Abstract
Nucleotide excision repair (NER) in yeast is effected by the concerted action of a large complex of proteins. Recently, we identified a stable subcomplex containing the yeast Rad7 and Rad16 proteins. Here, we report the identification of autonomously replicating sequence binding factor 1 (ABF1) as a component of the Rad7/Rad16 NER subcomplex. Yeast ABF1 protein is encoded by an essential gene required for DNA replication, transcriptional regulation, and gene silencing. We show that ABF1 plays a direct role in NER in vitro. Additionally, consistent with a role of ABF1 protein in NER in vivo, we show that certain temperature-sensitive abf1 mutant strains that are defective in DNA replication are specifically defective in the removal of photoproducts by NER and are sensitive to killing by ultraviolet (UV) radiation. These studies define a novel and unexpected role for ABF1 protein during NER in yeast.
Keywords: DNA repair, Saccharomyces cerevisiae, ABF1, Rad7, Rad16
Autonomously replicating sequence (ARS) binding factor 1 (ABF1) is an abundant DNA-binding protein encoded by an essential gene in Saccharomyces cerevisiae (Rhode et al. 1992). ABF1 recognizes a specific DNA sequence motif present at numerous sites in the yeast genome, including ARS's, promoter elements and mating-type silencing sequences (Rhode et al. 1992). Genetic analyses have demonstrated that ABF1 protein plays roles in ARS activity during DNA replication, transcriptional activation, and transcriptional silencing of HM mating-type loci (Diffley and Stillman 1989; Rhode et al. 1989; Walker et al. 1989).
ARS's contain multiple functional elements (Marahrens and Stillman 1992; Rao et al. 1994; Rao and Stillman 1995). Element A is required for binding of the origin recognition complex (ORC) (Marahrens and Stillman 1992; Rao et al. 1994; Rao and Stillman 1995), whereas element B1 participates in ORC–DNA interactions. Element B3 has been identified as the ABF1-binding site (Marahrens and Stillman 1992; Rao et al. 1994; Rao and Stillman 1995). Additionally, temperature-sensitive abf1 mutants have been shown to be defective in DNA replication (Rhode et al. 1992), providing direct genetic evidence for a role of ABF1 protein in the initiation of DNA replication.
With respect to its role in transcription, ABF1-binding sites have been identified in the promoters of genes and mutational analysis has revealed both positive and negative regulatory effects on gene expression (Brand et al. 1987; Buchman and Kornberg 1990; Della Seta et al. 1990). Transcriptional silencing of mating-type loci in yeast is effected by localized alterations in chromatin structure (Rine and Herskowitz 1987). Silencing also requires the presence of cis-acting elements (which include an ARS), trans-acting factors such as histones H3 and H4, RAP1 protein, the Sir proteins, and ABF1 (Klar et al. 1979; Feldman et al. 1984; McNally and Rine 1991). A direct role for ABF1 protein in silencing has been established by the isolation of temperature-sensitive alleles of ABF1 that are specifically defective in this modality (Loo et al. 1995).
In contrast to most genes required for the process of nucleotide excision repair (NER) in yeast, the Rad7 and Rad16 proteins appear to play a specialized role in this process. They are required specifically for the removal of pyrimidine dimers from transcriptionally repressed regions of the genome, (such as the HM mating-type loci), but not from their transcriptionally activated homologs (Terleth et al. 1990; Verhage et al. 1994). Additionally, Rad7 and Rad16 are required for NER of the nontranscribed strand of transcriptionally active genes, but not for repair of the transcribed strand of such genes (Terleth et al. 1990; Verhage et al. 1994). The precise role of Rad7 and Rad16 proteins during NER of these structurally distinctive regions of the genome is not clear, nor have higher eukaryotic homologs of these proteins been identified. Paetkau et al. (1994) reported an interaction of Rad7 protein with Sir3, a protein involved in silencing of mating-type loci. However, current evidence suggests that Rad7 is not directly involved in gene silencing, and there are no indications that factors required for gene silencing are also involved in NER.
Another feature that distinguishes the RAD7 and RAD16 genes from most other genes involved in NER in yeast is that deletion of either of these genes confers only moderate sensitivity to ultraviolet (UV) radiation (Terleth et al. 1990). Furthermore, extracts of rad7 and rad16-deletion mutants are not defective in the early event of damage-specific DNA incision during NER in vitro or in vivo (Reed et al. 1998). However, such mutants are severely deficient (but not totally defective) in post-incision oligonucleotide excision and repair synthesis of DNA in vitro (Reed et al. 1998). These observations have led to the suggestion that the Rad7 and Rad16 proteins play a role in the displacement (excision) of oligonucleotide fragments generated by the bimodal incision reaction. This process may in turn be directly coupled to dissociation of the bound NER machinery from DNA and/or to chromatin modulation during NER. In this regard, it is of interest that Rad16 is a member of the SWI/SNF superfamily of ATPases, each of which is believed to be a subunit of a chromatin-modulating complex (Bang et al. 1992).
Here we report an unexpected role for ABF1 protein in NER in yeast. We show that ABF1 protein copurifies with the yeast Rad7 and Rad16 proteins [which form a stable complex both in vivo and in vitro (Wang et al. 1997; Reed et al. 1998) through multiple fractionation steps. A mutant strain depleted of ABF1 protein by the N-degron strategy (Dohmen et al. 1994) is both sensitive to killing following exposure to UV radiation, and defective in NER in vitro. Furthermore, defective NER in vitro can be specifically complemented with purified recombinant ABF1 protein. Finally, a subset of conditional-lethal abf1 mutants shown previously to be defective in DNA replication are now shown to be sensitive to killing by UV light, but not to agents that produce DNA damage that is repaired by pathways distinct from NER. Furthermore, we demonstrate that the UV radiation sensitivity of these mutants directly correlates with an inability to remove UV radiation-induced lesions in DNA.
