Over the past two decades, there have been tremendous advances in understanding the mechanisms by which the heart adapts to hemodynamic stress, producing either adaptive or maladaptive remodeling. Whereas, twenty years ago cardiomyopathies were only described by their physiologic consequences: dilation and systolic dysfunction, hypertrophy or restriction and diastolic dysfunction (or a combination), we have now developed a sophisticated understanding of the alterations in cell signaling and cell and tissue remodeling underlying these physiologic phenotypes. With the current revolution in gene sequencing technology, we are also rapidly advancing our understanding of the underlying genetic basis of these cellular events.
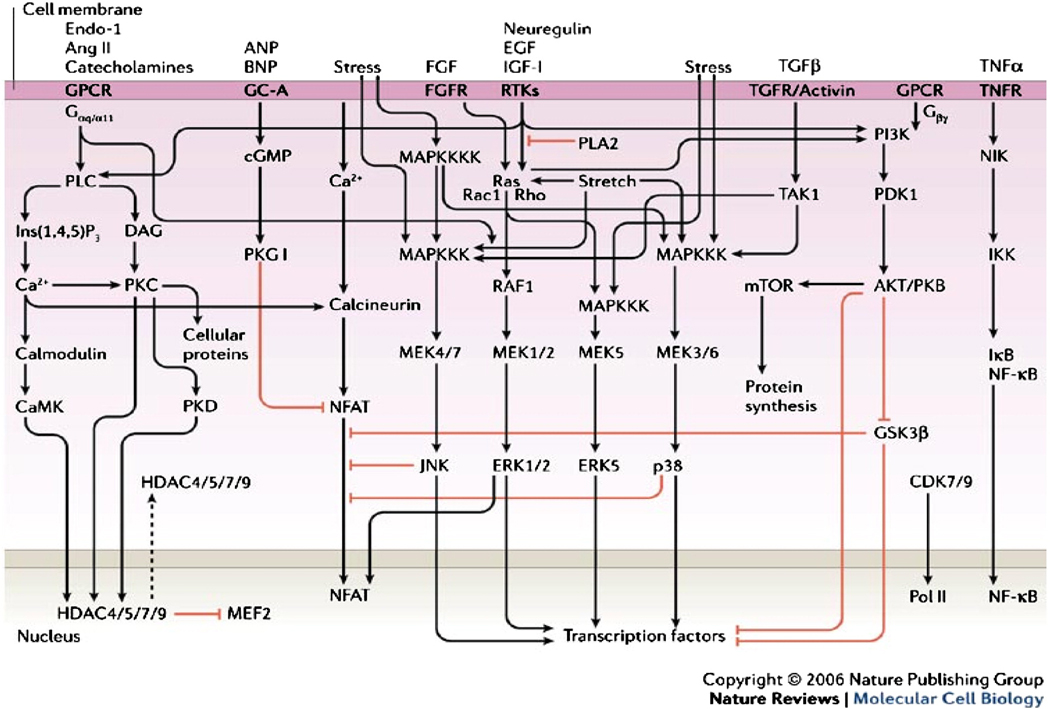
Pressure overload hypertrophy has been one of the principal models by which the pathophysiology of heart failure has been studied. Multiple signaling pathways have been identified as regulating the hypertrophic response, including G protein coupled receptors (endothelin, angiotensin and catecholamine), stress receptors, receptor tyrosine kinases (neuregulin, extracellular growth factor, insulin-like growth factor), the transforming growth factor/activin family, and tumor necrosis factor (Figure 1) (1). However, many of these studies have been performed using isolated cells that have been transfected with supra-physiologic levels of the protein of interest to increase the gain of the read-out system, and the relevance of these in vitro findings to human disease is often unclear. In a review entitled “Crosstalk and specificity in signaling: are we crosstalking ourselves into general confusion,” Dumont et al. reviewed the literature for a two year period for evidence of cross-signaling between four key signal transduction cascades, uncovering up to 760 potential signaling interactions (2). Clearly, this level of crosstalk (“everything does everything”) is inconsistent with the known specificity of action of extracellular signals.
Figure 1.
Signaling pathways regulating cardiac hypertrophy. From Ref. 1.
