc finger transcription factors rationally designed to recognize a mutated binding site in 5′HS3. Several binary systems for regulating gene expression have been described, for instance, the yeast GAL4/UAS, the bacterial tet-repressor/operator, and steroid-inducible systems (Ornitz et al. 1991; Gossen et al. 1995; No et al. 1996; Wang et al. 1997). Furthermore, an engineered zinc finger domain recognizing a unique region of the BCR–ABL fusion oncogene has been shown to impair the expression of this oncogene in transformed cells after transient transfection in culture (Choo et al. 1994). Our data show that engineered zinc finger transcription factors are particularly useful tools for the study of tissue-specific activation of gene expression in transgenic mice.
Transcription factors with altered binding specificities
Previously, it has been shown that the DNA-binding specificity of the Drosophila fushi tarazu (ftz) protein can be altered by changing a single amino acid in its homeodomain. This was used to demonstrate direct autoregulation through binding of the ftz mutant to mutant DNA target sites in a regulatory element of the ftz gene, establishing that this in vivo approach can be used to distinguish between activities of homeodomain proteins with very similar binding site specificities (Schier and Gehring 1992). Most transcription factors are members of families classified by highly conserved DNA-binding domains. Members of such families recognize very similar or even identical DNA sequences and are often found in the same cell types. This suggests that family members may have overlapping arrays of target genes, and in many cases it has proved impossible to show which factor activates which gene. 5′HS3 of the β-globin LCR is a good example of this problem. The core fragment contains three GATA footprints interspersed by three footprints over G boxes; fp 1–3 of the core are minimally required for activity in transgenic mice (Philipsen et al. 1990, 1993). Because GATA-1 is the only GATA family member present at late stages of erythroid differentiation, point mutagenesis and in vivo footprinting analyses have demonstrated convincingly that GATA-1 interacts with the GATA sites of fp 1 and 3 in vivo (Strauss and Orkin 1992; Philipsen et al. 1993). In contrast, the G boxes of fp 2 are bound in vitro by a number of different proteins belonging to the Sp1 family (Philipsen et al. 1990, 1993). These proteins have very similar binding sites, and no mutations specific for individual factors could be made (Philipsen et al. 1993). To resolve this issue, we have introduced compensatory mutations in the zinc finger domains of the Sp1 family members Sp1 and EKLF designed to interact with a mutated G box in fp 2. Our data show that mEKLF activates the mutated 5′HS3 fragment in binary transgenic mice by binding directly to the fp 2 mutant in vivo. In contrast, mSp1 does not act as an activator in this context, even though it interacts specifically with the fp 2 mutant in vitro and is a functional transactivator through this binding site in the context of the promoter of the OVEC vector. We conclude that specific properties of EKLF are required for activation of 5′HS3. Our data do not rule out that other factors are also capable of activating 5′HS3. Possible candidates are other Sp1 family members such as BKLF and Sp3 (Hagen et al. 1992; Crossley et al. 1996), or the recently described multitype zinc finger protein Friend of GATA-1 (Tsang et al. 1997). It remains possible that such proteins function as activators of 5′HS3 through direct interaction with the wild-type fp 2 sequence. Furthermore, the reporter genes are present as multicopy concatemers, and thus synergism among the multiple insertions might occur. Therefore, it will be of interest to test the activity of mutant binding sites/mutant transcription factor combinations in the structural context of single copy integrants of the complete human β-globin locus (Strouboulis et al. 1992).
It is likely that replacement of fp 2 with a completely artificial binding site will have a stronger effect on the remaining activity of fp 1–3 (Philipsen et al. 1993). The construction of novel zinc finger domains recognizing the artificial site would be required to activate such a mutant reporter gene. Recently, it has been shown that the specificity of zinc finger domains can be changed efficiently by successive rounds of affinity selection through randomization of the recognition helix of each finger (Greisman and Pabo 1997). Thus, a second generation of mutant Sp1 family members could be designed that interacts with a completely arbitrary sequence in fp 2, resulting in more stringent activator/reporter combinations.
EKLF and activation of the β-globin locus
In this paper we have used binary transgenic mice to show that EKLF is an activator of the LCR that binds directly to fp 2 of 5′HS3. EKLF appears to function in a specific combination with GATA-1 bound at fp 1 and 3, consistent with earlier results obtained with point mutations and deletions in 5′HS3 (Philipsen et al. 1993; Pruzina et al. 1994). Thus, we have identified an in vivo binding site for EKLF in the β-globin LCR.
