Abstract
Transcription of the P1 promoter of the Escherichia coli proP gene, which encodes a transporter of osmoprotectants, is strongly induced by a shift to hyperosmotic media. Unlike most other osmotically regulated promoters, the induction occurs for a brief period of time, corresponding to the replacement of intracellular K+ glutamate with osmoprotecting compounds. This burst of proP transcription is correlated with the osmolarity-dependent binding of the cAMP receptor protein CRP to a site within the proP P1 promoter. We show that CRP–cAMP functions as an osmotically sensitive repressor of proP P1 transcription in vitro. Binding of CRP to the proP promoter in vivo is transiently destabilized after a hyperosmotic shift with kinetics that correspond to the derepression of transcription, whereas Fis and Lac repressor binding is not osmotically sensitive. Similar osmotic regulation of proP P1 transcription by the CRP* mutant implies that binding of cAMP is not responsible for the unusual osmotic sensitivity of CRP activity. Osmotic regulation of CRP activity is not limited to proP. Activation of the lac promoter by CRP is also transiently inhibited after an osmotic upshift, as is the binding of CRP to the galΔ4 P1 promoter. These findings suggest that CRP functions in certain contexts to regulate gene expression in response to osmotic changes, in addition to its role in catabolite control.
Keywords: CRP–cAMP, osmotic gene regulation, protein–DNA interactions, ProP transporter, lac promoter
The majority of genes in Escherichia coli encode products that serve to protect cells from various environmental stresses. One such regulon responds to changes in medium osmolarity. When cells are subjected to an osmotic upshift, the intracellular concentration of K+ immediately increases by active transport, followed by a rapid accumulation of glutamate as the primary counter ion by de novo synthesis (Measures 1975; Epstein 1986; Larsen et al. 1987; Dinnbier et al. 1988; Cayley et al. 1991; McLaggan et al. 1994). Intracellular K+ in E. coli can approach concentrations as high as 1 m shortly after a hyperosmotic shift (Richey et al. 1987; Dinnbier et al. 1988). After a relatively short period of time, more biologically compatible osmoprotecting compounds such as glycine betaine (N,N,N-trimethylglycine), proline, and trehalose replace K+ glutamate. Through this process E. coli cells maintain a relatively constant turgor pressure on exposure to hyperosmotic conditions. Similar response mechanisms occur in many other organisms including a variety of bacteria, plants, and animals (Yancey et al. 1982; Galinski 1995; Kempf and Bremer 1998; Wood 1999).
During the course of the osmotic response in E. coli, a set of genes are specifically induced (Csonka and Epstein 1996). One gene whose transcription is strongly induced and has an important role in mediating protection to osmotic upshifts is proU (Lucht and Bremer 1994; Gowrishankar and Manna 1996). ProU is a high-affinity transporter for proline, glycine betaine, and other osmoprotecting compounds and is a member of the ABC superfamily of transporters. ProP is a second, lower-affinity transporter of a similar set of compounds, which has recently been shown to be important for colonization of the urinary tract by uropathogenic E. coli (Cairney et al. 1985; Gowrishankar 1986; Grothe et al. 1986; May et al. 1986; Culham et al. 1998). ProP is a multiple-spanning transmembrane protein that is a member of a superfamily of transporters and catalyzes a broad-specificity osmoprotectant-proton symport reaction (Culham et al. 1993; Farwick et al. 1995). Earlier studies concluded that ProP was primarily regulated by post-translational mechanisms as its activity is strongly activated upon osmotic upshift (Dunlap and Csonka 1985; Milner et al. 1988). This stimulation of transport activity can be reproduced using purified ProP reconstituted in proteoliposomes (Racher et al. 1999).
Recent studies have shown that ProP synthesis is also highly regulated (Mellies et al. 1995; Xu and Johnson 1995; see also Cairney et al. 1985; Gowrishankar 1986; Jovanovich et al. 1988). proP is transcribed from two primary promoters, P1 and P2 (Fig. 1). We show here that the P1 promoter is strongly, but only transiently, induced by a shift to high-osmolarity media. The proP P2 promoter is transiently activated as cells growing in rich medium begin to enter stationary phase. This narrow window of expression as a function of growth phase is regulated by the combined activities of two transcription factors: the RpoS ς factor, whose levels increase as cells enter stationary phase, and Fis, whose levels decrease as cells enter stationary phase. The presence of ProP enables cells in stationary phase to rapidly respond to an osmotic shock.
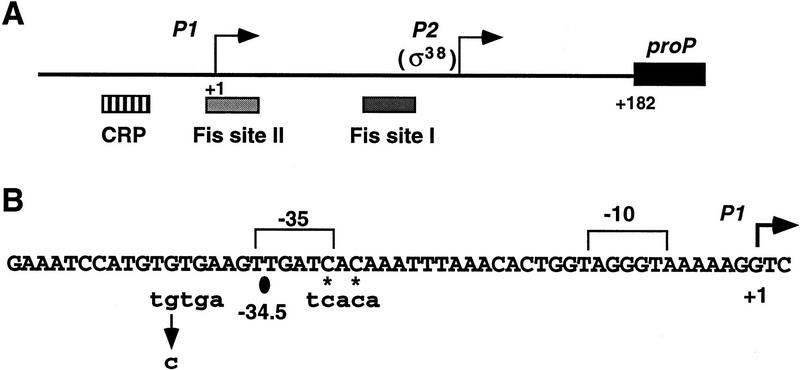
Figure 1.
The proP control region. (A) Schematic representation of the proP promoter region depicting the two promoters. P1 promoter activity is controlled by medium osmolarity and negatively regulated by CRP (Culham et al. 1993; Mellies et al. 1995, this paper). P2 promoter activity requires ς38 plus Fis binding at site I and is consequently activated in late exponential growth (Xu and Johnson 1995, 1997a). The weaker Fis-binding site II has a small negative effect on transcription from P1. (B) Sequence of the proP P1 promoter with the match to the CRP consensus noted. The G-to-C mutation strongly reduces CRP binding and leads to constitutive expression of the P1 promoter. The asterisks denote the positions of the guanines on the bottom strand that are protected from DMS reactivity by CRP–cAMP (Xu and Johnson 1997b).
A variety of mechanisms appear to be operating to control gene expression in enteric bacteria in response to osmotic upshifts, though precisely how increases in medium osmolarity are transduced to selective changes in gene expression are not known. One mechanism involves a two-component regulatory system in which the osmotic sensor EnvZ controls the phosphorylation state of the transcriptional regulator OmpR (Pratt and Silhavy 1995). A second general mechanism of osmotic control is via RpoS. RpoS levels increase primarily by post-transcriptional mechanisms upon a shift to hyperosmotic conditions, which lead to increased transcription of RpoS-dependent promoters (Hengge-Aronis et al. 1993; Lange and Hengge-Aronis 1994). Osmotic induction of the proP P2 promoter is probably due to increased RpoS levels (Xu and Johnson 1995). Finally, proU synthesis is derepressed by an osmotic upshift in part by release of the abundant nucleoid-associated protein H-NS from an extended regulatory sequence located downstream from its promoter (Lucht and Bremer 1994; Gowrishankar and Manna 1996; Jordi et al. 1997).
In this paper, we report that a sequence-specific binding protein, the cAMP receptor protein (CRP), whose primary function is to control transcription as a function of catabolite conditions (de Crombrugghe et al. 1984; Kolb et al. 1993), can also function as an osmoregulator of gene expression. At the proP P1 promoter, CRP is functioning as an osmotically sensitive repressor, and at the lac promoter, CRP is functioning as an osmotically sensitive activator. This newly discovered activity for CRP may have broad regulatory implications, given the large number of genes under its control.
Results
Transient osmotic induction of transcription from the proP P1 promoter
A modest induction of the proP P1 promoter by increases in medium osmolarity has been reported (Mellies et al. 1995). We have further investigated the osmotic regulation of P1 transcription by following the kinetics of transcription after different levels of hyperosmotic shock. As elaborated below, we find that transcription from the P1 promoter is undetectable in cells growing in a low-osmolarity medium but that the promoter is rapidly and strongly induced after a hyperosmotic shift. However, the promoter is only induced for a short period of time as transcription decreases to a low level after the initial burst.
Activation of proP P1 transcription as a function of hyperosmotic shifts was initially assayed using a proP–lacZ gene fusion on a λ prophage. A fis mutant strain was used to abolish expression from the downstream P2 promoter because P2 promoter activity is dependent on Fis (Xu and Johnson 1995). Similar results were obtained in an rpoS instead of a fis mutant background, which also eliminates P2 expression (data not shown; Mellies et al. 1995; Xu and Johnson 1995). NaCl was added at different concentrations to cells growing in LBN [Luria broth (LB) without added NaCl], and cell growth and β-galactosidase activity were measured at various times. As shown in Figure 2A, ProP–LacZ activity increases sharply after the addition of 0.3–0.5 m NaCl. However, there is little increase in overall activity after 30 min, implying that nascent synthesis is shut off. After addition of 0.5 m NaCl, cell growth lags for ∼30 min and then resumes. Cell growth is further retarded at 0.6 m NaCl with a corresponding lag in proP–lacZ induction. Upon addition of 0.7 m NaCl, cell growth ceases and only a slow induction is observed over the course of several hours. These experiments indicate that maximum induction of the P1 promoter in cells growing in complex media occurs after addition of 0.5–0.6 m NaCl and corresponds to a 500-fold increase in LacZ activity after 30 min.