Results
ABF1 copurifies with Rad7 and Rad16 proteins
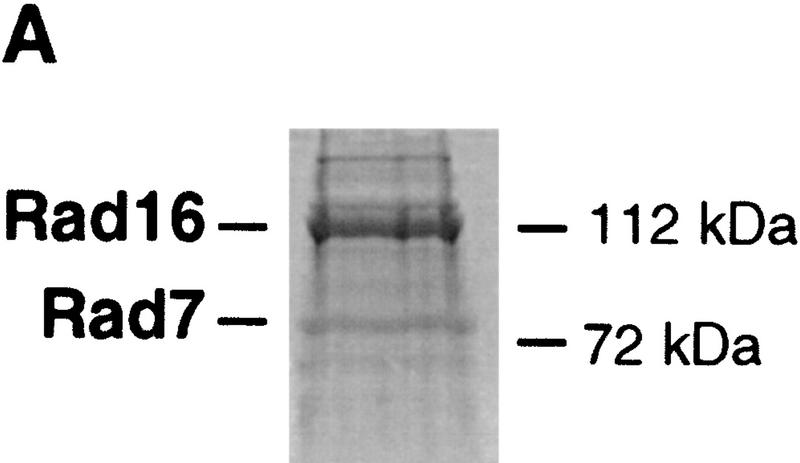
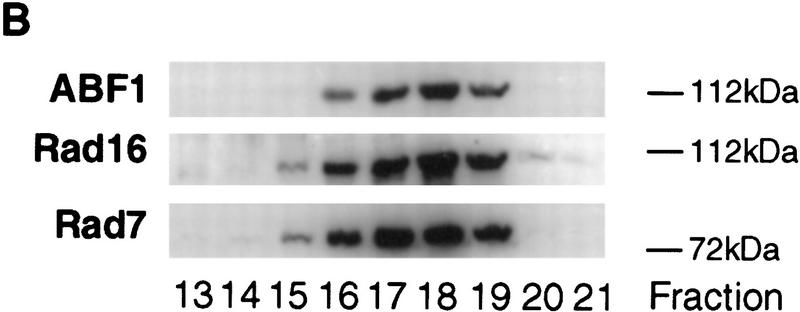
Recently, we reported the purification of a Rad7/Rad16-containing protein complex (Reed et al. 1998). Gel filtration of the purified complex revealed an apparent molecular mass of ∼300 kD (Reed et al. 1998), significantly larger than the summed molecular masses of Rad7 and Rad16 (∼190 kD). In the present studies, electrophoretic analysis of the purified complex revealed that the larger of two bands identified (∼112 kD), known by immunoblotting to contain Rad16 protein, stained more intensely than the Rad7 band (∼75 kD) (Fig. 1A). Tandem mass spectrometry analysis of the 112-kD band revealed the presence of ABF1 protein in addition to the previously identified Rad16 protein (see Materials and Methods). We confirmed this result by immunoblotting with affinity-purified ABF1 antibody [see Materials and Methods (Diffley and Stillman 1988, 1989), which does not cross react with Rad16 protein (Fig. 1B)].
Figure 1.
(A) Polyacrylamide gel stained with Coomassie blue shows the presence of Rad16 (∼112 kD) and Rad7 (∼75 kD) proteins. The purification protocol and gel electrophoresis procedures are as described previously (Reed et al. 1998). The presence of these proteins was confirmed by Western blotting and by tandem mass spectrometry (see Materials and Methods). Note that the intensity of the 112-kD band is greater than that of the 75-kD band. (B) Western blot analysis shows cofractionation of purified ABF1, Rad16, and Rad7 proteins through a glycerol gradient as described (Reed et al. 1998). Antibodies to Rad7 and Rad16 were prepared as described (Reed et al. 1998). Antibodies to ABF1 protein are described in Materials and Methods.
Details of the purification of Rad7/Rad16 complex have been reported previously (Reed et al. 1998). Briefly, we His-tagged Rad7 protein and purified it on a nickel column as the first fractionation step. It should be noted that the His-tagged Rad7 strain expresses Rad7 at similar levels to the wild-type strain from which it was created. All Rad7 and Rad16 protein was immobilized on the nickel matrix and the two proteins copurified in subsequent purification steps. In contrast, we noted that ∼70% of the ABF1 protein was in the flow-through fraction from the nickel column. Hence, we estimate that only ∼30% of the cellular ABF1 protein is associated with Rad7 and Rad16.
Depleting ABF1 in cells results in UV radiation sensitivity and defective NER
ABF1 protein is encoded by an essential gene for which a limited repertoire of conditional-lethal mutants are available (Rhode et al. 1992). Therefore, we investigated the functional significance of its physical association with the Rad7/Rad16 complex using an experimental approach that facilitates the temperature-dependent degradation of selected gene products in living yeast cells (the N-degron strategy) (Dohmen et al. 1994). Fusing a temperature-dependent degradation signal to ABF1 generated a mutant strain designated abf1td, in which ABF1 protein could be selectively inactivated, thus obviating the need to laboriously screen for informative conditional-lethal mutants.
