Introduction
During the past decade, DNA research has advanced from human genome sequencing to the mapping of genetic variations among individuals.1 In recent years, these interindividual genetic variations have been associated with and used to identify potential for disease, drug response, and adverse reactions.1 Pharmacogenetic research findings have already been applied to varying degrees in several clinical fields and are expected to support a further shift toward a more personalized, less empirical approach to health care.2 However, we are only beginning to understand the impact that molecular diagnostics and targeted therapies will have on patient treatment, clinical outcomes, and cost-effectiveness.1 Despite significant advances, many scientific, economic, educational, legal, and commercial barriers impede the translation of pharmacogenomic research findings into clinical practice.3
The History of Pharmacogenomics
One of the most impressive scientific advances during the past decade was the sequencing of the human genome by the Human Genome Project.2 The complete human genome sequence was released in April 2003 to coincide with the 50th anniversary of the research publication announcing the discovery of the DNA double helix.4 Since then, advances in laboratory technology, computing, and bioinformatics have allowed genetic research to grow exponentially.4 Consequently, genetic research has since shifted from relatively rare monogenic diseases to more common and genetically complex diseases, such as cancer, cardiovascular and psychiatric disorders, and diabetes.4 These diseases are not only more prevalent; they also affect public health to a greater degree, since they are responsible for the majority of disease-related mortality and morbidity.4 Genetic research also now explores the role that RNA, proteins, and metabolites play in disease etiology.4
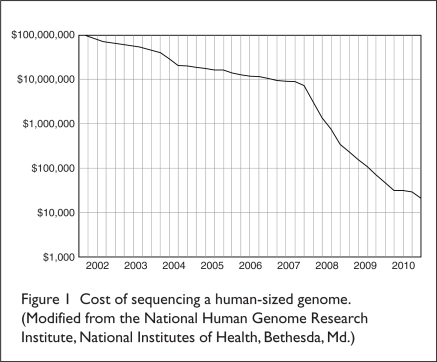
DNA sequencing has also been applied to the study of genetically influenced variations in drug response, or “pharmacogenomics.” The FDA has defined pharmacogenomics as “the study of variations of DNA and RNA characteristics as related to drug response,” whereas “pharmacogenetics” is “the study of variations in DNA sequence as related to drug response.”2 More specifically, pharmacogenomics evaluates molecular determinants at the genome-, transcriptome-, and proteome-wide levels, whereas pharmacogenetics involves limited and specific genetic markers.5 The application of pharmacogenetics to the clinical management of an individual is referred to as “personalized medicine” (PM).2 The goal of PM is to use an individual’s genetic data to avert preventable diseases or to choose a treatment that has the highest probability of being safe and efficacious according to a person’s genetic makeup.2 As is true for most technologies, the cost of DNA sequencing has dropped substantially over the years (Figure 1), increasing the likelihood that PM will become much more prevalent in the future.2
Figure 1.
Cost of sequencing a human-sized genome.
(Modified from the National Human Genome Research Institute, National Institutes of Health, Bethesda, Md.)
The Promise of Pharmacogenomics
As genomic medicine advances, there is hope that genetic biomarkers will encourage movement away from empirical and population-based treatment approaches to those that are stratified according to better patient outcomes, thus ending a “trial-and-error” approach to therapy.4,6–8 It is estimated that only 50% of patients respond positively to their medications.5,9 Therefore, half of the patient population might not be properly medicated or might experience therapeutic delays if they need to change medications because of a lack of efficacy.5
Adverse drug reactions (ADRs) are also unpredictable, even among individuals who are receiving the same therapeutic regimen.5 Because ADRs are a leading cause of death in the U.S., this unpredictability represents a significant safety risk.5 Identification of genetic factors that may predispose a patient to an ADR would be greatly helpful in preventing such reactions.5,7 Genetic testing may also be useful for predicting drug interactions. Estimates reveal that between 20% and 25% of drugs are metabolized, at least in part, by the cytochrome P450 (CYP) isoenzyme 2D6 (CYP2D6).3 Patients with multiple copies of the CYP2D6 gene may therefore be rapid metabolizers of these drugs and may not achieve therapeutic plasma levels at the usual drug dose.3,7 Conversely, subjects who have few functional CYP2D6 genes may be slow metabolizers, causing drug levels to exceed the therapeutic range.3,7
It is also expected that molecular-screening tests will be able to identify many diseases at earlier stages, when these conditions may be preventable, treatable, or curable.10 Early DNA testing may become key in disease-prevention strategies, since genetic information is accessible long before many disease processes begin.2,7 For example, detection of an elevated cholesterol level by a laboratory test may indicate a risk for the future development of heart disease.2 However, a genetic assay might identify a variant in the LDLR gene before cholesterol levels become elevated.2 Genetic testing may therefore be more powerful than traditional phenotypic tests, since this DNA mutation can be identified long before cholesterol levels start to rise.2
Genetic testing may also be more cost-effective than phenotypic tests, because delayed diagnosis often leads to increased morbidity and a need for more expensive medical procedures.2 Advanced genomic monitoring assays may also provide physicians with an improved ability to detect the likelihood of disease recurrence after successful treatment.8,10 In addition, genomic-based prognostic tests might be capable of assessing the risk of disease progression, and the results, therefore, may be able to inform decisions about whether to use adjuvant chemotherapy in cancer treatment.10
It has become widely accepted that genetically or molecularly targeted diagnostic tests and therapies are required to further drive the progress in treating many diseases, particularly cancer.8,10 Although pharmaceutical companies were initially reluctant to apply pharmacogenomics or biomarkers to drug development, this is now a pervasive strategy.8–10 Pharmacogenetic testing is used in phase 2A and 2B clinical trials in order to facilitate the development of new molecules and to reduce the associated risks and costs.5,7,8 In order to develop therapeutics that are targeted toward an individual’s genetic makeup, it is also necessary to develop predictive diagnostic genetic tests, known as “companion diagnostics,” along with new drug candidates.5,10 Companion diagnostics are a critical and necessary complement to targeted drug therapies, since they enable biomarker-stratified patient subsets to be correlated with therapeutic outcomes.10 Pharmacogenetic testing may also benefit pharmaceutical companies by identifying patients who will safely respond to medications that are approved but aren’t often prescribed because of toxicity and inefficacy issues, potentially expanding market share.8
