Abstract
Replication of the Escherichia coli chromosome is initiated at a unique site, oriC. Concurrent initiation occurs at all oriC sites present in a cell once, and only once, per cell cycle. A mechanism to ensure cyclic initiation events was found operating through the chromosomal site, datA, a 1-kb segment located at 94.7 min on the genetic map that titrates exceptionally large amounts of the bacterial initiator protein, DnaA. A strain lacking datA grew normally but exhibited an asynchronous initiation phenotype as a result of extra initiation events. This mutant phenotype was suppressed by DnaA-titrating plasmids. Furthermore, mutations in a 9-bp DnaA-binding sequence (the DnaA box) in datA were enough to induce the mutant phenotype. Thus, datA is a novel chromosomal element that appears to adjust a balance between free and bound DnaA for a single initiation event at a fixed time in the bacterial cell cycle. Titration of DnaA to newly duplicated datA during oriC sequestration, which is mediated by hemimethylated GATC sequences in oriC and the SeqA protein, would contribute to prevention of reinitiations when oriC is desequestered.
Keywords: datA, DnaA, initiation, oriC, replication
Chromosomal replication in Escherichia coli is normally initiated at a unique site, oriC, and proceeds bidirectionally (Bird et al. 1972; Marsh and Worcel 1977). In the initiation reaction, DnaA protein plays a key role. It binds to five 9-bp sequences, termed DnaA boxes, within oriC and, assisted by protein HU or integration host factor (IHF), causes unwinding of the strands in the region containing three AT-rich 13-mer repeats. DnaB helicase and other replisome components are loaded onto this open complex to start DNA replication (for review, see Skarstad and Boye 1994; Messer and Weigel 1996).
The initiation reaction is precisely regulated such that it takes place at a fixed time in the bacterial cell cycle. Control of DnaA protein activity seems to be most crucial to the timing of initiation (Løbner-Olesen et al. 1989; Atlung and Hansen 1993), but the molecular basis of this regulation is largely unknown. In a steady-state culture, intervals between initiation events in a cell are equal to the doubling time of cell number. Irrespective of growth rate, the time required for a round of replication and subsequent cell division is constant, being about 60 min at 37°C. As a consequence, in rapidly growing cells, new rounds of replication initiate while previous rounds are still in progress. Under such conditions, all origins fire essentially synchronously and, therefore, cells always contain 2n and 2n+1 origins (where n is a positive integer; Skarstad et al. 1986). This distribution enables partitioning of equal numbers of chromosomes into two daughter cells when cells divide.
In addition, each copy of oriC fires once, and only once, per cell cycle. A mechanism called sequestration is involved in preventing secondary initiations, which could occur immediately after primary initiations when initiation potential is still high. There are 11 GATC sequences in the 245-bp minimal oriC region that are sites for methylation by DNA adenine methyltransferase (Dam methylase; Zyskind and Smith 1992). Newly replicated GATC sites remain hemimethylated until Dam methylase transfers methyl groups to these sites. The time required for conversion of hemimethylated GATC sequences in oriC and in the promoter region of the dnaA gene (PdnaA), which also contains several GATC sequences, to the fully methylated state is much longer than that for other GATC sequences in the genome (Campbell and Kleckner 1990). The prolonged hemimethylated state is assumed to be caused by specific binding (sequestration) of these sites to the membrane (Ogden et al. 1988; Campbell and Kleckner 1990). Under such circumstances, oriC and PdnaA are protected from further methylation by Dam methylase, and reinitiation is blocked (Russell and Zinder 1987; Campbell and Kleckner 1990; Landoulsi et al. 1990). A gene, seqA, is involved in this process (Lu et al. 1994; von Freiesleben et al. 1994). Purified SeqA protein has a strong affinity for hemimethylated DNA. The binding, however, is specific neither to oriC nor to PdnaA (Brendler et al. 1995; Slater et al. 1995), suggesting involvement of other factor(s) for specific sequestration. SeqA also binds fully methylated oriC DNA in a specific manner (Slater et al. 1995). This binding may be involved in the timing of initiation.
To prevent reinitiation, initiation potential, which most likely implies active DnaA concentration, must fall to a level that can no longer cause initiation from oriC before the end of sequestration. Duplication of oriC itself could contribute to this requirement. oriC and PdnaA are located close on the chromosome (about 40 kb apart) and, therefore, the periods of sequestration of these two sites overlap most of the time. Transcription from PdnaA is transiently blocked during this period (Campbell and Kleckner 1990; Theisen et al. 1993). This block should also contribute to lowering the initiation potential, although it is not essential inasmuch as initiation synchrony is maintained when the dnaA gene is expressed constitutively at a level controlled by the lac promoter (Løbner-Olesen et al. 1989).
We have recently discovered a novel chromosomal site that titrates unusually large amounts of DnaA protein (Kitagawa et al. 1996). A single copy of this site, named datA (DnaA titration), titrates eightfold more DnaA molecules in vivo than a region spanning oriC and the mioC promoter, despite its twofold higher Kd value. The datA locus spans about 950 bp between the glyVXY and amiB-mutL operons at 94.7 min on the genetic map and could be replicated during oriC sequestration. In this study we have examined the role of datA in the control of initiation of replication. The data indicate that datA is a crucial cis element for regulated initiation. DnaA titration to datA appears to be a unique system that keeps a balance between DnaA and DNA concentrations and enables cell cycle-coupled synchronous initiation of replication. Sequestration and DnaA titration by datA are separate mechanisms, and each operates independently; presumably, the latter works after the former to assure the single initiation event in the cell cycle.
Results
A datA null mutant grows normally
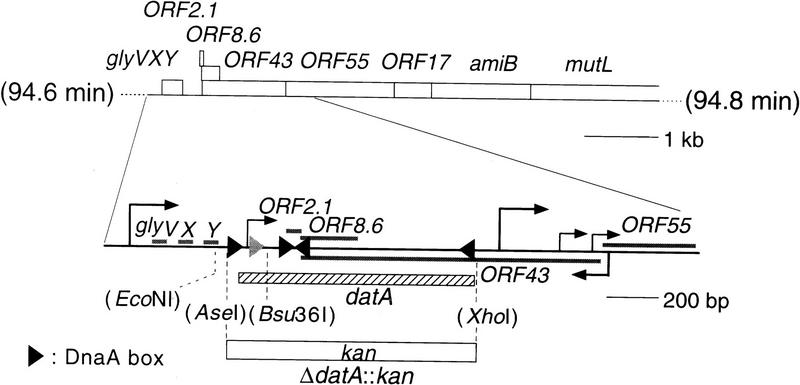
A deletion mutant of datA was constructed by replacing the 962-bp AseI–XhoI fragment (Kitagawa et al. 1996) containing the whole datA region by the kanamycin-resistance gene (Fig. 1). The mutation (ΔdatA::kan) was made in a recD strain and was introduced by P1 transduction without any problems into several other strains that differ in genetic backgrounds. The presence of a plasmid containing the AseI–XhoI fragment in a recipient cell (W3110) did not affect the efficiency of P1 transduction, suggesting that datA is dispensable for cell growth. In another experiment, the null allele was first integrated into the chromosome by homologous recombination such that direct repeats of mutant and wild-type alleles were separated by a plasmid vector sequence carrying the sacB gene (Slater and Maurer 1993; Ohmori et al. 1995). When the recombined segregants retaining only one allele were selected as sucrose-resistant colonies, 17 of 96 clones contained the mutant allele. Although this is half of the value expected from the length of homology available for the two required recombination events (1 kb at the left and 2 kb at the right), the results seemed to indicate that datA is a nonessential locus, inasmuch as all eight tested clones had expected mutant restriction patterns when examined by Southern hybridization.
Figure 1.
Location of datA and ΔdatA::kan mutation on the E. coli chromosome. ORFs are indicated with molecular masses (kD) of their products. In the enlarged map, the extent of the ORFs are indicated by bars; those above the line read to the right and the one below the line to the left. Bent arrows are initiation sites of transcription. Five arrowheads denote DnaA boxes. The second DnaA box, which was inactivated by the datA mutation is gray. The datA region is indicated by a thick striped bar. The region substituted by the gene conferring kanamycin resistance (kan) is also shown.
