Abstract
Background & objectives:
The greater tendency to diabetes in Indians may be due to genetic factors in addition to environment and diet. CD36, a class B scavenger cell surface receptor mediates internalization of oxidized low density lipoprotein (Ox-LDL) leading to the formation of macrophage foam cells. CD36 deficiency is related to phenotypic expression of the metabolic syndrome, frequently associated with atherosclerotic cardiovascular diseases resulting in raised levels of glucose thereby contributing to type 2 diabetes (T2DM). Therefore, the association of human CD36 gene mutation to T2DM needs investigation. We undertook this study to investigate CD36 gene status in north Indian subjects by screening for the deletion of exons 3, 4 and 5 and certain polymorphisms.
Methods:
Clinical characteristics were compared between 300 T2DM patients and 100 healthy controls. Deletion analysis was carried out for exons 3, 4 and 5 of CD36 gene in 300 T2DM patients using PCR and agarose gel electrophoresis. Genotype analysis for two polymorphisms 478C>T and delAC in exons 4 and 5 respectively was carried out using PCR-RFLP method.
Results:
Biochemical parameters such as fasting and post-prandial glucose levels, total cholesterol, LDL-cholesterol and blood pressure were slightly raised in the T2DM patients when compared with controls with lowered HDL-cholesterol. No exonic deletion was observed in the 300 patients and 100 controls screened. All individuals were found to be homozygous (CC and -/-) for the two polymorphisms studied.
Interpretation & conclusions:
Although no exonic deletion was found in T2DM patients, our study suggests that all 15 exons need to be screened for mutations which lead to CD36 deficiency. Genotyping studies of the two SNPs in the CD36 gene confirmed the absence of exons 4 and 5 deletion. This is perhaps the first report from India suggesting that CD36 is one of the several important genes that need to be explored in relation to T2DM.
Keywords: Biochemical characteristics, CD36 gene, deletion, exons, type 2 diabetes
Type 2 diabetes mellitus (T2DM) is the common form of diabetes accounting for 90 per cent of cases worldwide and is continuously increasing. Over the past 30 years, the status of diabetes has changed from being considered as a mild disorder of the old age to one of the major causes of morbidity and mortality affecting the young and middle aged people. It is estimated that approximately 285 million people worldwide, or 6.6 per cent in the age group 20-79 yr, will have diabetes by the end of 2010, 70 per cent of whom live in low- and middle-income countries. This number is expected to increase by more than 50 per cent in the next 20 years if preventive programmes are not put in place. Some 438 million people, or 7.8 per cent of the adult population, are projected to have diabetes by 20301. The largest increases will take place in the regions dominated by developing economies as is evident from the alarming number of diabetic patients in India1,2.
T2DM occurs primarily due to a ‘high blood sugar level’ resulting in hyperglycaemia, glycosuria, dyslipidaemia, high levels of low density lipoproteins (LDL) and low levels of high density lipoproteins (HDL). If remained uncontrolled, T2DM may lead to various complications such as abdominal obesity, hypertension, atherosclerosis, stroke, coronary heart disease, etc. However, the mechanisms involved in determination of the risk of developing diabetes are still not known clearly. It has been established that T2DM has a genetic component and several candidate genes are responsible for inducing susceptibility to this disease such as adiponectin gene (ADIPOQ), TCF7L20, β3 adrenergic receptor (β3AR) gene and many more3–8. There have been reports of a gene coding for a macrophage receptor, CD36 which has shown association with hypertension and metabolic syndrome in the Caucasian and African-American populations, respectively9,10.
CD36 is a 88 kDa integral transmembrane protein, one of the most important molecules found on the surface of many cells in vertebrates and has the ability to endocytose oxidized LDL (OxLDL)11,12. Several studies suggested the role of CD36 as an important regulator of the metabolic pathways involved in insulin resistance13–15. The pathophysiology of human CD36 deficiency in metabolic syndrome and atherogenesis has been explained16. Highly oxidized lipoproteins can be rapidly taken up by macrophages to give rise to foam cells, cholesterol engorged cells that are hallmark of early atherosclerotic lesions. The increased fatty acid availability can induce insulin resistance if the capacity of adipose tissue to store triglycerides and/or that of muscle to oxidize FA is exceeded17. A recent report has shown that alteration in CD36 levels might be involved in the development of diet-induced insulin-resistance in mice18.
