Abstract
The human immunodeficiency virus type 1 (HIV-1) Tat protein strongly and specifically stimulates transcription elongation from the HIV-1 LTR and provides an important in vitro model system to study this process. Here we use protein-affinity chromatography to identify cellular factors involved in transcription elongation. A Tat-affinity column bound one transcription factor, Tat–SF1, efficiently and selectively. Tat–SF1 was identified originally as a Tat-specific coactivator, but we show it is a general transcription elongation factor. Our results also reveal the existence of an ATP-inactivatable general elongation factor (AIEF) required for Tat–SF1 activity and for which Tat can substitute functionally.
Keywords: HIV-1, Tat, Tat–SF1, transcription, elongation factor
Transcription in eukaryotic cells requires the assembly of an elaborate multisubunit preinitiation complex, followed by initiation, elongation, and termination. Although it was thought originally that regulation occurs primarily at the level of initiation, it is now recognized that some genes are regulated, at least in part, at the transcription elongation step (for review, see Bentley 1995). Recent studies suggest that regulation of elongation is probably of general importance in eukaryotic gene expression, especially in higher organisms (for review, see Bentley 1995; Li and Green 1996a). Compared to our understanding of transcription initiation, much less is known about the factors and mechanisms involved in transcription elongation.
The human immunodeficiency virus-1 (HIV-1) Tat protein is a potent activator of the HIV-1 long terminal repeat (LTR) and functions by binding to an RNA element, Tar, in the nascent transcript (for review, see Cullen 1993; Jones and Peterlin 1994). Tat can also stimulate transcription when directed to the promoter through a heterologous DNA-binding domain (Southgate and Green 1991), suggesting that it may be functionally similar to typical activators that target promoters by sequence-specific DNA binding. Based on both in vivo and in vitro evidence, HIV-1 Tat stimulates transcription at the level of elongation (Kao et al. 1987; Laspia et al. 1989; Marciniak et al. 1990; Feinberg et al. 1991; Marciniak and Sharp 1991) and thus has provided a valuable system to study this process.
We investigate cellular factor(s) that are required for general and Tat-directed transcription elongation. Our results reveal a new general elongation factor, Tat–SF1.
Results and Discussion
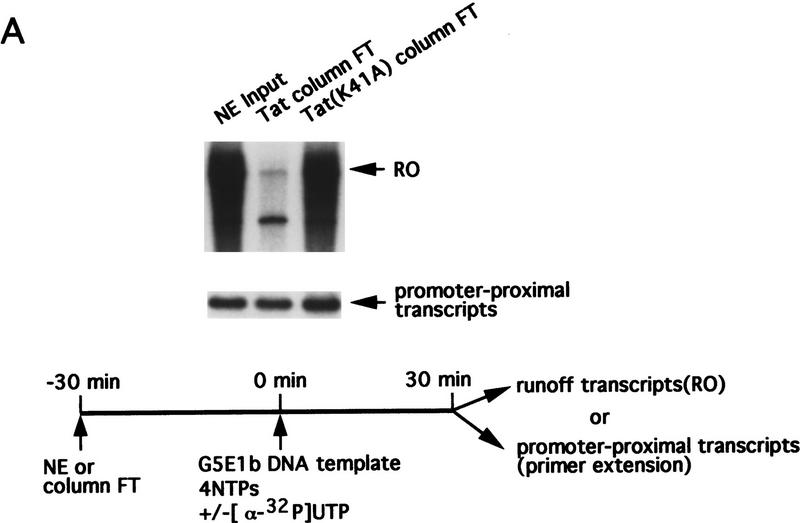
To identify cellular factors with which Tat interacts, a HeLa cell nuclear extract was chromatographed on a protein-affinity column containing immobilized wild-type Tat. A Tat activation-defective mutant, Tat(K41A), was used as a control. Flowthrough (FT) fractions were tested initially in a standard run-off (RO) transcription assay using the generic DNA template, G5E1b. Figure 1A shows that the crude nuclear extract and the Tat mutant protein-affinity column FT supported efficient transcription elongation as evidenced by synthesis of an ∼1600-nucleotide RO transcript. In contrast, the amount of 1600 nucleotide RO transcript synthesized in the FT of the wild-type Tat protein-affinity column was decreased dramatically. To determine whether the transcription defect was at the level of initiation or elongation, we performed primer-extension analysis using a ‘promoter-proximal’ primer located near the transcription start-site. Figure 1A shows that the FT fractions from the wild-type and mutant Tat columns, and the crude nuclear extract, all gave rise to a similar amount of this promoter-proximal primer-extension product. We conclude that the Tat-affinity column depleted a factor(s) involved specifically in transcription elongation.
Figure 1.