Targeted inactivation of the EKLF gene in mice results in lethality around E14–15 caused by severe anemia due to failure to express the β-globin gene at appropriate levels (Nuez et al. 1995; Perkins et al. 1995). The expression of other erythroid-specific genes examined, including the α-like and the embryonic/fetal β-like globins, is not affected showing that EKLF is essential for the activation of the adult β-globin promoter (Nuez et al. 1995; Perkins et al. 1995; Wijgerde et al. 1996). In addition, we have demonstrated recently that the expression of 5′HS3–lacZ transgenes requires EKLF (Tewari et al. 1998), and DNase I hypersensitive site analysis of the human β-globin locus in the EKLF null background showed strongly reduced sensitivity at the β-globin promoter and moderately reduced sensitivity at 5′HS3 (Wijgerde et al. 1996). These data agree well with our observation that direct binding of mEKLF to the mutant fp 1–3 sequence results in elevated reporter gene expression and increased DNase I hypersensitivity at fp 1–3. We conclude that direct binding of EKLF is implicated in chromatin remodeling over 5′HS3, the only LCR fragment that appears to have an intrinsic dominant chromatin opening activity (Ellis et al. 1996).
Deletion of 5′HS3 in the human or the mouse locus does not inactivate the LCR completely (Hug et al. 1996; Milot et al. 1996b). Deletion of 5′HS3 in the endogenous mouse locus results in a 30% reduction of β gene expression in adult mice (Hug et al. 1996). Natural mutations in the CACC box of the β-globin promoter result in a less severe reduction of β gene expression than observed in EKLF knockout mice (Thein 1993; Perkins et al. 1996; Wijgerde et al. 1996). Thus, it appears that EKLF sites are necessary in both the promoter and the LCR and that these sites act independently of each other. Remarkably, these interactions occur in a different context of other factors, and it is of interest to note that EKLF is not required for the activation of the γ-globin promoter in fetal liver cells, whereas it is essential for β-globin promoter activity in the same cells (Perkins et al. 1996; Wijgerde et al. 1996). Thus, we suggest that direct binding of EKLF to both the LCR and the β promoter is mandatory for high level transcription of the adult β-globin gene.
Outlook
Direct binding of EKLF appears to be pivotal for the function of 5′HS3 and the β-globin promoter. Therefore, mutation of both elements should result in a reporter gene that is expressed at extremely low levels in the absence of the appropriate mEKLF. Rendering mEKLF steroid dependent by fusing it to a steroid–receptor ligand binding domain would give an additional level of control to study the kinetics of gene activation in erythroid cells. The feasibility of steroid-inducible target gene expression in binary transgenic mice has been demonstrated (Wang et al. 1997). Inducible activation of LCR-dependent gene expression provides an in vivo system that can be used to address different aspects of gene expression, for instance, recruitment of RNA polymerase II to the promoter, interactions between the LCR and the promoter through DNA looping, and the potential link between transcriptional activation and DNA replication (for review, see Milot et al. 1996a; Ptashne and Gann 1997). We are currently exploring these issues in the context of the complete human β-globin locus (Strouboulis et al. 1992).
Materials and methods
DNA constructs and transgenesis
Mutations were introduced in the fp 1–3 region of 5′HS3 as described (Philipsen et al. 1993), and linked to the HpaI–EcoRV fragment of the human β-globin gene. The microinjection fragments were released from vector sequences by digestion with EcoRV, gel purified and used for transgenesis (Kollias et al. 1986). Founder transgenics were either dissected at E13.5 (Philipsen et al. 1990, 1993; Talbot et al. 1989) or bred to obtain lines. Intactness, mosaicism, and copy numbers of the transgenes were analyzed by Southern blotting (Philipsen et al. 1993).
To change the binding specificity of fingers 1 and 3 of EKLF and Sp1, the amino acid substitutions indicated in Figure 2A were introduced by PCR-mediated mutagenesis; the sequence of the cloned PCR products was verified by dideoxy sequencing (Sambrook et al. 1989); the mutated proteins were designated mEKLF and mSp1. For transfection experiments in CHO cells, cDNAs encoding Sp1, mSp1, and mEKLF were cloned in the NotI site of the expression vector pRc/CMV (Invitrogen). Control and mutant binding sites were cloned in the SalI site of the OVEC vector (Westin et al. 1987). The integrity and orientation of the cloned oligonucleotides was determined by dideoxy sequencing. For erythroid-specific expression, the EKLF, mEKLF, Sp1, and mSp1 cDNAs were cloned in the vector pEV3 (Needham et al. 1992), resulting in EKLF–pEV, mEKLF–pEV, Sp1–pEV, and mSp1–pEV. Microinjection fragments for transgenesis were obtained by digestion with AatII and Asp718.