CD36 gene has 15 exons extending over 32 kb on chromosome 7q11.219,20. It has been reported that genetic factors like single nucleotide polymorphisms (SNPs) in the CD36 gene are significantly associated with T2DM10,21–23 while exonic deletions such as exons 4, 9, 11, 12 and 13 in the CD36 gene lead to its deficiency24–26. Deletion of exons 3, 4 and 5 in CD36 gene has been studied in the Japanese population24 while significant alteration in the activity of CD36 gene has been shown to occur due to small nucleotide deletions of 10, 12, 16 and 29 bps9,25. Exon 3 contains the last 89 nucleotides of the 5’-untranslated region, encodes the N-terminal cytoplasmic and transmembrane domains. Deletion of the two non-coding exons 1 and 2 along with third exon results in no expression of CD36 protein. Exons 4 and 5 encode amino acids 41-94 and 94-143, respectively which constitute potential N-glycosylation sites. The skipping of these exons results in a defect of the ligand binding site of CD36 protein20.
CD36 deficiency can be divided into two subgroups, type I in which neither platelets nor monocytes/macrophages express CD36, and in type II monocytes/macrophages express CD36 but platelets do not. Two important mutations in CD36 gene responsible for CD36 deficiency are SNP T/C at nt 478 of CD36 (Exon 4) and a dinucleotide deletion (delAC in Exon 5)27–29. The 478C>T polymorphism (proline-90 serine) predominates in the type I and type I1 CD36 deficiency24,28 via defects in post-translational modification, while the dinucleotide deletion causes a frameshift mutation leading to the appearance of a translation stop codon and a marked reduction in the level of CD36 mRNA.
The role of CD36 gene in lipid metabolism and T2DM susceptibility prompted us to take up this study on CD36 gene status in the north Indian population. We screened the deletion of exons 3, 4 and 5 and the polymorphisms viz. 478C>T SNP in exon 4 and a delAC in exon 5 of the CD36 gene.
Material & Methods
Sample collection: The study protocol was approved by the institutional ethical committee of Chatrapati Sahuji Maharaj Medical University (CSMMU), Lucknow, and the study was performed during March-November, 2009. Written informed consent was obtained from all participants. Blood samples were collected randomly from 300 patients (173 males and 127 females) from the Diabetic Clinic of Medicine OPD, CSMMU, Lucknow and their clinical details, family history of diabetes and associated complications were recorded. Control samples (n=100) were collected from healthy staff members and students of University of Lucknow. The screening and management of patients were done as per American Diabetes Association guidelines30. Criteria for patients’ participation included fasting plasma glucose (FPG) ≥126 mg/dl or a 2 h blood glucose level of ≥200 mg/dl after a 75 g oral glucose load (oral glucose tolerance test). Individuals with a history of major illness were excluded. Normal individuals were included on the basis of normal FPG of 70-109 mg/dl or random plasma glucose (RPG) 110-199 mg/dl.