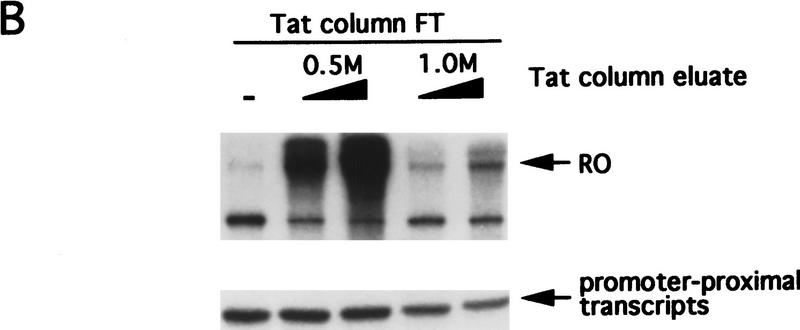
A Tat-affinity column depletes a factor(s) required for transcription elongation. Nuclear extract was chromatographed on a Tat-affinity column, or a control column containing the Tat mutant, Tat(K41A). FT fractions were collected, and bound proteins eluted with 0.5 m or 1.0 m KCl. (A) Activities of the nuclear extract input and the FT fractions were tested in a standard RO transcription assay or by primer-extension. The RO transcripts are shown at top; the primer-extension products at bottom. (B) The Tat protein-affinity column eluates (5 or 10 μl) were tested for their ability to complement the Tat protein-affinity column FT to support transcription. RO transcripts and primer-extension products are shown.
Figure 1B shows that a 0.5 m eluate of the Tat protein-affinity column complemented the FT fraction, whereas the 1.0 m eluate had little effect. Primer-extension analysis revealed that the 0.5 m eluate had no significant effect on transcription initiation. We conclude that the 0.5 m eluate of the Tat protein-affinity column contains a general transcription elongation factor(s) that binds to the Tat-activation domain.
To determine whether the activity bound to the Tat-affinity column corresponded to a known transcription component, we carried out immunoblot analysis using antibodies directed against various transcription-initiation factors [TFIIB, TFIID (TBP subunit), TFIIF (RAP30 subunit), TFIIH (ERCC3 subunit) (Drapkin et al. 1994)], elongation factors [SII, Elongin (SIII) (Elongin B subunit) (Aso et al. 1995; Garrett et al. 1995), ELL (Shilatifard et al. 1996), P-TEFb (CDK9 subunit) (Zhu et al. 1997)], RNA polymerase II, and Tat–SF1 (Zhou and Sharp 1996), a putative Tat-specific cofactor. Figure 2 shows that