Abstract
A variety of both Gram-positive and Gram-negative bacteria produce large quantities of indole as an intracellular signal in microbial communities. Biosynthesis of indole is well-studied, and while carbon sources and amino acids are important environmental cues for indole production in Escherichia coli, other environmental factors affecting indole production for this strain are less clear. This study demonstrates that the environmental cue pH is an important factor for indole production that further controls biofilm formation of E. coli. Moreover, E. coli produced a higher level of extracellular indole in the presence of the antibiotics ampicillin and kanamycin, and the increased indole enhanced cell survival during antibiotic stress. Additionally, we found here that temperature is another important factor for indole production; E. coli produces and accumulates a large amount of indole at 50°C, even at low cell densities. Overall, our results suggest that indole is a stable biological compound, and E. coli may utilize indole to protect itself against other microorganisms.
Keywords: indole, signal molecule, Escherichia coli, environmental factors, antibiotics
1. INTRODUCTION
In nature, bacteria are most commonly found in complex communities (Vendeville et al., 2005). Many bacteria have developed their ability to sense the local environment, such as the nutritional limitation, their population, the presence of toxic chemicals from other bacteria and host signals. Therefore, it is important to coordinate their behavior in order to adapt and survive in environmental niches. For example, bacteria have developed signaling systems, such as intercellular signaling systems (Waters and Bassler, 2005; Keller and Surette, 2006) and diverse two-component regulatory systems (Mitrophanov and Groisman, 2008) to synchronize cellular behavior.
Among intercellular signal molecules, indole is often produced by a variety of both Gram-positive and Gram-negative bacteria (to date, more than 85 species) (Lee and Lee, 2010). Importantly, recent studies have demonstrated many biological functions of indole, such as drug resistance in E. coli (Hirakawa et al., 2005), plasmid stability in E. coli (Chant and Summers, 2007), virulence control in pathogenic E. coli (Anyanful et al., 2005; Hirakawa et al., 2009), and biofilm formation in E. coli (Di Martino et al., 2003; Lee et al., 2007a) as well as in Vibrio cholerae (Mueller et al., 2009). Interestingly, indole controls phenotypes of other bacteria that cannot produce indole. For example, indole decreases the growth of fungus Aspergillus niger (Kamath and Vaidyanathan, 1990), increases drug resistance in Salmonella enterica (Nikaido et al., 2008), and attenuates virulence in Pseudomonas aeruginosa (Lee et al., 2009). Moreover, indole significantly affects gene expression for human epithelial cells leading to tighter cell-junctions and increases in beneficial cytokines (Bansal et al., 2010).
An early report showed that Bacillus coli (Escherichia coli) and Asiatic cholera (Vibrio cholerae) produced indole during a stationary cell growth phase in 1897 (Smith, 1897). Indole biosynthesis in E.coli has been investigated over many decades (Newton and Snell, 1965; Botsford and DeMoss, 1971; Yanofsky et al., 1991). In E. coli, indole is produced by tryptophanase (TnaA; EC 4.1.99.1) that can reversibly convert tryptophan into indole, pyruvate, and ammonia (Newton and Snell, 1965) in the tryptophan pathway in E. coli (Pittard, 1996; Lee et al., 2007a). E. coli uses several mechanisms (repression, transcription attenuation, and feedback inhibition) to regulate the expression of the tryptophan (trpABCDE) and tna operons (tnaCAB) in tryptophan metabolism (Yanofsky et al., 1991; Gong and Yanofsky, 2002; Lee et al., 2007a). Environmental conditions and composition of media critically influence the level of extracellular indole. For example, the extracellular indole concentration is cell population density-dependent in E. coli (Wang et al., 2001; Kobayashi et al., 2006). Extracellular indole reached 0.5 mM in a rich medium in the stationary phase so that it has been called a stationary-phase signal molecule (Wang et al., 2001; Kobayashi et al., 2006). In addition, glucose repressed indole biosynthesis (John and Wyeth, 1919) due to catabolic repression of tnaA (Botsford and DeMoss, 1971). Different carbon sources also influence the accumulation of extracellular indole (John and Wyeth, 1919; Botsford and DeMoss, 1971). Additionally, temperature and pH affect indole biosynthesis in E. coli. A low pH inhibited indole production in E. coli (John and Wyeth, 1919), and TnaA was one of the most induced proteins at pH 9.0 (Blankenhorn et al., 1999). Gene expression of tnaAB was induced in E. coli by temperature shifting from 30°C to 43°C (Li et al., 2003), and E. coli lost the ability of indole biosynthesis at 44.5°C (Bueschkens and Stiles, 1984).
Although the importance of indole signaling in microbial communities (Lee and Lee, 2010) as well as in the human immune system (Wikoff et al., 2009; Bansal et al., 2010) has been recently demonstrated, environmental factors that contribute to and regulate indole production remain poorly defined. Recently, several transcriptomic studies provide informative data on the expression of tnaA in response to various environmental cues in E. coli (Table 1). Hence, the overall objective of this study is to identify the important environmental factors that control indole production as well as cell growth to better understand indole signaling in microbial communities.
Table 1.