Difficulties Translating Pharmacogenomic Data Into Clinical Practice
Discovery of a biomarker is only the first step in the long and complex process toward its translation into clinical practice.6 So far, the translation of pharmacogenomic discoveries into clinical practice has been surprisingly disappointing.3 In fact, many genetic biomarkers have not advanced much further beyond identification.6 This lack of progress may result, in part, from the failure to partially or fully replicate research identifying genetic biomarker associations, an issue that is not uncommon in genetic research.6 Overestimation of the magnitude of an effect can encourage the use of underpowered sample sizes, which often leads to replication failure.6 This failure to replicate pharmacogenomic research findings makes it difficult to establish the clinical validity of a biomarker and can lead to skepticism, confusion, criticism, and ultimately failure to accept a test.6
The impact of environmental factors can also complicate the ability to replicate pharmacogenomic research.4 It has been estimated that only 10% to 15% of genetic biomarkers have a direct impact on drug response.3 Instead, drug response phenotypes are more commonly influenced by a complex interplay between environmental, genetic, and gene–environment interactions.3,5 For instance, it is known that tumor-associated inflammatory responses can down-regulate CYP3A-mediated drug metabolism, thus contributing to drug clearance variability and toxicity of docetaxel (Taxotere, Sanofi-aventis; Docefrez, Sun) in cancer patients.3 In addition, drug interactions can influence drug response and can often explain why a phenotype does not accurately reflect a genotype for drug metabolism.3,5 Only fragmentary information is known regarding how the interplay between genetics and the environment influences pharmacological response.3 These complex factors highlight the need for prescriptions that are personalized to consider phenotypic, environmental, and genetic data in order to significantly reduce therapeutic failures and ADRs.5
Multiple genes can also have an impact on the predictive value of a genetic biomarker. Historically, many known pharmacogenetic traits were attributed to single nucleotide polymorphisms (SNPs), variations in alleles at a single gene locus that produced clearly discernible phenotypes.3 However, it is now thought to be likely that most drug–response phenotypes result from variations at multiple gene loci.3,4 Pharmacogenetic tests that evaluate only a few genes overlook the contributions of other genetic variations, thereby reducing the predictive value and applications for the test.3 Pharmacogenetic tests could therefore be more clinically applicable if they included a comprehensive survey of the human genome and considered the multigenic nature of many drug disposition and response phenotypes.3 It is expected that next-generation, whole-genome sequencing will be capable of investigating large genes for diagnostic purposes, a development that has the potential to significantly advance the adoption and clinical utility of pharmacogenomics.2 However, the challenge of identifying the contributions of many variations in multiple genes, and then translating this information into a predictive test, is formidable.3
A further complication is the lengthy and extensive investigation that is required to clinically verify genetic risk factors that are suspected of affecting drug pharmacokinetics and pharmacodynamics.3 With only 3% of published clinical data in this field focusing on phase 2 studies and beyond, there is a lack of evidence-based guidelines for many pharmacogenetic applications.4 In addition, some biomarker tests are in need of phase 3 and 4 research to evaluate whether recommended guidelines have been successful in reducing morbidity and mortality, such as testing for HLA-B*5701 and HLA-B*1502 in patients with human immunodeficiency virus (HIV) or HER2/neu in patients with breast cancer (see Table 1, page 419).4
Table 1.
List of Selected Clinically Valid Pharmacogenetic Biomarkers and Level of Recommendation For Related Drugs in the Context of FDA-Approved Drug Labels
| Pharmacogenetic Marker* | Representative Drug | Disease | Test Name† |
|---|---|---|---|
| CCR5 expression +++ | Maraviroc | HIV infection | Trofile |
| c-KIT expression + | Imatinib | Gastrointestinal stromal tumor | DakoCytomation c-Kit pharmDx |
| CYP2C9 variants; VKORC1 variants ++ | Warfarin | Thromboembolism | Verigene Warfarin Metabolism Nucleic Acid Test |
| CYP2C19 variants + | Voriconazole | Fungal infection | Roche Amplichip CYP450 test |
| CYP2D6 variants + | Atomoxetine, fluoxetine | Attention-deficit hyperactivity disorder, depression, obsessive-compulsive disorders | Roche AmpliChip CYP450 test |
| DPD deficiency + | Capecitabine, 5-FU | Colorectal cancer | TheraGuide 5-FU |
| EGFR expression + | Erlotinib | Non–small-cell lung cancer | DakoCytomation EGFr pharmDx |
| EGFR expression and K-RAS mutation +++ | Cetuximab, panitumumab | Colorectal cancer | DakoCytomation EGFr pharmDx and Nucleotide sequencing-high-resolution melting (HRM) analysis |
| G6PDH deficiency + | Primaquine | Malaria | Glucose-6-phosphate dehydrogenase screening |
| G6PDH deficiency ++ | Rasburicase | Hyperuricemia | Glucose-6-phosphate dehydrogenase screening |
| HER2/NEU overexpression+++ | Trastuzumab | Breast cancer | HercepTest |
| HLA-B*1502 ++ ‡ | Carbamazepine, phenytoin | Epilepsy | HLA typing |
| HLA-B*5701 ++ | Abacavir | HIV infection | HLA typing |
| NAT variants + | Isoniazid, rifampin | Tuberculosis | Genelex |
| Ph1 chromosome + | Busulfan | Chronic myelogenous leukemia | BCR/ABL test |
| Ph1 chromosome +++ | Dasatinib, imatinib | Acute lymphoblastic leukemia | BCR/ABL test |
| PML/RAR gene expression + | Tretinoin | Acute promyelocytic leukemia | PML/RARα quantitative real-time polymerase chain reaction |
| TPMT variants ++ | Azathioprine, 6-MP, thioguanine | Acute lymphocytic leukemia | Prometheus TPMT Genetics |
| UGT1A1 variants + | Nilotinib | Chronic myelogenous leukemia | Invader UGT1A1 Molecular Assay |
| UGT1A1 variants ++ | Irinotecan | Colorectal cancer | Invader UGT1A1 Molecular Assay |
ABL = Abelson; BCR = breakpoint cluster region; CCR = chemokine (C-C motif) receptor; c-KIT = v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; DPD = dihydropyrimidine dehydrogenase; EGFR = epidermal growth factor receptor; 5-FU = 5-fluorouracil; G6PDH = glucose-6-phosphate dehydrogenase; HER = human epidermal growth factor receptor; HLA = human leukocyte antigen; 6-MP = 6-mercaptopurine; NAT = N-acetyltransferase; Ph1 = Philadelphia; PML/RAR = promyelocytic leukemia/retinoic acid receptor; TPMT = thiopurine S-methyltransferase; UGT1A1 = uridine diphosphate glucuronosyltransferase 1A1; VKORC1 = vitamin K epoxide reductase complex subunit 1.