of these factors, only Tat–SF1 was both depleted significantly from the Tat-affinity column FT and present at high concentration in the 0.5 m column eluate, which contained the elongation activity. The Tat-affinity column also bound several general transcription factors to a lesser extent, in particular, TFIIF and the cyclin-dependent kinase/P-TEFb subunit, CDK9, which have both been implicated in Tat function previously (Kato et al. 1992; Moncebo et al. 1997; Zhu et al. 1997; Gold et al. 1998; Wei et al. 1998).
Figure 2.
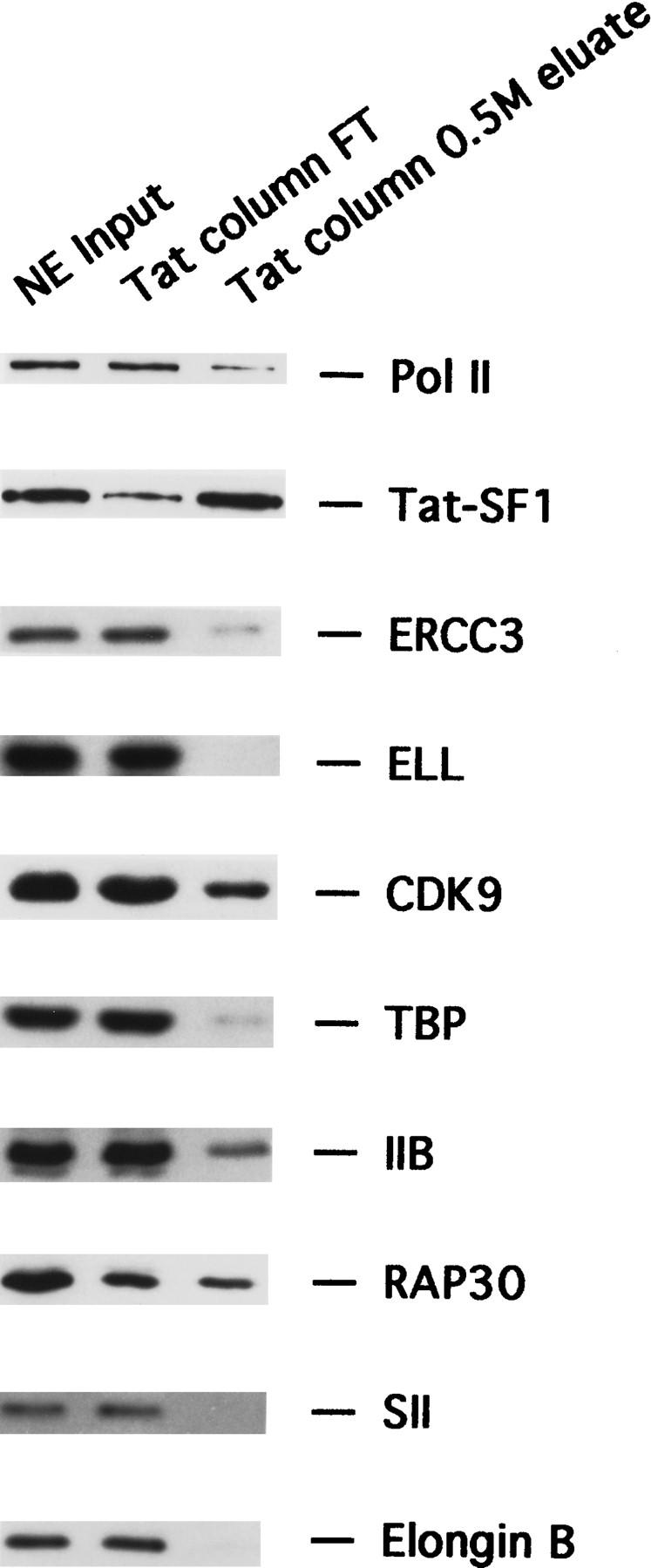
Selective depletion of Tat–SF1 by a Tat-affinity column. The presence of the known general transcription components in the nuclear extract input, Tat-affinity column flow through and the 0.5 m eluate was analyzed by immunobloting.
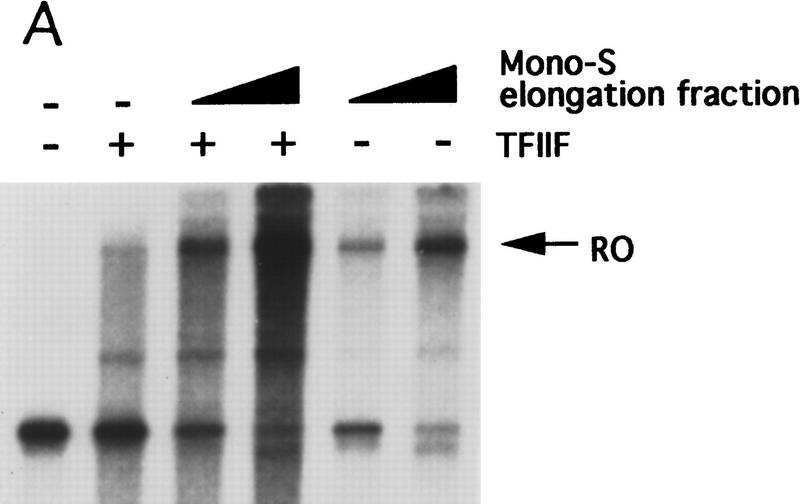
To identify the general elongation factor retained on the Tat-affinity column, the 0.5 m column eluate was subjected to biochemical fractionation (see Materials and Methods). Figure 3A shows that the final fraction complemented the Tat-affinity column FT to support efficient elongation. Significantly, the immunoblot experiment of Figure 3B shows that this fraction contained high levels of Tat–SF1 but no other known transcription components. Because TFIIF was depleted modestly by the Tat-affinity column and has been implicated in transcription elongation previously (for review, see Bentley 1995; Reine et al. 1996), it was also tested. Figure 3A shows that TFIIF increased transcriptional activity of the Tat-affinity column FT (lane 2) and synergized with Tat–SF1 (lanes 3,4). However, whereas TFIIF stimulated production of all transcripts, Tat–SF1 appeared to stimulate exclusively production of full-length transcripts.
Figure 3.