Transcriptomic data for gene expression of tnaA in wild-type E. coli with various environmental factors. Each comparison was made with its own control condition in the same reference. Studies with mutant strains are not included here.
| Factors | Conditions | Fold change of tnaA |
Reference |
|---|---|---|---|
| Culture time | 15 h suspension cells vs. 4 h suspension cells in LB |
−588 | (Domka et al., 2007) |
|
| |||
| Glucose | No glucose vs. 0.4% glucose in LB | 100 | (Gosset et al., 2004) |
|
| |||
| Carbon sources |
Glycerol vs. glucose in a minimal medium | 5 | (Liu et al., 2005) |
| Succinate vs. glucose in a minimal medium | 5 | (Liu et al., 2005) | |
| Acetate vs. glucose in a minimal medium | 13 | (Liu et al., 2005) | |
|
| |||
| Amino acids | Alanine vs. glucose in a minimal medium | 6 | (Liu et al., 2005) |
| Proline vs. glucose in a minimal medium | 12 | (Liu et al., 2005) | |
| 50 μg/mL L-tryptophan for 60 min in a minimal medium with 0.2% glucose |
18 | (Khodursky et al., 2000) | |
|
| |||
| Acid | pH 5.0 vs. pH 7.0 in LB | −14 | (Maurer et al., 2005) |
|
| |||
| Base | pH 8.7 vs. pH 5.0 in LB | 16 | (Maurer et al., 2005) |
| 10 mM trimethylamine N-oxide vs. none in LB |
15 | (Bordi et al., 2003) | |
|
| |||
| Pressure | 0.1 Mpa vs. 30 MPa in LB | −25 | (Ishii et al., 2005) |
|
| |||
| Oxidative stress |
35 ppm paraquat vs. none for 60 min in LB | 20 | (Kim et al., 2005) |
| 1 mM hydrogen peroxide vs. none in LB | 18 | (Zheng et al., 2001) | |
|
| |||
| Heavy metals |
1.2 mM cadmium (II) vs. none in LB | −15 | (Brocklehurst and Morby, 2000) |
| 4 mM nickel (II) vs. none in LB | −15 | (Brocklehurst and Morby, 2000) | |
|
| |||
| Aeration | Aerobic vs. anaerobic for 60 min in a minimal medium with 30 mM glucose |
7 | (Patridge and Ferry, 2006) |
| Aerobic vs. anaerobic in a minimal medium with 0.1% glucose |
−2 | (Kang et al., 2005) | |
2. MATERIALS AND METHODS
2.1. Bacterial strains, materials and growth rate measurements
E. coli K-12 BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBA-DAH33 ΔrhaBADLD78) and its isogenic tnaA mutant (Baba et al., 2006) that does not produce indole (Lee et al., 2008) were used. Luria-Bertani (LB) (Sambrook et al., 1989) was used as a basic medium for growth unless indicated. Indole, ampicillin, kanamycin, chloramphenicol, N-hexanoyl-DL-homoserine lactone, N-(3-oxotetradecanoyl)-L-homoserine lactone, N-(3-oxododecanoyl)-L-homoserine, N-(3-oxooctanoyl)-L-homoserine lactone, paraquat, cadmium chloride, sodium dodecyl sulfate (SDS), and 4-(dimethylamino)-benzaldehyde were purchased from Sigma-Aldrich Co. (Missouri, USA). Hydrogen peroxide (H2O2), HCl, NaOH, NaCl, FeCl3, ethanol, methanol, and dimethyl sulfoxide (DMSO) were purchased from Duksan Pure Chemical Co. (Ansan, Korea). Bacterial strains were initially streaked from −80°C glycerol stocks on LB plates, and a fresh single colony was inoculated into LB medium (25 mL) in 250-mL flasks and routinely cultured at 250 rpm at 37°C unless indicated. Overnight cultures were diluted 1:100 using LB medium. For cell growth measurements, the optical density was measured at 600 nm (OD600) with a spectrophotometer (UV-160, Shimadzu, Japan). When the value of OD600 was above 0.7, culture sample was diluted to fit in a linear range of 0.2 and 0.7. The specific growth rates were determined by measuring OD600 and calculated by using the linear portion of the natural logarithm of OD600 versus time. In order to measure cell viability and cell number, diluted cells were enumerated with LB agar plates.
2.2. Indole assays
Extracellular and intracellular indole concentrations were measured as indicated previously using Kovac’s reagent (Kawamura-Sato et al., 1999). Briefly, Kovac’s reagent (0.4 mL) was mixed with supernatants (1 mL) of bacterial cultures. The reaction mixture was diluted 1:10 in HCl-amyl alcohol solution (30 mL of HCl and 90 mL of amyl alcohol), and the absorbance of the mixture was measured at 540 nm with a spectrophotometer (Shimadzu UV-160). For the intracellular indole concentrations, cells were completely lysed with 1% SDS and 0.2% NaOH by mixing well (Sambrook et al., 1989). In addition, the spectrophotometric indole assay was corroborated with reverse-phase HPLC (Lee et al., 2007a) using a 100 × 4.6 mm Chromolith Performance RP-18e column (Merck KGaA, Darmstadt, Germany) and elution with H2O-0.1% (v/v) trifluoroacetic acid and acetonitrile as the mobile phases at a flow rate of 0.5 mL/min (50:50). Under these conditions, the retention time and the absorbance maximum was 5.1 min/271 nm for indole.
2.3. Crystal-violet biofilm assay
A static biofilm formation assay was performed in 96-well polystyrene plates (Fisher Scientific, Pittsburg, USA) as previously reported (Pratt and Kolter, 1998). Briefly, cells were inoculated at an initial turbidity at 600 nm of 0.05 for 24 h without shaking at 30°C and 37°C. Cell density (turbidity at 620 nm) and total biofilm (absorbance at 540 nm) were measured using crystal violet staining. Total biofilm was normalized by bacterial growth for each condition. Each data point was averaged from at least twelve replicate wells (six wells from each of two independent cultures).