FDA (2009). Table of valid genomic biomarkers in the context of approved drug labels. Available at: www.fda.gov. Accessed June 20, 2011.
The names of specific pharmacogenetic tests are provided for information purposes only as examples of available tests and do not constitute an endorsement of any particular test or vendor.
For patients with Asian ancestry.
FDA classification: + = for information only; ++ = recommended; +++ = required.
From Gervasini G, et al. Eur J Clin Pharmacol 2010;66(8):755–774.3 (With kind permission from Springer Science & Business Media.3)
Further investigation is also needed to verify pharmacogenetic testing that is used to determine dose and patient response to warfarin.5 Although this practice is somewhat routine and an algorithm even exists for this test, there is still concern regarding its validity and reliability.4 To aid in resolving such issues, the Centers for Disease Control and Prevention (CDC), Office of Public Health Genomics, has sponsored the ACCE Model Project (analytic validity, clinical validity, clinical utility and associated ethical, legal and social implications [ELSI]) to create a process for evaluating emerging genetic tests.6,10,11 The aim of this project was to develop a model system for assembling, analyzing, disseminating, and updating existing data on the safety and effectiveness of DNA-based genetic tests and algorithms.11 The process includes collecting, evaluating, interpreting, and reporting data on genetic testing in a format that allows policymakers to have access to current and reliable information.10,11
Some Clinically Significant Pharmacogenetic Findings
Pharmacogenetic testing has been adopted to varying degrees in several clinical fields.2 A discussion of some of these applications follows.
Oncology
Cancer varies both phenotypically and genetically, even among patients with identical types and stages of disease.5 Many targeted cancer therapies are active against cell–surface receptors or downstream effector molecules, so that mutations in signaling pathway components can influence drug sensitivity and resistance.10 One major theme emerging from pharmacogenomic research in oncology is that such mutations can inform treatment decisions and predict patient outcomes.10 Significant advances in cancer tissue analysis techniques and patient stratification have also occurred in the past decade.5 Much of this progress has been based on the identification of molecular features that determine tumor classification, prognosis, targeted treatments, and treatment response.10 Diagnostic pharmacogenetic tests for some cancers have been developed and are now readily available.5
The availability of tests for nonheritable, somatic cell DNA mutations is rapidly increasing. The best known of these is the HER2 receptor gene amplification test, which is used to guide breast cancer treatment with trastuzumab (Herceptin, Genentech).2,5 HER2 (human epidermal growth factor receptor 2) is overexpressed in approximately one quarter of patients with breast cancer.5 Overexpression of the HER2 oncogene has been found to correlate with increased tumor formation and metastasis, a poor prognosis, and resistance to chemotherapy.5 Trastuzumab treatment is considered only when the patient has HER-positive 3 cancer, defined by very high levels of HER2 protein in the tumor.5 Pharmacogenetic testing has become an integral part of the breast cancer treatment with trastuzumab, since variability in HER2 receptor gene expression aids in determining degree of patient response.5
Pharmacogenomics has also been responsible for significant advances in treating lung cancer. Erlotinib (Tarceva, OSI/Genentech) and gefitinib (Iressa, AstraZeneca) are tyrosine kinase inhibitors (TKIs) designed to target the epidermal growth factor receptor (EGFR), which has been shown to influence predisposition to lung cancer.5 A recent East Asian study investigated the role of an EGFR mutation as a predictor for improved progression-free survival (PFS) with gefitinib treatment compared with carboplatin–paclitaxel therapy.5 Results indicated that the response to gefitinib was almost entirely limited to the mutation-positive group, whereas mutation-negative patients benefited more from chemotherapy.5 A European study also screened patients with non–small-cell-lung cancer (NSCLC) for EGFR mutations to identify those who were most likely to respond to erlotinib treatment.5
Two colorectal cancer treatments, cetuximab (Erbitux, Bristol-Myers Squibb/ImClone) and panitumumab (Vectibix, Amgen), are also directed against the EGFR.5 Mutations in K-ras are thought to activate the Ras/Raf/MAPK pathway independent of EGF binding and to block the activity of EGFR inhibitors.5 A relationship between K-ras mutations and survival was identified in metastatic colorectal cancer patients treated with cetuximab, showing that the presence of a K-ras mutation was an independent predictor for shorter progression-free and overall survival.5,8 A similar relationship between K-ras mutation and lack of response to panitumumab has also been demonstrated.5 In addition to K-ras status, other molecular markers for cetuximab and panitumumab efficacy are being investigated.5 For example, increases in EGFR gene copy number have also been shown to correlate with tumor response rate.5
Cardiology
Pharmacogenomic research in cardiology lagged in the 1990s but has grown quickly in recent years.5 In particular, promising discoveries have been made regarding two anti-thrombotic drugs, warfarin(Coumadin, Bristol-Myers Squibb; Jantoven, Upsher-Smith) and clopidogrel (Plavix, Bristol-Myers Squibb/Sanofi-aventis).5 Newer anticoagulant agents have been introduced to the market, such as dabigatran etexilate mesylate (Pradaxa, Boehringer Ingelheim), which was approved by the FDA in October 2010.12 However, the oral coumarin anticoagulants (OCAs) warfarin, acenocoumarol, and phenprocoumon have been the standard treatment for thromboembolic disorders for more than 60 years.5 Despite their efficacy, these drugs have a narrow therapeutic window and pose a high risk of major bleeding, especially during the initial phase of treatment.5 There is also substantial individual variation in response to OCAs, depending on the patient’s age, sex, body mass index, smoking, vitamin K intake, and concomitant drug therapy, thereby requiring frequent monitoring and dosage adjustment.5 Research during the past decade has found that interindividual differences in OCA dose response are also significantly influenced by genetic variations in two enzymes: CYP2C9, which metabolizes OCAs, and VKORC1, the target for these two drugs.5 Variations in the CYP2C9*2 and *3 alleles decreased CYP2C9 enzymatic activity and inhibit the metabolism of OCAs, whereas the VKORC1-1639G>A polymorphism influences pharmacodynamic response to coumarins.5