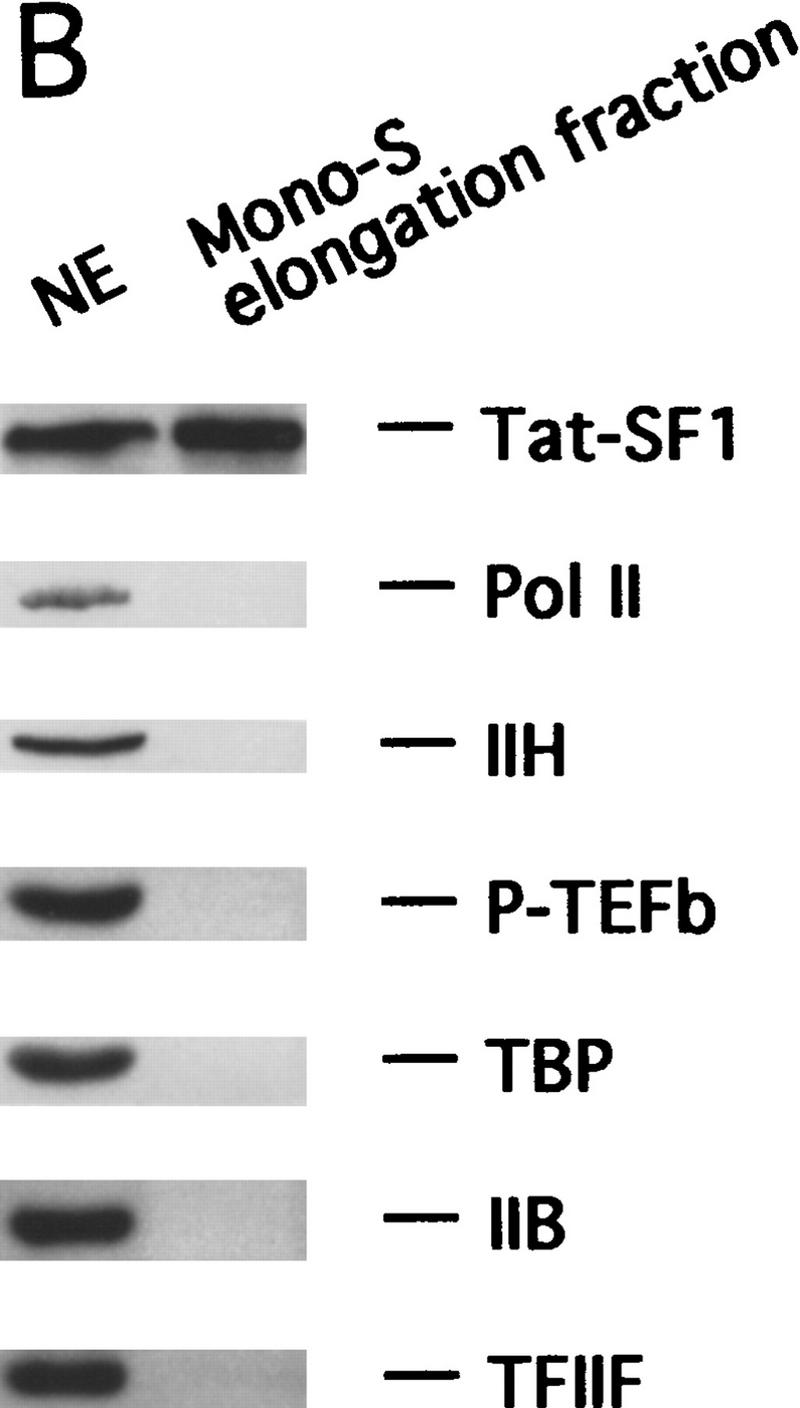
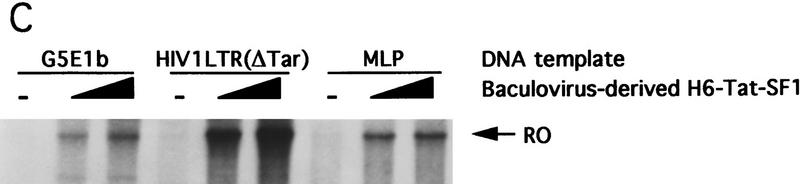
Tat–SF1 is a general transcription elongation factor. (A) The elongation activity present in the 0.5 m Tat-affinity column eluate was partially purified as described in Materials and Methods. Peak fraction containing this factor from the final Mono S column (2, 4 μl) and a TFIIF-containing fraction (2 μl) were added individually or together to the Tat-affinity column FT. Run-off transcripts on the G5E1b template are shown. (B) The Mono S fraction containing the elongation activity was analyzed by immunoblotting using antibodies against the indicated factors. (C) The same as A except that baculovirus-derived Tat–SF1 was used. The DNA templates are indicated.
The finding that Tat–SF1 may be a general transcription elongation factor was unexpected because it was proposed originally to be a Tat-specific coactivator (Zhou and Sharp 1995, 1996). To confirm this conclusion, we expressed Tat–SF1 in a baculovirus system. Figure 3C shows that following addition to the Tat-affinity column flow through, the baculovirus-derived Tat–SF1 stimulated transcription elongation efficiently from three different DNA templates. Based on these results, we conclude that Tat–SF1 is a general transcription-elongation factor.
Tat–SF1 was identified originally as a Tat-specific coactivator (Zhou and Sharp 1995, 1996) and therefore its depletion was expected to abolish Tat-directed transcription activation. To confirm this prediction, we used a previously described Tat-transactivation assay that involves an initial preincubation of the nuclear extract (or fractions thereof) with ATP (Marciniak and Sharp 1991). Figure 4A shows, as expected, that the flow through of a Tat-affinity column failed to support Tat-activated transcription, whereas efficient activation by Tat was observed with the crude nuclear extract input and the flow through of the Tat-mutant column. Figure 4B shows that the Tat-affinity column 0.5 m eluate complemented the ability of the Tat-affinity column flow through to support Tat-directed transcription activation, which as expected, was observed with the wild-type but not mutant [HIV-1LTR(ΔTar)] LTR template. Figure 4C shows that addition of TFIIF to the Tat-affinity column flow through increased the overall production of transcripts from both the wild-type and mutant LTR templates but was not required for activation by Tat. In contrast, Tat–SF1 complemented the Tat-affinity column flow through to support synthesis of full-length transcripts from the HIV LTR but not HIV-1LTR(ΔTar) DNA template (Figure 4B,C). These results indicate that Tat–SF1 is required for activation by Tat, consistent with previous studies (Zhou and Sharp 1995, 1996).
