The worlds of RNA and DNA are not entirely separate. They frequently come together when RNA/DNA hybrids form during replication and transcription often inducing genome instability. Topoisomerases (Top) play an important role in preventing RNA/DNA annealing during transcription1–3, while the resolution of these hybrids by ribonucleases H (RNase H) helps to maintain genome integrity. In the report by Kim et al. (page 123), Top1, one of the two types of topoisomerases, was found to not only prevent R-loop formation but also to participate in the removal of abundant, persistent rNMPs incorporated during DNA replication , and in the process– assaulting genome stability. As RNA polymerases traverse DNA during transcription, distortions of DNA occur in front and behind the polymerase causing torsion that compacts the DNA in front (positive supercoiling) and relaxes the DNA behind (negative supercoiling)4. Topoisomerases must perform molecular gymnastics to restore normal DNA topology5. When Top1 is defective, nascent RNA can anneal to the underwound DNA forming R-loops (FIGURE). Similarly, R-loops can form when genes are transcribed at high rates1 or if the proteins that normally interact with the nascent RNA are defective2;3. In most cases the formation of R-loops has deleterious consequences, because the displaced DNA strand is left susceptible to breakage leading to transcription associated mutation (TAM)6;7 or transcription associate recombination (TAR). But what can come to the rescue when Top1 fails to prevent R-loop formation? RNases H can by cleaving the RNA strand of the R-loop thus restoring the original dsDNA form1–3.
Figure.
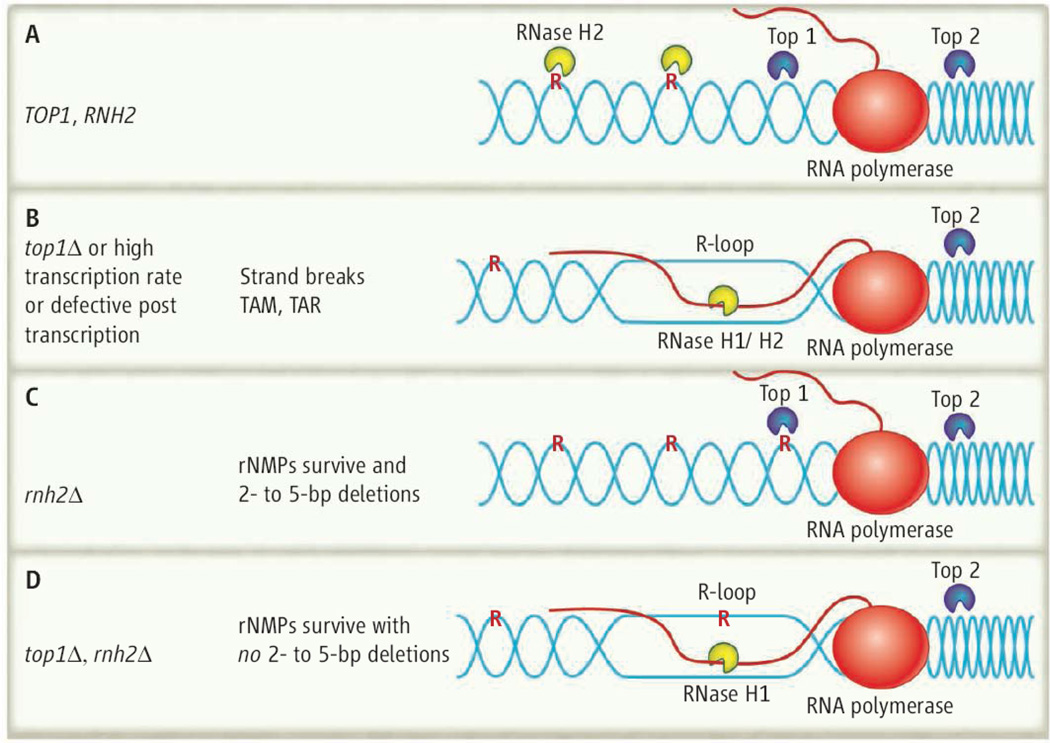
On top. (A) In yeast wild-type cells (TOP1, RNase H2), R-loop formation is prevented when Top2 relieves the positive supercoiling ahead of the transcribing RNA polumerase, and Top1 relieves the negative supercoiling behind. RNase H2 cleaves rNMPs incorporated into DNA, which leads to their replacement with dNMP. (B) In top1Δ mutants, RNA/DNA hybrids are formed and processed by RNase H1/H2, and rNMP removal is initiated by RNase H2. Mutations can result by TAM and TAR. (C) in rnh2Δ cells, Top1 will encounter and cleave rNMPs that have not been removed because RNase H2 is missing, which produces 2- to 5-bp deletions. (D) In the double mutant top1Δ rnh2Δ, RNA in R-loops may be cleaved by RNase H1, but rNMPs will remain in the DNA. DNA is blue, RNA is red, and single rNMPs in DNA are noted as red R.
There are two types of RNases H and both can process R-loops (fig)8. In Escherichia coli, deleting the genes encoding RNase HI and TopI results in synthetic lethality. In Saccharomyces cerevisiae, top1Δ strains are viable when only one RNase H (either RNase H1 or RNase H2) is absent, but when all three enzymes (Top1, RNase H1 and RNase H2) are missing, strains are unable to grow9. Not only are RNases H and Top1 linked through transcription and R-loop formation/removal, Kim et al. made the intriguing observation that these two types of enzymes are involved in processing of single ribonucleotides embedded in DNA, a recently described replication misincorporation error.