Discovery of these two polymorphisms caused the FDA to revise the warfarin drug label to include pharmacogenetic information in 2007.5 The revisions indicate that CYP2C9 and VKORC1 genotyping can assist in optimizing warfarin dosing and that lower doses may be best for patients with the identified genetic variations in one or both of these enzymes.5 However, although several pharmacogenetic-based dosing algorithms that incorporate CYP2C9 and VKORC1 genotype information have been proposed for warfarin, medical societies such as the American College of Chest Physicians have not yet changed their guidelines because of a lack of sufficient data from prospective randomized studies.5 For this reason, large randomized clinical trials are currently planned or under way in order to further determine the influence of pharmacogenetic-guided OCA dosing on treatment outcomes.5
Clopidogrel is currently the standard of care for acute coronary syndrome;5 it is indicated for patients undergoing percutaneous coronary interventions with or without stenting, the reduction of atherothrombotic events in patients with recent stroke or myocardial infarction, and diagnosed peripheral arterial disease.5 Nonresponsiveness to clopidogrel is widely reported—approximately 25% of patients taking it experience a subtherapeutic antiplatelet response that is associated with an increased risk of recurrent ischemic events.5 Growing evidence indicates that the response to clopidogrel might be determined by the CYP2C19 genotype.5 Specifically, the CYP2C19*2 allele impairs CYP2C19 function, which causes a marked decrease in platelet response to clopidogrel.5 Consequently, in May 2009, the FDA revised the drug’s label to include the impact of the CYP2C19 genotype on the drug’s pharmacodynamics and clinical response.5 Recently, a novel allelic variant, CYP2C19*17, was also discovered.5 It increases the transcriptional activity and enzymatic activity of CYP2C19, and with an observed prevalence of 30% or less, this variation is quite common in Caucasian populations.5
Psychiatry
Recently, several genome-wide association studies have identified genetic variants that provide new insights into possible molecular targets for antipsychotic and antidepressant agents.5 Typical antipsychotic medications exert effects on components of the dopamine pathway.5 Published studies have reported a significant association between polymorphisms in dopamine receptor genes DRD2 and DRD3 and response outcomes.5 With respect to atypical antipsychotic agents, pharmacogenetic studies have reported an association between the serotonin receptor genes HTR2A and HTR2C and response outcomes.5 In addition, depression studies have identified treatment outcome associations for genes in the serotonergic and noradrenergic systems.5 Specifically, significant associations have been reported for polymorphisms of the 5-HTTLPR serotonin transporter (SLC6A4) gene as well as for the HTR2A and HTR1A serotonin receptor genes.5
Genome-wide association studies to identify the genetic determinants of lithium response have also been undertaken.5 The phenotypic response to lithium, an ion with antisuicidal and mood-stabilizing effects, is complex, and its mechanism of action is unclear.5 However, many studies have implied that genes that encode for components of the inositol pathway may be involved in lithium’s mechanism of action.5 Various research papers have also reported associations between lithium response and the 5-HTTLPR polymorphism of the SLC6A4 serotonin transporter gene.5
Many pharmacogenetic studies in psychiatry have also produced intriguing results regarding genes encoding for phase 1 metabolic enzymes.5 Most psychiatric drugs are metabolized by CYP 450 isoenzymes.5 Specifically, antidepressants and antipsychotic agents are metabolized mainly by the CYP2D6, CYP1A1, CYP3A4, CYP2C9, and CYP2C19 isoenzymes.5 A number of studies report that CYP2D6 polymorphisms predict metabolism and side effects for risperidone (Risperdal, Janssen) but do not predict response to this psychotropic medication.5 Genotyping the CYP2D6 gene may therefore assist health professionals in identifying patients who need to be monitored for risperidone serum levels and ADRs.5
Although variants in gene encoding for the P450 isoenzyme CYP1A2 have been associated with decreased drug metabolism, these polymorphisms do not seem to affect response to clozapine, a CYP1A2 substrate.5 A number of findings have also demonstrated that genetic variants in the gene encoding for CYP2D6 correlate with serum levels of the antidepressants venlafaxine (Effexor, Wyeth/Pfizer), nortriptyline (e.g., Pamelor, Aventyl), and paroxetine (Paxil, GlaxoSmithKline; Pexeva, Noven).5 Depressed patients with a duplication of the gene for CYP2D6 have been found to be ultra-metabolizers of nortriptyline and fail to respond to treatment.5 Conversely, subjects with two nonfunctional copies of the gene for CYP2D6 were shown to be poor metabolizers of tricyclic antidepressants and had elevated plasma levels of these drugs.5
Infectious Disease