Figure 4.
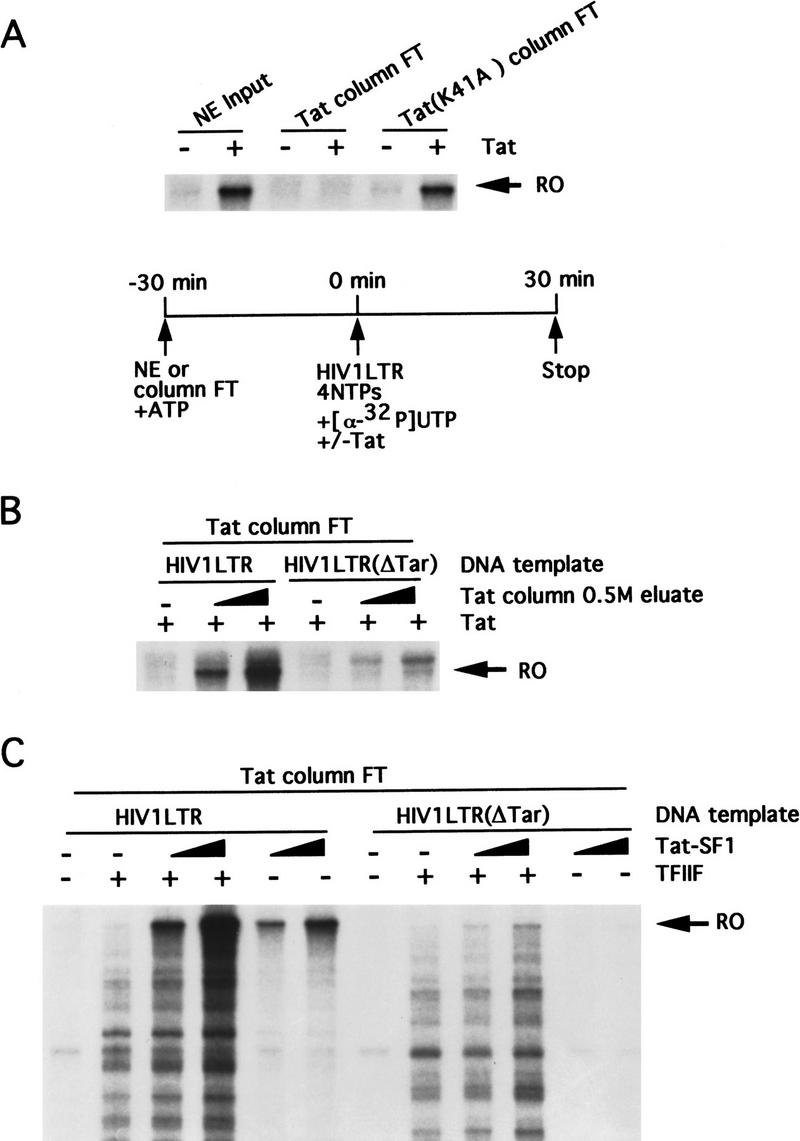
Tat–SF1 is required for transcription activity by Tat. Affinity chromatography was performed as described in Fig. 1. (A) The nuclear extract input and the FT fractions from the Tat protein-affinity column and the control column were tested in a Tat-transactivation assay as shown schematically. (B) The 0.5 m eluate from the Tat protein-affinity column (0, 5, or 10 μl) was tested for complementation of the Tat protein-affinity column FT for activation by Tat. (C) The same as B except that TFIIF (2 μl) and Tat–SF1 (2, 4 μl) from the Mono S column were added.