The ΔdatA::kan mutation did not cause any altered physiological characteristics in several genetic backgrounds. Doubling times of the mutants were similar to those of wild-type cells when grown in minimal media or in broth at temperatures between 20°C and 43°C. Cell size and morphology were indistinguishable from wild-type cells. In addition, the ΔdatA::kan mutation did not affect temperature sensitivity of growth of strains carrying dnaA5, dnaA204, or dnaA508. Also, the datA deletion did not affect growth of himA::Tn10, dam-13::Tn9, or a seqA null mutant. Strains containing the datA deletion did not exhibit any significant change in the total cellular amount of DnaA protein as measured by an immunoblotting assay (data not shown).
Overinitiation occurs and initiation synchrony is disturbed in ΔdatA::kan mutant
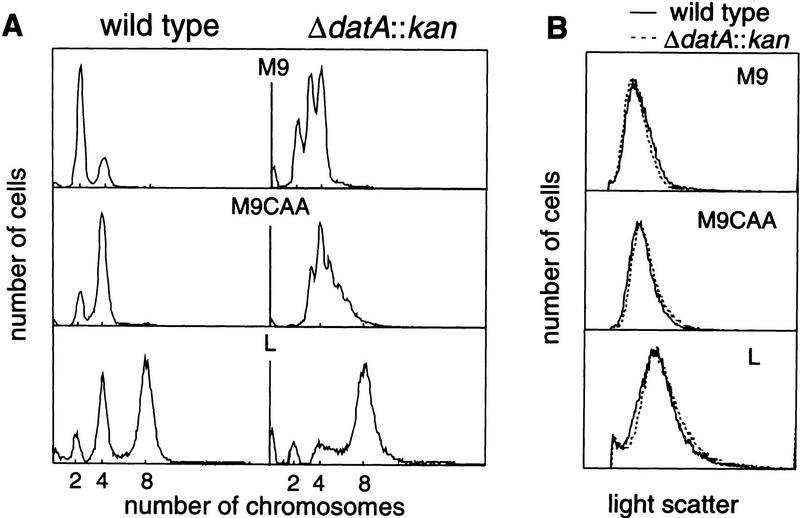
A drastic alteration in phenotype caused by ΔdatA::kan was revealed by flow cytometry. In the experiments shown in Figure 2A, distribution of the number of chromosomes per cell was measured after treating exponentially growing cells with rifampicin and cephalexin. These drugs block initiation of replication and cell division, respectively. After cultivation for enough time to complete ongoing replication, the number of chromosomes per cell normally distributes to 2n and 2n+1, where n = 0 or a positive integer, indicating that replication begins at the same time in the cell cycle at all origins present in a cell (Skarstad et al. 1986). Wild-type cells (W3110) exhibited the expected synchronous initiation pattern. The average number of chromosomes per cell, which is equivalent to the number of oriCs present in a cell at the time of addition of the drugs, increased upon increase of the cell growth rate, but no detectable peak of cellular chromosome numbers deviating from 2n appeared under these conditions (Fig. 2A, Table 1).
Figure 2.
Asynchronous and extra initiations in the ΔdatA:: kan strain. Cells growing exponentially in the indicated medium were treated with rifampicin and cephalexin for six generations, and run-out DNA histograms (A) or light-scatter histograms (B) were obtained by flow cytometry. Both W3110 (wild type) and RSD448 (ΔdatA::kan) had the same doubling time of 72 min in M9 medium, 50 min in M9CAA medium, and 28 min in L broth. Run-out DNA histograms measure the distribution of chromosome numbers per cell, which is equal to the number of origins per cell present at the time of addition of rifampicin and cephalexin. Light-scatter histograms measure cell size distribution.
Table 1.
DNA replication phenotypes of wild-type and ΔdatA::kan mutant
| Strain/genotypea
|
Medium
|
Origin/cellb
|
Asynchrony indexc
|
oriC/terd
|
DNA (nmoles/OD650)e
|
|
|---|---|---|---|---|---|---|
| − drugs
|
+ drugs
|
|||||
| Wild type | M9 | 2.4 ± 0.1 | <0.1 | 1.3 ± 0.1 | 23.0 ± 0.9 | 21.8 ± 1.3 |
| ΔdatA::kan | M9 | 3.3 ± 0.1 | 0.54 | 1.7 ± 0.1 | 22.4 ± 0.8 | 34.8 ± 2.4 |
| Wild type | M9CAA | 3.3 ± 0.3 | <0.1 | 1.5 ± 0.1 | 21.8 ± 2.4 | 21.5 ± 2.1 |
| ΔdatA::kan | M9CAA | 4.8 ± 0.8 | 2.1 | 2.0 ± 0.1 | 24.4 ± 2.9 | 29.6 ± 2.0 |
| Wild type | L | 6.1 ± 0.5 | <0.1 | 2.9 ± 0.3 | 32.5 ± 1.8 | 34.3 ± 0.9 |
| ΔdatA::kan | L | 7.2 ± 0.2 | 0.13 | 3.6 ± 0.1 | 34.6 ± 0.9 | 41.2 ± 2.4 |
Wild type is W3110. ΔdatA::kan is RSD448 (=W3110 ΔdatA::kan).
Calculated from the data of flow cytometry of cells treated with rifampicin and cephalexin.
Ratio of frequencies of three- and five- through seven-chromosome cells to two-, four- and eight-chromosome cells [(f3 + f5 + f6 + f7)/(f2 + f4 + f8)] in rifampicin- and cephalexin-treated culture.
Determined by Southern hybridization with exponentially growing cells.
Determined by the colorimetric assay using diphenylamine. (− drugs) Measured with exponentially growing cells; (+ drugs) measured with cells treated with rifampicin and cephalexin.
In contrast to the clear synchrony in wild-type cells, initiations were asynchronous in RSD448, a ΔdatA::kan derivative of W3110, in all media. In the M9 medium, 35% of cells contained three chromosomes. In the M9 medium supplemented with 0.2% casamino acids (M9CAA), all possible numbers of chromosomes between four and eight were present. The extent of asynchrony was reproducibly highest in M9CAA (Fig. 2A, Table 1), and, in RSD448 grown in L broth, the asynchrony of initiation was modest.
Another remarkable change caused by the datA deletion was an increase in the number of oriCs per cell as compared with wild type. Origin numbers in the mutant measured as chromosome numbers after treatment with the drugs were 20% to 45% higher than that of wild type (Table 1). The increase was caused by increased initiation frequency and was not attributable to inhibition of cell division, inasmuch as the mutant cells were not elongated (Fig. 2B). Initiation frequency in the ΔdatA:: kan mutant relative to wild-type cells was again highest in M9CAA (Table 1). Higher DNA concentration in the mutant after drug treatment was confirmed by a chemical assay with diphenylamine reagent. Overinitiation was also inferred from the increased oriC/ter ratio in the mutant measured by Southern hybridization (Table 1).
The above results indicate that datA is essential to prevent overinitiation and asynchronous initiation. The fact that origin number per cell was increased in datA− strains despite their cell size being indistinguishable from wild-type cells implies that primary initiations took place at an initiation mass (the cell mass at initiation divided by the number of origins) smaller than that of wild-type cells. Thus, datA has another role in the control of the timing of primary initiations.