Pharmacogenomic research has also assumed an important role in infectious disease.5 A genetic biomarker for abacavir (Ziagen, Viiv) hypersensitivity syndrome has been identified and can prevent potentially life-threatening complications from this human immunodeficiency virus (HIV) treatment.2 A lower frequency of abacavir sensitivity syndrome had initially been observed in populations with African ancestry, and a higher risk was seen in families, which suggested a genetic component.2 Abacavir hypersensitivity syndrome has since been linked to a major histocompatibility complex (MHC) class I allele, HLA B*5701.2,5 A pharmacogenetic test is now available to screen Caucasians, the “at-risk” group, for this HLA marker.10
The acceptance of the HLA-B*5701 allele as a pharmacogenetic marker for abacavir hypersensitivity is one of the few examples of the rapid evolution of a genetic biomarker from research tool to clinical use.10 This evolution was driven by strong clinical utility and alignment of stakeholder interests.10 One study reported that screening for the HLA-B5701 allele has reduced abacavir hypersensitivity syndrome reactions to less than 1%, compared with 4% to 8% before HLA testing was routinely performed.5
The c.516G/T variant in the CYP2B6 gene has also been identified as a potential pharmacogenetic marker for ADRs in patients who have been treated with efavirenz (Sustiva, Bristol-Myers Squibb).6 Furthermore, nucleotide substitutions in genes encoding for the organic anion transporter 1, or multidrug-resistant protein 2 or 4, have been associated with an increased risk of kidney tubulopathy in patients receiving tenofovir disoproxil fumarate (Viread, Gilead), a nucleotide analogue used in HIV therapy.5 Certain polymorphisms, such as the c.3435C/T variation in the MDR1 gene, can also be used to predict antiretroviral therapy response.5
The therapeutic management of infectious diseases has been challenged by antibiotic resistance, which is mainly a result of improper prescribing and use of antimicrobials.5 Personalized medicine (PM) for infectious diseases is a developing concept in which molecular biology tools are used to provide more rapid, informative, and accurate diagnostic assays, enabling more effective treatment.5 Over the past decade, several companies have developed various nucleic acid testing assays for the direct detection of viral pathogens and some resistant bacteria in clinical samples.5 These new technologies offer faster diagnosis and will likely slowly replace classical phenotypic methods of identifying and determining antimicrobial susceptibility patterns for microbes.5 These novel, rapid molecular diagnostic tools will provide clinicians with real-time, crucial clinical information that should greatly improve the management of microbial and viral infections.5
Some Currently Available Pharmacogenetic Tests
A number of pharmacogenetic tests are commercially available or are being performed in selected laboratories.5 A handful of these protein- and DNA-based tests have been approved by the FDA for in vitro diagnostic testing.5 Early tests tended to identify a single genetic mutation to predict a patient’s risk for disease; however, newer tests can evaluate thousands of genes and dozens of genetic variations.13
One of the first test kits available was HercepTest (Dako), which was approved in 2001 by the Center for Devices and Radiological Health (CDRH) for detecting the overexpression of HER2 protein in breast cancer tissue.5,7 Similar tests that measure HER2 gene copy number using fluorescence in situ hybridization (FISH) are also available.5 Complex multigene products for breast cancer diagnosis are also now emerging, such as the FDA-approved, 70-gene-based MammaPrint (Agendia).2 This test is designed to stratify patients with early-stage breast cancer into low- and high-risk groups to aid in long-term management decisions.2
In 2005, the FDA approved the AmpliChip CYP450 Test (Roche Molecular Systems), the first pharmacogenetic test for genotyping 27 CYP2D6 and 3 CYP2C19 alleles that are associated with different drug-metabolizing phenotypes.5 The test is used to predict the metabolic rate for drugs that are substrates of CYP isoenzymes 2D6 and 2C19.5,7 Another test, the DMET Plus Panel (Affymetrix), covers an even wider range of genetic variations that influence drug metabolism, including common and rare SNPs, insertions, deletions, trialleles, and copy number variants, many of which are not assayed by conventional pharmacogenetic methods.5 The DMET Plus Panel identifies 1,936 drug metabolism biomarkers present in 225 genes, including all of those that the FDA has included in drug labels.5 The panel can identify common genetic variants with allelic frequencies of approximately 20%, along with absorption, distribution, metabolism, and excretion (ADME) markers that have allelic frequencies below 9%.5
Pharmacogenetic tests that identify predictors of ADR susceptibilities to antipsychotic pharmacotherapies have also been developed.5 The PhyzioType (Genomas) system employs 384 SNPs from 222 genes as well as a biostatistical algorithm.5 Genomas is waiting for a patent and FDA approval for this test.5 The PGxPredict:clozapine test (PGxHealth) detects a nucleotide polymorphism in the HLA-DQB1 gene.5 This test predicts the likelihood of clozapine-induced agranulocytosis and helps to determine risk–benefit balance for clozapine treatment.5 Laboratory tests that detect genetic factors that may influence psychotropic pharmacodynamics (such as genetic variants in the HTR2A, HT2RC, and 5-HTT genes that predict clozapine response) can also now be performed.5
Other pharmacogenetic tests that can detect the HLA-B*1502 allele for carbamazepine (e.g., Carbatrol, Tegretol, Epitol)–induced Stevens–Johnson syndrome, are being performed in some laboratories.5 This test is recommended by the FDA for patients of Asian descent, based on the finding that the incidence of this reaction is 10 times higher in this population.5,9 Some additional pharmacogenetic tests are presented in Table 1.3
Obstacles to the Translation Of Pharmacogenomics Into Clinical Practice
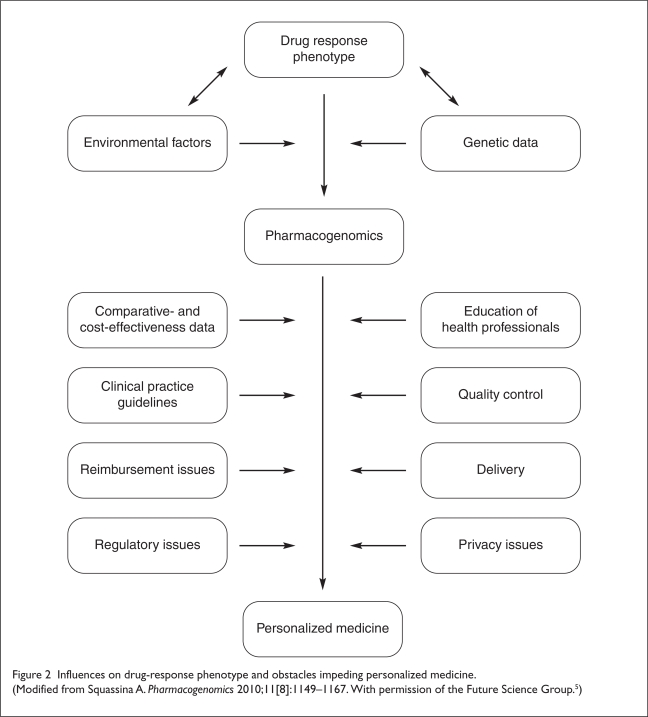
Many significant scientific, economic, educational, legal, and commercial barriers impede the translation of pharmacogenomics into clinical practice (Figure 2, page 420).3,9 These are discussed next.