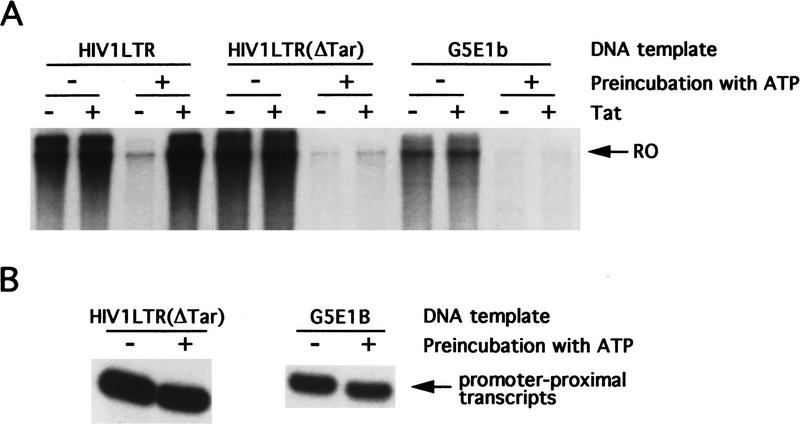
The results of Figure 3 indicate that Tat–SF1 can stimulate elongation in the standard nuclear extract but following preincubation with ATP the ability of Tat–SF1 to simulate elongation required Tat (Fig. 4). Figure 5A shows that preincubation of the nuclear extract reduced production of the 1600-nucleotide run-off transcript drastically without affecting initiation (Figure 5B). Therefore, the preincubation step inactivated an elongation activity, which we refer to as ATP-inactivatable elongation factor (AIEF). AIEF is a general elongation factor because the effect of preincubation was observed with several different DNA templates tested. Taken together, our results suggest that general transcription elongation requires both Tat–SF1 and AIEF. Because Tat can activate transcription in an ATP-preincubated extract that lacks functional AIEF, we conclude that Tat can substitute for AIEF on the HIV LTR.
Figure 5.
Preincubation of nuclear extract with ATP inactivates an elongation factor, AIEF. Nuclear extract was preincubated in the presence or absence of ATP. Tat and DNA templates were added as indicated. (A) RO transcripts; (B) primer-extension products.
The intrinsic elongation efficiency of RNA polymerase II is relatively low and is increased by a set of elongation factors including SII, SIII (elongin), ELL and p-TEFb, and transcription initiation factors TFIIF and TFIIH (for review, see Bentley 1995; Reine et al. 1996). Here we have shown that Tat–SF1 is also a general elongation factor. Further studies will be required to determine how Tat–SF1 stimulates elongation. Because Tat–SF1 does not affect promoter-proximal transcription, and is required for Tat function, we speculate that it increases the processivity of the transcription complex. Tat–SF1 is a member of a family of factors that include EWS, TLS/FUS, Drosophila SARFH, and hTAFII68, all of which share a similar RNA binding motif. Accumulating evidence suggests that these factors are involved in transcription (Bertolotti et al. 1996 and references therein). However, further studies are needed to define the relationship between these factors and determine their roles in transcription elongation.
Previous studies have shown that Tat–SF1 is essential for Tat-transactivation (Zhou and Sharp 1995, 1996), and a role for TFIIF in Tat function has also been suggested (Kato et al. 1992). Our finding that a Tat-affinity column can selectively and efficiently bind Tat–SF1, and to a lesser extent TFIIF, is consistent with these previous reports. We have shown that Tat–SF1 is required for Tat function and thus is also an essential Tat-specific coactivator as reported previously (Zhou and Sharp 1995, 1996). However, whereas the previous studies suggested that Tat–SF1 was Tat-specific, instead we find it is a general elongation factor. This apparent discrepancy can be explained readily by considering the assays used in the different studies. The Tat-transactivation assay used in this and the previous studies (Zhou and Sharp 1995, 1996) involves preincubation of nuclear extract with ATP. We have shown that this preincubation inactivates a general elongation activity, AIEF. Because AEIF is a general elongation factor, following ATP-preincubation elongation becomes inefficient; under these conditions, efficient elongation occurs only on the HIV LTR in the presence of Tat. These results indicate that Tat can substitute functionally for AIEF and further suggest that Tat–SF1 cooperates normally with AIEF to stimulate transcription elongation of cellular genes. AIEF could correspond to one of the previously identified elongation factors, but based upon several criteria, including its chromatographic properties, we think it is most likely a novel activity (X.-Y. Li and M.R. Green, unpubl.).
In addition to Tat–SF1, several other factors have been implicated in Tat-transactivation (Herrmann and Rice 1995; Parada and Roeder 1996; Cujec et al. 1997; Garcia-Martines et al. 1997; Moncebo et al. 1997; Suñé et al. 1997; Zhu et al. 1997; Wei et al. 1998; Wu-Baer et al. 1998). Of these, TFIIH and P-TEFb have been studied most extensively (Parada and Roeder 1996; Cujec et al. 1997; Garcia-Martines et al. 1997; Moncebo et al. 1997; Zhu et al. 1997; Wei et al. 1998). Both TFIIH and P-TEFb can phosphorylate the CTD of RNA polymerase II, a process thought to be stimulated by Tat. CTD phosphorylation is believed to be important during promoter clearance and required to counteract the effect of elongation inhibitory factors (for review, see Jones 1997). However, CTD phosphorylation may not be sufficient for Tat-transactivation: Tat-transactivation can be detected under conditions in which the CTD phosphorylation is unaffected (Keen et al. 1997). CTD phosphorylation may also be required to recruit elongation factors to the elongation complex, and our results raise the possibility that Tat–SF1 is one such a factor. Significantly, Tat–SF1 can associate with and be phosphorylated by P-TEFb (Zhou et al. 1998).