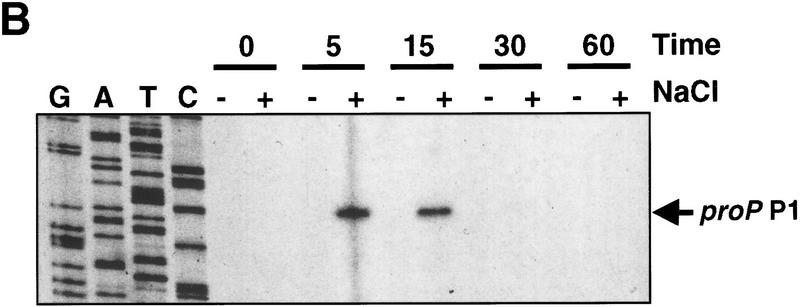
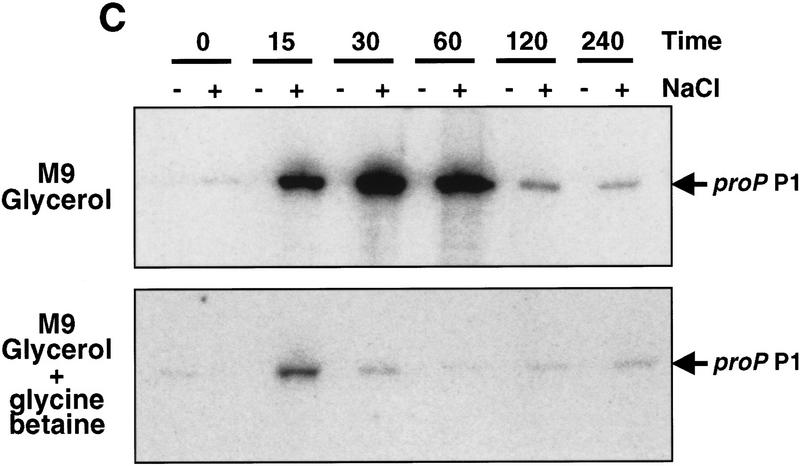
Figure 2.
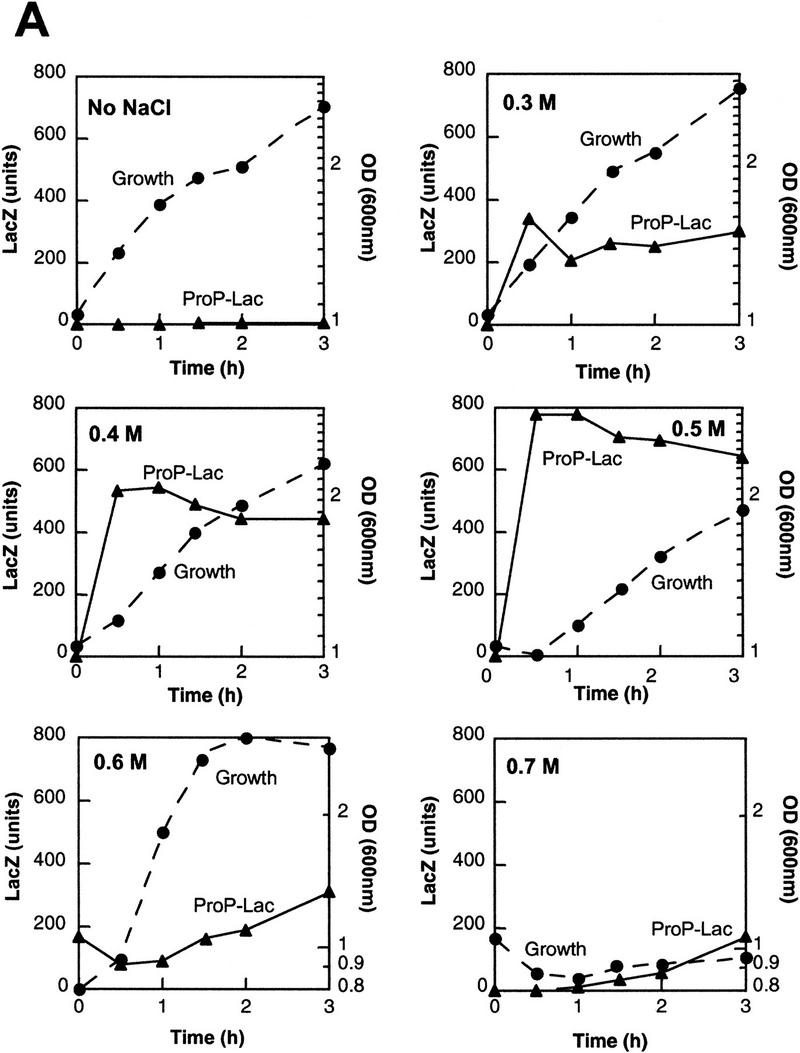
Osmotic induction of proP P1. (A) β-Galactosidase activities programmed by a proP–lacZ protein fusion in rich media during different levels of osmotic induction. RJ4373 was grown in LBN, and NaCl was added to a final concentration of 0.3–0.7 m as shown. The absence of Fis in RJ4373 insures that transcription initiates from the P1 promoter only. β-Galactosidase activities (▴) and cell growth (OD600, ●) were measured at the indicated times after addition of NaCl. β-Galactosidase activities are expressed as Miller units without normalization to cell densities (units per ml culture) to reflect synthesis rates. (B) Primer extension analysis of proP P1 RNA after addition of 0.5 m NaCl to CAG4000 growing in LBN. Times (min) after addition of NaCl (+) or LBN (−) are given at top. The sequence ladder was generated using the same primer used for the primer extension of the RNA. (C) Osmotic induction of proP P1 in minimal media. One-tenth volume of M9 plus glycerol containing 4 m or no NaCl was added to cultures of CAG4000 growing in M9 plus glycerol with or without 1 mm glycine betaine. RNA was extracted from cells collected at the indicated times after salt addition and subjected to primer extension as in B. The two panels represent images taken from one gel at the same exposure and thus are directly comparable.
To more directly measure the transient nature of osmotic induction of proP P1, RNA was isolated from cells growing in LBN after addition of 0.5 m NaCl and subjected to primer extension (Fig. 2B). RNA initiated at proP P1 was undetectable prior to osmotic induction but was maximal just 5 min after upshift. proP mRNA remained high at 15 min but was barely detectable after 30 min. Levels of proP P1 initiated RNA 5 min after salt addition were >100-fold over the noninduced control.
Osmotic induction of proP P1 was also measured in M9 plus glycerol medium in the presence or absence of glycine betaine. Optimal induction of the proP–lacZ fusion was observed after addition of 0.4 m NaCl (data not shown). Figure 2C shows a time course of RNA initiated from the proP P1 promoter after addition of 0.4 m NaCl. RNA levels increased about 400-fold 30 min after salt addition and then declined to a low level. When glycine betaine was added together with the NaCl, transcription reached only 6% of the maximal level obtained without the osmoprotectant and declined after 15 min. A decrease in osmotic induction of proP by glycine betaine has also been noted by Mellies et al. (1995). Thus, the availability of an osmoprotecting compound such as glycine betaine in the media results in the rapid shut off of proP P1 transcription and can therefore explain the transient nature of proP P1 transcription observed by cells growing in LB after osmotic induction. The shut-off of proP P1 transcription seen in the synthetic media is probably due to de novo synthesis of osmoprotecting compounds.
CRP–cAMP is an osmotically sensitive repressor of the proP P1 promoter in vitro
We previously showed that CRP–cAMP binds to a site within the proP P1 promoter region centered at −34.5 and functions as a repressor of P1 transcription both in vivo and in vitro (Xu and Johnson 1997b). Loss of CRP–cAMP binding either by a Δcrp or Δcya mutation or by a point mutation within the CRP-binding site led to constitutive expression of proP. In the following experiment, we wished to determine whether CRP–cAMP could function as an osmotically-sensitive repressor in vitro, which would provide evidence that CRP–cAMP binding directly mediates the osmotic control of the P1 promoter. In vitro transcription on supercoiled plasmid DNA was performed in the presence and absence of CRP–cAMP at different K+ glutamate concentrations. As shown in Figure 3A, CRP–cAMP was an effective repressor at 0.1 m and 0.2 m K+ glutamate. At 0.3 m K+ glutamate a small amount of P1 transcription was detected in the presence of CRP–cAMP, and above 0.4 m K+ glutamate CRP–cAMP was no longer an effective repressor. Transcription of P1 remained high with up to 0.6 m K+ glutamate in the reaction; RNA synthesis at 0.6 m K+ glutamate was 40% greater than at 0.1 m K+ glutamate in the absence of CRP–cAMP. Transcription of the plasmid-encoded rnaI gene was unaffected by CRP–cAMP, as expected, and decreased somewhat at the higher salt concentrations.