Cell transfections
CHO cells (0.5 × 106) were transfected using the calcium phosphate precipitation method (Graham and Van der Eb 1973) with 3 μg of OVEC test vector, 0.15 μg of OVEC–REF vector, and 0.1, 0.5, or 2 μg of the appropriate pRc/CMV expression vector, supplemented with pTZ to a total amount of 18 μg of DNA. The cells were harvested after 72 hr and used to prepare RNA (Antoniou 1991).
MEL cells (25 × 106) were transfected by electroporation with 25 μg of pEV3 derivatives linearized with ScaI. After selection for G418 resistance, erythroid differentiation was induced by the addition of dimethylsulfoxide (2% vol/vol) to the media (Antoniou 1991). After 4 days of induction, the cells were harvested and protein extracts were made according to Scholer et al. (1989).
Fetal liver protein and RNA samples
Fetal livers were dissected from E13.5 fetuses and frozen immediately in liquid nitrogen. RNA was isolated from frozen tissue samples as described (Antoniou 1991; Philipsen et al. 1993). Protein was extracted according to Scholer et al. (1989) and stored at −80°C. Western blotting was performed as described, using 20 μg of protein per lane (Marin et al. 1997). Rabbit polyclonal antibodies recognizing EKLF and Sp1 were used in 1:3000 and 1:1000 dilutions, respectively. As a loading control, the blots were reprobed with a rat monoclonal antibody directed against GATA-1 (N6, cat. no. sc-265, Santa Cruz Biotechnology) used at 3 ng/μl.
Expression of the human β-globin transgene was quantitated by S1 nuclease analysis, using a probe specific for human β-globin mRNA, and probes detecting mouse β-major or mouse α-globin mRNA as the internal control (Antoniou 1991; Philipsen et al. 1993).
Gel retardation analysis
Oligonucleotide probes covering fp 2 were labeled and annealed according to Wall et al. (1988). Double-stranded competitor oligonucleotides were added at the amounts indicated prior to the addition of extract to the reactions. Polyclonal antibodies recognizing Sp1, Sp3, and EKLF were used at 1:160, 1:320, and 1:40 dilutions in the binding reactions, respectively. Incubation was for 30 min at room temperature. The samples were loaded on 4% polyacrylamide (PAA)/0.5× TBE gels and run at 300 V for 3 hr at 4°C. Other details are described in Wall et al. (1988). The sequences of the oligonucleotides used are as follows (only the sense strand is shown): fp 2 control, 5′-AGGTTCCAGT GAGTTTGGGGTGGGGTCAGTG-3′; fp 2 mutant, 5′-AGGTTCCAGTGAGTTTGTGGTGGTGTCAGTG-3′; and Sp1, 5′-ATAGTCCCGCCCCTAACTCCGCCCAT-3′. Bases different from the wild-type sequence of fp 2 are underlined; bases different between control fp 2 and mutant fp 2 are underlined and in boldface type.
Primary transcript in situ hybridization
E13.5 livers from individual fetuses were disrupted in 150 μl of PBS. Twenty microliters of this cell suspension was spotted onto a polylysine-coated slide and fixed in 4% formaldehyde, 5% acetic acid, for 20 min at room temperature. The slides were then washed three times for 10 min in PBS and stored in 70% ethanol at −20°C. Two sets of probes were used for in situ hybridization. Four biotinylated oligonucleotides were used to detect human β-globin transcription. This probe mixture is directed against intron 1; two of the oligonucleotides overlap partially with exon sequences and thus cytoplasmic β-globin mRNA can also be detected (Dillon et al. 1997). Three DIG-labeled mouse α-globin intron-specific oligonucleotides were used to reveal the erythroid cells (Tewari et al. 1996). Hybridizations were done as described by Tewari et al. (1996) and Wijgerde et al. (1995). For antibody detection, the Tyramid amplification system of Dupont NEN was used (Raap et al. 1995). For quantitation, at least 300 cells were scored for each slide. The reproducibility of these data fell within a 10% error margin.
The following oligonucleotides were used for in situ hybridizations: Human β-globin, 5′-CTGTCTCCACATGCCCAGTTTCTATGGTCTCCTTAAACCTGTCTTGTAA-3′, 5′-GGGTGGGAAAATAGACCAAAGGCAGAGAGAGTCAGTGCCTATCAGAAAC-3′, 5′-AGGGCAGTAACGGCAGACTCTCCTCAGGAGTCAGGT-3′, and 5′-ATAACAGCATCAGGAGGGACAGATCCCCAAAGGACTCA-3′; Mouse α-globin, 5′-CACAGAAAAGCATAGTTAGAAGCGCCCACTGAGCGAGTGCCAGGTCC-3′, 5′-AGCCCTTCCTAGGGGCCCAGATGCCGCCTGCCAGGTCCC-3′, and 5′-GCTCCCCTTCCTGGGACCACTATGTCCCTGCCTTGGGCACGAGGACCC-3′.