Figure 2.
Influences on drug-response phenotype and obstacles impeding personalized medicine.
(Modified from Squassina A. Pharmacogenomics 2010;11[8]:1149–1167. With permission of the Future Science Group.5)
Lack of Comparative- and Cost-Effectiveness Data
Evidence that the cost of pharmacogenetic testing is justified by clinical outcome is lacking and needs to be generated, ideally through comparative effectiveness research.5,9 Unfortunately, although the number of published reports regarding the cost-effectiveness of pharmacogenetic biomarkers has been increasing, the data are still scarce and often inadequate, incomplete, or contradictory, or fail to demonstrate cost savings.6 Because of rising constraints on health care expenditures, it is essential that pharmacogenomic studies, particularly randomized controlled trials, be designed to include a thorough cost-effectiveness analysis.6 Such an analysis is likely to demonstrate that pharmacogenetic testing leads to improved clinical care but is cost-effective only for certain genetic marker tests, diseases, and treatments.5,8 Those who design cost-effectiveness studies must also consider the expense for the computational resources, storage, and interpretation that will be required to process, analyze, and save the generated genetic data.2,14
Need for Education and Clinical Practice Guidelines
Educational programs and guidelines for the utilization of pharmacogenetic data in clinical decision-making also need to be developed and disseminated.14 Medical professionals have acknowledged that they lack the training to apply pharmacogenetics in clinical practice.15 A 2008 survey (sponsored by the American Medical Association and Medco Health Solutions) of more than 10,000 physicians found that 98% of respondents were aware that a patient’s genetic profile could influence response to drug therapy.15 However, only 10% of the surveyed physicians felt that they were adequately informed to apply pharmacogenetic information to clinical decision-making.15 Furthermore, only 26% of survey takers reported that they had received pharmacogenomic training during medical school or postgraduate training.15 Predictably, physicians who considered themselves to be well informed were twice as likely to order pharmacogenetic tests.15
Studies also show that patients expect health care professionals to competently explain and interpret pharmacogenetic test results.5 Governmental regulations that may prohibit direct-to-consumer access to genetic tests are reasonable only if health care providers are adequately trained to counsel patients.16 In an effort to correct this knowledge gap, the International Society of Pharmacogenomics has requested that deans of education at medical schools include pharmacogenomic training in the core medical curriculum.5 This request was made in order to prevent physician education from creating a bottleneck in the implementation of PM.5
The lack of clinical practice guidelines and algorithms is an additional barrier to the translation of pharmacogenetics into clinical practice.2,6,9 Guidelines need to be robust, up to date, and consistent but also flexible so that they can be quickly revised to reflect new research findings.11 In order to facilitate developments in this field, regulatory agencies such as the FDA and the European Medicines Agency have published guidance documents.6,17 Clinical guidelines define only best medical practices and are often insufficient to change clinical practice.6 Therefore, multipronged approaches may be essential, such as regulatory changes that are coordinated with changes to clinical guidelines.6
Reimbursement Policies
Payer reimbursement policies exert a great influence on the diffusion of pharmacogenetic tests into clinical practice.5 Currently, diagnostic tests in the U.S. are reimbursed according to Current Procedural Terminology (CPT) codes and Medicare clinical laboratory-fee and physician-fee schedules.10 However, different models have been proposed to pay for pharmacogenomic testing, including value-based pricing, pay-for-performance, or even “money-back guarantees.”1
Experts have suggested that changes in the Centers for Medicare and Medicaid Services (CMS) reimbursement policies for pharmacogenetic tests are critical to the more widespread diffusion of this technology into clinical practice.5 However, such changes are not likely to occur soon.5 Although the FDA revised the warfarin label to include information about the influence of the CYP2C9 and VKORC1 genes in 2007,5 the CMS has approved Medicare reimbursement for pharmacogenetic warfarin tests only when they are performed during a prospective randomized controlled study that meets certain criteria.5 The CMS policy is based on regulations that state “tests for screening purposes ... performed in the absence of signs, symptoms, complaints, or personal history of disease or injury are not covered, except as explicitly authorized by statute.”5
Insurers have been reluctant to reimburse for expensive diagnostics and therapeutics because they don’t have data demonstrating that disease prevention is cost-effective.8,9 It has therefore been predicted that equitable payer reimbursement will occur only when public and private insurers acknowledge that targeted patient therapies can produce cost savings.8,10 However, some recent positive insurer reimbursement decisions regarding targeted therapies have been based on a pay-for-performance model that considered clinical and cost-effectiveness data and thereby recognized the economic value of molecular diagnostics.10 However, some studies have shown that cost-effectiveness analyses don’t always influence reimbursement.7 Instead, the two most important factors influencing reimbursement identified were the strength of clinical evidence and endorsement by professional guidelines.10 However, the disparity between the cost of randomized clinical trials needed to clinically validate pharmacogenetic tests and the traditionally low rates of reimbursement for diagnostics poses challenges.10 This disparity can potentially threaten the motivation of test developers, manufacturers, and laboratories to develop or offer pharmacogenetic tests.11
Regulatory Issues
By including pharmacogenetic information in an increasing number of drug labels, the FDA has been one of the main proponents of PM.6 However, the regulation of pharmacogenetic tests and targeted therapies by two separate centers within the FDA is an impediment.1 The Center for Drug Evaluation and Research (CDER) regulates targeted therapies, whereas the Center for Devices and Radiological Health (CDRH) regulates molecular diagnostics.1 The FDA has agreed that the regulatory oversight of pharmacogenetic tests needs to be re-examined and that clear guidance on new biomarkers is forthcoming.1,9 In a 2010 address, FDA Commissioner Margaret Hamburg identified three areas for improving the regulatory oversight of pharmacogenetics: (1) the need for a more flexible regulatory path and product approval process that adapts to genomic and clinical data; (2) collaboration between government regulatory and research agencies; and (3) transparency between the government, industry, and patient communities to maximize the safety and efficacy of genetic diagnostics and therapeutics.9