Figure 3.
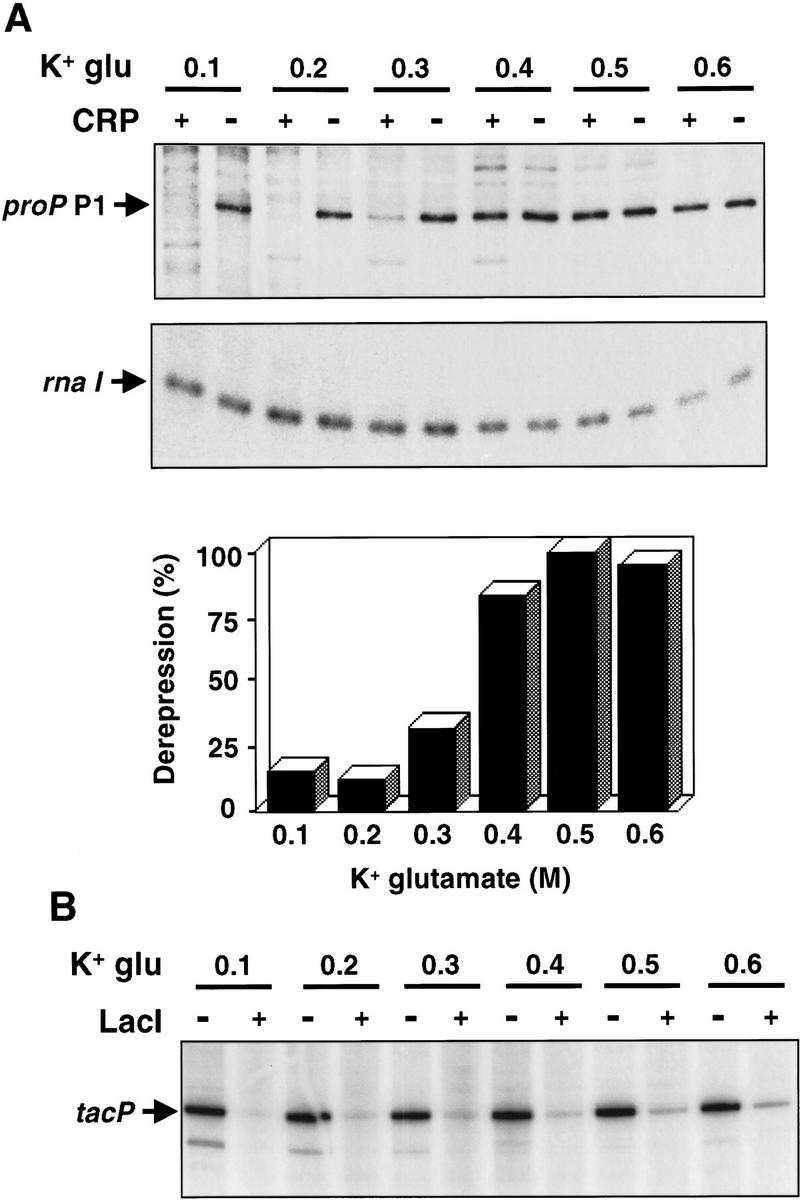
In vitro repression of transcription by CRP and Lac repressor. (A) In vitro single-round transcription reactions were performed using the proP substrate pRJ4069 in the presence (+) or absence (−) of CRP–cAMP and in the presence of 0.1–0.6 m K+ glutamate as denoted. The portions of the gel containing the P1 transcript and the vector rrn1 transcript are shown. The bar graph depicts the levels of the proP P1 transcript synthesized in the presence versus the absence of CRP–cAMP at each K+ glutamate concentration from the data shown above. (B) A similar set of reactions were performed using the substrate pKK223-3 containing the tac promoter in the presence (+) or absence (−) of Lac repressor.
For comparison, in vitro transcription reactions were performed on the tac promoter in the presence or absence of the Lac repressor under different K+ glutamate concentrations. In contrast to CRP–cAMP, the Lac repressor functioned effectively at K+ glutamate concentrations up to 0.6 m (Fig. 3B); transcript levels obtained with 0.1 or 0.6 m K+ glutamate in the presence of repressor were ≤10% of the no-repressor control. The tac promoter also remained active at high K+ glutamate concentrations.
In vivo binding of CRP–cAMP to the proP P1 promoter is osmotically sensitive
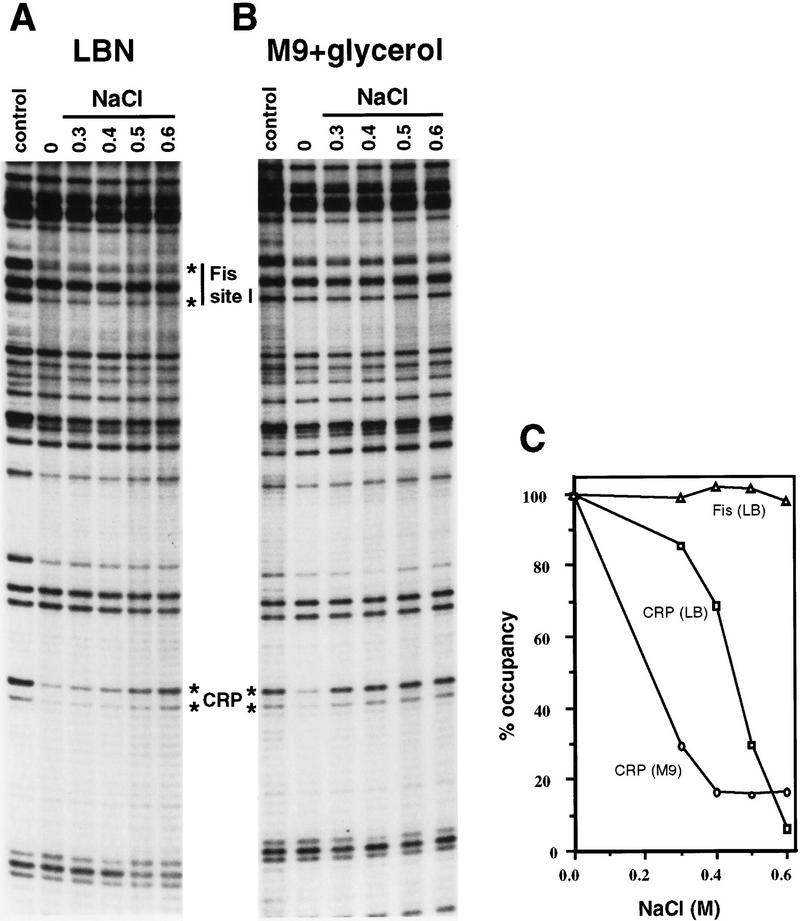
For CRP–cAMP to function as an osmoregulator of proP transcription in vivo, its binding to the P1 promoter would be expected to be osmotically sensitive. The binding of CRP–cAMP after an osmotic upshift was measured by dimethylsulfate (DMS) footprinting in growing cells where molecular-crowding forces caused by the loss of free water are operating to stabilize protein–DNA interactions (Record et al. 1998a,b). CRP protects the guanines at −28 and −30 (weakly) on the bottom strand of the P1 promoter (Xu and Johnson 1997b). Figure 4A shows the protection patterns of these guanines 15 min after the addition of varying amounts of NaCl to cells growing in LBN. The CRP site is well protected from DMS modification in the unshocked cells (0 NaCl) as compared with the control that was treated with DMS after DNA isolation (control). After addition of 0.3 m NaCl, there was a small decrease in CRP–cAMP binding, and occupancy of the CRP site became increasingly reduced with increasing NaCl such that at 0.5 m NaCl there was almost a 70% loss of binding (Fig. 4C). After addition of 0.6 m NaCl, essentially no CRP–cAMP binding was detected. Osmotic shock had no effect on the methylation patterns of a plasmid containing a point mutation at −41 that abolishes CRP binding (Xu and Johnson 1997b). The −28 and −30 guanines were equally methylated in either shocked or unshocked conditions (data not shown), confirming that the effects seen with the wild-type sequence were mediated by CRP–cAMP binding.
Figure 4.