DNase I hypersensitivity assay
Ten E13.5 livers were used to isolate nuclei followed by treatment with increasing amounts of DNase I at 37°C (Ellis et al. 1996). DNA was purified, digested with NcoI, and Southern blotted. A 0.44-kb HpaI–AccI fragment from the β-globin promoter was used as a probe to detect the hypersensitive site at fp 1–3. A 0.6-kb BamHI–SacI fragment of the mouse α1 globin gene, detecting a 0.85-kb NcoI fragment, was used as a control probe. Quantitation was done on a PhosphorImager with ImageQuant software (Molecular Dynamics).
Acknowledgments
We thank Drs. Jim Kadonaga, Jim Bieker, and Guntram Suske for the gift of Sp1 cDNA and EKLF and Sp1/Sp3 antibodies, respectively. We are indebted to Dr. Albert Brinkman and Cor Berrevoets for help with the transfection assays. R.T., F.L. and M.W. were supported by a Netherlands Organisatie voor Wetenschappelijk Orderzoek (The Netherlands) grant to F.G. T.dW. was supported by Therexsys (UK).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact
Footnotes
E-MAIL vangeest@ch1.fgg.eur.nl; FAX 31-10-436 0225.
References
- Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: Identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou M. Induction of erythroid-specific expression in murine erythroleukemia (MEL) cell lines. Methods Mol Biol. 1991;7:421–434. doi: 10.1385/0-89603-178-0:421. [DOI] [PubMed] [Google Scholar]
- Choo Y, Klug A. A role in DNA binding for the linker sequences of the first three zinc fingers of TFIIIA. Nucleic Acids Res. 1993;21:3341–3346. doi: 10.1093/nar/21.15.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Y, Sanchez-Garcia I, Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc Natl Acad Sci. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- Ellis J, Tanun KC, Harper A, Michalovich D, Yannoutsos N, Philipsen S, Grosveld F. A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human β-globin locus control region. EMBO J. 1996;15:562–568. [PMC free article] [PubMed] [Google Scholar]
- Feng WC, Southwood CM, Bieker JJ. Analyses of β-thalassemia mutant DNA interactions with erythroid Kruppel-like factor (EKLF), an erythroid cell-specific transcription factor. J Biol Chem. 1994;269:1493–1500. [PubMed] [Google Scholar]
- Fraser P, Hurst J, Collis P, Grosveld F. DNase I hypersensitive sites 1, 2 and 3 of the human β-globin dominant control region direct position-independent expression. Nucleic Acids Res. 1990;18:3503–3508. doi: 10.1093/nar/18.12.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- Grosveld F, Dillon N, Higgs D. The regulation of human globin gene expression. Bailliere’s Clin Hematol. 1993;6:31–55. doi: 10.1016/s0950-3536(05)80065-4. [DOI] [PubMed] [Google Scholar]
- Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug BA, Wesselschmidt RL, Fiering S, Bender MA, Epner E, Groudine M, Ley TJ. Analysis of mice containing a targeted deletion of β-globin locus control region 5′ hypersensitive site 3. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc Natl Acad Sci. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GH. Determination of the base recognition positions of zinc fingers from sequence analysis. EMBO J. 1992;11:4507–4517. doi: 10.1002/j.1460-2075.1992.tb05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kingsley C, Winoto A. Cloning of GT box-binding proteins: A novel Sp1 multigene family regulating T-cell receptor gene expression. Mol Cell Biol. 1992;12:4251–4261. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollias G, Wrighton N, Hurst J, Grosveld F. Regulated expression of human A γ-, β-, and hybrid γ β-globin genes in transgenic mice: manipulation of the developmental expression patterns. Cell. 1986;46:89–94. doi: 10.1016/0092-8674(86)90862-7. [DOI] [PubMed] [Google Scholar]
- Letovsky J, Dynan WS. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989;17:2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Knight JD, Jackson SP, Tjian R, Botchan MR. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991;65:493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milot E, Fraser P, Grosveld F. Position effects and genetic disease. Trends Genet. 1996a;12:123–126. doi: 10.1016/0168-9525(96)30019-x. [DOI] [PubMed] [Google Scholar]
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, Deboer E, Langeveld A, Tanun K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996b;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- Narayan VA, Kriwacki RW, Caradonna JP. Structures of zinc finger domains from transcription factor Sp1. Insights into sequence-specific protein-DNA recognition. J Biol Chem. 1997;272:7801–7809. doi: 10.1074/jbc.272.12.7801. [DOI] [PubMed] [Google Scholar]