2.4. Scanning electron microscopy (SEM)
In order to investigate the impact on cell morphology upon addition of ampicillin and indole, SEM was used by modifying the protocol (Hossain et al., 1996). Briefly, E. coli cells were cultured in LB medium at 37°C for 6.5 h, and cells were directly fixed by adding glutaraldehyde (2.5% final) and formaldehyde (2% final) and incubated at 4°C overnight. Fixed cells were collected by filtering with a 0.45 μm nylon filter (Nalgene, New York, USA) with vacuum. The filter contained cells was cut into 0.5 × 0.5 mm squares and washed with 0.2 M sodium phosphate buffer before fixating for 90 min with osmium solution (containing 1.5 mL of sodium phosphate buffer 0.2 M, 3 mL of 2% OsO4 and 3mL deionized water). Then, samples were washed and dehydrated by successive 10 min incubations in 50% ethanol, 70% ethanol, 80% ethanol, 90% ethanol, and 95% ethanol followed by two successive incubations for 20 min in 100% ethanol. After dehydrating, the filters with cells were incubated in isoamyl acetate for 20 min and dried with critical-point dryer (HCP-2, Hitachi, Japan). The nylon filters were affixed to SEM stubs and coated with white gold for 200 seconds using ion sputtering (E-1030, Hitachi). Specimens were examined with the SEM S-4100 (Hitachi). The voltage was set at 15 kV and viewed at a magnification from x 2,000 to x15,000.
3. RESULTS
The main purpose of this research was to determine key environmental factors affecting the production of extracellular indole in E. coli. Environmental conditions such as pH, antibiotics, temperature, heavy metals, oxidative reagents, osmolarity, solvents, and quorum-sensing molecules (acylhomoserine lactones) from other bacteria were tested. Since indole signaling shows different responses at different temperatures (Lee et al., 2008), both a human body temperature (37°C) and a low temperature (25°C or 30°C) were investigated with various conditions.
3.1. pH controls indole production and biofilm formation
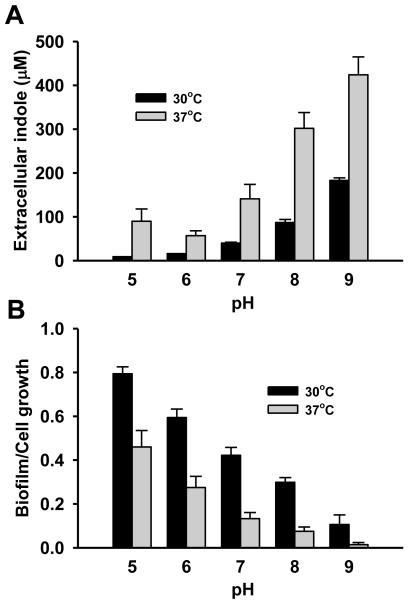
Low pH (pH 4) inhibits indole production (John and Wyeth, 1919) while high pH (pH 9) induces the expression of tnaA significantly in E. coli (Blankenhorn et al., 1999; Stancik et al., 2002; Yohannes et al., 2004). Hence, different pHs (4, 5, 6, 7, 8, 9, and 10 adjusted with 37% HCl and 5 N NaOH which caused the total volume change less than 0.2 %) were tested to measure the production of extracellular indole at the same cell turbidity of 1.5 at 30°C and 37°C, respectively. Growth rates of E. coli were similar between pH 5 and pH 9 as 0.83 ± 0.01 h−1 at pH 5, 1.01 ± 0.01 h−1 at pH 6, 1.04 ± 0.02 h−1 at pH 7, 0.87 ± 0.01 h−1 at pH 8, and 0.70 ± 0.08 h−1 at pH 9, respectively at 30°C and 1.37 ± 0.02 h−1 at pH 5, 1.51 ± 0.04 h−1 at pH 6, 1.50 ± 0.02 h−1 at pH 7, 1.35 ± 0.01 at pH 8, and 1.21 ± 0.04 h−1 at pH 9, respectively at 37°C, while cell growth of E. coli was much slow at pH 4 and 10 so that data at pH 4 and 10 were excluded in the comparison. The initial pH (between pH 5 and pH 9) was not changed until the cell turbidity of 1.5 at which the production of indole was measured.
Fig. 1A clearly shows that low pH inhibits indole production while high pH increases indole production both at 30°C and at 37°C. The results match well with the previous transcriptomic data (Table 1) in which tnaA was repressed under acidic conditions while tnaA was induced with a basic condition. It was also observed that indole production was higher at 37°C than at 30°C for all tested pHs.
Fig. 1.
Effect of pH on extracellular indole production (A) and biofilm formation after 24 h (B) in E. coli in LB medium. pH (5, 6, 7, 8, and 9) was adjusted with 37% HCl and 5 N NaOH and cells were cultured at 30°C and 37°C. Extracellular indole was measured at a cell turbidity (optical density at 600 nm) of 1.5. The pH at the cell turbidity of 1.5 was almost same as the initial pH. Total biofilm (OD540) was normalized by cell growth (OD620) for each condition. Each data point was averaged from at least two independent cultures and one standard deviation is shown.