In vivo binding of CRP after addition of different concentrations of NaCl. (A) DMS footprinting reactions on cells growing in LBN and subjected to increasing concentrations of NaCl as denoted for each lane. (B) DMS footprinting reactions on cells growing in M9 plus glycerol and subjected to increasing concentrations of NaCl as in A. The cells were treated with DMS 15 min (A) or 30 min (B) after addition of NaCl. Media without NaCl was added to the cells labeled 0. The control lanes represent DMS reactions performed on plasmids after isolation and thus no cellular proteins are present. The locations of the guanines at −28 and −30 on the bottom strand that are protected from DMS modification in vitro by purified CRP or Fis protein at site I are designated with the asterisks. Fis-mediated protections in cells growing in M9 plus glycerol are weak because of the low Fis concentrations present under these growth conditions. (C) Graph depicting the percent occupancy of the CRP-binding site, as determined from the level of protection of the −28 guanine, in cells growing in LB and M9 plus glycerol. Occupancy of Fis site I is also shown from cells growing in LB after the different salt additions. The level of protection measured at 0 NaCl is defined as 100% occupancy relative to the control (0% occupancy).
CRP–cAMP binding after addition of different concentrations of NaCl to cells growing in M9 + glycerol was also determined (Fig. 4B,C). CRP–cAMP binding was found to be more sensitive to osmotic upshift in the synthetic media as compared to LB. Addition of just 0.3 m NaCl was sufficient to release about 70% of CRP. The greater sensitivity of CRP–cAMP binding to increases in medium osmolarity in minimal media correlates with the greater induction at lower osmolarity in minimal as compared to complex media.
For comparison, in vivo DNA binding of two other regulatory proteins was measured after osmotic upshift of cells growing in LBN. The Fis protein binds to a site centered 81 bp downstream of the CRP binding site in the proP regulatory region and functions as a positive activator of P2 promoter transcription. As shown in Figure 4A, Fis binding was unaffected by the same osmotic upshift that released CRP. Lac repressor binding to the primary O1 and weaker O3 secondary operators was also assayed. Repressor binding to both operators was unaffected by osmotic upshifts up to 0.6 m NaCl (Fig. 5A). Moreover, the lacL8UV5 promoter remained efficiently repressed after addition of 0.5 m NaCl under the same conditions where the proP P1 promoter was induced 500-fold (Fig. 5B, see also Richey et al. 1987).
Figure 5.
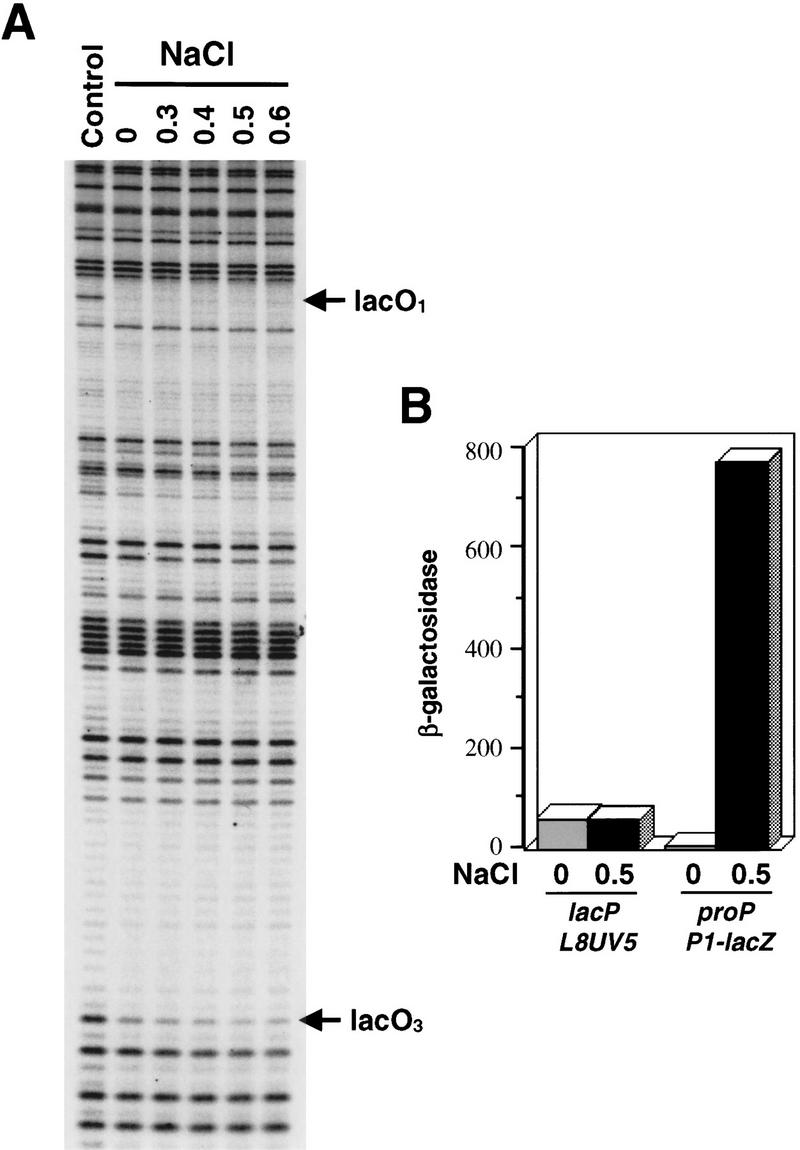
Absence of osmotic sensitivity of Lac repressor binding and activity. (A) In vivo DMS footprinting reactions on cells growing in LBN and subjected to increasing concentrations of NaCl. This experiment was performed identically to Fig. 4A except that the cells contained pRZ4004, which carries the wild-type lac promoter region. (B) Lac repressor activity after an osmotic upshift. RJ3355 (lacI+PL8UV5 Z+Y+) was grown in LBN. β-Galactosidase (Miller units) was assayed 30 min after addition of no NaCl (control) or 0.5 m NaCl. For comparison, induction of β-galactosidase by the proP–lac fusion in RJ3265 under identical conditions is shown.
Release of CRP–cAMP from the proP promoter is transient and independent of genomic context
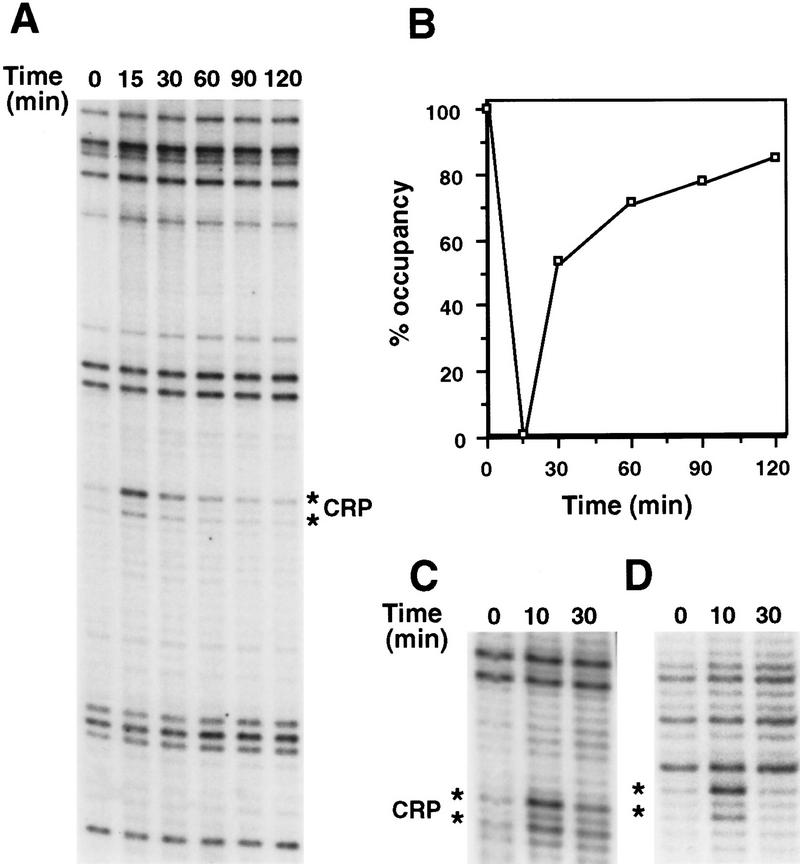
Because induction of the proP P1 promoter only occurs for a short time after cells are exposed to an osmotic upshift, we measured the in vivo occupancy of the proP CRP site as a function of time after addition of 0.5 m (data not shown) and 0.6 m (Fig. 6A,B) NaCl to cells growing in LBN. The CRP site was sensitive to DMS modification 15 min after salt addition as observed previously but became increasingly resistant to DMS modification after 30 min, indicating rebinding of CRP–cAMP. The kinetics of CRP–cAMP binding during the course of adaptation to an osmotic upshift is thus consistent with the transient nature of the transcriptional derepression at proP P1.
Figure 6.
Kinetics of DNA binding by CRP after osmotic upshift. (A) In vivo DMS footprinting of cells growing in LBN immediately prior to time = 0 or at various times after NaCl addition as denoted. The locations of the guanines that are diagnostic for CRP binding are shown by the asterisks. (B) Graph depicting the occupancy of the CRP site at different times after osmotic upshift. The level of protection immediately preceding the osmotic upshift (time 0) was set at 100% occupancy and the amount of reactivity at 15 min was set at 0% occupancy. (C) Binding to the isolated proP CRP-binding site. In vivo DMS footprinting was performed on cells carrying pRJ1662 immediately prior to time 0, 10 min, or 30 min after addition of 0.6 m NaCl. (D) Binding to the CRP site in the galP1Δ4 promoter. Cells containing pRJ1663 were treated as in C.