Correlation between the ΔdatA mutant phenotype and loss of DnaA-titrating activity
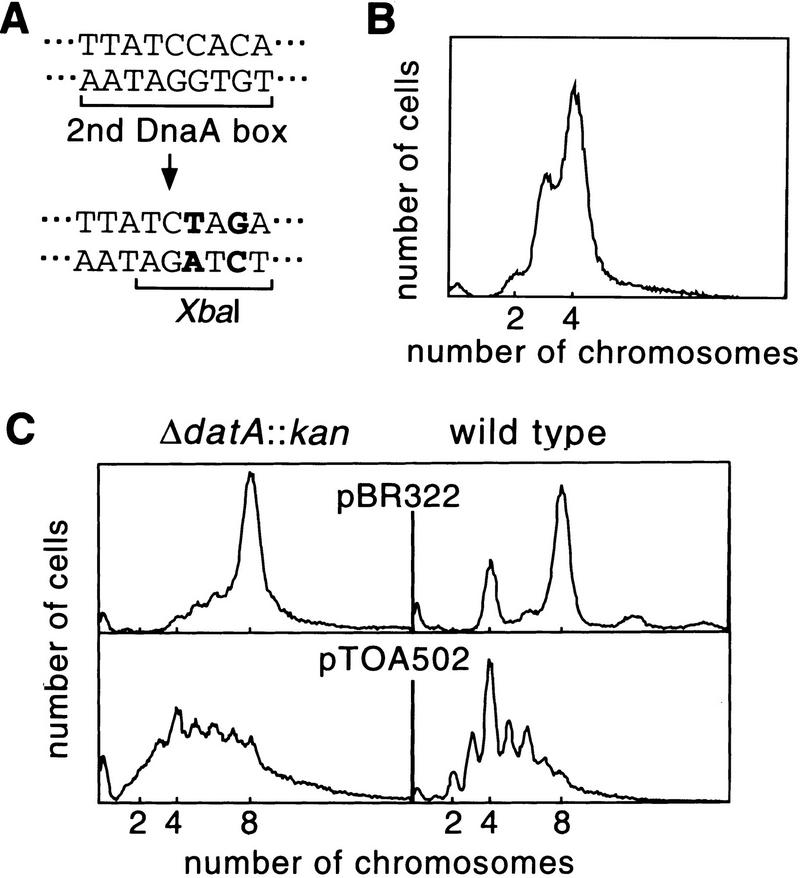
For the DnaA-titrating activity of datA, a region including the second DnaA box is essential (Kitagawa et al. 1996). Therefore, we introduced base substitutions into this DnaA box on the chromosome of wild-type strain W3110 to examine its role in initiation control. Two bases were substituted to create a XbaI site (designated as the datA1 mutation) as shown in Figure 3A. No other sequence alteration was made on the chromosome. The DNA histogram in Figure 3B indicates that the datA1 mutation was enough to bring about the mutant phenotype observed with RSD448. The inability of datA1 to titrate DnaA was demonstrated in vivo by measuring expression of the mioC–lacZ fusion gene which is repressed by DnaA (Kitagawa et al. 1996) (Table 2). pMW119-EX, carrying wild-type datA on an EcoNI–XhoI fragment (Fig. 1) of the pSC101-based vector pMW119, derepressed the fusion gene on pRML1. This derepression was not observed with pMW119-EX2, in which the second DnaA box is destroyed by the datA1 mutation. Levels of β-galactosidase activity decreased upon induction of DnaA protein from pBRCDI1 (Table 2), confirming that the activity responded to changes of cellular DnaA concentration.
Figure 3.
Fluorescence (DNA) histograms of strains disturbed in DnaA titration. (A) The datA1 mutation. (B) Run-out DNA histogram of RSD561 (W3110datA1) grown in M9 medium. The experiment was performed as described in Figure 2. (C) Run-out DNA histogram of RSD448 (pBR322), RSD448 (pTOA502), W3110 (pBR322), and W3110 (pTOA502). Cells grown in L broth supplemented with 40 μg/ml ampicillin were treated as described in Fig. 2. pTOA502 is a pBR322 derivative carrying the DnaA boxes of oriC region (see text).
Table 2.
Derepression of MioC–β-galactosidase by DnaA protein
| Plasmidsa
|
IPTG
|
β-galactosidase activity (units)
|
|---|---|---|
| pRML1, pMW119 | − | 9.4 ± 0.3 |
| pRML1, pMW119-EX | − | 20.8 ± 2.9 |
| pRML1, pMW119-EX2 | − | 9.4 ± 0.4 |
| pRML1, pBRCDI1, pMW119 | − | 7.0 ± 0.2 |
| pRML1, pBRCDI1, pMW119 | + | 0.7 ± 0.1 |
| pRML1, pBRCDI1, pMW119-EX | − | 11.4 ± 0.4 |
| pRML1, pBRCDI1, pMW119-EX | + | 2.0 ± 0.3 |
| pRML1, pBRCDI1, pMW119-EX2 | − | 8.1 ± 0.4 |
| pRML1, pBRCDI1, pMW119-EX2 | + | 1.7 ± 0.3 |
pRML1 contains a mioC–lacZ fusion gene downstream of the mioC promoter, which is under control of the DnaA protein. pBRCDI1 carries the dnaA gene under control of the tac promoter and the lacIq gene. pMW119-EX and pMW119-EX2 contain the EcoNI–XhoI fragment (Fig. 1) carrying wild-type datA and the datA1 mutation, respectively, on a pMW119 vector.
There are two small open reading frames (ORF2.1 and ORF8.6) and a portion of ORF43 in datA (Fig. 1). All of these ORFs appear to be expressed (Kitagawa et al. 1996). Although nonessential for growth, it was possible that the absence of the products of these genes causes unregulated initiation in the ΔdatA::kan mutant. However, exhibition of the mutant phenotype by the datA1 mutation outside these genes argues against this possibility. Expression of both ORF2.1 and ORF8.6 depends on transcription from a promoter just upstream of the second DnaA box (Fig. 1). The activity of this promoter was measured as the activity of chloramphenicol acetyl transferase (CAT) with a promoter assay vector pROAKK1 (Table 3). pROAKK-EB carrys the EcoNI–Bsu36I fragment (Fig. 1) on pROAKK1 in an orientation in which the promoter directs transcription of the cat gene. The datA1 mutation carried on pROAKK-EB2 did not cause any inhibition of this transcription. Some enhancement observed is in accordance with the negative role of DnaA on this transcription (Kitagawa et al. 1996). Thus, absence of ORF products of datA is not likely to be the cause of aberrant initiation control in datA::kan cells. Although less likely, however, the possibility that overexpression of the ORF products causes the same phenotype as that exhibited in their absence cannot be eliminated.
Table 3.
Effect of datA1 mutation on the promoter activity in datA
| Plasmida
|
CAT activity (units)
|
|---|---|
| pROAKK1 | 0 |
| pROAKK-EB | 193 ± 20 |
| pROAKK-EB2 | 550 ± 80 |
pROAKK-EB and pROAKK-EB2 contain the EcoNI–Bsu36I fragment (Fig. 1) with wild-type sequence and the datA1 mutation, respectively, on a promoter assay vector, pROAKK1, in an orientation that directs transcription of the cat gene on the vector from the promoter for ORF2.1 and ORF8.6.
A correlation between the DnaA-titrating activity and the activity that suppresses overinitiation was inferred further from experiments with the plasmid pTOA502, which is totally unrelated to datA. This plasmid carries five DnaA boxes in oriC and one in the mioC promoter region on pBR322 but is defective in oriC function because of a deletion in the AT-rich 13-mer region at the left end of oriC. In datA::kan cells carrying pTOA502, initiation frequency was remarkably reduced compared with control cells carrying pBR322 (Fig. 3C). Average chromosome number per cell in this strain was even lower than that in wild-type cells carrying pBR322. pTOA502 also reduced initiation frequency in wild-type cells. Furthermore, initiation synchrony was severely disturbed not only in mutant but also in wild-type cells carrying pTOA502. In these cells, reduced levels of free DnaA protein might have caused inefficient and asynchronous initiations. Other factors that may bind to the defective oriC would also contribute to these phenotypes.
The above results strongly suggest that initiation control is disturbed in datA mutants by the loss of DnaA-titrating activity. Although a defective oriC-carrying plasmid, pTOA502, was used in the experiment in Figure 3C to avoid possible effects of titration of replication proteins other than DnaA, essentially the same results were obtained with strains carrying an intact oriC–pBR322 chimeric plasmid, pTOA50 (Kano et al. 1991; data not shown). On the other hand, the presence of plasmid vectors including mini-F-based pKV713, pSC101-based pMW119, and pBR322 did not affect initiation of replication of host chromosomes (data not shown). All these plasmids carry DnaA box(es) in their origin region (Fuller et al. 1984). Futhermore, introduction of minichromosomes did not affect the initiation synchrony of host chromosome replication (Skarstad 1988). Therefore, a large increase of DnaA-titrating activity beyond a certain level appears to be necessary to disturb the initiation control.
Effect of datA sequence carried on plasmids
To examine whether the overinitiation and initiation asynchrony that occurred in the ΔdatA::kan mutant could be suppressed by the datA sequence in trans, various plasmids were introduced into RSD448. Both of these phenotypes were suppressed by pKV713-EX, which carries the EcoNI–XhoI fragment (Fig. 4). Therefore, the datA sequence does not need to be located on the chromosome for controlled initiation. Neither pKV713-EX2 bearing the datA1 mutation nor the vector pKV713 alone could suppress the mutant phenotype.
Figure 4.