Fortunately, developments in murine genetic manipulation have made possible the examination of the role of individual proteins in vivo by selectively ablating (knockout) or increasing (transgenic) expression of individual genes. Early experiments with gene knockouts were often complicated by the lack of tissue specificity, e.g. whole-body deletion of a particular receptor does not separate the role of that receptor in myocytes vs. endothelial cells, and these differences were often dramatic (RxR knockout). Furthermore, many gene knockouts resulted in embryonic or early neonatal demise, preventing the examination of their effects on cardiovascular physiology.
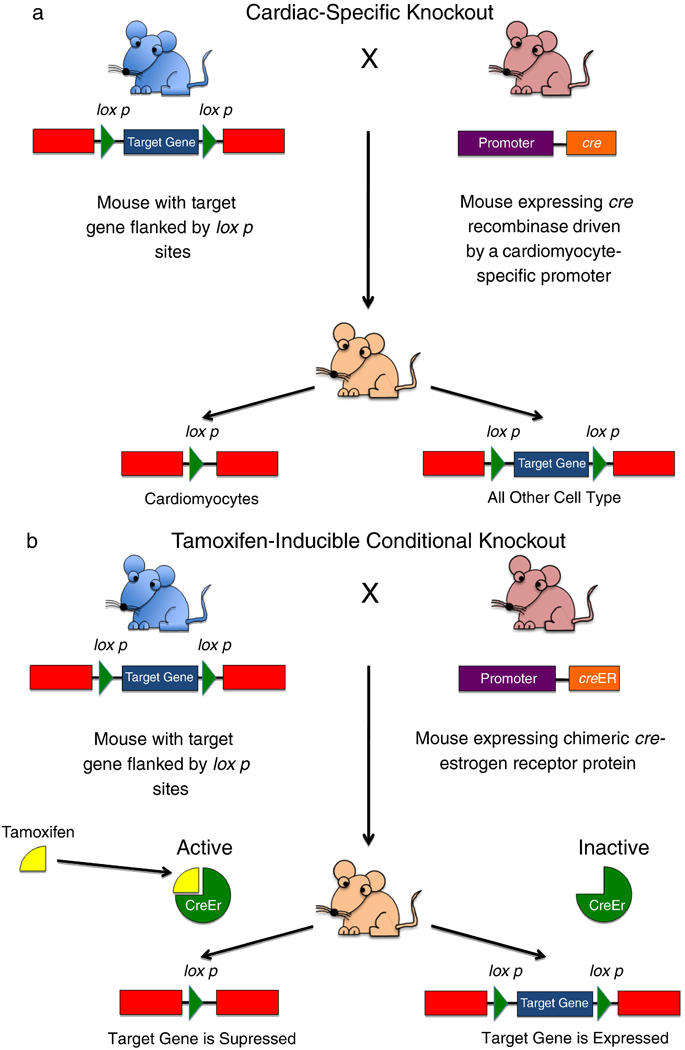
Two major advances have significantly improved our ability to manipulate the murine genome (reference here). To solve temporal issues and avoid embryonic lethality, conditional knockouts have been developed, where expression of a gene is placed under control of an external modifier, usually a drug such as tamoxifen or doxycycline, which can be administered to the animal to turn expression of the target gene on or off (Figure 2). To solve spatial issues of gene expression, tissue-specific transgenic techniques have been developed, where expression of a specific gene can be down- or upregulated in a particular cell type (e.g. myocytes or vascular smooth muscle cells) using tissue-specific promoters.
Figure 2.
Advances in murine genetic manipulation.