To address whether the genomic context of the CRP binding site within the proP P1 promoter was causing CRP–cAMP binding to be osmotically sensitive, we measured binding to a synthetically derived sequence representing the proP P1 CRP site plus the flanking 13 bp (−55 to −14 in Fig. 1). The isolated proP CRP site was efficiently bound in cells growing in LBN, but protection was lost 10 minutes after addition of 0.6 m NaCl (Fig. 6C). After 30 min, binding was largely restored. Thus, the osmotic sensitivity of CRP–cAMP binding does not require an intact overlapping RNA polymerase-binding site or some other feature of the proP sequence outside of the 42-bp cloned segment.
The osmotic sensitivity of CRP binding to DNA is not mediated by changes in cAMP binding
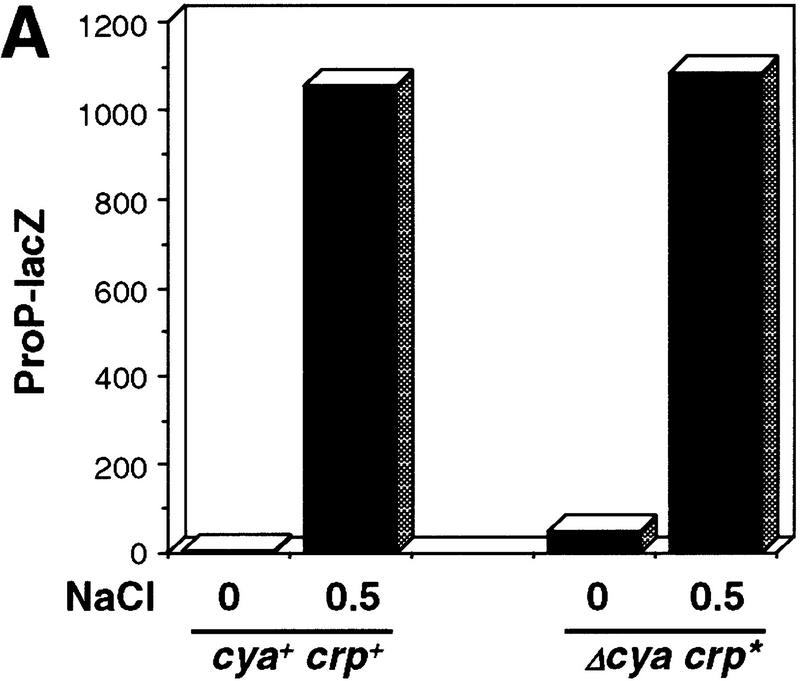
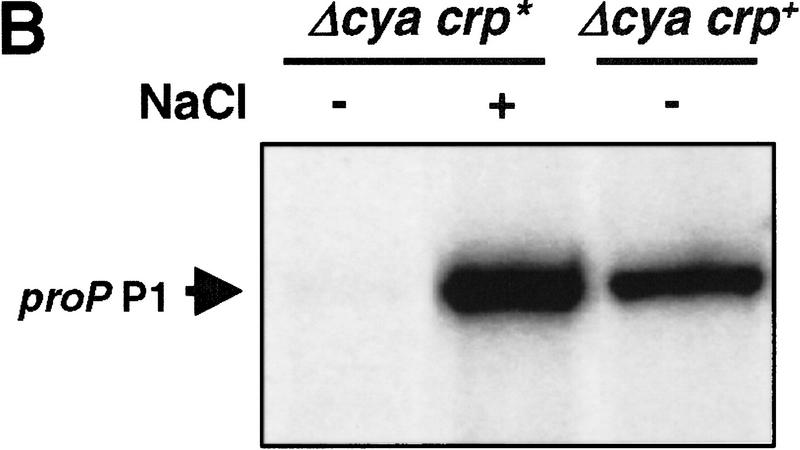
The presence of cAMP is required for repression by wild-type CRP as proP P1 transcription is constitutively expressed in a Δcya crp+ mutant (Fig. 7B; data not shown). We wondered if the osmotically sensitive DNA binding by CRP–cAMP could reflect a salt sensitivity of cAMP binding to CRP, which would thereby affect DNA binding. To determine whether cAMP binding was important for the osmotic regulation of proP P1 transcription, a CRP* mutant that could bind DNA in the absence of cAMP was assayed for osmotic control. The Δcya crp* mutant responded to an osmotic upshift in a manner similar to wild-type (cya+ crp+) as measured by either induction of proP–lacZ (Fig. 7A) or direct RNA analysis of the proP P1 message (Fig. 7B).
Figure 7.
Osmotic regulation of proP P1 by the cAMP-independent CRP* mutant. (A) Bar graph of β-galactosidase activities (Miller units) assayed immediately before or 30 min after addition of 0.5 mM NaCl to RJ3265 (cya+ crp+ proP-104–lacZ fis::str/spc) or RJ3372 (Δcya crp* proP-104–lacZ fis::str/spc) growing in LBN. (B) Primer extension assays of proP P1 mRNA. RNA was isolated from IT1131 (Δcya crp*) or IT1002 (Δcya crp+) growing in LBN (−) immediately before or 10 min after addition of 0.5 m NaCl (+). Time-course experiments showed that proP P1 mRNA in IT1131 decreased to basal levels 30 min after osmotic induction (data not shown).
CRP–cAMP activation of lacP and binding to galP1Δ4 is osmotically sensitive
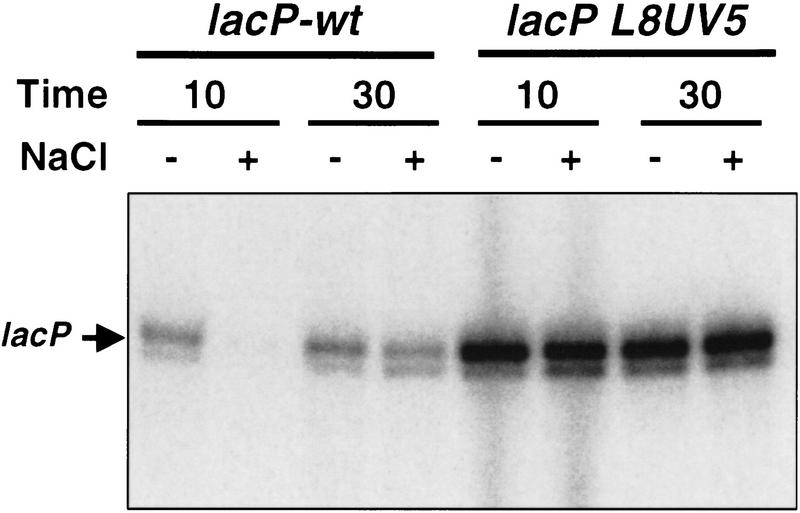
CRP is a positive and negative regulator of many different genes in E. coli. We wondered if its osmotically sensitive activity is unique for proP or may occur in other genetic contexts. Osmotic sensitivity of CRP–cAMP-mediated activation of the lac promoter was assessed by comparing activated transcription of the CRP-dependent wild-type lac promoter with the CRP-independent lacL8UV5 promoter after addition of 0.5 m NaCl to cells growing in LBN. Transcription from the lacP+ promoter was reduced by 95% 10 min after the osmotic upshift, whereas the lacL8UV5 promoter was unchanged (Fig. 8). After 30 min, transcription from lacP+ was restored. Thus, activation of the lac promoter by CRP–cAMP appears transiently sensitive to an osmotic upshift. We were unable to directly measure binding of CRP–cAMP to the lac promoter site by in vivo DMS footprinting (e.g., Fig. 5), probably because of its relatively weak affinity to the nonconsensus lacP site.
Figure 8.
Osmotic sensitivity of CRP-mediated activation of the lac promoter. Primer extension analysis of transcription initiated at the lac promoter by the CRP-dependent wild-type lac promoter (RJ3354) or the CRP-independent lacL8UV5 promoter (RJ3355). Cells were grown in LBN + IPTG and 0.5 m NaCl was added to a portion of the culture. RNA was extracted 10 min and 30 min after salt addition and subjected to primer extension.
CRP–cAMP binding to the consensus site located at −37.5 in the galP1Δ4 promoter (Bell et al. 1990) was assayed by in vivo DMS footprinting after an osmotic upshift. As shown in Figure 6D, CRP-mediated protection at this site was lost 10 min after addition of 0.5 m NaCl to cells growing in LBN but was restored after 30 min. Thus, binding of CRP–cAMP to the galP1Δ4 site was osmotically sensitive in a transient manner that resembles the proP site. CRP–cAMP functions to negatively regulate this promoter; however, we were only able to demonstrate a small increase in β-galactosidase activity by galP1Δ4–lacZ after addition of 0.5 m NaCl. The lack of strong derepression in this case may reflect an intrinsic sensitivity of the promoter to high osmolarity. In support of this idea, initial experiments indicate that the otherwise constitutive activity of galP1Δ in a cya mutant is transiently inhibited upon an osmotic upshift.