A more effective regulatory system will also be necessary to navigate the complex issues surrounding patient and kin protection with respect to genetic data ownership, a discussion that has already been initiated.14 Although historically, commercial test kits have mostly been the focus of FDA oversight, the agency has initiated a public dialogue regarding the development of a consistent, reasonable, and fair approach to all genetic tests, including those performed in a laboratory.13
Quality Control
In order for molecular diagnostics to be successfully implemented, guidelines that emphasize best practices with respect to technical performance, analytical validation, clinical interpretation, and proficiency need to be established.9,10 However, establishing quality assurance and proficiency guidelines at a pace that matches biomarker discovery and development is a significant challenge.10 In fact, the current lack of standards for high-quality specimens and assays has become one of the most significant barriers to progress in cancer research.10
Ideally, diagnostic providers should collaborate with the medical community and global proficiency testing organizations (Quality Control for Molecular Diagnostics, United Kingdom National External Quality Assessment Service, and NordiQC) to ensure that interpretation training and proficiency-testing programs for molecular diagnostics are in place.10 Pathologists will also be expected to include molecular diagnostic results in surgical pathology reports and therefore should also establish quality-control processes that ensure efficacy and accuracy.10
Delivery
It has been shown that point-of-care testing greatly improves clinical decision-making and outcomes.2 Ideally, pharmacogenetic tests will also be performed on a point-of-care basis, enabling health practitioners to make on-the-spot decisions regarding drug choice and dose.2 One current obstacle to performing point-of-care pharmacogenetic testing is the need for polymerase chain reaction (PCR) analysis.2 However, it is predicted that this obstacle may be overcome as alternatives to DNA amplification are developed.2 For example, development of third-generation DNA sequencers that can analyze single molecules, therefore circumventing the need for cloning or amplification, is now being discussed.2
Pharmacogenetic discoveries also need to be adapted into diagnostic technologies that are analytically accurate, reproducible, reliable, cost-effective, and compatible with sample types that are obtainable in routine clinical practice.10,14 Molecular technologies must also deliver diagnostic reports within a clinically useful turnaround time.14 Further, pharmacogenetic tests performed in large academic laboratories often use nonstandardized assays.10 This is an additional reason that pharmacogenetic testing needs to progress to a point where it is standardized and can be applied within a routine clinical setting.10
Privacy
Because pharmacogenetic testing identifies an individual’s disease risk, it inspires questions regarding privacy, genetic discrimination, and eligibility for health insurance or employment.5,9 The fact that data sharing is considered essential to pharmacogenomic research further complicates this issue.4 To illustrate, a typical question on an informed consent form for a pharmacogenomics study might request permission to store a participant’s genetic information and material along with phenotypic data.4 Permission to access an individual’s electronic patient record might also be requested in order to acquire future phenotypic data.4 Although genetic research presently occurs within a university medical center or a research institute, collaboration among research groups also makes it likely that genetic samples, information, or both, will be sent elsewhere.4 The samples or data might also be used to investigate traits that the participant doesn’t know about, since they were not defined in the original research study description.4
The level of genetic sample confidentiality in pharmacogenomic studies ranges from anonymous to identified.4 Although unidentified samples are the most private, researchers may discourage this choice, because unidentified data are considerably less valuable with respect to verifiability and follow-up.4 In addition, few collected samples are truly anonymous.4 Rapid advances in technology make it possible to generate, store, and share highly specific participant-unique data.4 This increases the likelihood that “anonymous” genetic data could be linked to a particular person.4 Ironically, this was illustrated when the genome of Dr. James Watson, a discoverer of the DNA double helix, was sequenced.4 He agreed to release his genetic information to public databases except for information regarding apolipoprotein E (Apo E), a protein associated with late-onset Alzheimer’s disease.4 However, even with this information excluded, Dr. Watson’s genotype could be imputed with more that 99% certainty because of linkage disequilibrium between polymorphisms flanking Apo E.4
In 2008, in order to encourage U.S. citizens to participate in genetic research and testing, Congress passed the Genetic Information Nondiscrimination Act (GINA),4,9 which provides citizens with protection against the misuse of genetic information by employers and insurers.9 Specifically, GINA prohibits insurers from using genetic information to determine underwriting decisions and employers from using these data to make selections regarding who gets hired, fired, or promoted.9 This legislation also prevents health insurers from requesting applicants to submit to genetic testing before they are granted coverage.9 However, some experts argue that because employers are still permitted to request access to health records of potential employees or their medical examination findings, the value of GINA is mainly symbolic.4 Other areas in which GINA is said to fall short include the fact that this legislation protects only individuals with a genetic predisposition and not a diagnosed disease, and it doesn’t prohibit long-term-care and life insurers from using genetic information to select plan participants.9
Sources of Impetus
Although many barriers currently interfere with the translation of pharmacogenomics into clinical practice, there are also some sources of impetus. A brief discussion regarding several of these influences follows.