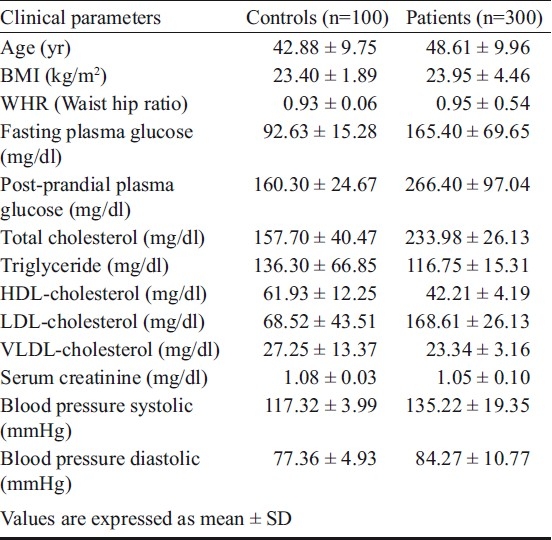
Biochemical parameters: Two ml blood samples were equally distributed in two vials, one ml in 0.5M EDTA and the other in a plain vial for DNA extraction and biochemical estimations respectively. Serum was collected from the blood in plain vials after centrifugation for 10 min at 2655 g at 4°C. Estimations of plasma glucose (mg/dl), serum insulin (mg/dl) and lipid profile (total serum cholesterol, TC; High density lipoprotein-cholesterol, HDL-C and serum triglycerides, TG) were done using commercially available Ecoline kits (Merck, India) by double beam spectrophotometer (Shimadzu, Japan) at 550 nm (TGL-C), 510 nm (S. creatinine), 500 nm (TC) and 560 nm (HDL-C). Height, weight and waist circumference were measured to calculate body mass index (BMI) and waist hip ratio (WHR). Systolic and diastolic (disappearance of Korotkoff sound, phase V) blood pressures were measured in the sitting position with an appropriately sized cuff after a 5 min rest. Clinical details of patients and control subjects were recorded (Table I).
Table I.
Clinical parameters of T2DM patients and control subjects
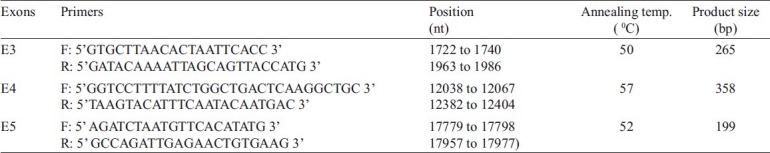
DNA extraction and deletion analysis: DNA was extracted from blood samples of T2DM patients using the salting out method31 with slight modifications. Lysis was followed by proteinase K buffer (0.375M NaCl, 0.012M EDTA) and SDS (10%) treatment. After frothing, cold 5M NaCl was added followed by phenol-chloroform extraction. DNA was checked on 0.8-1 per cent agarose gel. The quantity of DNA was estimated using double beam UV-visible spectrophotometer and quality was checked by measuring A260/A280. Exons 3 (nt 1722 to 1986), 4 (nt 12038 to 12404) and 5 (nt 17779 to 17977) of CD36 gene were amplified by polymerase chain reaction (PCR) using respective primers using Master Cycler ep Gradient (Eppendorf, USA)24. The primer sequences and PCR conditions are shown in Table II. PCR was performed for 30 cycles using 0.5U Taq polymerase, 10 pmol/μl of each primer, and 200μM dNTP in 25μl reaction volume. The PCR products were checked on 1.5 per cent agarose gel along with 50 and 100 bp markers. The gels were documented and analyzed.
Table II.
Primer sequences, nucleotide (nt) positions, PCR conditions and product sizes of exons
Genotype analysis: Two polymorphisms in CD36 gene responsible for exon deletions viz. 478C>T in exon 4 and a dinucleotide deletion (delAC) in exon 5 were analyzed in T2DM patients and control subjects by polymerase chain reaction and restriction fragment length polymorphisms (PCR-RFLP)24,27–29. The primers used were as shown in Table IIand the restriction enzymes used were Cfr131 and BoxI, identified by NEB cutter software (www.tools.neb.com/NEB cutter 2) for 478C>T and delAC respectively. The PCR amplicons and digested products were resolved on 10 per cent polycrylamide gels (PAGE), stained with ethidium bromide and documented in Gel Doc (Vilber Lourmat, France).
Statistical analysis: Statistical analysis was applied to biochemical data using SPSS software (www.spss.com) (v15.0). Mean ± SD (standard deviation) of all clinical parameters and diabetic duration was calculated in each age group (≤40, 41-59 and ≥ 60 yr) as well as in BMI groups (<18.5; 18.5-25 and >25 kg/m2). P values were calculated by 2x2 contingency table using paired t-test. All P values were two sided and P<0.05, was considered significant.