Discussion
The proP P1 promoter is rapidly, but only transiently, induced by osmotic upshifts (Mellies et al. 1995; this paper). In low-osmolarity medium, this promoter is strongly repressed by the CRP protein (Xu and Johnson 1997b). We show here that the osmotic regulation of proP P1 correlates with CRP–cAMP binding to DNA. In low-osmolarity medium, the crp site at proP is fully occupied, and transcription from this promoter is very low. After a hyperosmotic shift, CRP is released from this site, and the proP1 promoter is derepressed. Induction ratios as measured by direct RNA analysis or ProP–LacZ fusions exceed 100-fold and thus are in the same range as reported for the proU promoter (Csonka 1989; Booth and Higgins 1990; Lucht and Bremer 1994).
Osmotic induction of the proP P1 promoter is transient
An important difference between the proP P1 promoter and the proU or other previously characterized osmotically activated promoters is that induced transcription from proP P1 only occurs for a brief period of time. This is most clearly illustrated by direct RNA analysis where peak transcription occurs just 5 min after an upshift in LB and transcript levels decrease dramatically after 15 min. The transient nature of the response probably reflects the rapid replacement of K+ glutamate with osmoprotecting solutes such as glycine betaine (Fig. 2C) and related compounds present in complex media and the de novo synthesis of compounds such as trehalose in minimal media (Dinnbier et al. 1988; McLaggan et al. 1994; Record et al. 1998a). The kinetics of induction are consistent with K+ or possibly glutamate being the primary signal to which the promoter is responding. K+ glutamate has also been proposed to be the signal controlling osmotic induction of proU (Booth and Higgins 1990; Prince and Villarejo 1990), but the high levels of proU transcription that are maintained over a long period of time in response to a hyperosmotic shift argue that other signals must also be operating for this promoter.
CRP is an osmotically sensitive repressor of proP
We have shown previously that CRP–cAMP represses P1 transcription in vivo (Xu and Johnson 1997b) and in vitro by binding to the CRP-binding site centered at −34.5 with respect to the transcription start site. In this work we have demonstrated that CRP–cAMP can directly function as an osmotically sensitive repressor of the P1 promoter in vitro. Whereas the presence of CRP–cAMP inhibited in vitro transcription by purified Eς70 at K+ glutamate concentrations up to 0.3 m, repression was lost at ≥0.4 m K+ glutamate. By contrast, the Lac repressor effectively inhibited transcription up to 0.6 m K+ glutamate. In vitro studies with the proU promoter have demonstrated that the repressing activity of the nucleoid-associated protein H-NS, which is responsible for a large part of the osmotic control of proU transcription, is inhibited above 0.3 m K+ glutamate (Ueguchi and Mizuno 1993).
In vivo binding studies demonstrated that CRP binding to proP was unusually salt sensitive. CRP was released from its binding site immediately after an osmotic upshift but then rebounded with kinetics that approximated proP P1 transcriptional activity. On the other hand, Fis binding to a high affinity site within the proP regulatory region was unaffected. Likewise, Lac repressor remained efficiently bound to both the strong O1 and weak O3 sites in the lac regulatory region and effectively inhibited transcription after an osmotic upshift (Richey et al. 1987; this paper).
The transient reduction in the in vivo occupancy of the proP crp site with increasing medium osmolarity qualitatively correlates with the magnitude and kinetics of induction of P1 transcription observed at different levels of osmotic upshifts. For example, after a 0.4 m upshift in LB about 30% of the CRP site becomes accessible to DMS modification (Fig. 4), and proP P1 transcription is induced to 70% of its maximal value as assayed by LacZ fusions (Fig. 2). The greater apparent transcriptional derepression relative to the decrease of binding to the CRP site can be explained by considering that a competition exists between RNA polymerase and the CRP–cAMP repressor proteins at the promoter. At low K+ glutamate concentrations, CRP–cAMP is tightly bound to the promoter effectively preventing RNA polymerase binding or activity. At intermediate K+ glutamate concentrations, the binding equilibrium of CRP–cAMP is shifted such that over the course of a 2-min DMS reaction, the guanines become partially methylated. Under these conditions RNA polymerase can successfully compete with CRP to form transcriptionally active complexes. Thus, a precise quantitative correspondence between the degree of CRP occupancy measured by a 2-min DMS reaction and the extent of osmotic induction measured by ProP–LacZ fusion activity may not be expected. To test the relationship between repressor binding and promoter derepression in an unrelated system, we measured Lac repressor binding and transcriptional derepression in the presence of different concentrations of IPTG at the lacL8UV5 promoter in vitro. Purified Lac repressor was added to reaction mixes that included an end-labeled 203-bp DNA fragment containing the lacL8UV5 promoter and various amounts of IPTG. A portion was removed and subjected to DNase I footprinting to measure repressor occupancy, and RNAP plus NTPs was added to the remainder to assay transcription. In a manner similar to what was observed with CRP–cAMP at the proP P1 promoter, IPTG concentrations that led to only a partial loss of repressor binding generated near fully derepressed transcription (data not shown).
How is CRP binding to DNA regulated by osmolarity?
Increasing salt concentrations generally have a destabilizing effect on protein–DNA interactions in vitro, and this has been specifically reported for CRP [Fried and Stickle 1993; but note that chloride salts, which strongly inhibit CRP–DNA interactions (J.Xu, unpubl.; see Leirmo et al. 1987), were used in this study]. However, protein–DNA interactions in vivo have been found to be highly tolerant to osmotic upshifts (Record et al. 1998b). Indeed, we have directly shown in this paper that binding of Fis and the Lac repressor to their specific sites on DNA is not detectably altered after addition of up to 0.6 m NaCl to the media. It has been proposed that macromolecular crowding caused by a reduction of cytoplasmic volume stabilizes protein–DNA interactions (Cayley et al. 1991). A reduction of water activity, generated by the addition of neutral solutes such as sucrose or glycerol, has been shown to stabilize CRP–cAMP binding to linear fragments in vitro (Vossen et al. 1997). It is therefore surprising that CRP–cAMP binding to specific sites within the regulatory regions of proP, galP1, pmelRcon (data not shown), and by inference lacP, appears so sensitive to changes in medium osmolarity in vivo.
There are several possibilities that can be considered to account for the unique sensitivity to high intracellular K+ glutamate by CRP–cAMP. One is that osmotically induced changes in DNA structure could influence DNA binding by CRP–cAMP, as has been proposed for osmotic control of the proU system (Jordi et al. 1995). As discussed above, such changes would have to be highly specific for CRP–DNA interactions and not reflect special features of the surrounding DNA or the overall genetic context (native, λ prophage, or plasmid). Moreover, release of CRP by a hyperosmotic shift was not accompanied by a significant change in plasmid supercoiling; 10 min after addition of 0.5 m NaCl the change in the linking number was measured to be ≤−1 (data not shown). In addition, CRP binding to the proP site displayed a similar osmotic sensitivity in vivo when the DNA was relaxed (ΔLK of +10 relative to the untreated cells) by prior treatment with norfloxacin (data not shown).
The structure of the CRP–cAMP–DNA complex does not reveal any obvious feature that would cause it to be more osmotically sensitive than other protein–DNA complexes such as Fis or Lac repressor (Kaptein et al. 1990; Schultz et al. 1991; Lewis et al. 1996; Pan et al. 1996; Parkinson et al. 1996). A unique feature of CRP is that at least one molecule of cAMP must be bound within the dimer interface to promote the appropriate conformational change with the DNA-binding region to enable DNA binding. Each of the cAMP molecules are bound to the CRP dimer by a set of molecular contacts that include two direct and one indirect electrostatic interactions (McKay et al. 1982). Thus, a possible model explaining the osmotic sensitivity of DNA binding could be reduced binding of cAMP at high ionic conditions. However, the near-normal osmotic control by the cAMP-independent crp* mutant in the absence of adenylate cyclase, together with the relative insensitivity of cAMP binding to CRP measured in vitro with increasing salt concentration (Takahashi et al. 1980), argues against such a model. The normal osmotic control by the CRP* mutant also rules out the possibility that changes in cAMP levels are responsible for the osmotically sensitive binding. On the other hand, the allosteric change modulated by cAMP, which controls DNA binding by CRP may be osmotically sensitive but not in a manner influenced by the crp* mutation used. Such a model is attractive because it explains the unique osmotic sensitivity of CRP in comparison to other DNA-binding proteins.
A general role for CRP in osmoregulation?