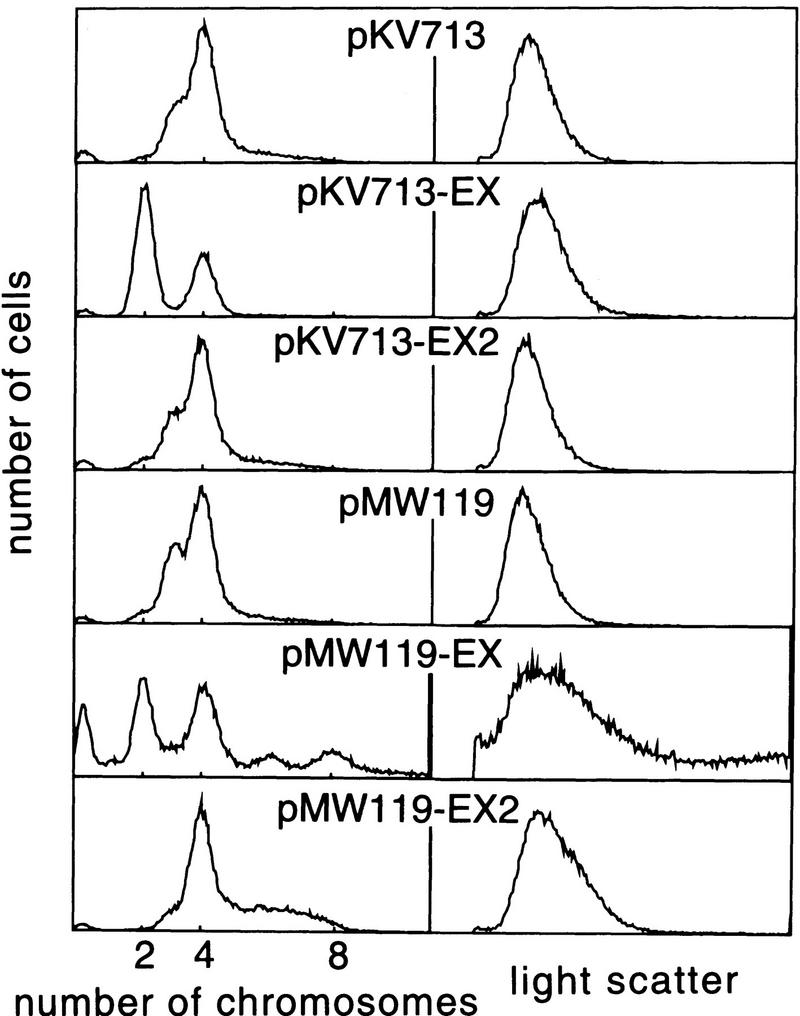
Fluorescence (DNA) and light-scatter histograms of RSD448 carrying various plasmids. RSD448 (ΔdatA::kan) cells transformed with the indicated plasmids were grown to log phase in M9 medium supplemented with 20 μg/ml ampicillin and analyzed as described in Fig. 2. pKV713 and pMW119 are mini-F- and pSC101-based vectors, repectively. Fragments EX and EX2 are EcoNI–XhoI fragments (Fig. 1), carrying wild-type datA and the datA1 mutation, respectively.
Remarkably, the fluorescence profile was strongly affected by plasmid copy number. pKV713-EX, which gave optimum suppression, is a mini-F-based plasmid and is assumed to be present at a copy number of one to two molecules per cell grown in M9 minimal medium. On the other hand, when datA was introduced into RSD448 on a pSC101-based vector pMW119, which is present at a copy number severalfold higher than pKV713, a significant number of cells contained over four chromosomes, although two-chromosome cells appeared with concomitant disappearance of three-chromosome cells as expected from suppression of the asynchrony phenotype. It turned out that this was attributable to the irregular cell size distribution as revealed by light scatter (Fig. 4); the larger number of chromosomes was contained in elongated cells as revealed by the fluorescence-light scatter diagram (data not shown). Inhibition of cell division became more significant when datA was present on a high-copy number plasmid such as pBR322 (data not shown). Growth of cells with datA on such a plasmid was severely slowed. We assume that too much titration of DnaA to excess datA sequences decreases cellular DnaA concentration and brings about division inhibition as has been observed when the cellular DnaA level is limited (Løbner-Olesen et al. 1989).
Initiation time in the cell cycle
Variation of origin numbers per cell during the cell cycle was investigated by use of synchronized cells of ON338 (B/rF26ΔdatA::kan). Newborn cells were collected by elution of growing cells from a nitrocellulose membrane filter (baby machine) and cultivated in test tubes at 37°C (Helmstetter et al. 1992). Origin numbers per cell were measured at various times by flow cytometry after treatment with rifampicin and cephalexin (Fig. 5). In accordance with the doubling time of 40 min under the growth conditions, a cyclic change of two- and four-origin cells was observed at this interval. The initiation event causes an increase of four-origin cells and a concomitant decrease of two-origin cells, while the opposite effects are caused by cell division. Two- and four-origin cells amounted to about 80% of the population under the experimental conditions with ON338. This suggests that the majority of the initiation events occur regularly at normal intervals in mutant cells.
Figure 5.
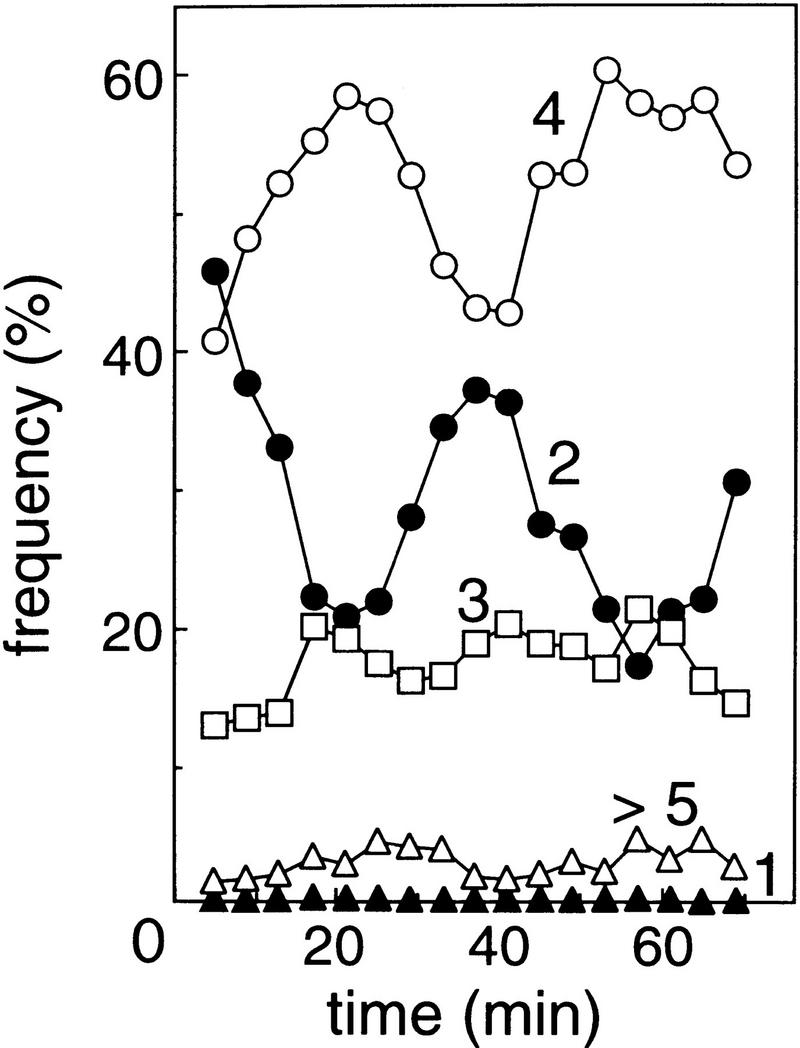
Change of origin number per cell during the cell cycle. Newborn cells of ON338 (B/rF26ΔdatA::kan) in M9 medium were collected using the baby machine technique and grown as batch cultures at 37°C. At the indicated times, samples were treated with rifampicin and cephalexin, and run-out DNA histograms were obtained by flow cytometry. The frequency of origin numbers was determined by measuring the area of each peak. Origin numbers are indicated on the graph.
Most of the remaining cells consisted of three-origin cells, which were typical to the datA mutant. The proportion of three-origin cells remained roughly constant throughout the cell cycle and did not decrease during the period of cell division (25–45 min and 55–70 min in Fig. 5). It seems that three-origin cells initiate replication to form four-origin cells before they divide, as there were no detectable one-origin cells. Decrease in the fraction of three-origin cells should be balanced by initiations in cells with two- or four-origins; the former forming three origins and the latter forming five or more origins and dividing soon after the initiations to produce three-origin cells. The period during which these extra initiations appeared to occur coincided with the period that included cell division and also overlapped with the time during which oriC is expected to be freed from sequestration after normally scheduled initiation. Because of the broad time range for initiation and division as well as the minor population of cells containing other than two or four origins, it is possible that, in certain population of cells, extra initiations occur at any time and those cells have irregular division intervals. However, because the cell size distribution of datA mutants was indistinguishable from that of wild-type cells (Fig. 2B; data not shown), the most reasonable explanation of the data would be that reinitiations occurred during the period between the end of sequestration and the next initiation event.