(a) Generation of a cardiac-specific gene knockout. The myocardium is a complex tissue, consisting of many cell types (myocytes, fibroblasts, vascular smooth muscle cells, vascular endothelial cells). Often it is desirable to remove a gene from one cell type only, to differentiate the role of each cell type in contributing to a specific cardiac pathology. This technology relies on the use of a DNA recombinase known as Cre (for cyclization recombination). This enzyme recognizes a specific 34 base pair-long site in DNA known as the loxP site. Cre induces a recombination event between two loxP sites, cutting out any DNA between them. One mouse is generated with the target gene of interest flanked by two loxP sites; this is known as a “floxed” gene (blue mouse). A second mouse is generated expressing Cre under the control of a cardiac-specific promoter, e.g. the α-myosin heavy chain (α-MHC) promoter (pink mouse). When these two mice are crossbred, their cells will contain both transgenes, however, Cre will be expressed only in cardiomyocytes, thus only in cardiomyocytes will the target gene be removed by the recombination event between the two loxP sites. All other cells will express that gene normally. (b) Generation of a conditional knockout. Sometimes it is desirable to remove a gene in a specific time frame. This is especially important when deletion of a gene from conception results in embryonic lethality, whereas deletion in the adult heart does not. In this case, a mouse is created with a floxed target gene (blue mouse). This mouse is crossbred to a mouse where Cre is fused to a mutated estrogen receptor which has lost its ability to bind endogenous hormones, but which can still bind tamoxifen. In the absence of tamoxifen, cre is maintained in an inactive state. However, when the mouse is given tamoxifen, it binds to the estrogen receptor which allows Cre to migrate to the nucleus and remove the target gene.
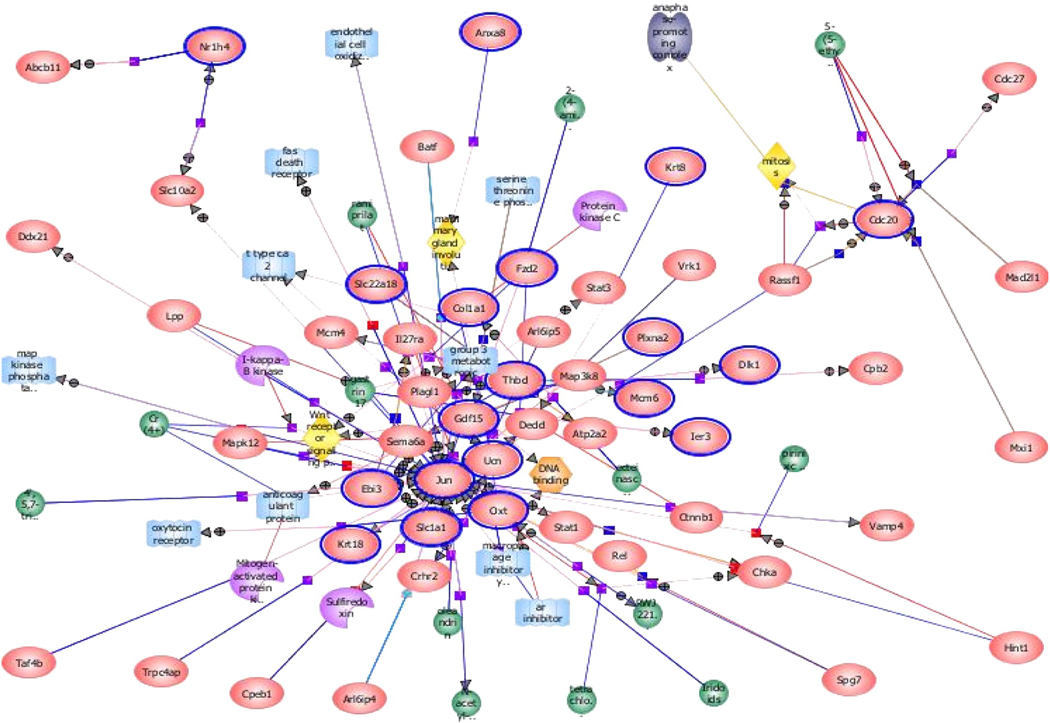
Using increasingly sophisticated bioinformatics tools, we are now able to map genome-wide alterations associated with cardiac stressors such as pressure overload. Within 2 hours of aortic banding, over 500 transcripts are unregulated and 700 downregulated and by 10 days, nearly 800 are upregulated and 1200 are downregulated (3). However, the pathophysiologic relevance of each of these gene chip results remains in question. For one, the choice of controls is critical for these studies, given that the stress of sham surgery alone can result in over 500 gene expression alterations in the mouse. Advances in bioinformatics technology, such as pathway analysis, allows us to place each of these gene alterations within relevant signaling cascades (metabolism, apoptosis, cell growth, etc.) to further enhance our understanding of which pathways are truly relevant and which are merely bystanders (Figure 3).