CRP is one of the most common regulators of gene expression in enteric bacteria. Its primary regulatory function is to control transcription in response to glucose levels. The generality of the osmoregulatory activity of CRP remains to be fully determined. We have shown that osmotically sensitive regulation by CRP–cAMP is not limited to repression at the proP locus as activation of lacP is also transiently sensitive to osmotic upshifts. Binding of CRP–cAMP to the galP1Δ4 promoter is also osmotically sensitive, but this promoter is not significantly derepressed by an osmotic upshift. Thus, even though CRP–cAMP binding at a particular promoter may be affected by changes in medium osmolarity, other controlling features or the intrinsic properties of the promoter itself may mitigate a corresponding change in transcription. Although transcription of only a subset of CRP-regulated genes may be osmoresponsive, the potential for a relatively global osmoregulatory effect mediated by CRP may have important physiological consequences. It is important to emphasize that the response at different loci would be expected to occur only for a short period of time immediately after an osmotic upshift. This is a critical time for cell survival as the intracellular environment is undergoing enormous changes as the cell adapts to maintain a constant turgor pressure.
Materials and methods
Strains and plasmids
Unless otherwise noted, all in vivo experiments were performed in derivatives of CAG4000 (E. coli MG1655 ΔlacX74). RJ4373 contains a single copy prophage of λproP104Δ1–lacZ in CAG4000 fis::kan-767 (Xu and Johnson 1997b). RJ3265 is CAG4000 fis::spc/str-985 containing the proP-104 lacZ gene fusion inserted within the proP locus (Xu and Johnson 1995). RJ3354 and RJ3355 are CAG4000 with an F′proAB lacZYA episome containing the wild-type or L8UV5 lac promoter, respectively. IT1002 (W3110 Δcya::cat crp+ lacI::Tn10) and IT1131 (IT1002 crp*T28K A144T) were from H. Aiba (Tagami et al. 1995). RJ3372 is RJ3265 Δcya::cat crp*T28K A144T.
The duplex oligonucleotide with top strand sequence d[AACGAAATCCATGTGTGAAGTTGATCACAAATTTAAACACTG] representing the proP CRP-binding site was cloned into the SmaI site of pUC18 to give pRJ1661. The EcoR1–HindIII fragment containing the CRP-binding site from pRJ1661 was then subcloned into pBR322 to give pRJ1662. The galP1Δ4 promoter region was cloned into pBR322 to give pRJ1663 by transferring an EcoR1–HindIII fragment from pRW50-galP1Δ4 (Bell et al. 1990, provided by S. Busby).
β-galactosidase and in vivo RNA assays
For assays performed in rich media, overnight cultures were diluted 1:100 into LBN and grown for 3 hr at 37°C with shaking. LB containing 5 m NaCl, or no NaCl for the control, was added to give the appropriate final concentration of NaCl and incubation was continued. For assays performed in minimal media, overnight LB cultures were diluted 1:100 into M9 + 0.4% glycerol and grown until an OD600 of 1.2 was reached prior to addition of 4 m NaCl in M9 + glycerol to the desired final concentration. β-Galactosidase assays were performed on duplicate cultures and the average was determined (Miller 1992). RNA was isolated from 10 ml of culture by the hot phenol method (Case et al. 1988). An oligonucleotide complementary to +173 to +196 of P1-initiated RNA was used for primer extension of proP mRNA and the lacZ universal primer was used for analysis of lacP mRNA. Ten micrograms of total RNA was used for primer extension in all cases.
In vivo DMS footprinting
DMS reactions were performed essentially as described (Sasse-Dwight and Gralla 1990; Xu and Johnson 1997b) on CAG4000 containing the following plasmids: pRJ4039 for analysis of the proP region (Xu and Johnson 1995), pRJ1662 for analysis of the isolated proP crp site, and pRJ1663 for analysis of the galP1Δ4 promoter region. Lac repressor binding to the lac operators was probed in RJ7065 (CAG4000 pRZ4004 F′proAB+ lacIsqZu118 fzz::Tn5). pRZ4004 is a derivative of pBR322 that contains the HaeIII 203-bp lacP fragment (W. Reznikoff, University of Wisconsin, Madison). Ten ml of culture grown as described above were incubated with 5 mm DMS for 3 min at 37°C. The culture was then diluted into 2 volumes of ice-cold LB + 1 m β-mercaptoethanol and the cells were collected by centrifugation. The cells were washed once, and plasmid DNA was isolated using a Qiagen kit. The DNA was treated with piperidine and purified by a G50 spin column prior to primer extension. The sequences of the primers used were as follows: proP P1 nucleotides −103 to −85 for detection of the proP crp site in pRJ4039, lacZ nucleotides −117 to −99 for detection of Lac repressor binding in pRZ4004, and pBR322 nucleotides 4333–4351 for detection of CRP binding in pRJ1662 (proP) and pRJ1663 (galP). Primer extension products were electrophoresed on sequencing gels and bands indicative of protein binding, along with nearby reference bands whose levels of protection did not change, were quantitated using a PhosphorImager. The values obtained from the reference bands were used to adjust for minor loading variations.
In vitro transcription
In vitro single-round transcription reactions were performed as described previously (Xu and Johnson 1997a). pRJ4069 was used for the proP template and contained P1 sequences from −113 to +196 upstream of the rrnB terminator region (Xu and Johnson 1997b). pKK223-3 (Pharmacia) was used for tacP transcription. Supercoiled plasmid substrates were prepared by 2 CsCl-ethidium bromide bandings. Plasmid DNA (0.1 pmole) was incubated with 1 pmole of CRP plus 100 μm cAMP or 1 pmole of Lac repressor in 50 mm Tris-HCl (pH 8.0) buffer containing 0.1–0.6 m K+ glutamate, 3 mm magnesium acetate, 0.1 mm EDTA, 1 mm DTT, and 50 μg/ml BSA per ml for 10 min at room temperature. RNA polymerase (0.5 pmole) was added, and the reaction incubated at 37°C for 15 min followed by the addition of NTPs, including 10 μCi of [α-32P]UTP and 60 μg of heparin per ml in a final volume of 50 μl. The elongation reaction was terminated after 6 min and prepared for electrophoresis on 8% polyacrylamide-7 m urea gels as described (Xu and Johnson 1997a). Lac repressor protein was purified by M. Haykinson of this laboratory from a strain containing pUC18 with the lacIq gene (pRJ1655). RNA polymerase was purified by the method of (Burgess and Jendrisak 1975) from RJ4128 (fis::spc-985 rpoS::Tn10). CRP was a gift from J. Krakow (Hunter College, New York, NY).
Acknowledgments
We thank J. Krakow for purified CRP, S. Busby and H. Aiba for strains, and Sarah McLeod for advice throughout the work. This work was supported by grant GM38509 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rcjohnson@mednet.ucla.edu; FAX (310) 206-5272.