Since indole inhibited the biofilm formation of E. coli BW25113 strain (Lee et al., 2007a) and pH significantly changes indole production (Fig. 1 A), the effect of pH was investigated for biofilm formation in E. coli BW25113 (Fig. 1B). Biofilm formation was clearly decreased at alkali conditions (pH 8 and 9) due to the high level of indole accumulation. To confirm the biofilm reduction by indole, when indole (0.5 mM) was exogenously added in the medium at pH 5, the biofilm formation/cell was significantly decreased as 0.29 ± 0.06 at 30°C and 0.27 ± 0.09 at 37°C. This result supports the previous result that indole decreases biofilm formation in nonpathogenic E. coli as well as pathogenic E. coli O157:H7 (Lee et al., 2007b; Lee et al., 2007a; Lee et al., 2008).
3.2. Antibiotics induce indole production
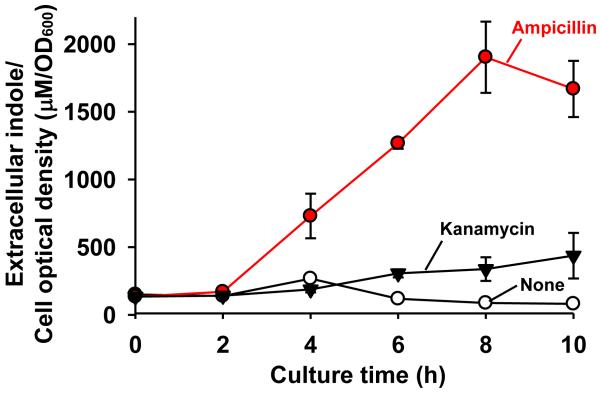
Since indole increased drug resistance in E. coli by inducing multidrug export genes (Hirakawa et al., 2005), we investigated the effect of antibiotics on indole production. Antibiotics (bactericidal ampicillin of the β-lactam class of antibiotics, bactericidal kanamycin of the aminoglycosides, and bacteriostatic chloramphenicol) at sub-inhibitory concentrations were added in the beginning of each cell culture and indole production and cell growth were measured at 25°C and 37°C. E. coli produced significantly more extracellular indole/cell density (22-fold) in the presence of ampicillin (2 μg/mL) (Fig. 2); 1900 ± 260 μM indole/cell OD with ampicillin vs. 88 ± 3 μM indole/cell OD for the control at 8 h. Similarly, kanamycin (2 μg/mL) also enhanced indole production/cell density by 4-fold with 338 ± 87 μM indole/cell OD compared to the control at 8 h (Fig. 2). However, bacteriostatic chloramphenicol did not affect the indole production (data not shown). Additionally, the effect of antibiotics at a low temperature, such as 25°C, was similar to the results at 37°C (data not shown). Therefore, E. coli produced more indole signal in the presence of the bactericidal antibiotics, ampicillin and kanamycin, at both 25°C and 37°C.
Fig. 2.
Effect of antibiotics on extracellular indole production/cell growth in E. coli. Sub-inhibitory concentrations of ampicillin (2 μg/mL) and kanamycin (2 μg/mL) were added at the beginning of culturing in LB medium at 37°C. Each data point was averaged from at least four independent cultures and one standard deviation is shown.
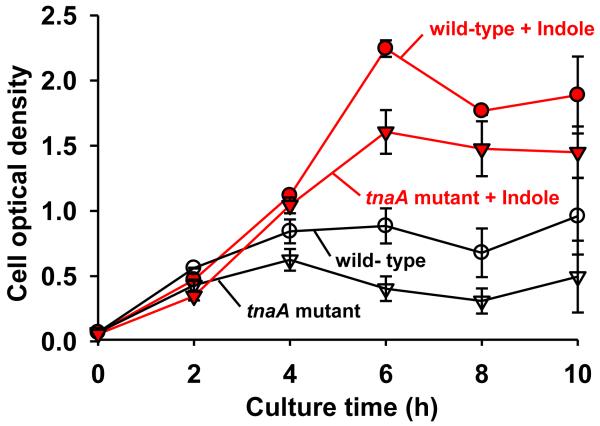
Since indole induced the expression of multidrug exporter genes in E. coli and increased cell survival rate in the presence of several drugs, such as rhodamine 6G, sodium dodecyl sulfate, carbenicillin (Hirakawa et al., 2005), we studied the effect of indole on cell growth in the presence of antibiotics. As expected, the addition of indole (1 mM) to both E. coli K-12 BW25113 wild-type and tnaA knock mutant(non-indole producing strain (Lee et al., 2008)) increased cell growth 2- to 5-fold in the presence of ampicillin at 8 h (Fig. 3). Apparently, cell growth of the tnaA mutant was more sensitive against ampicillin than the wild-type, and the effect of indole on cell growth was more significant (5-fold) in the tnaA mutant than the wild-type (Fig. 3). Also the exogenous addition of indole (1 mM) to the tnaA mutant complemented the effect of indole on cell growth in the presence of ampicillin (Fig. 3). Therefore, we conclude that E. coli produces a high amount of indole against antibiotics, which leads to enhanced cell survival.
Fig. 3.
Effect of indole on cell growth in the presence of ampicillin. Sub-inhibitory concentration of ampicillin (2 μg/mL) was added at the beginning of culturing E. coli K-12 BW25113 and E. coli K-12 BW25113 ΔtnaA (indole deficient mutant (Lee et al., 2008)) in LB medium at 37°C. Exogenous indole (1 mM) was added to test the effect of indole. Each data point was averaged from four independent cultures and one standard deviation is shown.