Figure 3.
Pathway analysis of altered gene expression during pressure overload hypertrophy. This type of analysis examines groups of genes which are altered during a particular cardiovascular stressor. If multiple components of a signaling pathway are all up- or down-regulated in concert, the liklihood that pathway plays an important role in pathophysiology is enhanced. (Data courtesy of Dr. Sushma Reddy).
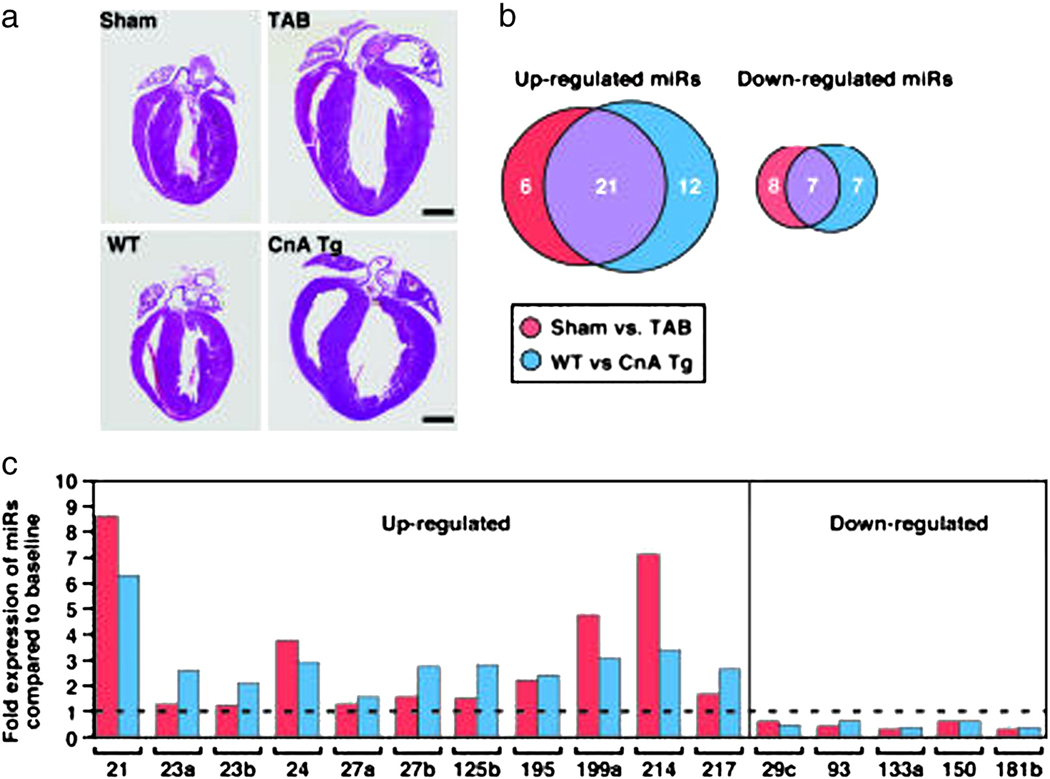
However, gene expression analysis is only a part of the picture of the adaptation to hypertrophy and the progression to heart failure. Most of the key signaling proteins involved in these events are regulated at the post-translational level, e.g. by phosphorylation (which alters their activity) or ubiquitination (which alters their targeting for proteasomal degradation) (4, 5). The role of histone modification (6, 7, 8) and microRNAs in the hypertrophic response are additional examples of the complexity of cellular regulation of cardiac structure and function (Figure 4) (9, 10).
Figure 4.
Epigentic control in the heart: the role of microRNAs (miRs) in hypertrophy and heart failure. In this study, van Rooij and colleagues examined the expression of a panel of miRs in mice after aortic banding and in transgenic mice overexpressing the protein calcineurin, both of which induce hypertrophy (A). There were some changes in miR expression that were unique to either condition, however the majority overlapped between the two hyperterophy-inducing stimuli (B). The specific miRs that were up- or down-regulated are shown in C. From Ref. 9.