Results & Discussion
The average age of the patients was 48.61±9.96 yr and their fasting and post-prandial glucose levels were 165.40 ± 69.65 and 266.40 ± 97.04 mg/dl, respectively. The average systolic BP was slightly elevated (135.22 ± 19.35 mmHg) and the mean diastolic pressure was almost normal (84.27 ± 10.77 mmHg). Total cholesterol (233.98 ± 26.13 mm/dl) and LDL-C (168.61 ± 26.13 mmHg) levels were slightly raised and HDL-C was low (42.21 ± 4.19 mmHg). However, no significant difference was observed in BMI, WHR, triglycerides and serum creatinine levels between the T2DM and control groups (Table I).
The PCR products of exons 3 (265 bp), 4 (358 bp) and 5 (199 bp) are shown in Fig. 1. Exonic deletion was observed neither in patients nor controls. In our study population all individuals (patients and controls) were found to be homozygous ‘CC’ and -/- for 478C>T and delAC polymorphisms respectively (Fig. 2).
Fig. 1.
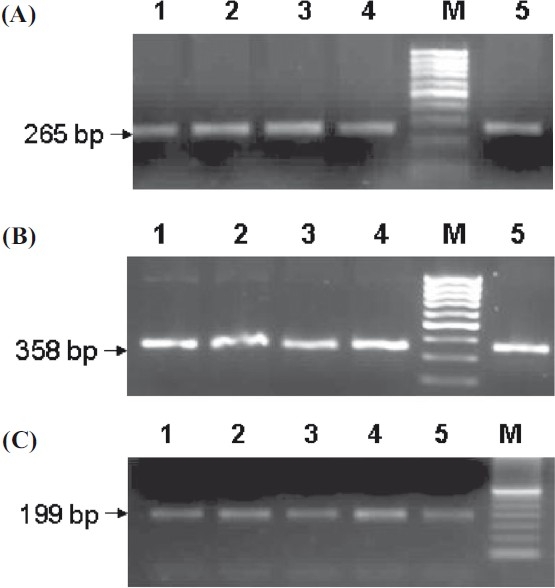
Agarose gels showing PCR products of (A) Exon 3, (B) Exon 4, (C) Exon 5, Lane M:100 and 50 bp ladder.
Fig. 2. (A).
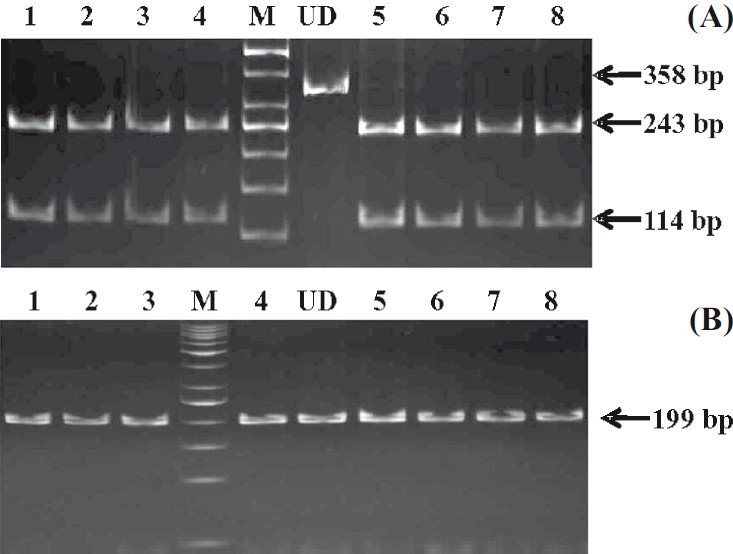
Gel showing 478C>T SNP in exon 4, lane M, 50 bp ladder; lane UD, undigested PCR product; lanes 1-4, 5-8, PCR products digested with Cfr131 showing CC genotype. (B). Gel picture of delAC in exon 5, lane M, 50 bp ladder; lane UD undigested PCR product; lanes 1-3, 4, 5-8, Box I digested PCR products showing -/- genotype.