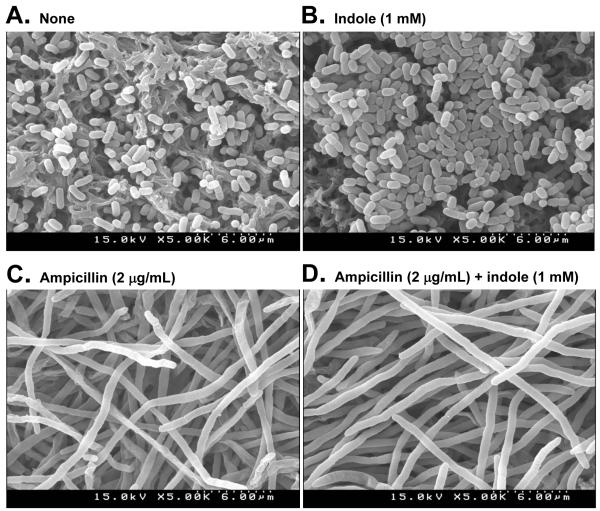
To better understand the effect of indole on cell survival in the presence of antibiotic ampicillin, cell morphology was examined using SEM because indole could inhibit cell division in E. coli (Chant and Summers, 2007; Lee et al., 2008). Addition of ampicillin blocks cell division and made cells elongated (Fig. 4) which matches with the previous result (Comber et al., 1975). However, the addition of indole (1 mM) in the presence and absence of ampicillin does not affect cell morphology. Hence, indole did not prevent the inhibition of cell division by antibiotics.
Fig. 4.
Effect of indole and ampicillin on cell morphology of E. coli. For SEM analysis, cells were grown in LB medium in the absence of exogenous indole (A), with 1 mM indole (B), with 2 μg/mL ampicillin (C), or with 2 μg/mL ampicillin and 1 mM indole (D). Cells were cultured in LB medium at 37°C for 6.5 h and were directly fixed by adding glutaraldehyde and formaldehyde and filtered with 0.45 μm Nylon filter.
3.3. High temperature induces indole production
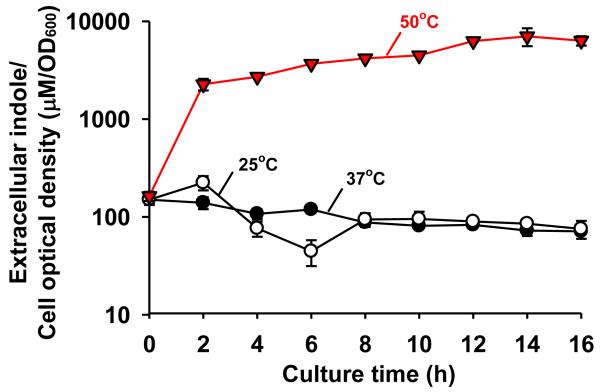
Since temperature is an important factor in indole cell signaling (Lee et al., 2008) and thermal destruction of E. coli is important in the food industry (Juneja et al., 1997), the effect of temperature (25, 37, 42, and 50°C) on indole production was investigated. Maximal indole accumulation at a low temperature (25°C) and at a high temperature (42°C) was similar to that at 37°C (data not shown). For example, extracellular indole accumulation was reached maximally at about 500 μM and maintained at this concentration at 25°C and 37°C. However, at 50°C, E. coli quickly started to produce indole within 2 hours although E. coli could not grow at 50°C (Fig. 5). The indole concentration increased to 7020 ± 1420 μM indole/cell OD after 14 h although the optical cell density of E. coli was 120 times lower than that at 37°C. Therefore, the extracellular indole/cell density at 50°C was 89-fold higher than that at 37°C (79 ± 2 μM indole/cell OD) at 14 h. This experiment was conducted six times to confirm the result. Therefore, we sought to find some mechanism of this phenomenon. Initially, cell survival was measured. The cell survival rates were maintained more than 73% for more than 10 hours (98 ± 1% at 1 h, 93 ± 1% at 2 h, and 73 ± 1% at 10 h, respectively), which indicates the small number of live E. coli produced a large amount of indole at 50°C. Additionally, the intracellular indole concentration of the inoculum was measured because intracellular indole from dead (lysed) cells could affect the level of extracellular indole. However, the intracellular indole concentration of the inoculum was an insignificant 0.25 ± 0.07 μM. Therefore, we concluded that a small number of E. coli (optical cell density of 0.05 at 600 nm that is about 4 × 107 cells/mL) produce more than 300 μM extracellular indole at 50°C. This result is quite different from the previous report that E. coli lost the ability of indole biosynthesis at 44.5°C (Bueschkens and Stiles, 1984).
Fig. 5.
Effect of temperature on extracellular indole production/cell growth in E. coli. Cells were cultured in LB medium at 25°C, 37°C, and 50°C. Each data point was averaged from at least four independent cultures and one standard deviation is shown.
Notably, indole was quickly produced for the first two hours at 50°C and stably accumulated even at a low cell density (Fig. 5). The result supports the previous transcriptomic study that the gene expression of tnaAB is induced in E. coli by temperature shifting from 30°C to 43°C (Li et al., 2003). Hence, we hypothesized that E. coli may enhance indole production at high temperatures, which may lead to a higher survival rate at high temperatures. In order to test the impact of heat shock, the culture temperature was shifted from 37°C to 50°C at the optical density of 1.5. However, the final accumulation of indole with the temperature shift is similar to that without the temperature shift (data not shown). Additionally, the effect of indole on cell growth at a high temperature was investigated by adding indole (1 mM) at the beginning of the cell culture and culturing at 46°C. However, indole (1 mM) did not increase cell growth of E. coli at 46°C (data not shown). Therefore, it appears that a high temperature enhances indole production, which does not increase cell growth. The genetic mechanism of this phenomenon has not been resolved yet.