It has been reported previously that Tat can stimulate transcription by counteracting the inhibitory effect of sodium citrate, which apparently involves TFIIH (Parada and Roeder 1996). Our work further implicates an ATP-inactivatable general elongation factor, AIEF. Collectively, these various studies suggest that Tat functions through multiple factors. Future experiments will be required to define the precise roles of these factors in general and Tat-directed transcription elongation.
Materials and methods
Plasmids
pGEXFH–Tat and pGEXFH–Tat(K41A) contain, respectively, the DNA fragments encoding the HIV-1 Tat protein (amino acids 1–86) and its mutant, Tat(K41A), fused in-frame to the GST–Flag–HMK sequence in pGEX2T(128/129). The plasmids p(−119)HIV-1LTRCAT, p(−119)HIV-1LTRCAT(ΔTar) (Southgate and Green 1991), and pG5E1bCAT (Lillie and Green 1989) have been described previously.
Preparation of GST–Tat and GST–Tat(K41A)
The procedure was the same as described previously with modifications (Li and Green 1996b). Briefly, 20 μm ZnCl2 was added to the bacterial culture prior to induction by IPTG. The cell lysate was prepared in a lysis buffer [20 mm Tris-Cl (pH 8.0), 1 m NaCl, 20 μm ZnCl2, 0.1% NP-40, 1 mm DTT, and protease inhibitors]. After batch incubation, glutathione-agrose beads containing bound GST-fusion proteins were transferred to a column, and washed extensively first with the lysis buffer and then with buffer B [20 mm HEPES (pH 8.0), 300 mm KCl, 20 mm ZnCl2, 10% glycerol, 0.1% NP-40, 0.5 mm DTT, and 0.5 mm PMSF]. The proteins were eluted with buffer B containing glutathione and dialyzed extensively against buffer B.
Tat protein-affinity chromatography
The Tat protein-affinity column and the control column contained, respectively, GST–Tat and GST–Tat(K41A) immobilized on glutathione–agarose beads at a concentration of ∼3 mg/ml. The columns, with a volume of 1 ml each, were washed extensively first with the lysis buffer and then with buffer D [20 mm HEPES (pH 8.0), 0.2 mm EDTA, 0.5 mm DTT, and 0.5 mm PMSF] containing 0.1 m KCl and 20 μm ZnCl2. Nuclear extract (3 ml) was passed through the columns at a flow rate of 1.5 column vol/hr. The columns were washed with buffer D containing 0.1 m KCl, and bound protein eluted with buffer D containing 0.5 m and 1.0 m KCl. The eluted fractions were dialyzed against buffer D containing 0.1 m KCl.
Partial purification of Tat-binding factors
The 0.5 m eluate from the Tat-affinity column, derived from 150 ml of nuclear extract, was dialyzed in buffer E (20 mm HEPES, 10% glycerol, 0.2 mm EDTA, 0.03% NP-40, 0.5 mm DTT, and 0.5 mm PMSF) containing 0.1 m KCl. The dialyzed sample was applied to a 1-ml Resource Q column (Pharmacia) equilibrated in 0.1 m buffer E, and proteins were fractionated using 20 ml of buffer E with 0.1–0.6 m KCl. The fractions from the gradient were dialyzed in 0.1 m buffer E and analyzed for transcription activity in reconstitution experiments. The active fractions were pooled and loaded onto a 1-ml Mono S column (Pharmacia) equilibrated in 0.1 m buffer E, and fractionated using 20 ml of buffer E with 0.1–0.6 m KCl, and dialyzed in 0.1 m buffer E.
Run-off transcription assays
The DNA templates including pHIV-1LTRCAT, pHIV-1LTRCAT(ΔTar), and pG5E1bCAT were linearized with HpaI, and consequently the full-length run-off transcripts are ∼1600 nucleotides. A typical transcription reaction contained 7.5 μl of nuclear extract or flow through from the Tat-affinity or control column, 0.25 μg of DNA template, 600 μm of ATP, GTP, and CTP, 20 μm UTP, 1 μCi (800 mCi/mmole) [α-32P]UTP (NEN) in the reaction buffer (20 mm HEPES, 75 mm KCl, 6%–13% glycerol, 20 μm ZnCl2, 0.01% NP-40, 40 ng pGEM3, 1.0 mm DTT, and 0.5 mm PMSF). GAL4-AH was added to transcription reactions using G5E1bCAT as template. Where indicated, 50 ng Tat or its mutant Tat(K41A), purified from Escherichia coli as a GST fusion-protein, was also added. The nuclear extract was preincubated for 30 min at 30°C in the reaction buffer in the presence or absence of 300 μm ATP before transcription was started by addition of DNA template and nucleotide triphosphates including [α-32P]UTP. ATP was added during preincubation in the Tat-transactivation assay but not in standard transcription assay. Transcription reactions were allowed to proceed for 30 min. Transcripts were purified and analyzed on a 5% denaturing polyacrylamide gel containing 8 m urea.