- Needham M, Gooding C, Hudson K, Antoniou M, Grosveld F, Hollis M. LCR/MEL: A versatile system for high-level expression of heterologous proteins in erythroid cells. Nucleic Acids Res. 1992;20:997–1003. doi: 10.1093/nar/20.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- No D, Yao TP, Evans RM. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: Targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Perkins ND, Agranoff AB, Pascal E, Nabel GJ. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- Perkins AC, Gaensler KM, Orkin SH. Silencing of human fetal globin expression is impaired in the absence of the adult β-globin gene activator protein EKLF. Proc Natl Acad Sci. 1996;93:12267–12271. doi: 10.1073/pnas.93.22.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S, Talbot D, Fraser P, Grosveld F. The β-globin dominant control region: Hypersensitive site 2. EMBO J. 1990;9:2159–2167. doi: 10.1002/j.1460-2075.1990.tb07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S, Pruzina S, Grosveld F. The minimal requirements for activity in transgenic mice of hypersensitive site 3 of the β globin locus control region. EMBO J. 1993;12:1077–1085. doi: 10.1002/j.1460-2075.1993.tb05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzina S, Antoniou M, Hurst J, Grosveld F, Philipsen S. Transcriptional activation by hypersensitive site three of the human β-globin locus control region in murine erythroleukemia cells. Biochim Biophys Acta. 1994;1219:351–360. doi: 10.1016/0167-4781(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Raap AK, van de Corput MP, Vervenne RA, van Gijlswijk RP, Tanke HJ, Wiegant J. Ultra-sensitive FISH using peroxidase-mediated deposition of biotin- or fluorochrome tyramides. Hum Mol Genet. 1995;4:529–534. doi: 10.1093/hmg/4.4.529. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schier AF, Gehring WJ. Direct homeodomain-DNA interaction in the autoregulation of the fushi tarazu gene. Nature. 1992;356:804–807. doi: 10.1038/356804a0. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Balling R, Hatzopoulos AK, Suzuki N, Gruss P. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 1989;8:2551–2557. doi: 10.1002/j.1460-2075.1989.tb08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjottem E, Andersen C, Johansen T. Structural and functional analyses of DNA bending induced by Sp1 family transcription factors. J Mol Biol. 1997;267:490–504. doi: 10.1006/jmbi.1997.0893. [DOI] [PubMed] [Google Scholar]
- Strauss EC, Orkin SH. In vivo protein-DNA interactions at hypersensitive site 3 of the human β-globin locus control region. Proc Natl Acad Sci. 1992;89:5809–5813. doi: 10.1073/pnas.89.13.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouboulis J, Dillon N, Grosveld F. Developmental regulation of a complete 70-kb human β-globin locus in transgenic mice. Genes & Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- Talbot D, Collis P, Antoniou M, Vidal M, Grosveld F, Greaves DR. A dominant control region from the human β-globin locus conferring integration site-independent gene expression. Nature. 1989;338:352–355. doi: 10.1038/338352a0. [DOI] [PubMed] [Google Scholar]
- Tewari R, Gillemans N, Harper A, Wijgerde M, Zafarana G, Drabek D, Grosveld F, Philipsen S. The human β-globin locus control region confers an early embryonic erythroid-specific expression pattern to a basic promoter driving the bacterial lacZ gene. Development. 1996;122:3991–3999. doi: 10.1242/dev.122.12.3991. [DOI] [PubMed] [Google Scholar]
- Tewari R, Gillemans N, Wijgerde M, Nuez B, von Lindern M, Grosveld F, Philipsen S. Erythroid Krüppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the β-globin locus control region. EMBO J. 1998;17:2334–2341. doi: 10.1093/emboj/17.8.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein SL. β-thalassaemia. Bailliere’s Clin Hematol. 1993;6:151–175. doi: 10.1016/s0950-3536(05)80069-1. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu CN, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Wall L, deBoer E, Grosveld F. The human β-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes & Dev. 1988;2:1089–1100. doi: 10.1101/gad.2.9.1089. [DOI] [PubMed] [Google Scholar]
- Wang YL, Demayo FJ, Tsai SY, Omalley BW. Ligand-inducible and liver-specific target gene expression in transgenic mice. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- Westin G, Gerster T, Mueller MM, Schaffner G, Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- Wijgerde M, Gribnau J, Trimborn T, Nuez B, Philipsen S, Grosveld F, Fraser P. The role of EKLF in human β-globin gene competition. Genes & Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]