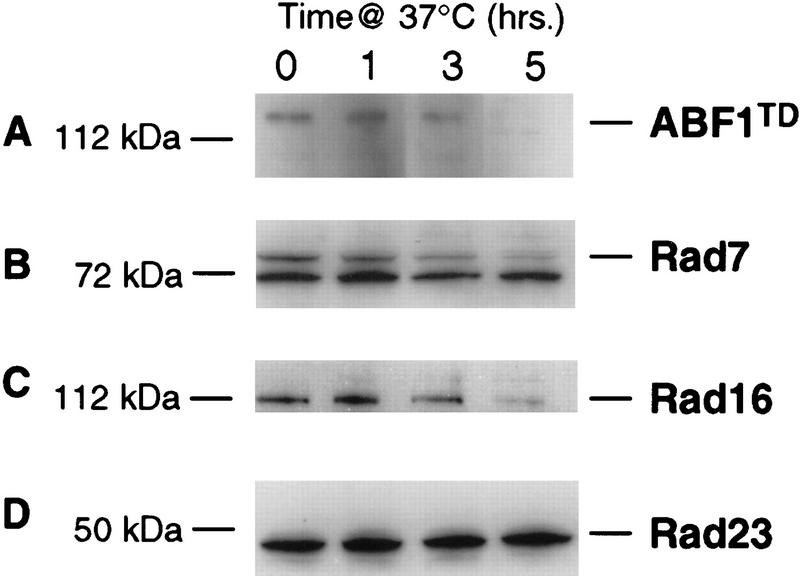
As shown in Figure 2, ABF1 protein was depleted progressively from extracts during 1–5 hr of incubation at the restrictive temperature (37°C). Rad7 and Rad16 proteins were also degraded under these conditions, but at a significantly reduced rate compared with ABF1 protein (Fig. 2). The time required for steady-state levels of ABF1, Rad7, and Rad16 proteins to decrease by 50% under these conditions (measured by quantitative densitometry scanning) was calculated at 2, 3.5, and 4 hr, respectively. Such degradation was not observed in wild-type strains at 37°C (data not shown). Additionally, other NER proteins such as Rad23 (Fig. 2), as well as several cross-reacting proteins (Fig. 2b), showed no obvious degradation over a 5-hr period at 37°C. The codegradation of Rad7 and Rad16 with ABF1 provides supporting evidence that the three proteins exist in a complex in vivo.
Figure 2.
An N-degron strain was constructed that expresses a temperature-degradable ABF1TD fusion protein (Dohmen et al. 1994). When this strain was maintained at 37°C for varying periods of time, there was a progressive loss of the ABF1TD fusion protein as monitored by Western blotting with monoclonal antibodies to an HA epitope in the protein (A). Rad7 (B) and Rad16 protein (C) were also degraded, but the rate of this degradation was significantly slower than that of the ABF1TD fusion protein. Rad23 protein (D) and various other proteins that cross-reacted with the antibodies (e.g., the lower band in B) did not show detectable degradation. Antibodies to Rad23 protein were prepared as described (Wang et al. 1997).
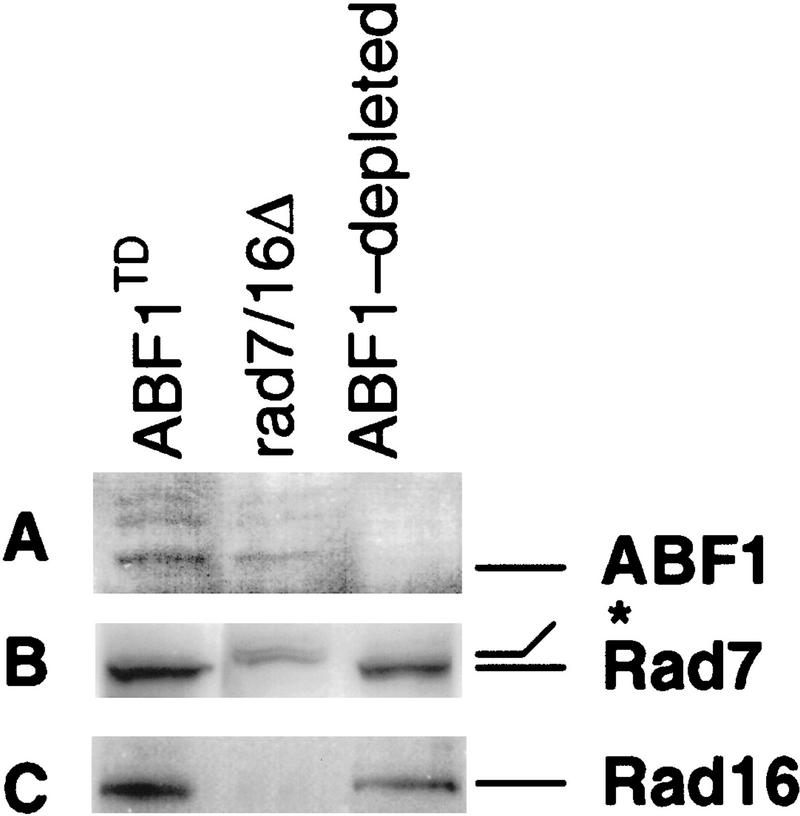
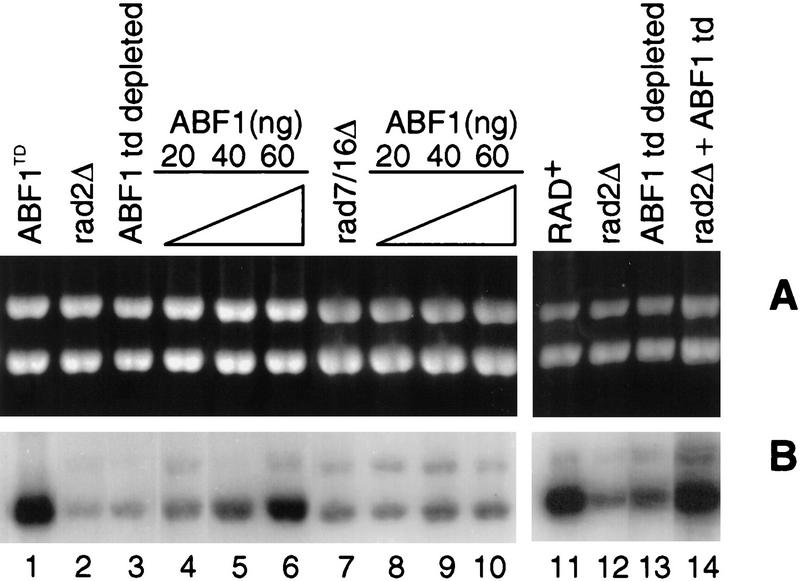
To correlate the temperature-dependent degradation of ABF1 protein with defective NER, we prepared whole-cell extracts (WCE) from the abf1td strain held at the restrictive temperature (37°C) for 4 hr. Under these conditions, ABF1 protein was depleted (Fig. 3A), but Rad7 protein (Fig. 3B) and Rad16 protein (Fig. 3C) were still readily detectable by immunoblotting. We tested the NER activity of this extract and showed it to be severely deficient compared with extracts prepared from the abf1td strain maintained at the permissive temperature (23°C), but comparable with a known NER-defective rad2 mutant (Fig. 4, cf. lanes 1–3 and lanes 11–13). Defective NER in the ABF1-depleted extract was complemented by mixing with an extract from the rad2 mutant (Fig. 4, cf. lanes 11–14), demonstrating that the extract was not nonspecifically inactivated. Furthermore, the addition of purified recombinant ABF1 protein expressed in baculovirus (see Materials and Methods) and hence free of any yeast Rad7 and Rad16 proteins, restored repair synthesis in abf1td mutant extracts (Fig. 4, lanes 4–6), but not in rad2 mutant extracts (data not shown). Purified ABF1 protein had no stimulatory effect on repair synthesis in extracts of a wild-type strain (data not shown). Finally, purified ABF1 did not complement defective repair synthesis in either rad7 or rad16 single-mutant extracts (data not shown), nor in rad7/rad16 double-mutant extracts (Fig. 4, lanes 8–10). These results demonstrate a direct role for ABF1 protein in NER in vitro.