Replication of datA during oriC sequestration is not essential for controlled initiation
Sequestration of oriC mediated by SeqA protein and hemimethylated GATC sequences in oriC has been known to be a mechanism preventing reinitiation that could occur just after initiation when the initiation potential is still high (for review, see Crooke 1995). Inasmuch as sequestration is assumed to continue for about one-third of the cell cycle after replication initiation at the oriC located at 84.5 min on the genetic map, the datA locus at 94.7 min could be replicated during the sequestration period. Titration of DnaA to the newly replicated datA locus should contribute to lowering the initiation potential before the end of sequestration.
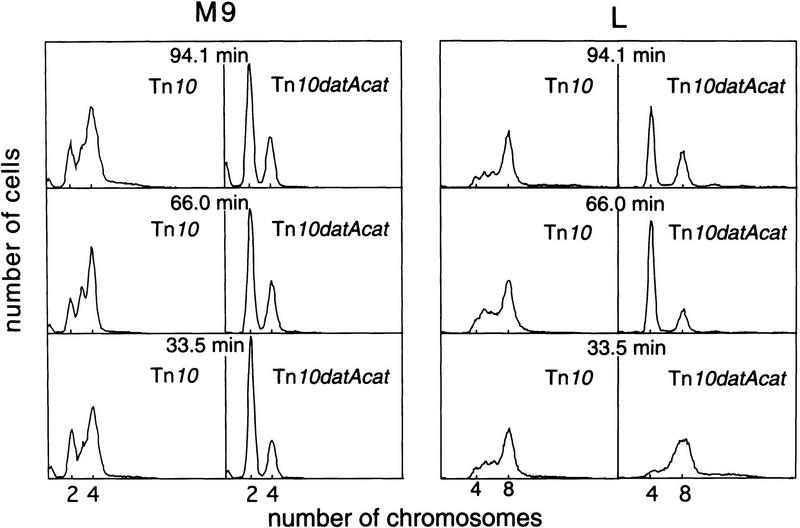
If titration of DnaA to the newly replicated datA locus during the sequestration period is essential for preventing reinitiation that appeared to occur after release of the sequestered oriC from the cellular membrane (Fig. 5), movement of the datA locus to positions that are replicated after the end of sequestration should permit some reinitiation reactions to occur prematurely. To construct strains carrying datA at different loci, we made use of strains that were derived from the wild-type strain MG1655 and have Tn10 located at various positions around the E. coli chromosome (Singer et al. 1989). Ten such strains were chosen that have Tn10 insertions ∼10 min apart on the genetic map. A portion of the Tn10 of each strain was replaced with the wild-type datA sequence ligated to the cat gene, and the original datA sequence was deleted. Chromosome ploidy of these strains was analyzed after treatment with rifampicin and cephalexin. In M9 minimal medium, all 10 strains carrying datA at different positions did not exhibit any significant alterations in the chromosome number distribution as compared with the wild type. Three representatives are shown in Figure 6 together with their datA− controls. Therefore, replication of datA during oriC sequestration is not required to prevent reinitiation.
Figure 6.
Effect of the chromosomal location of datA on the frequency and synchrony of initiation. Derivatives of the wild-type strain MG1655 devoid of datA at the original locus and instead carrying Tn10 or Tn10datA cat at indicated chromosomal loci were grown in M9 medium or L broth and analyzed as described in Fig. 2.
When the same 10 strains were grown in L broth, datA sequences translocated at all positions except for one eliminated the reinitiation reactions. The only exception that could not suppress the mutant phenotype was a strain carrying datA at 33.5 min (Fig. 6). Because this site is very close to the replication terminus (ter site), the inability to suppress reinitiations might be attributable to a reduced dosage of datA relative to the DnaA amounts, which is increased in fast-growing cells (see Discussion).
Discussion
Initiator titration model and datA
Our results suggest that datA titrates a large amount of the initiator protein DnaA, which consequently reduces free DnaA concentration below a level that provokes overinitiations. It has generally been assumed that initiation potential accumulates to a maximum level before initiation, then drops to a level that can no longer trigger initiation during the period of oriC sequestration on a membrane (Campbell and Kleckner 1990; for review, see Crooke 1995). The initiation potential most likely implies active DnaA concentration. In the computer-simulated initiator titration model, Hansen et al. (1991b) postulated that, after initiation, accumulated DnaA protein is titrated to newly replicated DnaA boxes on the chromosome during oriC sequestration. Our data provide evidence of the operation of a DNA-binding mechanism of the initiator protein in the control of initiation frequency. Induction of overinitiation by deleting the datA locus implies that DnaA titration to datA is essential for the chromosome number control.
Although many DnaA-binding sites are present on the E. coli chromosome (Messer and Weigel 1996; Roth and Messer 1998), which would contribute to lowering the initiation potential, datA appears to be prominent in its DnaA-titrating activity in vivo (Kitagawa et al. 1996). Severe growth inhibition observed with cells carrying datA on high-copy-number plasmids was accompanied neither by pTOA502-carrying cells nor by cells carrying an oriC–pBR322 chimeric plasmid, which has a higher copy number as a result of replication from oriC. Mutations in datA on a high-copy-number plasmid that recovered normal cell growth simultaneously reduced DnaA-titrating activity of datA (data not shown). Recently, in an in vitro assay, five high-affinity binding sites for DnaA were found on the chromosome besides oriC (Roth and Messer 1998). One of them was datA, and four others were located away from oriC. It would be interesting to know their capacity of DnaA titration in vivo and the phenotype of strains with deletions of these sites.
datA as a reservoir of DnaA
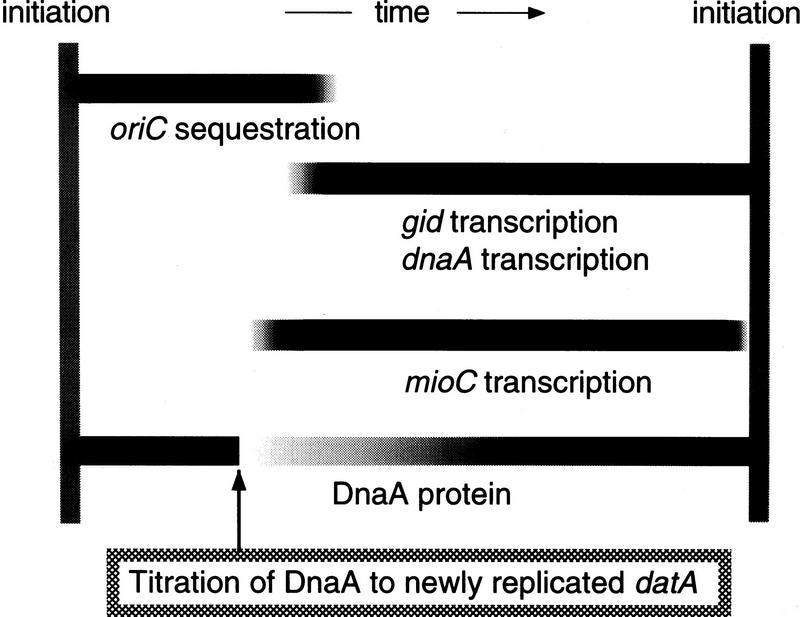
Despite the high level of DnaA titration to datA in vivo, the dissociation constant (Kd) of DnaA binding to datA is twofold higher than that to oriC as measured in vitro (Kitagawa et al. 1996). The lower affinity and higher capacity of DnaA binding relative to oriC might suggest the role of datA as a reservoir for DnaA molecules. DnaA boxes R1, R2, and R4, but not R3, in oriC are bound by DnaA throughout most of the cell cycle, and it is suggested that further aggregation of DnaA molecules to cover DnaA box R3 may be necessary to effect initiation (Samitt et al. 1989; Cassler et al. 1995). The concentration of free DnaA molecules would be kept to a very low level in wild-type cells, mainly because of titration to the datA reservoir, throughout most of the cell cycle (Fig. 7). Accumulation of free DnaA above a critical level to trigger initiation would be achieved just before initiation as inferred from the increased mRNA level at this time (Campbell and Kleckner 1990; Theisen et al. 1993). After initiation, duplicated oriC is sequestered to the membrane, and reinitiation is blocked during sequestration. The capacity of the datA reservoir is increased by duplication during oriC sequestration. Accumulated DnaA would be temporarily titrated to the datA reservoir and would be immediately retrieved by the DnaA boxes R1, R2, and R4 in oriC after desequestration.