CD36, being an important receptor molecule for modified lipoproteins, plays an important role in the regulation of lipid metabolism. Studies have shown its involvement in diverse disorders such as insulin resistance, dyslipidaemia, hyperlipidaemia, atherosclerosis32,33 and T2DM21,22. Lipid abnormalities in CD36 deficiency might depend on the presence of diabetes since the total cholesterol and triglyceride levels in diabetic CD36 deficient patients were higher than in control subjects and non-diabetic CD36-deficient patients33. Several kinds of CD36 gene mutations have been reported in CD36-deficient patients25. A dinucleotide (AC) deletion in exon 5 of CD36 gene led to a frameshift mutation resulting in a stop codon24 while the insertion of nucleotide A at position 1159 led to the abnormal splicing in mRNA resulting in skipping of exons 4, 9, 11 in type 1 CD36 deficient subjects26. However, our study did not show deletion of exons 3, 4 and 5 of the CD36 gene in north Indian T2DM patients as well as controls. All individuals tested for 478C>T and delAC polymorphisms were ‘CC’ and -/- homozygotes respectively. This suggests that in our population CD36 exonic deletion is not prevalent; as a result deficiency of CD36 protein is not evident.
CD36 expression in monocytes is upregulated by oxidized low density lipoprotein (Ox-LDL), whose levels increase in case of T2DM, hyperglycaemia and related atherosclerosis15. CD36 deficiency can promote defective insulin action (resistance) and disordered fatty acid metabolism in spontaneous hypertension34.
Although the molecular aspects of CD36 and its complications are not very clear, there have been attempts to study its polymorphisms and their association with T2DM. CD36 genotype was identified as a fundamental determinant of myocardial long chain fatty-acid uptake13,35,36. We have recently reported that one of the several SNPs in the CD36 gene (rs1761667, G>A) shows a significant association with T2DM in the north Indian population37. This provides a lead for the CD36 genotypes and their association with T2DM. Our findings are in support of the data obtained from other populations such as the -178 A/C SNP promoter mutation in the CD36 gene in the French population21,22 and a CD36 nonsense mutation in caucasians21. It appears that CD36 variants vary in different racial and ethnic populations. The CD 36 gene polymorphisms in selected north Indian population in this study did not show any variation but since the CD36 molecule has a definite role to play in T2DM, it will be interesting to determine the genetic variants at the CD36 locus and their association with insulin sensitivity, diabetes, and atherosclerosis in our population.
Acknowledgments
Authors acknowledge the financial support by Department of Biotechnology, New Delhi. The first author (SG) acknowledges the Junior Research Fellowship from Rajiv Gandhi National Fellowship, RGNF, UGC, New Delhi. Authors are also thankful to the technicians at the Diabetes Clinic, CSMMU, Lucknow.
References
- 1.International Diabetes Federation Diabetes Atlas. [accessed on September 10, 2010]. Available from: http://atlas.idf-bxl.org/content/diabetes .