As pediatric cardiologists our view of hypertrophy and heart failure is also a little different than that of our adult colleagues. We routinely care for children with mild to moderate degrees of increased afterload (e.g. pulmonic or aortic stenosis), in whom long-standing cardiac hypertrophy is not inevitably associated with heart failure. The murine models which have been the mainstay of the above research induce hypertrophy with an aortic band which acutely increases left ventricular pressure by 50–100% and results in heart failure within 1–2 weeks. The relevance of these models to children with congenital heart disease (or even to adults with stable aortic stenosis or systemic hypertension) is still unclear. Although the literature has neatly divided hypertrophy into physiologic (i.e. the athletic heart) vs. pathologic (i.e. that induced by pressure overload), the question remains as to whether all pathologic hypertrophy is deleterious in the long run?
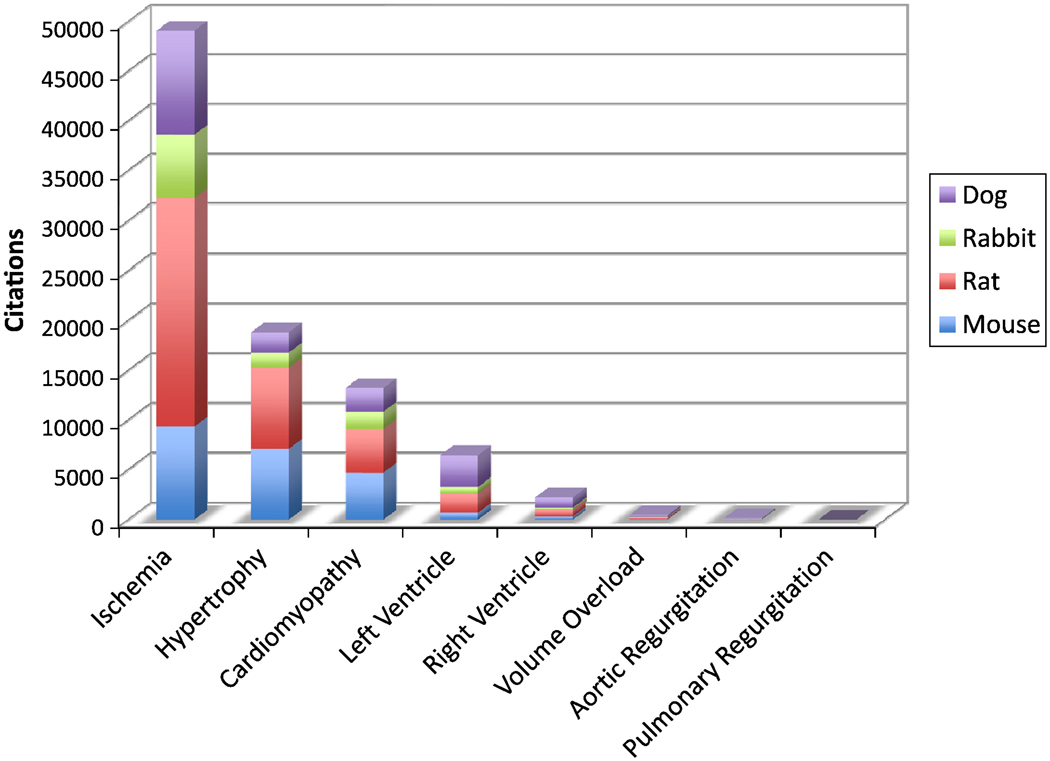
Additionally, for pediatric cardiologists it is often the right ventricle, not the left, which is at risk for failure in our patients. Despite surgical repair or palliation, many patients with congenital heart disease are left with residual alterations in either volume or pressure loading (e.g. tetralogy of Fallot, pulmonary atresia, truncus arteriosus). Those with systemic right ventricles (l-transposition of the great arteries, hypoplastic left heart and other single ventricle conditions) are also at increased risk of heart failure. Yet despite considerable data on the molecular events of left ventricular remodeling and failure, little is known about these events in the right ventricle (Figure 5). There are few models of right ventricular pressure overload (11, 12). Although many of the molecular events associated with right ventricular afterload stress are similar to those of the left ventricle, there are several key differences in both gene and micro RNA expression which may reveal unique pathways by which the right ventricle adapts to stress. The development of models of right ventricular volume overload (13), simulating the condition of many patients after right ventricle-pulmonary artery conduit insertion, will be a key next step to increase the relevance of these basic investigations to children with congenital heart disease.