3.4. Effects of acylhomoserine lactones, osmolarity, aeration, metals, solvents, and oxidative stresses
E. coli increased indole production against antibiotics originated from other microorganisms (Fig. 2) and SdiA (a LuxR homologue) of E. coli recognizes acylhomoserine lactones (AHLs) from other bacteria (Lindsay and Ahmer, 2005) and binds AHLs (Yao et al., 2006), and AHLs decrease the biofilm formation of E. coli (Lee et al., 2007a). Hence, we hypothesized that AHLs may affect indole production. However, four kinds of AHLs (N-hexanoyl-DL-homoserine lactone, N-(3-oxotetradecanoyl)-L-homoserine lactone, N-(3-oxododecanoyl)-L-homoserine, and N-(3-oxooctanoyl)-L-homoserine lactone) up to 10 μM did not change the accumulation of extracellular indole as well as cell growth either at 30°C or at 37°C (data not shown).
Since various environmental stimuli, such as osmolarity, aeration, metals, and solvents, affect AI-2 production in chemostat cultures (DeLisa et al., 2001), the effects of these stimuli were investigated. Osmolarity, adjusted by the addition of sodium chloride (0, 2, 10, and 40 g per liter in NaCl-free LB medium) did not much affect indole production (data not shown). Anaerobic conditions created by sealing culture flasks from the beginning of culturing also did not change the indole production (data not shown). The addition of ferrous chloride (III) (up to 10 mg/L) did not change the indole production as well as cell growth. Also, the addition of cadmium chloride (II) (up to 150 mg/L) and ethanol (5 v/v%) did not alter the final indole production (0.5 mM), although both cadmium chloride and ethanol inhibited cell growth significantly (less than 1.0 of optical density at 600 nm after 10 h). These results indicate that the enhanced indole production seen with high pH, antibiotics, and temperature does not always occur under the inhibition of cell growth.
Since the previous transcriptomic data showed that oxidative stresses (1 mM hydrogen peroxide (Zheng et al., 2001) and 35 ppm paraquat (Kim et al., 2005)) induced tnaA (Table 1), oxidative stress was investigated. However, unexpectedly, hydrogen peroxide up to 5 mM and paraquat up to 200 ppm did not alter indole production and cell growth (data not shown). These results suggest that oxidative stresses cause induction of tnaA gene expression but do not led to the accumulation of extracellular indole.
DISCUSSION
Bacteria can sense environmental cues, regulate their overall gene expression, and control their phenotypes. As a well-known metabolite, indole has recently shown diverse biological roles as an intercellular signal in microbial communities (Lee and Lee, 2010). Our current study demonstrates that a variety of environmental cues play an important role in the accumulation of extracellular indole in E. coli and can lead to significant increases in indole production per cell.
Carbon sources (John and Wyeth, 1919; Botsford and DeMoss, 1971) amino acids (Newton and Snell, 1965), and growth status (Wang et al., 2001; Kobayashi et al., 2006) are important factors for indole biosynthesis and TnaA activity for E. coli. Several reports also indicated that pH is important factor for indole production (John and Wyeth, 1919). It was even proposed that TnaA counteracted alkaline stress, such as trimethylamine N-oxide because an indole-mutant (ΔtnaLAB) showed low survival in trimethylamine N-oxide (Bordi et al., 2003). We clearly demonstrated that E. coli produces more indole at high pH, which further impacts biofilm formation (Fig. 1). These results support the hypothesis that E. coli may turn off tnaA in the acidic stomach and turn it on in the basic gut (Lee et al., 2007a).
This study demonstrated for the first time that E. coli has the ability to enhance the accumulation of indole against antibiotics (Fig. 2) and the enhanced indole production increases cell survival (Fig. 3). Although highly speculative, these results suggest that E. coli may utilize indole to compete against other microorganisms that could produce antibiotics. For example, the β-lactam antibiotic penicillin may be derived from Penicillium fungi (Fleming, 1929) and bactericidal kanamycin may be isolated from Streptomyces kanamyceticus (Garrod, 1981). In natural environments, E. coli may encounter these antibiotic-producing microorganisms outside animal host. It was previously reported that indole increases drug resistance by inducing intrinsic xenobiotic exporter genes (mdtEF and acrD) in E. coli, where indole acts via two-component signal transduction systems (BaeSR and CpxAR) (Hirakawa et al., 2005). Similar to E. coli, non-indole producing bacteria, such as S. enterica (Nikaido et al., 2008) and P. aeruginosa (Lee et al., 2009) also increased drug resistance in the presence of indole. The results indicate that the presence of environmental signal indole could seriously affect the drug resistance of pathogenic bacteria.
The degradation of indole is possible because various oxygenases from non-indole producing bacteria, plants, and animals may transform indole (Lee and Lee, 2010). Hence, non-indole producing bacteria may have acquired some defense against indole. For example, P. aeruginosa, could rapidly decrease the level of extracellular indole (Lee et al., 2009). Furthermore, previous results showed that indole from E.coli diminished Pseudomonas virulence by repressing quorum sensing regulated genes encoding the mexGHI-opmD multidrug efflux pump, phz operon, pqs operon, pch operon, and pvd operon while AHLs induce these genes (Lee et al., 2009). Therefore, it appears that signaling interference for indole and defense against indole could be widespread in microbial communities.