Figure 3.
Western blot analysis shows that when the abf1td mutant strain was maintained at an elevated temperature, ABF1 protein in extracts competent for NER is depleted (A). The protein was detectable in an extract prepared from the abf1td strain maintained at the permissive temperature and in a rad7/rad16 double-deletion mutant strain (A). Under these conditions, Rad7 protein was detected in extracts of the abf1td strain but not in extracts of the rad7/rad16 double-deletion mutant (B). The center lane in B was deliberately overloaded to demonstrate the absence of Rad7 protein. The band migrating at a higher molecular mass than Rad7 (*) is a nonspecific cross-reacting species in this strain. (C) Similar data for Rad16 protein.
Figure 4.
NER was measured by monitoring repair synthesis of a plasmid substrate carrying defined base damage (bottom bands) relative to the levels of nonspecific DNA synthesis in a larger control plasmid (top bands). The details of this assay are as described (Wang et al. 1997). (A) Ethidium-stained gels indicating the relative amounts of DNA loaded. (B) An autoradiograph of the gel. ABF1+ extracts (ABF1TD) prepared from the abf1td strain at the permissive temperature support robust repair synthesis (lane 1). However, such synthesis is defective in a rad2 deletion mutant known to be required for NER (lanes 2,12), and in extracts of a rad7/rad16 double-deletion mutant (lane 7). Defective or deficient NER is also observed in extracts of a strain expressing temperature-degradable ABF1 protein (lanes 3,13). Defective NER is complemented by mixing rad2 mutant and ABF1-depleted (by temperature degradation) extracts (lane 14), or by adding increasing amounts of purified ABF1 protein to the ABF1-depleted extracts (lanes 4–6). Addition of ABF1 protein has no effect on the level of repair synthesis in the rad7/rad16 deletion mutant (lanes 8–10).
DNA-binding domain mutants of ABF1 are sensitive to UV radiation
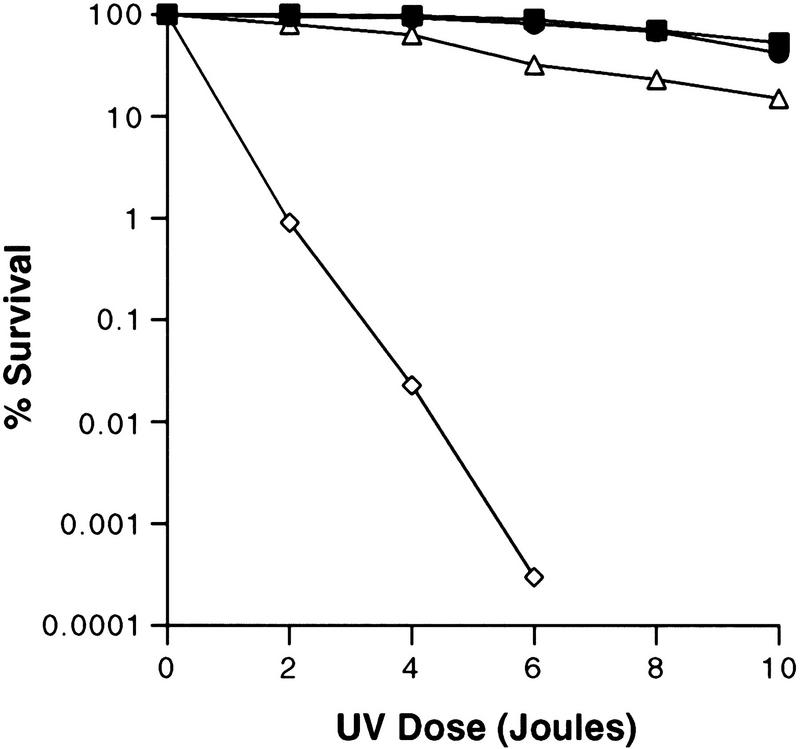
Conditional-lethal abf1 mutants have been isolated in two independent laboratories on the basis of screens for defective transcriptional silencing (Loo et al. 1995) and defective DNA replication (Rhode et al. 1992). Sensitivity of the former mutants to UV radiation was examined at nonpermissive and permissive temperatures for silencing. No increased UV-radiation sensitivity was observed under the latter conditions (data not shown), suggesting that the silencing function of ABF1 protein does not play a role in NER. In contrast, when we examined conditional-lethal mutants obtained from a screen for defective DNA replication, a strain designated abf1-1 with a mutation in the DNA-binding domain of ABF1 protein (Rhode et al. 1992), was clearly sensitive to UV light at the