References
- Bell A, Gaston K, Williams R, Chapman K, Kolb A, Buc H, Minchin S, Williams J, Busby S. Mutations that alter the ability of the Escherichia coli cyclic AMP receptor protein to activate transcription. Nucleic Acids Res. 1990;18:7243–7250. doi: 10.1093/nar/18.24.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR, Higgins CF. Enteric bacteria and osmotic stress: Intracellular potassium glutamate as a secondary signal of osmotic stress? FEMS Microbiol Rev. 1990;6:239–246. doi: 10.1111/j.1574-6968.1990.tb04097.x. [DOI] [PubMed] [Google Scholar]
- Burgess RR, Jendrisak JJ. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Cairney J, Booth IR, Higgins CF. Salmonella typhimurium proP gene encodes a transport system for the osmoprotectant betaine. J Bacteriol. 1985;164:1218–1223. doi: 10.1128/jb.164.3.1218-1223.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case CC, Roels SM, Gonzalez JE, Simons EL, Simons RW. Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. Gene. 1988;72:219–236. doi: 10.1016/0378-1119(88)90147-3. [DOI] [PubMed] [Google Scholar]
- Cayley S, Lewis BA, Guttman HJ, Record MT., Jr Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J Mol Biol. 1991;222:281–300. doi: 10.1016/0022-2836(91)90212-o. [DOI] [PubMed] [Google Scholar]
- Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN, Epstein W. Osmoregulation. In: Neidhardt FC, et al., editors. Eschericia coli and Salmonella: Cellular and molecular biology. Washington, D.C.: ASM; 1996. pp. 1210–1223. [Google Scholar]
- Culham DE, Lasby B, Marangoni AG, Milner JL, Steer BA, van Nues RW, Wood JM. Isolation and sequencing of Escherichia coli gene proP reveals unusual structural features of the osmoregulatory proline/betaine transporter, ProP. J Mol Biol. 1993;229:268–276. doi: 10.1006/jmbi.1993.1030. [DOI] [PubMed] [Google Scholar]
- Culham DE, Dalgado C, Gyles CL, Mamelak D, MacLellan S, Wood JM. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology. 1998;144:91–102. doi: 10.1099/00221287-144-1-91. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B, Busby S, Buc H. Cyclic AMP receptor protein: Role in transcription activation. Science. 1984;224:831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]
- Dinnbier U, Limpinsel E, Schmid R, Bakker EP. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of growing cells of Escherichia coli K-12 to elevated sodium chloride concentrations. Arch Microbiol. 1988;150:348–357. doi: 10.1007/BF00408306. [DOI] [PubMed] [Google Scholar]
- Dunlap VJ, Csonka LN. Osmotic regulation of L-proline transport in Salmonella typhimurium. J Bacteriol. 1985;163:296–304. doi: 10.1128/jb.163.1.296-304.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein W. Osmoregulation by potassium transport in Escherichia coli. FEMS Microbiol Rev. 1986;39:73–78. [Google Scholar]
- Farwick M, Siewe RM, Kramer R. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J Bacteriol. 1995;177:4690–4695. doi: 10.1128/jb.177.16.4690-4695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried MG, Stickle DF. Ion-exchange reactions of proteins during DNA binding. Eur J Biochem. 1993;218:469–475. doi: 10.1111/j.1432-1033.1993.tb18398.x. [DOI] [PubMed] [Google Scholar]
- Galinski EA. Osmoadaptation in bacteria. Adv Microb Physiol. 1995;37:272–328. [PubMed] [Google Scholar]
- Gowrishankar J. ProP-mediated proline transport also plays a role in Escherichia coli osmoregulation. J Bacteriol. 1986;166:331–333. doi: 10.1128/jb.166.1.331-333.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar J, Manna D. How is osmotic regulation of transcription of the Escherichia coli proU operon achieved? A review and a model. Genetica. 1996;97:363–378. doi: 10.1007/BF00055322. [DOI] [PubMed] [Google Scholar]
- Grothe S, Krogsrud RL, McClellan DJ, Milner JL, Wood JM. Proline transport and osmotic stress response in Escherichia coli K-12. J Bacteriol. 1986;166:253–259. doi: 10.1128/jb.166.1.253-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R, Lange R, Henneberg N, Fischer D. Osmotic regulation of RpoS-dependent genes in Escherichia coli. J Bacteriol. 1993;175:259–265. doi: 10.1128/jb.175.1.259-265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordi BJ, Owen-Hughes TA, Hulton CS, Higgins CF. DNA twist, flexibility and transcription of the osmoregulated proU promoter of Salmonella typhimurium. EMBO J. 1995;14:5690–5700. doi: 10.1002/j.1460-2075.1995.tb00256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordi BJ, Fielder AE, Burns CM, Hinton JC, Dover N, Ussery DW, Higgins CF. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J Biol Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- Jovanovich SB, Martinell M, Record MT, Jr, Burgess RR. Rapid response to osmotic upshift by osmoregulated genes in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988;170:534–539. doi: 10.1128/jb.170.2.534-539.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein R, Lamerichs RM, Boelens R, Rullmann JA. Two-dimensional NMR study of a protein-DNA complex. Lac repressor headpiece-operator interaction. Biochem Pharmacol. 1990;40:89–96. doi: 10.1016/0006-2952(90)90183-l. [DOI] [PubMed] [Google Scholar]
- Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. The cellular concentration of the ς S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes & Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- Larsen PI, Sydnes LK, Landfald B, Strom AR. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: Betaines, glutamic acid, and trehalose. Arch Microbiol. 1987;147:1–7. doi: 10.1007/BF00492896. [DOI] [PubMed] [Google Scholar]
- Leirmo S, Harrison C, Cayley DS, Burgess RR, Record MT., Jr Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- Lucht JM, Bremer E. Adaptation of Escherichia coli to high osmolarity environments: Osmoregulation of the high-affinity glycine betaine transport system proU. FEMS Microbiol Rev. 1994;14:3–20. doi: 10.1111/j.1574-6976.1994.tb00067.x. [DOI] [PubMed] [Google Scholar]
- May G, Faatz E, Villarejo M, Bremer E. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol Gen Genet. 1986;205:225–233. doi: 10.1007/BF00430432. [DOI] [PubMed] [Google Scholar]
- McKay DB, Weber IT, Steitz TA. Structure of catabolite gene activator protein at 2.9-Å resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J Biol Chem. 1982;257:9518–9524. [PubMed] [Google Scholar]
- McLaggan D, Naprstek J, Buurman ET, Epstein W. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J Biol Chem. 1994;269:1911–1917. [PubMed] [Google Scholar]
- Measures JC. Role of amino acids in osmoregulation of non-halophilic bacteria. Nature. 1975;257:398–400. doi: 10.1038/257398a0. [DOI] [PubMed] [Google Scholar]
- Mellies J, Wise A, Villarejo M. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J Bacteriol. 1995;177:144–151. doi: 10.1128/jb.177.1.144-151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Milner JL, Grothe S, Wood JM. Proline porter II is activated by a hyperosmotic shift in both whole cells and membrane vesicles of Escherichia coli K12. J Biol Chem. 1988;263:14900–14905. [PubMed] [Google Scholar]
- Pan CQ, Finkel SE, Cramton SE, Feng JA, Sigman DS, Johnson RC. Variable structures of Fis-DNA complexes determined by flanking DNA-protein contacts. J Mol Biol. 1996;264:675–695. doi: 10.1006/jmbi.1996.0669. [DOI] [PubMed] [Google Scholar]
- Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RE, Berman HM. Structure of the CAP-DNA complex at 2.5 angstroms resolution: A complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Silhavy TJ. Porin regulation of Escherichia coli. In: Hoch JA, Silhavy TJ, editors. Two component signal transduction. Washington, D.C.: ASM; 1995. pp. 105–127. [Google Scholar]
- Prince WS, Villarejo MR. Osmotic control of proU transcription is mediated through direct action of potassium glutamate on the transcription complex. J Biol Chem. 1990;265:17673–17679. [PubMed] [Google Scholar]
- Racher KI, Voegele RT, Marshall EV, Culham DE, Wood JM, Jung H, Bacon M, Cairns MT, Ferguson SM, Liang WJ, et al. Purification and reconstitution of an osmosensor: Transporter ProP of Escherichia coli senses and responds to osmotic shifts. Biochemistry. 1999;38:1676–1684. doi: 10.1021/bi981279n. [DOI] [PubMed] [Google Scholar]
- Record MT, Jr, Courtenay ES, Cayley DS, Guttman HJ. Responses of E. coli to osmotic stress: Large changes in amounts of cytoplasmic solutes and water. Trends Biochem Sci. 1998a;23:143–148. doi: 10.1016/s0968-0004(98)01196-7. [DOI] [PubMed] [Google Scholar]
- ————— Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments. Trends Biochem Sci. 1998b;23:190–194. doi: 10.1016/s0968-0004(98)01207-9. [DOI] [PubMed] [Google Scholar]
- Richey B, Cayley DS, Mossing MC, Kolka C, Anderson CF, Farrar TC, Record MT., Jr Variability of the intracellular ionic environment of Escherichia coli. Differences between in vitro and in vivo effects of ion concentrations on protein-DNA interactions and gene expression. J Biol Chem. 1987;262:7157–7164. [PubMed] [Google Scholar]
- Sasse-Dwight S, Gralla JD. Footprinting protein-DNA complexes in vivo. Methods Enzymol. 1990;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: The DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Tagami H, Inada T, Kunimura T, Aiba H. Glucose lowers CRP* levels resulting in repression of the lac operon in cells lacking cAMP. Mol Microbiol. 1995;17:251–258. doi: 10.1111/j.1365-2958.1995.mmi_17020251.x. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Blazy B, Baudras A. An equilibrium study of the cooperative binding of adenosine cyclic 3′,5′-monophosphate and guanosine cyclic 3′,5′-monophosphate to the adenosine cyclic 3′,5′-monophosphate receptor protein from Escherichia coli. Biochemistry. 1980;19:5124–5130. doi: 10.1021/bi00563a029. [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Mizuno T. The Escherichia coli nucleoid protein H-NS functions directly as a transcriptional repressor. EMBO J. 1993;12:1039–1046. doi: 10.1002/j.1460-2075.1993.tb05745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen KM, Wolz R, Daugherty MA, Fried MG. Role of macromolecular hydration in the binding of the Escherichia coli cyclic AMP receptor to DNA. Biochemistry. 1997;36:11640–11647. doi: 10.1021/bi971193e. [DOI] [PubMed] [Google Scholar]
- Wood JM. Osmosensing by bacteria: Signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Johnson RC. Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J Bacteriol. 1995;177:5222–5231. doi: 10.1128/jb.177.18.5222-5231.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Activation of RpoS-dependent proP P2 transcription by the Fis protein in vitro. J Mol Biol. 1997a;270:346–359. doi: 10.1006/jmbi.1997.1133. [DOI] [PubMed] [Google Scholar]
- ————— Cyclic AMP receptor protein functions as a repressor of the osmotically inducible promoter proP P1 in Escherichia coli. J Bacteriol. 1997b;179:2410–2417. doi: 10.1128/jb.179.7.2410-2417.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: Evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]