- 2.Mohan V, Radhika G, Vijayalakshmi P, Sudha V. Can the diabetes/cardiovascular disease epidemic in India be explained, at least in part, by excess refined grain (rice) intake? Indian J Med Res. 2010;131:369–72. [PubMed] [Google Scholar]
- 3.Herder C, Rathmann W, Strassburger K, Finner H, Grallert H, Huth C, et al. Variants of the PPARG, IGF2BP2, CDKAL1, HHEX, and TCF7L2 genes confer risk of type 2 diabetes independently of BMI in the German KORA studies. Horm Metab Res. 2008;40:722–26. doi: 10.1055/s-2008-1078730. [DOI] [PubMed] [Google Scholar]
- 4.Lee YH, Kang ES, Kim SH, Han SJ, Kim CH, Kim HJ, et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet. 2008;53:991–8. doi: 10.1007/s10038-008-0341-8. [DOI] [PubMed] [Google Scholar]
- 5.Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes. 2008;57:2226–33. doi: 10.2337/db07-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Li H, Loos RJ, Yu Z, Ye X, Chen L, et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57:2834–42. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena M, Banerjee M. Diabetes: History, prevalence, insulin action and associated genes. J Appl Biosci. 2008;34:139–51. [Google Scholar]
- 8.Iwai N, Tago N, Yasui N, Kokubo Y, Inamota N, Tomoike H, et al. Genetic analysis of 22 candidate genes for hypertension in the Japanese population. J Hypertens. 2004;22:1119–26. doi: 10.1097/00004872-200406000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Ma X, Bacci S, Mlynarski W, Gottardo L, Soccio T, Menzaghi C, et al. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum Mol Genet. 2004;13:2197–205. doi: 10.1093/hmg/ddh233. [DOI] [PubMed] [Google Scholar]
- 10.Love-Gregory L, Sherva R, Sun L, Wasson J, Schappe T, Doria A, et al. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum Mol Gene. 2008;17:1695–704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noushmehr H, D’Amico E, Farilla L, Hui H, Wawrowsky KA, Mlynarski W, et al. Fatty acid translocase (FAT/CD36) is localized on insulin-containing granules in human pancreatic beta-cells and mediates fatty acid effects on insulin secretion. Diabetes. 2005;54:472–81. doi: 10.2337/diabetes.54.2.472. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg IJ, Eckel RH, Abumrad NA. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J Lipid Res. 2009;50(Suppl):S86–90. doi: 10.1194/jlr.R800085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyaoka K, Kuwasako T, Hirano K, Nozaki S, Yamashita S, Matsuzawa Y. CD36 deficiency associated with insulin resistance. Lancet. 2001;357:686–87. doi: 10.1016/s0140-6736(00)04138-6. [DOI] [PubMed] [Google Scholar]
- 14.Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J Clin Invest. 2002;109:1381–9. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handberg A, Levin K, Hojlund K, Beck-Nielsen H. Identification of the oxidized low-density lipoprotein scavenger receptor CD36 in plasma: a novel marker of insulin resistance. Circulation. 2006;114:1169–76. doi: 10.1161/CIRCULATIONAHA.106.626135. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita S, Hirano K, Kuwasako T, Janabi M, Toyama Y, Ishigami M, et al. Physiological and pathological roles of a multi-ligand receptor CD36 in atherogenesis; insights from CD36-deficient patients. Mol Cell Biochem. 2007;299:19–22. doi: 10.1007/s11010-005-9031-4. [DOI] [PubMed] [Google Scholar]
- 17.Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev. 2002;23:201–9. doi: 10.1210/edrv.23.2.0461. [DOI] [PubMed] [Google Scholar]
- 18.Koonen DP, Sung MM, Kao CK, Dolinsky VW, Koves TR, Ilkayeva O, et al. Alterations in skeletal muscle fatty acid handling predisposes middle-aged mice to diet-induced insulin resistance. Diabetes. 2010;59:1366–75. doi: 10.2337/db09-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J Biol Chem. 1994;269:18985–91. [PubMed] [Google Scholar]