Figure 7.
Model for the control of DNA replication initiation in E. coli. Periods of oriC sequestration and transcription of the indicated genes are shown by horizontal bars. Relative concentrations of free DnaA molecules are expressed by the degree of shading. See text for details. Cell cycle-dependent transcription from the gid and mioC promoters is accommodated, inasmuch as they are implicated in the control of initiation of replication of minichromosomes (Theisen et al. 1993; Ogawa and Okazaki 1994). Involvement of transcription from these promoters for efficient replication from the chromosomal oriC is suggested only for replication that takes place under certain suboptimal conditions (de Wind et al. 1987; Bates et al. 1997).
The present study revealed that the location of datA is not crucial for its function (Fig. 6). Thus, tight regulation of free DnaA concentration coupled to replication is not essential to prevent reinitiations. Although nonessential, replication of datA during oriC sequestration would be favored to reduce free DnaA to a safer level to prevent reinitiation especially under circumstances in which the amount of DnaA protein is elevated, inasmuch as the strain carrying datA at the ter region could not suppress the mutant phenotype when grown in L broth (Fig. 6).
During the period of oriC sequestration, the dnaA promoter region is also sequestered and transcription from this promoter is blocked (Fig. 7; Campbell and Kleckner 1990; Theisen et al. 1993). This should help promptly reduce the level of free DnaA protein.
In datA mutants, the free DnaA level might be increased as a result of the absence of the datA reservoir, the DnaA box R3 in oriC would be easily occupied by such molecules, and aberrant initiations could occur. Furthermore, excess free DnaA could trigger primary initiations at an initiation mass smaller than that of wild-type cells.
Asynchrony phenotype and overinitiation
The mechanism that keeps initiation synchrony is not known, but several conditions are known to disturb this synchrony. Temperature-sensitive dnaA mutants were the first example (Skarstad et al. 1988). Cells carrying mutations at the ATP-binding site in the dnaA gene seem to initiate replication at random, even under permissive conditions. Disturbance of the synchrony was less significant in other dnaA mutants. It has been suggested that the activity of DnaA protein is not simply related to its ability to synchronize multiple initiations. In another case, oversupply of DnaA protein provoked asynchronous initiation (Boye et al. 1988; Atlung and Hansen 1993). In dam and seqA mutants, initiations are strikingly asynchronous, presumably because newly replicated origins are not sequestered (Boye and Løbner-Olesen 1990; Lu et al. 1994). Finally, mutations in the fis and him genes, which assist the initiator function of DnaA (Boye et al. 1993), and mutations affecting the global level of chromosomal supercoiling (von Freiesleben and Rasmussen 1992) are known to cause initiation asynchrony. Therefore, initiation synchrony seems to be based on a fine balance of several factors involved in the initiation of replication. Overinitiation could accompany the asynchrony phenotype in some, but not all cases. In such cases, the origin number per cell should be larger than that of wild-type cells. On the other hand, initiations triggered under reduced initiation capacity might be asynchronous. In such cases, the origin number per cell should be smaller than that of wild-type cells.
Formally, the asynchrony phenotype may be classified into two types. In the first type, initiation occurs at random because of a distortion in the balance of factors essential to keep the timing and synchrony of initiation. In the second type, initiation occurs synchronously but some origins are aborted immediately after initiation. In the case of datA mutant cells, an increased cellular concentration of free DnaA molecules seems to have provoked asynchronous extra initiations after the oriC desequestration. It is also possible, however, that the second type of asynchrony phenotype described above is involved in datA mutants.
As shown in Figure 2 and Table 1, there was a drastic effect of cell growth rate on the degree of overinitiation and asynchrony in datA null cells. However, there was no apparent relationship between these factors. In addition, overinitiation was not strictly correlated with asynchrony. Among the three cultures, overinitiation and initiation asynchrony were most prominent in cells grown in M9CAA medium with an intermediate growth rate. Although these results were reproducible, there is no good explanation for this observation. These parameters of initiation phenotype should depend not only on a balance between cellular concentrations of free DnaA molecules and DNA (DnaA boxes), but also on many other factors as discussed above. Therefore, it is possible that, in datA null cells, some other factors in addition to concentrations of DnaA and DNA may be involved in the differential effect of growth rate on the datA phenotype.
Location of datA on the chromosome did not affect the frequency and synchrony of initiation, except when placed at 33.5 min and cultured in L broth (Fig. 6). Therefore, at a fixed growth rate, the normal initiation phenotype is not affected by the time of temporal change of free DnaA concentration in the cell cycle, which is assumed to occur by titration of DnaA to newly replicated datA. This suggests that, in datA+ cells, there is a certain range of free DnaA levels that does not cause aberrant initiation. When cultured in L broth, cellular numbers of DnaA molecules and original datA sites closely located to oriC should increase to high levels (Hansen et al. 1991a). Therefore, movement of datA to ter site, where the gene dosage is minimum, may have increased free DnaA concentration enough to induce the datA mutant phenotype in this medium.
The effect of overproduction of DnaA from a lacP-controlled dnaA gene has been studied extensively, and it is known that overinitiation occurs at increased DnaA levels (Atlung and Hansen 1993). In addition, depending on the degree of overproduction, various levels of initiation asynchrony accompany the overinitiation. Atlung and Hansen (1993) found that the increased initiation did not lead to a large increase in the DNA concentration because of the slowdown of replication fork velocity, which, for unknown reasons, appears to occur mostly near oriC. Similar consequences seem to have occurred in the datA-deleted strain, inasmuch as DNA content was not increased (Table 1).
DnaA inactivation and DnaA titration to datA
In vitro, ATP-bound DnaA is slowly converted to an ADP-bound inactive form by an ATPase activity intrinsic to DnaA protein (Sekimizu et al. 1987). Recently, it was found that the β subunit of DNA polymerase III holoenzyme, accompanied by a partially purified protein, IdaB, accelerates hydrolysis of ATP bound to DnaA (Katayama et al. 1998). This activity of the β subunit is dependent on its assembly as a sliding clamp on DNA and is stimulated by DNA synthesis. Thus, DNA replication appears to be coupled to inactivation of DnaA. Furthermore, a mechanism of conversion of ATP-bound DnaA to an ADP-bound inactive form has been suggested in which the β subunit appears to be involved independently of replication (Katayama and Crooke 1995). Although their physiological significance remains to be established, these activities could also contribute to restrict the initiation event to occur once, and only once, per cell cycle. The two forms of DnaA (ATP bound and ADP bound) should not be discriminated by datA, inasmuch as the DNA binding of DnaA is not affected by bound adenine nucleotide (Sekimizu et al. 1987). Therefore, the β clamp-dependent inactivation of DnaA could cooperate with DnaA titration to datA to assure complete inhibition of reinitiation.
Materials and methods
Bacterial strains, plasmids, and media
Bacterial strains used in this study are listed in Table 4. RSD497 was constructed by transformation of DPB271 (recD) by pMW119-BBΔdatA::kan linearized with SpeI (Russell et al. 1989). pMW119-BB carries the chromosomal 4162-bp BamHI fragment containing the datA locus on pSC101-based plasmid pMW119 (purchased from Nippon Gene, Toyama, Japan). pMW119BBΔdatA::kan was derived from this plasmid by replacing a 962-bp Asel–XhoI datA fragment (Kitagawa et al. 1996) with the 1340-bp StuI kan fragment of pACYC177. Clones containing ΔdatA::kan on the chromosome were obtained by selecting kanamycin-resistant colonies and the correct replacement of the datA sequence was confirmed by genomic Southern hybridization.
Table 4.