Transcription initiation assay
Transcription reactions were carried out as described above without [α-32P]UTP. Primer-extension analysis was performed as described by Lin et al. (1988) using a primer corresponding to a sequence near the transcription start site.
Purification of H6–Tat–SF1
A DNA fragment encoding Tat–SF1 (Zhou and Sharp 1996) was cloned into the BglII-EcoRI site of the pBlueBacHis2B vector (Invitrogen). The resulting plasmid was cotransfected with linearized Bac-N-Blue DNA into SF9 cells. Recombinant virus obtained was used to express Tat–SF1 with a His-tag at the amino terminus in SF9 cells. The cells were harvested 60 hr postinfection, resuspended in lysis buffer [20 mm Tris-Cl (pH 8.0), 500 mm NaCl, 0.1% NP-40, 5 mm β-mercaptoethanol and protease inhibitors], and lysed by brief sonication. After it was cleared by centrifugation, the lysate was loaded onto a Ni2+-agarose column, which was then washed extensively with the lysis buffer containing 20 mm imidazole. H6–Tat–SF1 was eluted from the column with the lysis buffer containing 100 mm imidazole, and was dialyzed against buffer D containing 0.1 m KCl.
Acknowledgments
We thank D. Reinberg, R. Drapkin, R.C. Conaway, J.W. Conaway, P.A. Sharp, Q. Zhou, D. Price, and M. Sawadogo for providing valuable reagents, and L. Qiang for excellent technical assistance. M.R.G. is an investigator of the Howard Hughes Medical Institute. This work was supported by a grant from the National Institutes of Health to M.R.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Michael.Green@ummed.edu; FAX (508) 856-5473.
References
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Bentley DL. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L. hTAFII68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Does HIV-1 Tat induce a change in viral initation rights? Cell. 1993;73:417–420. doi: 10.1016/0092-8674(93)90126-b. [DOI] [PubMed] [Google Scholar]
- Cujec T, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D, Peterlin B. The HIV trans-activator Tat binds to the CDK-activating kinase (CAK) and activates the phosphorylation of the C-terminal domain of RNA polymerase II. Genes & Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R, Reardon JT, Ansari A, Huang J-C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcription elongation. Proc Natl Acad Sci. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martines L, Mavankal G, Neveu J, Lane W, Ivanov D, Gaynor R. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 1997;16:2836–2850. doi: 10.1093/emboj/16.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett KP, Aso T, Bradsher JN, Foundling SI, Lane WS, Conaway RC, Conaway JW. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc Natl Acad Sci. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MO, Yang X, Herrmann CH, Rice AP. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: Candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA. Taking a new TAK on Tat transactivation. Genes & Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- Jones KA, Peterlin BM. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Bochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM. Antitermination of transcription within the long terminal repeat of HIV by the tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kato H, Sumimoto H, Pognonec P, Chen CH, Rosen CA, Roeder RG. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factor. Genes & Dev. 1992;6:655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- Keen N, Churcher M, Karn J. Transfer of Tat and release of Tar RNA during the activation of the human immunodeficiency virus type-1 transcription elongation complex. EMBO J. 1997;16:5260–5272. doi: 10.1093/emboj/16.17.5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stablizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Li X-Y, Green MR. Transcription elongation and Cancer. Curr Biol. 1996a;6:943–944. doi: 10.1016/s0960-9822(02)00633-4. [DOI] [PubMed] [Google Scholar]
- ————— Intramolecular inhibition of activating transcription factor-2 function by its DNA-binding domain. Genes & Dev. 1996b;10:517–527. doi: 10.1101/gad.10.5.517. [DOI] [PubMed] [Google Scholar]
- Lillie JW, Green MR. Transcriptional activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lin Y-S, Carey MF, Ptashne M, Green MR. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Mancebo H, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes & Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Calnan BJ, Frankel AD, Sharp PA. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990;63:791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- Parada C, Roeder R. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxyl-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- Reine D, Conaway JW, Conaway RC. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Southgate CD, Green MR. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes & Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- Suñé C, Hayashi T, Liu Y, Lane WS, Young RA, Garcia-Blanco MA. CA150, a nuclear protein associated with the RNA polymerase II holoenzyme, is involved in Tat-activated human immunodeficiency virus type 1 transcription. Mol Cell Biol. 1997;17:6029–6039. doi: 10.1128/mcb.17.10.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang S-M, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to Tar-RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Sharp PA. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Tat–SF1: Cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science. 1996;274:605–610. doi: 10.1126/science.274.5287.605. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chen D, Pierstorff E, Lou K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Peery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M, Price D. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes & Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]