semipermissive temperature of 32°C (Fig. 5). The abf1-1 mutant strain was not abnormally sensitive to the DNA-damaging agents methylmethane sulfonate or hydrogen peroxide. Both of these agents are known to produce DNA damage that is repaired by pathways other than NER (Friedberg et al. 1995; data not shown). A second independently isolated mutant strain (abf1-2) shown to carry an identical mutation in the ABF1 DNA-binding domain (Rhode et al. 1992) was also abnormally sensitive to killing by UV light (data not shown). A third replication-defective strain (abf1-5), which carries a mutation outside of the DNA-binding motif and is not defective in binding the ABF1 recognition sequence (Rhode et al. 1992), was not abnormally sensitive to UV radiation (Fig. 5). These observations indicate an in vivo role of ABF1 in DNA repair and specifically implicate the DNA-binding domain of the protein in this process.
Figure 5.
UV-radiation sensitivity of the abf1-1 and abf1-5 mutants. Survival was measured by quantitating colony-forming ability as described (Huang et al. 1998) at the highest temperature at which the strains could grow (32°C for abf1-1 and 28°C for abf1-5). (█) Wild type at 32°C; (⋄) abf1-1 at 32°C; (▵) abf1-5 at 28°C; (●) wild type at 28°C.
DNA-binding domain mutants of ABF1 are defective in photoproduct removal in vivo
To directly examine the in vivo removal of DNA damage following exposure to UV radiation, we used a slot blot assay. This assay monitors the loss of cyclobutane pyrimidine dimer (CPD) and 6-4 photoproducts from genomic DNA with specific monoclonal antibodies raised against these lesions (Mori et al. 1991). Figure 6 shows the kinetics of the loss of 6-4 photoproducts in the abf1-1 (Fig. 6B) and abf1-5 (Fig, 6A) mutants and in the wild-type parental strain (Fig, 6A,B). The abf1-1 mutant is defective in the removal of these lesions compared with the wild-type parental strain at the semipermissive temperature of 32°C (Fig. 6B, cf. lanes 3 and 4). In contrast, both the abf1-5 strain and the wild-type parental strain showed a progressive loss of lesions as a function of time at this temperature (Fig. 6A, cf. lanes 3 and 4). At the permissive temperature of 23°C, lesion removal was observed in all strains. However, even under these conditions the kinetics of lesion removal in the abf1-1 strain was reduced compared with wild type (Fig. 6A and B, lanes 1 and 2).
Figure 6.
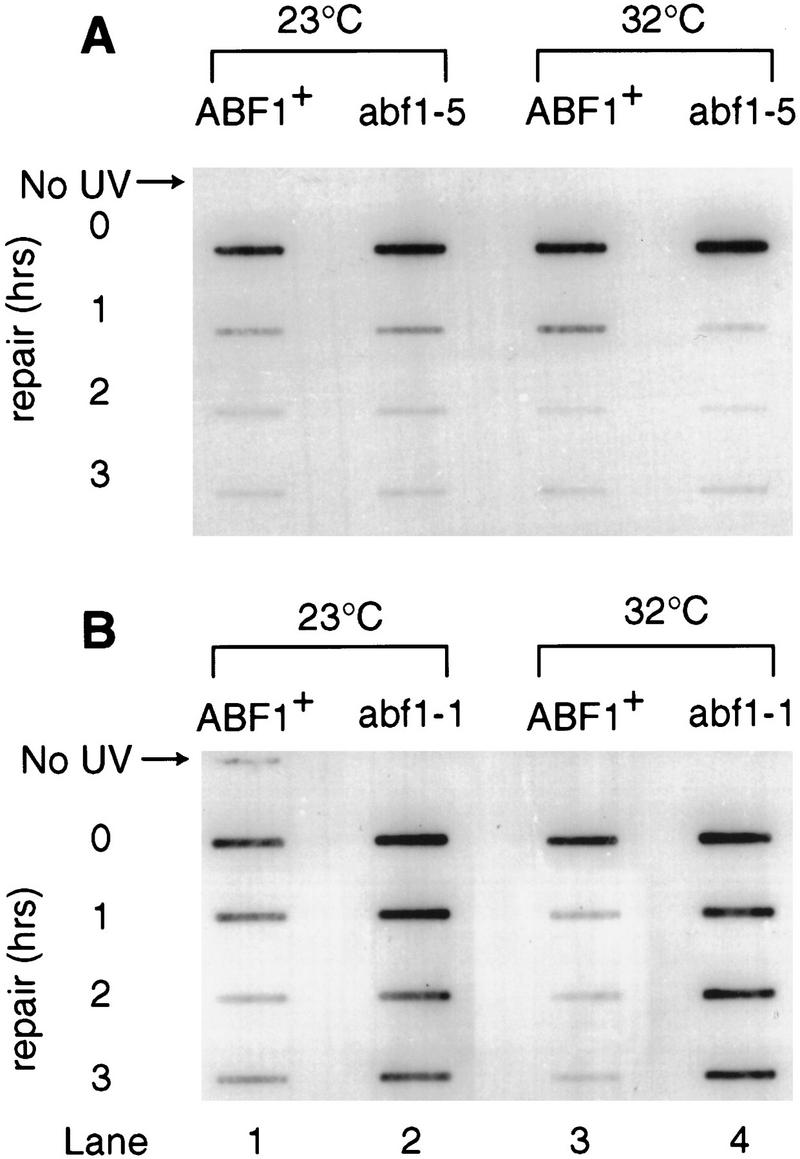
Kinetics of the loss of UV radiation-induced 6-4 photoproducts from genomic DNA. Removal of (6-4) photoproducts was measured by an immunological slot blot assay (see Materials and Methods). Monoclonal (6-4) photoproduct antibodies are used as a probe for the presence of these lesions in genomic DNA following exposure of the cells to UV and their subsequent incubation at the noted conditions to allow for DNA repair. Strains were maintained at the permissive (23°C) and semipermissive (32°C) temperatures (see Materials and Methods and Results for details).