Bacterial strains
| Strain
|
Genotype
|
Source or referencea
|
|---|---|---|
| DH5α | supE44 ΔλlacU169 (φ80 lacZ ΔM15) recA1 endA1 gyrA96 thi-1 relA1 | laboratory stock |
| DPB271 | recD::minitet | (1) |
| RSD497 | DPB271 ΔdatA::kan | this work |
| W3110 | IN(rrnD–rrnE)1 | laboratory stock |
| RSD448 | W3110 ΔdatA::kan | P1 (RSD497) × W3110 |
| RSD561 | W3110datA1 | this work |
| B/rF26 | thyA his | (2) |
| ON338 | B/rF26ΔdatA::kan | P1 (RSD497) × B/rF26 |
| CSH50 | araΔ(lac pro) strA thi | laboratory stock |
| Q358 | supE hsdR φ80r | laboratory stock |
| YMEL | F+mel-1 supF58 | laboratory stock |
| CAG18436 | MG1655zae502::Tn10 | (3) |
| CAG12021 | MG1655zae502::Tn10 | (3) |
| CAG12078 | MG1655zce726::Tn10 | (3) |
| CAG18459 | MG1655zde234::Tn10 | (3) |
| CAG12099 | MG1655zee3129::Tn10 | (3) |
| CAG18481 | MG1655zff208::Tn10 | (3) |
| CAG12168 | MG1655zgd210::Tn10 | (3) |
| CAG18452 | MG1655zhe3085::Tn10 | (3) |
| CAG18501 | MG1655zie296::Tn10 | (3) |
| CAG18427 | MG1655zje2241::Tn10 | (3) |
| RSD411 | zae502::Tn10datA cat ΔdatA::kan | this work |
| RSD412 | zbc3105::Tn10datA cat ΔdatA::kan | this work |
| RSD413 | zce726::Tn10datA cat ΔdatA::kan | this work |
| RSD414 | zde234::Tn10datA cat ΔdatA::kan | this work |
| RSD415 | zee3129::Tn10datA cat ΔdatA::kan | this work |
| RSD416 | zff208::TndatA cat ΔdatA::kan | this work |
| RSD417 | zgd210::Tn10datA cat ΔdatA::kan | this work |
| RSD418 | zhe3085::Tn10datA cat ΔdatA::kan | this work |
| RSD419 | zie296::Tn10datA cat ΔdatA::kan | this work |
| RSD420 | zje2241::Tn10datA cat ΔdatA::kan | this work |
| RSD421 | CAG18436ΔdatA::kan | P1 (RSD497) × CAG18436 |
| RSD422 | CAG12021ΔdatA::kan | P1 (RSD497) × CAG12021 |
| RSD423 | CAG12078ΔdatA::kan | P1 (RSD497) × CAG12078 |
| RSD424 | CAG18459ΔdatA::kan | P1 (RSD497) × CAG18459 |
| RSD425 | CAG12099ΔdatA::kan | P1 (RSD497) × CAG12099 |
| RSD426 | CAG18481ΔdatA::kan | P1 (RSD497) × CAG18481 |
| RSD427 | CAG12168ΔdatA::kan | P1 (RSD497) × CAG12168 |
| RSD428 | CAG18452ΔdatA::kan | P1 (RSD497) × CAG18452 |
| RSD429 | CAG18501ΔdatA::kan | P1 (RSD497) × CAG18501 |
| RSD430 | CAG18427ΔdatA::kan | P1 (RSD497) × CAG18427 |
(1) Russell et al. (1989); (2) Leonard and Helmstetter (1986); (3) Singer et al. (1989), NIG collection (National Institute of Genetics, Mishima, Japan).
In addition to transformation of a recD mutant by a linear DNA as described above, a method described by Ohmori et al. (1995) was performed with some modifications to make ΔdatA:: kan strains for assessing the requirement of datA for growth. The AseI–XhoI datA fragment in the 4162-bp BamHI fragment was replaced by ΔdatA::kan and cloned into pKH5002SB, a derivative of pKH5002 (Ohmori et al. 1995) in which the rpsL+ gene is replaced by the Bacillus subtilis sacB gene (Ried and Collmer 1987) as a negative selection marker. The sacB gene was obtained from pBIP (Slater and Maurer 1993). pKH5002 is able to replicate only in an rnhA− strain and, therefore, upon transformation of wild-type cells with pKH5002SB carrying the BamHI Δdat::kan fragment, the plasmid is integrated into the chromosome via homologous recombination to form a nontandem direct repeat in which wild-type and mutant alleles are separated by the vector sequence. The cointegrates are unstable, and resolution of the diploid structure tends to occur by a second homologous recombination event that accompanies deletion of either the intact or the kan-disrupted genes together with the vector sequence. Selection for sucrose resistance gives rise to the wild type (Amps Kans) and, if the datA locus is dispensable for growth, the disruptant (Amps Kanr) colonies. RSD561 was also constructed by this procedure. The datA1 mutation was introduced into pKH5002SB carrying the 4162-bp BamHI fragment by the method of Kunkel et al. (1987). Southern hybridization was employed to select clones with the datA1 mutation, which creates an XbaI site (Fig. 3A).
Strains RSD411–RSD420 were constructed by introducing the datA sequence ligated to the cat gene into the Tn10 resident on the chromosome of a series of 10 CAG strains (Table 4). Tn10s of these strains distribute on the chromosome roughly 10 min apart. The SphI fragment of Tn10 (8.6 kb) was cloned into pACYC177, and its BglII tetr fragment was replaced by the AseI–XhoI datA fragment ligated to the BsaAI cat fragment of pACYC184. Q358 harboring Tn10datA cat carried on pACYC177 was infected with λ::Tn10 (λb221 cI857 cIII163:: Tn10 Oam29), and the resulting lysate was used to infect λcI857 Sam7-lysogen of YMEL. The lysate was then spread on L plates containing 10 μg/ml chloramphenicol. λ::Tn10datA cat phages were recovered from the lysogen following heat induction. Ten CAG strains carrying Tn10 were infected with λ::Tn10datA cat at a m.o.i. of 1, and Tets Camr colonies were isolated. To confirm that Tn10datA cat is located at the original Tn10 position, P1vir grown on the parental Tn10 strains was used to transduce the candidate strains to Tetr. The Tn10datA cat was considered to be 100% linked to the original Tn10 when all of the 100 transductants scored were Cams. The datA at the original locus on chromosome in each strain was replaced with ΔdatA::kan by P1 transduction.
pRML1 contains a mioC–lacZ fusion gene downstream of the mioC promoter (Kitagawa et al. 1996) on a miniR1 plasmid pKN1562 (obtained from S. Yasuda, National Institute of Genetics, Mishima, Japan). pMW119-EX and pKV713-EX were constructed by inserting the EcoNI–XhoI datA fragment into the SmaI site of pMW119 and the EcoRI site of a mini-F plasmid pKV713 (obtained from C. Wada, Kyoto University, Japan), respectively. pROAKK-EB carries the EcoNI–Bsu36I fragment (Fig. 1) in pROAKK1 (Kitagawa et al. 1996). pKV713-EX2 and pMW119-EX2 carry the datA1 mutation in pKV713-EX and pMW119-EX, respectively. pROAKK-EB2 carries the datA1 mutation in pROAKK-EB. pBRCDI1 is a pBR322-based vector carrying the dnaA gene under the control of the tac promoter and the lacIq gene. In addition, this plasmid has the cat gene of pACYC184 in place of the bla gene. pTOA502 was derived from pTOA50 (Kano et al. 1991), which carries the chromosomal AatII–HaeII fragment spanning oriC and mioC on pBR322, by deleting the SmaI–BglII fragment.
Culture media were L broth (10 grams/liter Bacto-tryptone, 5 grams/liter Bacto-yeast extract, 5 grams/liter NaCl, pH 7.2), M9 medium (Miller 1972) containing 0.2% glucose or M9CAA medium, which is M9 medium supplemented with 0.2% casamino acids. When necessary, required amino acids (40 μg/ml except where indicated), thiamine (5 μg/ml) or thymine (20 μg/ml) were added. Appropriate antibiotics (50 μg/ml ampicillin, 40 μg/ml kanamycin, 10 μg/ml tetracycline or 15 μg/ml chloramphenicol) were added for plasmid-carrying cells.
Flow cytometry
Origin number per cell was determined as described (Løbner-Olesen et al. 1989). Exponentially growing cells were incubated with 300 μg/ml rifampicin and 12 μg/ml cephalexin for six generations, fixed with ethanol, stained with 90 μg/ml mithramycin and 20 μg/ml ethidium bromide, and analyzed by a flow cytometer (Bryte HS, Bio-Rad).
Synchronization by the baby machine technique
To obtain synchronous culture under steady-state growth conditions, newborn cells of ON338 were collected using the baby machine (Helmstetter et al. 1992). Cells were grown in M9 medium supplemented with 40 μg/ml histidine and 20 μg/ml thymine at 37°C. When the culture reached an optical density at 460 nm of 0.25, cells were collected onto the surface of a 142-mm diameter membrane filter (Millipore, type GS) coated with poly-d-lysine. The effluents containing the newborn cells were pooled in a test tube for 4 min consecutively and transferred to a shaking water bath at 37°C. Samples were taken at various times for flow cytometry.