Figure 5.
Review of Pub Med citations over the past five years shows a strong left-ventricular and ischemia dominance in choice of disease model. Notice the paucity of models of volume overload, one of the more common conditions in the pediatric heart failure population.
Although most of these models simulate the types of stresses induced by congenital heart disease (pressure or volume overload), many of the signaling pathways activated are identical to those in the primary cardiomyopathies (e.g. β-receptor downregulation, activation of the fetal gene program, and alterations in regulators of intracellular calcium), however, many key differences likely exist. Parallel studies using murine models of genetic dilated and hypertrophic cardiomyopathies will be necessary to understand their basic mechanisms and determine disease-specific therapeutic approaches. An exciting new technology in this area is the use of induced pluripotent stem (iPS) cells to study mechanisms of the genetic myopathies. Skin fibroblasts or fat cells are taken directly from patients with a genetic cardiomyopathy, reprogrammed into pluripotent stem cells, and then differentiated into cardiomyocytes. These heart muscle cells can then be studied in vitro to determine the effects of the specific genetic alteration on cell signaling, calcium handling, sarcomere function, and contractile properties (14).
Finally, our reliance on animal models can now be supplemented by a level of genetic analysis in humans that was heretofore impossible. Advances in whole genome sequencing, e.g. the use of microfluidic technology combined with shotgun sequencing (15), have reduced the time and cost of such sequencing to the point where studies in human populations will soon be feasible.
ACKNOWLEDGMENT
This work was supported by a grant (HL061535) from the NIH/NHLBI to Dr. Bernstein.
REFERENCES
- 1.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signaling patheways. Nature Reviews. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 2.Dumont JE, Pecasse F, Maenhaut C. Crosstalk and specificity in signaling: are we crosstalking ourselves into general confusion>. Cellular Signaling. 2001;13:457–563. doi: 10.1016/s0898-6568(01)00168-1. [DOI] [PubMed] [Google Scholar]
- 3.Zhao M, Chow A, Powers J, Fajardo G, Bernstein D. Microarray analysis of gene expression after transverse aortic constriction in mice. Physiol. Genomics. 2004;19:93–105. doi: 10.1152/physiolgenomics.00040.2004. [DOI] [PubMed] [Google Scholar]
- 4.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nature Reviews Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 5.Qillis MS, Townley-Tilson WH, Kang EY, Homeister JW, Patterson C. Sent to destroy: the ubiquiting proteasomal system regulates cell signaling and protein quality control in cardiovascular development and disease. Circ. Res. 2010;106:463–478. doi: 10.1161/CIRCRESAHA.109.208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latham JA, Dent SYR. Cross-regulation of histone modifications. Nature Structural and Molecular Biology. 2007;14:1017–1024. doi: 10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 7.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 8.Leach M, van der Harst P, de Boer RA. Pharmacoepigenetics in heart failure. Curr Heart Fail Rep. 2010;7:83–90. doi: 10.1007/s11897-010-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitahori K, He H, Kawata M, Cowan DB, Friehs I, Del Nido PJ, McGowan FX., Jr N Development of left ventricular diastolic dysfunction with preservation of ejection fraction during progression of infant right ventricular hypertrophy. Circ Heart Fail. 2009;2:599–607. doi: 10.1161/CIRCHEARTFAILURE.109.862664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urashima T, Zhao M, Wagner R, Fajardo G, Farahani S, Quertermous T, Bernstein D. Molecular and physiologic characterization of RV remodeling in a murine model of pulmonary stenosis. Am J Physiol. 2008;295:H1351–H1368. doi: 10.1152/ajpheart.91526.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reddy S, Zhao M, Fajardo G, Urashima T, Rajagopalan V, Bernstein D. Physiologic and molecular characterization of a murine model of pulmonary insufficiency. Chicago: AHA Scientific Sessions; 2010. [Google Scholar]
- 14.Liu J, Verma PJ, Evans-Galea MV, et al. Generation of Induced Pluripotent Stem Cell Lines from Friedreich Ataxia Patients. Stem Cell Rev. 2010 December; doi: 10.1007/s12015-010-9210-x. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 15.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]