Most temperature-sensitive mutations in yeast reflect amino acid changes that result in destabilization of proteins at elevated temperature. Such mutations often result in alterations of some (but not all) phenotypes even at the permissive temperature. For example, Rhode et al. (1992) reported that at the permissive temperature, the ABF1-1 mutant protein was not able to bind the ABF1-binding site (a requirement for DNA replication), yet the strain showed no obvious replication defect. Therefore, they suggested that the protein must bind its cognate site to some extent in vivo. We propose a similar explanation for our observations involving lack of sensitivity to UV radiation and defective NER of 6-4 photoproducts in vivo at the permissive temperature.
Essentially similar results were obtained when the removal of CPD was analyzed (data not shown). These observations correlate the UV sensitivity of the abf1-1 and abf1-2 mutants directly with an inability to remove UV radiation-induced lesions from their DNA.
Discussion
In the present study, we have demonstrated that ABF1 protein is a component of a subcomplex of the nucleotide excision repairosome. Our experiments show that in addition to its previously noted roles in DNA replication, transcription, and gene silencing, ABF1 protein is involved directly in the process of NER. ABF1 is therefore identified as a novel DNA repair protein.
The association of this protein with the Rad7/Rad16 protein subcomplex, which is required for events during NER that follow bimodal damage-specific incision of DNA, suggests that like Rad7 and Rad16 proteins, ABF1 subserves a role in specialized functions during NER. Our experiments demonstrate that abf1 mutants that harbor mutations in the DNA-binding domain of the protein, and hence are defective in their ability to bind the ABF1 DNA recognition sequence, are also UV-radiation sensitive and defective in NER of photoproducts. In contrast, abf1 mutants that are DNA-replication defective, but carry mutations in other regions of ABF1, are not sensitive to killing by UV radiation and are NER proficient.
A possible explanation for these findings is that the overall ability of the ABF1 protein to nonspecifically bind DNA is altered in the abf1-1 and abf1-2 mutants, resulting in defective DNA repair. A more intriguing scenario is that ABF1-binding sites play a specific role in NER similar to that during DNA replication. We estimate that there are ∼2000 ABF1-binding sites in the yeast genome. Conceivably, the efficiency of NER in yeast is influenced by the proximity of this process to such sites. In this regard, it is provocative that Rad16 is a member of the SWI/SNF family of DNA-dependent ATPases involved in chromatin remodeling.
Previous biochemical studies with a reconstituted in vitro NER assay have shown that Rad7 and Rad16 stimulate the NER reaction (Guzder et al. 1997). The authors note that the ATPase activity of Rad16 is attenuated in the presence of high levels of damage, and suggest that the complex plays a role in scanning for DNA damage. Future studies are necessary to elucidate the precise role of ABF1 protein and the ABF1 DNA-binding site in the process of NER.
Materials and methods
Yeast strains
The RAD+ strain W303-B (MATa, ade2-1, trp1-1, leu2-3, 112, his3-11, 15, ura3-1) and the isogenic strains MGSC104 (rad7::LEU2), MGSC97 (rad7::URA3), and W303236 (rad16::URA3) were generously supplied by Dr. Jaap Brouwer (University of Leiden, The Netherlands). Other strains used were SX46a (MATa, ade2, his3-532, trp1-289, ura3-52) and its isogenic partners MGSC 139 (rad14::LEU2) and SX46a (rad1::URA3), rad4-10 (MATa, rad4-10, ade2, his3-832, trp1-289). SX46A (ubr1::his3, ubr1::HIS3, isogenic to SX46A); SX46Aabf1td (abf1td, isogenic to SX46A); SX46A ubr1::HIS3, abf1td (abf1td, isogenic to ubr1::HIS3); YNN281 (MATa trp1D, his3D200, ura3-52, lys2-801, ade2-1, gal), JCA30 (YNN281, ABF1 HIS3); JCA31 (YNN281, abf1-1, HIS3; JCA32 (YNN281, abf1-2, HIS3); JCA35 (YNN281, abf1-5, HIS3). These strains were a generous gift from Judith Campbell (Caltech, CA). W303-1A (MATa, ade2-1, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100; JRY3765 (MATa, abf1-102, ade2, his3-11, 15, leu2-3, 112, lys2-801, ura3; JRY4996 (MATa1, abf1-101, ste14, ade2, leu2-3, 112, lys2-801, ura3, were a kind gift from Jasper Rine (University of California). Yeast strains abf1td and ubr1::abf1td were maintained at 23°C in YPD medium supplemented with 0.5 mm CuSO4. The abf1 mutant strains from Jasper Rine and Judith Campbell were maintained on YPD at the permissive temperature of 23°C. The remaining strains were maintained at 30°C in YPD medium.