DNA polymerase incorporation of rNTPs is affected by the high ratio of rNTPs/dNTPs in vivo, the inherent preference of each DNA polymerase for dNTPs, and their (its) ability to remove rNMPs once incorporated into DNA10. In vitro experiments suggest that more than 10,000 ribonucleotides are incorporated during the replication of the ~ 12.5 Mbp S. cerevisiae genome. The lack of abundant rNMPs in normal DNA implies that they are quickly and efficiently removed. RNase H2, an enzyme that can cleave single ribonucleotides in duplex DNA leaving a 5’ rNMP attached to the downstream DNA11, seems to initiate the process, which most likely continues on a pathway involving Fen1 nuclease, a structure-specific nuclease having an important role in the maintenance of genome stability12. In the absence of RNase H2, rNMPs persist in abundance in genomic DNA, and are associated with the accumulation of 2–5 bp deletions that may be due to removal of rNMPs in DNA by an alternative pathway. This second pathway does not require the mismatch repair system suggesting repair occurs outside of DNA replication13.
This rare type of mutation has been reported previously and requires both Top1 activity and high rates of transcription for their formation6;7. This prompted Kim et al. to assess the involvement of Top1 in generating 2–5 bp deletions in RNase H2-defective strains. They found that eliminating Top1 activity prevents the 2–5 bp deletions that occur when RNase H2 is absent, suggesting that the 2–5 bp deletions are a signature of Top1 cleavage of rNMPs. Such deletions in the CAN1 gene14 were observed by TAM and in RNase H2-defective strains, but concordance of deletions were not perfect. Only two of the three TAM deletion hotspots in the CAN1 gene were observed in rnh201Δ strains (one of the subunit-encoding genes of RNase H2).
Top1 may encounter ribonucleotides in DNA due to high transcription rates that prevent RNase H2 access, or when RNase H2 is defective. Kim et al. convincingly showed that if there is a ribonucleotide at the site of Top 1 cleavage, an irreversible ssDNA break occurs. Top1 of S. cerevisiae nicks and seals only one of the DNA strands with the active site tyrosine forming a phosphodiester bond with the 3’-phosphate of the DNA end. After hydrolysis, the phosphodiester bond between two dNMPs is restored, thus reforming the natural 3’-5’ connection. However, when Top1 cleaves between a ribo- and deoxynucleotide, the 2’-OH of the ribose may form a 2’-3-cyclic phosphate rather than forming the 3’-5’ phosphodiester, prohibiting religation producing a stable single-stranded break. Deletions of 2–5 bp occur in tandem repeat sequences in the DNA suggesting repairing the ssDNA break within a tandem repeat can induce realignment and removal of some of the repeat sequence. However, most of the rNMPs in DNA persist, indicating that removal of all rNMPs is not a normal function of Top1 (Fig), and may be detrimental inducing genome instability.
Short tandem nucleotide repeats and rNMPs in DNA are necessary but not sufficient for deletions. Not all such repeats are deletion hot spots and some hot spots are different in TAM and RNase H2-defective strains. What accounts for these findings? Are rNMPs misincorporated by DNA polymerases the sole targets for Top1 and if so, is the race between Top1 and RNase H2 affected by transcription rates? Not all rNMPs in DNA Top1 targets. How are the Top1-resistent rNMPs in RNase H2-deficient strains copied and/or removed?
Interestingly, mutations in human RNase H2 can cause a rare autoimmune disorder (Aicardi Goutieres Syndrome or AGS) in which individuals suffer severe neurological defects often leading to early childhood death15. Kim et al. suggest that the cumulative effects of deletions in AGS patients with altered RNase H2 could be due to deletions similar to those seen in S. cerevisiae rnh201Δ strains. It will be of considerable interest to see if 2–5 bp deletions are more frequent in AGS patients.
Reference List
- 1.Drolet M. Molecular Microbiology. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 2.Huertas P, Aguilera A. Molecular Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Manley JL. Genes & Development. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 4.Liu LF, Wang JC. Proceedings of the National Academy of Sciences. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champoux JJ. Annual Review of Biochemistry. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Lippert MJ, et al. Proceedings of the National Academy of Sciences. 2011;108:698–703. doi: 10.1073/pnas.1012363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Proceedings of the National Academy of Sciences. 2011;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cerritelli SM, Crouch RJ. FEBS Journal. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Hage A, French SL, Beyer AL, Tollervey D. Genes & Development. 2010;24:1546–1558. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nick McElhinny SA, Kissling GE, Kunkel TA. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nick McElhinny SA, et al. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi ML, Purohit V, Brandt PD, Bambara RA. Chemical Reviews. 2006;106:453–473. doi: 10.1021/cr040497l. [DOI] [PubMed] [Google Scholar]
- 13.Clark AB, Lujan SA, Kissling GE, Kunkel TA. DNA Repair. 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The CAN1 gene encodes an arginine permease which can import the lethal arginine analog - canavanine. Rare canavanine resistant mutations can be detected when yeast cells are grown in the presence of the arginine analog.
- 15.Crow YJ, et al. Nature Genetics. 2006;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]