Pharmacoeconomics
Rapidly increasing medical costs continue to outpace our projected ability to pay for health care.2 This trend has encouraged a greater focus on preventive medicine.2 Molecular diagnostics holds great promise for the prevention of disease, even in “low resource” settings.4 It likely will also be cost-effective for practicing physicians to utilize pharmacogenetic tests to optimize treatment selection and prevent ADRs.3 For example, a lack of therapeutic efficacy can be as costly as drug toxicity, so it would be economically advantageous to identify treatment responders prior to therapy.3
Although we are only starting to gain an understanding of how pharmacogenetic-guided therapy can affect costs and clinical outcomes, it seems highly likely that pharmacogenetic testing will reduce health care expenditures.3 Pharmacy benefit managers Medco and CVS Caremark have also advocated real-world pharmacogenetic comparative-effectiveness studies.1,8 These companies have a combined member base totaling more than 100 million Americans.1 With such an enormous public reach, they are well positioned to promote this research and to make pharmacogenetic testing available to a large population.1
Drug Label Revisions
To date, the FDA has approved more than 200 drug labels that include information regarding genetic biomarkers; this number has increased substantially over the past decade.3 However, in many cases, the drug labels provide this content for informational purposes only (see Table 1, page 419).3 Few labels recommend or require that biomarker testing be performed before a therapeutic decision is made.3,9
Despite this trend, the inclusion of pharmacogenetic information in drug labels by the FDA has still had a positive impact. For example, the FDA revision of the drug labels for warfarin and clopidogrel triggered additional research that will likely lead to further pharmacogenetic insights.1 The increasing inclusion of pharmacogenetic information in drug labels has also caused a surge in the number of pharmacogenetic tests available.3
Nonprofit Advocacy Groups
During the past few years, nonprofit foundations have also undertaken initiatives to promote the clinical implementation of PM.5 The Personalized Medicine Coalition is such an organization; it is composed of many pharmaceutical, biotechnology, diagnostic, and information technology companies; health care providers and payers; patient advocacy groups; industry policy organizations; academic institutions; and government agencies.5 This organization encourages the clinical use of molecular diagnostics and PM; provides opinion leadership and public education; and disseminates information to the media, government officials, and health care leaders.5
Conclusion
Pharmacogenomics is redefining how disease is diagnosed, classified, and treated. Pharmacogenetic testing is now essential to the development and clinical use of many molecular diagnostics and targeted therapies. However, the many scientific, economic, educational, legal, and commercial barriers that exist need to be overcome before the full potential of pharmacogenetics and PM is achieved.9 Awareness of, and attention to, these various challenges are essential in order to allow pharmacogenetic technologies to provide innovative clinical treatments and optimize patient outcomes. Continued research, as well as the participation of all involved stakeholders, will be necessary to overcome these formidable barriers.9
References
- 1.Freuh Felix W. Real-world clinical effectiveness, regulatory transparency, and payer coverage: Three ingredients for translating pharmacogenomics into clinical practice. Pharmacogenomics. 2010;11(5):657–660. doi: 10.2217/pgs.10.46. [DOI] [PubMed] [Google Scholar]
- 2.Trent RJ. Pathology practice and pharmacogenomics. Pharmacogenomics. 2010;11(1):105–111. doi: 10.2217/pgs.09.150. [DOI] [PubMed] [Google Scholar]
- 3.Gervasini G, Benítez J, Carrillo JA. Pharmacogenetic testing and therapeutic drug monitoring are complementary tools for optimal individualization of drug therapy. Eur J Clin Pharmacol. 2010;66(8):755–774. doi: 10.1007/s00228-010-0857-7. [DOI] [PubMed] [Google Scholar]
- 4.Vijverberg SJ, Pieters T, Cornel MC. Ethical and social issues in pharmacogenomics testing. Curr Pharm Des. 2010;16(2):245–252. doi: 10.2174/138161210790112700. [DOI] [PubMed] [Google Scholar]
- 5.Squassina A, Manchia M, Manolopoulos VG, et al. Realities and expectations of pharmacogenomics and personalized medicine: Impact of translating genetic knowledge into clinical practice. Pharmacogenomics. 2010;11(8):1149–1167. doi: 10.2217/pgs.10.97. [DOI] [PubMed] [Google Scholar]
- 6.Pirmohamed M. Acceptance of biomarker-based tests for application in clinical practice: Criteria and obstacles. Clin Pharmacol Ther. 2010;88(6):862–866. doi: 10.1038/clpt.2010.245. (online, October 27, 2010). [DOI] [PubMed] [Google Scholar]
- 7.Vogenberg FR, Barash CI, Pursel M. Personalized medicine, part 1: Evolution and development into theranostics. P&T. 2010;35(10):560–576. [PMC free article] [PubMed] [Google Scholar]
- 8.Vogenberg FR, Barash CI, Pursel M. Personalized medicine, part 3: Challenges facing health care plans in implementing coverage policies for pharmacogenomic and genetic testing. P&T. 2010;35(12):670–675. [PMC free article] [PubMed] [Google Scholar]
- 9.Vogenberg FR, Barash CI, Pursel M. Personalized medicine, part 2: Ethical, legal, and regulatory issues. P&T. 2010;35(11):624–642. [PMC free article] [PubMed] [Google Scholar]
- 10.Walk EE. Improving the power of diagnostics in the era of targeted therapy and personalized healthcare. Curr Opin Drug Discov Dev. 2010;13(2):226–234. [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention (CDC) Genetic testing: ACCE model system for collection, analyzing, and disseminating information on genetic tests. Available at: www.cdc.gov/genomics/gtesting/ACCE/FBR/index.htm. Accessed May 6, 2011. [Google Scholar]
- 12.FDA approves Pradaxa to prevent stroke in people with atrial fibrillation. Available at: www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm230241.htm. Accessed May 14, 2011.
- 13.FDA Direct-to-consumer genetic testing and consequences to the public. Available at: www.fda.gov/NewsEvents/Testimony/ucm219925.htm. Accessed May 6, 2011.
- 14.Taylor BS, Ladanyi M. Clinical cancer genomics: How soon is now? J Pathol. 2011;223(2):318–326. doi: 10.1002/path.2794. [DOI] [PubMed] [Google Scholar]
- 15.Prainsack B, Wolinsky H. Direct-to-consumer genome testing: Opportunities for pharmacogenomics research. Pharmacogenomics. 2010;11(5):651–655. doi: 10.2217/pgs.10.33. [DOI] [PubMed] [Google Scholar]
- 16.Ries NM, Castle D. Nutrigenomics and ethics interface: Direct-to-consumer services and commercial aspects. OMICS J Integr Biol. 2008;12(4):245–250. doi: 10.1089/omi.2008.0049. [DOI] [PubMed] [Google Scholar]
- 17.FDA Guidance on pharmacogenetic tests and genetic tests for heritable markers. Available at: www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm077862.htm. Accessed May 6, 2011.