UV radiation sensitivity
Sensitivity to killing by UV radiation was measured by growing strains at the appropriate temperature (see below), following inoculation of starter cultures (5 ml) with a single colony and growing to late-log phase. An aliquot from this culture was then used to inoculate 25 ml of YPD and the culture grown to mid-log phase prior to harvesting. Serial 10-fold dilutions were performed for each culture and cells were plated such that 200–300 colonies were expected to grow on unirradiated control plates. Irradiated plates were exposed to UV radiation from a germicidal UV lamp at a fluence of 1 J/m2 per sec. Plates were incubated at the appropriate temperature in the dark until colonies were large enough to be counted. All manipulations were carried out in safe light to avoid photoreactivation. Experimental conditions for the survival of the abf1td mutant strain were as follows. Cells were grown overnight in YPD supplemented with CuSO4 (5 mm) at 23°C. Following plating on YPD (supplemented with CuSO4) and UV irradiation, cells were placed at 32°C to determine their survival. For the temperature-sensitive abf1 mutants defective in silencing, cells were grown overnight and incubated on plates post-irradiation at 23°C. Conditions for the abf1 mutant alleles temperature sensitive for replication were as follows: strains were grown overnight at 23°C and subsequently incubated at 32°C or 28°C (abf1-1 and abf1-5, respectively) to determine their UV sensitivity.
Plasmid construction
Plasmid pabf1td was derived from plasmid pPW66R containing the URA3 gene and Pcup1-Ub-Arg-DHFRts-ha-cdc281-95. This plasmid [temperature-degradable (td) vector] was generously provided by Dr. Alexander Varshavsky (Caltech). The cdc281-95 fragment was removed from pPW66R by digesting with HindIII. A 300-bp fragment of the ABF1-coding region was amplified by PCR using the following primers: 5′TATAGAAGCTTCCATGGACAAATTAGTCGTGAATTATTATGA-3′ 5′-TATAAAGCTTAAAATGTTTGTTCCTATGCGAATTCTT-3′. The PCR product has HindIII sites engineered into both ends. Digesting the PCR product with HindIII enzyme generated an ABF1 fragment that could be cloned directly in frame in the td vector. The ligated product was used to transform Escherichia coli DH5 α cells and transformants were selected. The ABF1 fragment could be cloned in either orientation and clones were tested and selected for ABF1 fragments in the correct orientation for ABF1 expression.
Construction of temperature-degradable yeast abf1 mutant strain
The pabf1td plasmid was linearized with a partial digest with NcoI, which cleaves within the ABF1 fragment. A second NcoI site exists within the URA3 selectable marker on the pPW66R vector. Therefore, partial digest conditions were necessary to linearize the plasmid. Yeast strains SX46A and SX46A ubr1::HIS3 were subsequently transformed. Recombinants of URA3 prototrophs were selected and integration of the abf1td allele into the genome was confirmed by Southern blot analysis.
Preparation of WCE from the abf1td mutant strain
Strain abf1td was grown at the permissive condition (23°C) in YPD supplemented with 0.5 mm CuSO4 to mid-log phase and cells were collected by centrifugation. WCE containing levels of ABF1 produced at the permissive temperature were prepared as described previously (Wang et al. 1997). To deplete temperature-degradable ABF1 protein in vivo, the cells were resuspended in YPD medium prewarmed to 37°C in the absence of CuSO4 and incubated at 37°C for 1–5 hr prior to collection for preparation of WCE.
ABF1 protein and antisera
ABF1 protein was produced in a recombinant baculovirus vector. The ABF1 gene (Diffley and Stillman 1989) was cloned into the transfer vector pVL1392 and then inserted into BaculoGold baculovirus DNA (PharMingen). The resulting virus was plaque purified and ABF1 protein was purified from recombinant baculovirus-infected SF9 insect cells by a procedure similar to that described previously (Diffley and Stillman 1988). Anti-ABF1p antibody was affinity purified as described (Diffley and Stillman 1989).
In vitro NER
Yeast WCE capable of supporting NER were prepared as described previously (Wang et al. 1997). NER was monitored by measuring DNA repair synthesis in pUC18 containing N-acetyl-2-aminofluorene (AAF) adducts. AAF-modified DNA was prepared by treating pUC18 with AAF and purifying the DNA on a 5%–20% sucrose gradient. Plasmid DNA was recovered from extracts by restricting with HindIII enzyme, which linearizes the substrate and reveals a discrete band in a 1% agarose gel following electrophoresis. In the standard NER assay that measures repair synthesis, an undamaged DNA template is used as an internal control. Background incorporation of radioactivity (typically ∼5% of that in the damaged plasmid) is subtracted from that incorporated in the damaged template. DNA was visualized in gels under UV light after staining with ethidium bromide. Repair synthesis was visualized by autoradiography of dried agarose gels and quantitated by PhosphorImaging.
Identification of ABF1 protein by tandem mass spectrometry
Details of the analysis are available on request. Proteins were proteolytically trypsinized in the gel. Extracted peptides were purified on a reverse-phase microcolumn and eluted in 60% methanol/5% formic acid into a nanoelectrospray capillary. Tandem mass spectrometry was performed. From the 112-kD band, a peptide fragment (Mr = 1791.88) identified uniquely the tryptic peptide comprising amino acids 638–652 of the ABF1 protein (NEDDKLPHEVAEQLR). Two other ABF1 peptides were sequenced from the same digest (data not shown).