Temperature is another important cue for indole signaling (Lee et al., 2008) as well as for indole production (Fig. 5). tnaAB is induced in E. coli by temperature shifting from 30°C to 43°C (Li et al., 2003) and mutants that lack heat shock proteins IbpA and IbpB have elevated extracellular concentrations of indole (Kuczyńska-Wiśnik et al., 2009). The current study confirms that the indole production is significantly increased at high temperatures (Fig. 1 and Fig. 5). Moreover, E. coli produced indole even at 50°C, which was different from a previous result that E. coli lost the ability of indole production at 44.5°C after 6 days (Bueschkens and Stiles, 1984). The difference is probably because of different culture times (16 hrs at 50°C vs. 6 days at 44.5°C) and different cell lines (BW25113 vs. ATCC 11775). It was also previously reported that indole cell signaling for biofilm formation, antibiotic resistance, and cell division in E. coli occurs primarily at a low temperatures (Lee et al., 2008), while indole could control plasmid stability in E. coli (Chant and Summers, 2007) and attenuate the virulence in P. aeruginosa at 37°C (Lee et al., 2009). Hence, it appears that E. coli may utilize indole signaling in different ways depending on the environmental temperature.
Humans maintain a symbiotic relationship with their intestinal microbial flora over a long time, and some bacteria are crucial for nutrient assimilation and are beneficial for the immune system (Hooper and Gordon, 2001). A variety of intestinal bacteria produce a large quantity of indole in the animal gut (a weak basic condition) including the human intestine (DeMoss and Moser, 1969; Lee and Lee, 2010). A recent study suggested that indole could be important in the intestinal epithelial cells response to gastrointestinal tract pathogens because indole beneficially affected gene expression of human epithelial cells (Bansal et al., 2010). Moreover, a mass spectrometry-based metabolomics study demonstrated that the production of a powerful antioxidant compound indole-3-propionic acid in animal blood completely depended on indole-producing enteric bacteria (Wikoff et al., 2009). The result suggested that animals possibly utilize indole derivatives originated from gut microflora for their immune systems (Wikoff et al., 2009). The current study further suggests that the intestinal microbial flora may protect itself using indole against pathogenic bacteria.
Recently, the role of indole has been also studied in Vibrio cholerae in which indole decreases its biofilm formation while increasing its grazing resistance to the phagocytic eukaryote Dictyostelium discoideum, probably by inducing expression of virulence-associated secretion proteins (Mueller et al., 2009). Furthermore, more than 85 species of both Gram-positive and Gram-negative bacteria produce large quantities of extracellular indole in microbial communities (Lee and Lee, 2010). Because the habitats of these bacteria are diverse, the mechanisms of indole production are probably different among bacterial species. Hence, it would be interesting to investigate indole production in other indole-producing bacteria. Further studies are essentially required to understand why many bacterial species produce indole, how bacteria regulate indole signaling, and how other species react to environmental indole.
ACKNOWLEDGEMENTS
This research was supported by the Yeungnam University research grant (to J. Lee). T. H. Han was supported by the Brain Korea 21 Project from the Ministry of Education and Human Resources, Korea. T. Wood is the T. Michael O’Connor endowed chair and is also supported by the NIH (R01 GM089999).
REFERENCES
- Anyanful A, Dolan-Livengood JM, Lewis T, Sheth S, Dezalia MN, Sherman MA, et al. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol Microbiol. 2005;57:988–1007. doi: 10.1111/j.1365-2958.2005.04739.x. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenhorn D, Phillips J, Slonczewski JL. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J Bacteriol. 1999;181:2209–2216. doi: 10.1128/jb.181.7.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi C, Theraulaz L, Mejean V, Jourlin-Castelli C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol Microbiol. 2003;48:211–223. doi: 10.1046/j.1365-2958.2003.03428.x. [DOI] [PubMed] [Google Scholar]
- Botsford JL, DeMoss RD. Catabolite repression of tryptophanase in Escherichia coli. J Bacteriol. 1971;105:303–312. doi: 10.1128/jb.105.1.303-312.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst KR, Morby AP. Metal-ion tolerance in Escherichia coli: analysis of transcriptional profiles by gene-array technology. Microbiology. 2000;146:2277–2282. doi: 10.1099/00221287-146-9-2277. [DOI] [PubMed] [Google Scholar]
- Bueschkens DH, Stiles ME. Escherichia coli variants for gas and indole production at elevated incubation temperatures. Appl Environ Microbiol. 1984;48:601–605. doi: 10.1128/aem.48.3.601-605.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant EL, Summers DK. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol Microbiol. 2007;63:35–43. doi: 10.1111/j.1365-2958.2006.05481.x. [DOI] [PubMed] [Google Scholar]
- Comber KR, Osborne CD, Sutherland R. Comparative effects of amoxycillin and ampicillin in the treatment of experimental mouse infections. Antimicrob Agents Chemother. 1975;7:179–185. doi: 10.1128/aac.7.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisa MP, Valdes JJ, Bentley WE. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J Bacteriol. 2001;183:2918–2928. doi: 10.1128/JB.183.9.2918-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss RD, Moser K. Tryptophanase in diverse bacterial species. J Bacteriol. 1969;98:167–171. doi: 10.1128/jb.98.1.167-171.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003;49:443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. Br J Exp Pathol. 1929;10:226–236. [Google Scholar]
- Garrod LP. Churchill Livingstone. 1981. Antibiotic and Chemotherapy; p. 131. [Google Scholar]
- Gong F, Yanofsky C. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J Biol Chem. 2002;277:17095–17100. doi: 10.1074/jbc.M201213200. [DOI] [PubMed] [Google Scholar]
- Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH., Jr. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol. 2004;186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol. 2005;55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- Hirakawa H, Kodama T, Takumi-Kobayashi A, Honda T, Yamaguchi A. Secreted indole serves as a signal for expression of type III secretion system translocators in enterohaemorrhagic Escherichia coli O157:H7. Microbiology. 2009;155:541–550. doi: 10.1099/mic.0.020420-0. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Hossain MM, Nakayama H, Goto N. In vitro induction of apoptosis of developing brain cells by 5-azacytidine. Int J Dev Neurosci. 1996;14:11–17. doi: 10.1016/0736-5748(95)00084-4. [DOI] [PubMed] [Google Scholar]
- Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C. Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles. 2005;9:65–73. doi: 10.1007/s00792-004-0414-3. [DOI] [PubMed] [Google Scholar]
- John F, Wyeth S. The Effects of Acids, Alkalies, and Sugars on the Growth and Indole Formation of Bacillus coli. Biochem J. 1919;13:10–24. doi: 10.1042/bj0130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja VK, Snyder OP, Jr., Marmer BS. Thermal destruction of Escherichia coli O157:H7 in beef and chicken: determination of D- and z-values. Int J Food Microbiol. 1997;35:231–237. doi: 10.1016/s0168-1605(96)01237-8. [DOI] [PubMed] [Google Scholar]
- Kamath AV, Vaidyanathan CS. New pathway for the biodegradation of indole in Aspergillus niger. Appl Environ Microbiol. 1990;56:275–280. doi: 10.1128/aem.56.1.275-280.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Sato K, Shibayama K, Horii T, Iimuma Y, Arakawa Y, Ohta M. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol Lett. 1999;179:345–352. doi: 10.1111/j.1574-6968.1999.tb08748.x. [DOI] [PubMed] [Google Scholar]
- Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- Khodursky AB, Peter BJ, Cozzarelli NR, Botstein D, Brown PO, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Youn CH, Ahn JM, Gu MB. Screening of target-specific stress-responsive genes for the development of cell-based biosensors using a DNA microarray. Anal Chem. 2005;77:8020–8026. doi: 10.1021/ac0514218. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Hirakawa H, Hirata T, Nishino K, Yamaguchi A. Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol. 2006;188:5693–5703. doi: 10.1128/JB.00217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczyńska-Wiśnik D, Matuszewska E, Laskowska E. E. coli heat shock proteins, IbpA and IbpB, affect biofilm formation by influencing the level of extracellular indole. Microbiology. 2009 doi: 10.1099/mic.0.032334-0. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Lee J. Indole as an Intercellular Signal in Microbial Community. FEMS Microbiol rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007a;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol. 2007b;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Attila C, Cirillo SLG, Cirillo JD, Wood TK. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microbial Biotech. 2009;2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Zhang XS, Hegde M, Bentley WE, Jayaraman A, Wood TK. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- Li Y, Cole K, Altman S. The effect of a single, temperature-sensitive mutation on global gene expression in Escherichia coli. RNA. 2003;9:518–532. doi: 10.1261/rna.2198203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay A, Ahmer BMM. Effect of sdiA on biosensors of N-acylhomoserine lactones. J Bacteriol. 2005;187:5054–5058. doi: 10.1128/JB.187.14.5054-5058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Durfee T, Cabrera JE, Zhao K, Jin DJ, Blattner FR. Global transcriptional programs reveal a carbon source foraging strategy by Escherichia coli. J Biol Chem. 2005;280:15921–15927. doi: 10.1074/jbc.M414050200. [DOI] [PubMed] [Google Scholar]
- Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. Indole acts as an Extracellular Cue Regulating Gene Expression in Vibrio cholerae. J Bacteriol. 2009;191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton WA, Snell EE. Formation and Interrelationships of Tryptophanase and Tryptophan Synthetases in Escherichia coli. J Bacteriol. 1965;89:355–364. doi: 10.1128/jb.89.2.355-364.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido E, Yamaguchi A, Nishino K. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem. 2008;283:24245–24253. doi: 10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patridge EV, Ferry JG. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J Bacteriol. 2006;188:3498–3506. doi: 10.1128/JB.188.10.3498-3506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittard AJ. Biosynthesis of the Aromatic Amino Acids: The Tryptophan Pathway. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington DC, USA: 1996. pp. 458–484. [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Smith T. A Modification of the Method for Determining the Production of Indol by Bacteria. J Exp Med. 1897;2:543–547. doi: 10.1084/jem.2.5.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancik LM, Stancik DM, Schmidt B, Barnhart DM, Yoncheva YN, Slonczewski JL. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J Bacteriol. 2002;184:4246–4258. doi: 10.1128/JB.184.15.4246-4258.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- Wang D, Ding X, Rather PN. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol. 2001;183:4210–4216. doi: 10.1128/JB.183.14.4210-4216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C, Horn V, Gollnick P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J Bacteriol. 1991;173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Martinez-Yamout MA, Dickerson TJ, Brogan AP, Wright PE, Dyson HJ. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol. 2006;355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Yohannes E, Barnhart DM, Slonczewski JL. pH-dependent catabolic protein expression during anaerobic growth of Escherichia coli K-12. J Bacteriol. 2004;186:192–199. doi: 10.1128/JB.186.1.192-199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]