- 20.Rac ME, Safranow K, Poncyljusz W. Molecular basis of human CD36 gene mutation. Mol Med. 2007;13:288–96. doi: 10.2119/2006-00088.Rac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepretre F, Vasseur F, Vaxillaire M, Scherer PE, Ali S, Linton K, et al. A CD36 nonsense mutation associated with insulin resistance and familial type 2 diabetes. Hum Mutat. 2004;24:104. doi: 10.1002/humu.9256. [DOI] [PubMed] [Google Scholar]
- 22.Lepretre F, Linton KJ, Lacquemant C, Vatin V, Samson C, Dina C, et al. Genetic study of the CD36 gene in a French diabetic population. Diabetes Metab. 2004;30:459–63. doi: 10.1016/s1262-3636(07)70143-x. [DOI] [PubMed] [Google Scholar]
- 23.Corpeleijn E, van der Kallen CJ, Kruijshoop M, Magagnin MG, de Bruin TW, Feskens EJ, et al. Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with Type 2 diabetes mellitus and insulin resistance. Diabet Med. 2006;23:907–11. doi: 10.1111/j.1464-5491.2006.01888.x. [DOI] [PubMed] [Google Scholar]
- 24.Kashiwagi H, Tomiyama Y, Kosugi S, Shiraga M, Lipsky RH, Kanayama Y, et al. Identification of molecular defects in a subject with type I CD36 deficiency. Blood. 1994;83:3545–52. [PubMed] [Google Scholar]
- 25.Tanaka T, Nakata T, Oka T, Ogawa T, Okamoto F, Kusaka Y, et al. Defect in human myocardial long-chain fatty acid uptake is caused by FAT/CD36 mutations. J Lipid Res. 2001;42:751–9. [PubMed] [Google Scholar]
- 26.Hanawa H, Watanabe K, Nakamura T, Ogawa Y, Toba K, Fuse I, et al. Identification of cryptic splice site, exon skipping, and novel point mutations in type I CD36 deficiency. J Med Genet. 2002;39:286–91. doi: 10.1136/jmg.39.4.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Y, Taylor KT, Sobieski DA, Medved ES, Lipsky RH. Identification of a human CD36 isoform produced by exon skipping. Conservation of exon organization and pre-mRNA splicing patterns with a CD36 gene family member, CLA-1. J Biol Chem. 1994;269:6011–15. [PubMed] [Google Scholar]
- 28.Kashiwagi H, Tomiyama Y, Honda S, Kosugi S, Shiraga M, Nagao N, et al. Molecular basis of CD36 deficiency. Evidence that a 478C-->T substitution (proline90-->serine) in CD36 cDNA accounts for CD36 deficiency. J Clin Invest. 1995;95:1040–6. doi: 10.1172/JCI117749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanai H, Chiba H, Fujiwara H, Morimoto M, Abe K, Yoshida S, et al. Phenotype-genotype correlation in CD36 deficiency types I and II. Thromb Haemost. 2000;84:436–41. [PubMed] [Google Scholar]
- 30.American Diabetes Association. Screening for type 2 Diabetes. Diabetes Care. 2004;27:S11–4. doi: 10.2337/diacare.27.2007.s11. [DOI] [PubMed] [Google Scholar]
- 31.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation and lipid metabolism. J Clin Invest. 2001;108:785–91. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuhashi M, Ura N, Nakata T, Shimamoto K. Insulin sensitivity and lipid metabolism in human CD36 deficiency. Diabetes Care. 2003;26:471–4. doi: 10.2337/diacare.26.2.471. [DOI] [PubMed] [Google Scholar]
- 34.Pravenec M, Zidek V, Simakova M. Genetics of Cd36 and the clustering of multiple cardiovascular risk factors in spontaneous hypertension. J Clin Chem. 1999;103:1651–7. doi: 10.1172/JCI6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai M, Tanaka T, Kintaka T, Ikemoto T, Shimizu A, Kitaura Y. Genomic heterogeneity of type II CD36 deficiency. Clin Chim Acta. 2002;321:97–106. doi: 10.1016/s0009-8981(02)00102-x. [DOI] [PubMed] [Google Scholar]
- 36.Kintaka T, Tanaka T, Imai M, Adachi I, Narabayashi I, Kitaura Y. CD36 genotype and long-chain fatty acid uptake in the heart. Circ J. 2002;66:819–25. doi: 10.1253/circj.66.819. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee M, Gautam S, Saxena M, Bid HK, Agrawal CG. Association of CD36 gene variants rs1761667 (G>A) and rs1527483 (C>T) with Type 2 diabetes in north Indian population. Int J Diab Mellitus. 2010;2:179–83. [Google Scholar]