In vivo DNA repair in abf1 mutant alleles
In vivo DNA repair was measured by growing the abf1-1 and abf1-5 mutant strains and isogenic parental strains at a permissive temperature of 23°C, by inoculating a starter culture (5 ml) with a single colony and growing to late-log phase. An aliquot from this culture was then inoculated to 100 ml of YPD and the culture grown to mid-log phase prior to harvesting. Cells were diluted in PBS to a cell density of 2 × 107/ml. Prior to irradiation, samples were divided equally into two fractions and maintained for 3 hr at the permissive temperature of 23°C or the semipermissive temperature of 32°C for the abf1 mutant alleles. The strains were irradiated in dishes such that the depth of the cell suspension was <3 mm. Each strain was exposed to UV light at a dose of 20 J from a germicidal lamp at a fluence of 1 J/m2 per sec. An unirradiated control sample was prepared, and irradiated samples were returned to their respective temperatures in the dark to allow for a period of repair. An aliquot from each sample was taken directly after irradiation. All manipulations were carried out in safe light to avoid photoreactivation. Samples were taken at the times indicated and genomic DNA prepared by standard techniques. Equal amounts (5 μg) of DNA from each time point were applied to Hybond N plus transfer membrane via a slot-blot transfer apparatus. DNA was denatured and fixed on the membrane by adding NaOH to a final concentration of 0.4 m to each DNA sample. Removal of UV radiation-induced photoproducts from genomic DNA was examined by Western blotting with monoclonal CPD or 6-4 photoproduct antibodies (Mori et al. 1991).
Acknowledgments
We thank Judith Campbell and Jasper Rine for yeast strains, Patty Wendel for technical assistance and our laboratory colleagues for extensive discussions and review of the manuscript. Supported by research grant CA12428 from the U.S. public Health Service (ECF).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL friedberg.errol@pathology.swmed.edu; FAX (214) 648-4067.
References
- Bang DD, Verhage R, Goosen N, Brouwer J, van de Putte P. Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Micklem G, Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987;51:709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Buchman AR, Kornberg RD. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990;10:887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Seta F, Treich I, Buhler JM, Sentenac A. ABF1 binding sites in yeast RNA polymerase genes. J Biol Chem. 1990;265:15168–15175. [PubMed] [Google Scholar]
- Diffley JF, Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci. 1988;85:2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989;246:1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Feldman JB, Hicks JB, Broach JR. Identification of sites required for repression of a silent mating type locus in yeast. J Mol Biol. 1984;178:815–834. doi: 10.1016/0022-2836(84)90313-9. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA repair and mutagenesis. Washington, DC.: ASM Press; 1995. [Google Scholar]
- Guzder SN, Sung P, Prakash L, Prakash S. The DNA dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J Biol Chem. 1997;272:21665–21668. doi: 10.1074/jbc.273.11.6292. [DOI] [PubMed] [Google Scholar]
- Huang W, Feaver WJ, Tomkinson AE, Friedberg EC. The N-degron protein degradation strategy for investigating the function of essential genes: Requirement for replication protein A and proliferating cell nuclear antigen proteins for nucleotide excision repair in yeast extracts. Mutat Res. 1998;408:183–194. doi: 10.1016/s0921-8777(98)00031-7. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Fogel S, Lusnak K. Gene conversion of the mating-type locus in Saccharomyces cerevisiae. Genetics. 1979;92:777–782. doi: 10.1093/genetics/92.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Laurenson P, Foss M, Dillin A, Rine J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Nokane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cylcobutane pyrimidine dimer of (6-4) photoproduct from the same mouse immunized with UV irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- Paetkau DW, Riese JA, MacMorran WS, Woods RA, Gietz RD. Interaction of the yeast RAD7 and SIR3 proteins: Implications for DNA repair and chromatin structure. Genes & Dev. 1994;8:2035–2045. doi: 10.1101/gad.8.17.2035. [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SH, Boiteux S, Waters R. UV-induced endonuclease III-sensitive sites at the mating type loci in Saccharomyces cerevisiae are repaired by nucleotide excision repair: RAD7 and RAD16 are not required for their removal from HML alpha. Mol & Gen Genet. 1996;250:505–514. doi: 10.1007/BF02174039. [DOI] [PubMed] [Google Scholar]
- Reed SH, You Z, Friedberg EC. The yeast RAD7 and RAD16 genes are required for postincision events during nucleotide excision repair. In vitro and in vivo studies with rad7 and rad16 mutants and purification of a Rad7/Rad16-containing protein complex. J Biol Chem. 1998;273:29481–29488. doi: 10.1074/jbc.273.45.29481. [DOI] [PubMed] [Google Scholar]
- Rhode PR, Sweder KS, Oegema KF, Campbell JL. The gene encoding ARS-binding factor I is essential for the viability of yeast. Genes & Dev. 1989;3:1926–1939. doi: 10.1101/gad.3.12a.1926. [DOI] [PubMed] [Google Scholar]
- Rhode PR, Elsasser S, Campbell JL. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1064–1077. doi: 10.1128/mcb.12.3.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C, Schenk P, Poot R, Brouwer J, van de Putte P. Differential repair of UV damage in rad mutants of Saccharomyces cerevisiae: A possible function of G2 arrest upon UV irradiation. Mol Cell Biol. 1990;10:4678–4684. doi: 10.1128/mcb.10.9.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SS, Francesconi SC, Tye BK, Eisenberg S. The OBF1 protein and its DNA-binding site are important for the function of an autonomously replicating sequence in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2914–2921. doi: 10.1128/mcb.9.7.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wei S, Reed SH, Wu X, Svejstrup JQ, Feaver WJ, Kornberg RD, Friedberg EC. The RAD7, RAD16, and RAD23 genes of Saccharomyces cerevisiae: Requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products. Mol Cell Biol. 1997;17:635–643. doi: 10.1128/mcb.17.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]