Southern hybridization
Gene dosages of oriC and ter were measured by Southern hybridization (Ogawa and Okazaki 1994; Table 1). Total cellular DNA was digested with PstI before agarose gel electrophoresis. 32P-labeled probes were the AvaI oriC fragment (463 bp) and a 1.3-kb KpnI fragment at 35 min on the genetic map (ter region) obtained from the Kohara clone 306 (2B2; Kohara et al. 1987).
Miscellaneous procedures
Assay for β-galactosidase (Table 2) was performed as described (Miller 1972) using plasmid-carrying CSH50 cells grown to a log phase in M9 minimal medium supplemented with the required nutrients and appropriate antibiotics. IPTG was added at 0.5 mm where indicated, which elevated the cellular DnaA level 5- to 10-fold. The diphenylamine assay for DNA content of E. coli cells was as described (Burton 1956). Assay for CAT was as described (Kitagawa et al. 1996). One unit of the activity was defined as the amount that catalyzes synthesis of acetylated chloramphenicol in 30 min at 30°C.
Acknowledgments
We are grateful to C.E. Helmstetter for introduction of the baby machine technique and for the strain B/rF26. We thank R. Maurer, A. Nishimura, H. Ohmori, C. Wada, and S. Yasuda for plasmids, phage, and E. coli strains. We also thank B.E. Funnell and J. Zyskind for comments on the manuscript, and T. Okazaki for encouragement. This work was partly supported by grants in aid for Scientific Research on Priority Area from the Ministry of Education, Science, Sports, and Culture of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL h4485la@nucc.cc.nagoya_u.ac.jp; FAX 81 052 789 3001.
References
- Atlung T, Hansen FG. Three distinct chromosome replication states are induced by increasing concentrations of DnaA protein in Escherichia coli. J Bacteriol. 1993;175:6537–6545. doi: 10.1128/jb.175.20.6537-6545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DB, Boye E, Asai T, Kogoma T. The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli. Proc Natl Acad Sci. 1997;94:12497–12502. doi: 10.1073/pnas.94.23.12497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird RE, Louarn J, Martuscelli J, Caro L. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol. 1972;70:549–566. doi: 10.1016/0022-2836(72)90559-1. [DOI] [PubMed] [Google Scholar]
- Boye E, Løbner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- Boye E, Løbner-Olesen A, Skarstad K. Timing of chromosomal replication in Escherichia coli. Biochim Biophys Acta. 1988;951:359–364. doi: 10.1016/0167-4781(88)90107-8. [DOI] [PubMed] [Google Scholar]
- Boye E, Lyngstadaas A, Løbner-Olesen A, Skarstad K, Wold S. Regulation of DNA replication in Escherichia coli. In: Fanning E, Knippers R, Winnacker E-L, editors. DNA replication and the cell cycle. 43. Mosbacher Kolloquium. Berlin, Germany: Springer; 1993. pp. 15–26. [Google Scholar]
- Brendler T, Abeles A, Austin S. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acids. Biochem J. 1956;62:315–322. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–969. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- Cassler MR, Grimwade JE, Leonard AC. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995;14:5833–5841. doi: 10.1002/j.1460-2075.1995.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E. Regulation of chromosomal replication in E. coli: Sequestration and beyond. Cell. 1995;82:877–880. doi: 10.1016/0092-8674(95)90020-9. [DOI] [PubMed] [Google Scholar]
- de Wind N, Parren P, Stuitje RA, Meijer M. Evidence for the involvement of the 16kD gene promoter in initiation of chromosomal replication of Escherichia coli strains carrying a B/r-derived replication origin. Nucleic Acids Res. 1987;15:4901–4914. doi: 10.1093/nar/15.12.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Hansen FG, Atlung T, Braun RE, Wright A, Hughes P, Kohiyama M. Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1991a;173:5194–5199. doi: 10.1128/jb.173.16.5194-5199.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen FG, Christensen BB, Atlung T. The initiator titration model: Computer simulation of chromosome and minichromosome control. Res Microbiol. 1991b;142:161–167. doi: 10.1016/0923-2508(91)90025-6. [DOI] [PubMed] [Google Scholar]
- Helmstetter CE, Eenhuis C, Theisen P, Grimwade J, Leonard AC. Improved bacterial baby machine: Application to Escherichia coli K-12. J Bacteriol. 1992;174:3445–3449. doi: 10.1128/jb.174.11.3445-3449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano Y, Ogawa T, Ogura T, Hiraga S, Okazaki T, Imamoto F. Participation of the histone-like protein HU and of IHF in minichromosomal maintenance in Escherichia coli. Gene. 1991;103:25–30. doi: 10.1016/0378-1119(91)90386-p. [DOI] [PubMed] [Google Scholar]
- Katayama T, Crooke E. DnaA protein is sensitive to a soluble factor and is specifically inactivated for initiation of in vitro replication of the Escherichia coli minichromosome. J Biol Chem. 1995;270:9265–9271. doi: 10.1074/jbc.270.16.9265. [DOI] [PubMed] [Google Scholar]
- Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- Kitagawa R, Mitsuki H, Okazaki T, Ogawa T. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol Microbiol. 1996;19:1137–1147. doi: 10.1046/j.1365-2958.1996.453983.x. [DOI] [PubMed] [Google Scholar]
- Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: Application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Landoulsi A, Malki A, Kern R, Kohiyama M, Hughes P. The E. coli cell surface specifically prevents the initiation of DNA replication at oriC on hemimethylated DNA template. Cell. 1990;63:1053–1060. doi: 10.1016/0092-8674(90)90508-c. [DOI] [PubMed] [Google Scholar]
- Leonard AC, Helmstetter CE. Cell-cycle specific replication of Escherichia coli minichromosomes. Proc Natl Acad Sci. 1986;83:5101–5105. doi: 10.1073/pnas.83.14.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Løbner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57:881–889. doi: 10.1016/0092-8674(89)90802-7. [DOI] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N. SeqA: A negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Marsh RC, Worcel A. A DNA fragment containing the origin of replication of the Escherichia coli chromosome. Proc Natl Acad Sci. 1977;74:2720–2724. doi: 10.1073/pnas.74.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W, Weigel C. Initiation of chromosome replication. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 1579–1601. [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Ogawa T, Okazaki T. Cell cycle-dependent transcription from the gid and mioC promoters of Escherichia coli. J Bacteriol. 1994;176:1609–1615. doi: 10.1128/jb.176.6.1609-1615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden GB, Pratt MJ, Schechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988;54:127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Saito M, Yasuda T, Nagata T, Fujii T, Wachi M, Nagai K. The pcsA gene is identical to dinD in Escherichia coli. J Bacteriol. 1995;177:156–165. doi: 10.1128/jb.177.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried JL, Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57:239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- Roth A, Messer W. High-affinity binding sites for the initiator protein DnaA on the chromosome of Escherichia coli. Mol Microbiol. 1998;28:395–401. doi: 10.1046/j.1365-2958.1998.00813.x. [DOI] [PubMed] [Google Scholar]
- Russell CB, Thaler DS, Dahlquist FW. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Zinder ND. Hemimethylation prevents DNA replication in E. coli. Cell. 1987;50:1071–1079. doi: 10.1016/0092-8674(87)90173-5. [DOI] [PubMed] [Google Scholar]
- Samitt CE, Hansen FG, Miller JF, Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989;8:989–993. doi: 10.1002/j.1460-2075.1989.tb03462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. A collection of strains containing genetically linked alternating antibiotic resistant elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, Boye E. The initiation protein DnaA: Evolution, properties and function. Biochim Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, von Meyenburg K, Hansen FG, Boye E. Coordination of chromosome replication initiation in Escherichia coli: Effects of different dnaA alleles. J Bacteriol. 1988;170:852–858. doi: 10.1128/jb.170.2.852-858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S, Maurer R. Simple phagemid-based system for generating allele replacement in Escherichia coli. J Bacteriol. 1993;175:4260–4262. doi: 10.1128/jb.175.13.4260-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- Theisen PW, Grimwade JE, Leonard AC, Bogan JA, Helmstetter CE. Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol Microbiol. 1993;10:575–584. doi: 10.1111/j.1365-2958.1993.tb00929.x. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV. The level of supercoiling affects the regulation of DNA replication in Escherichia coli. Res Microbiol. 1992;143:655–663. doi: 10.1016/0923-2508(92)90060-2. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14:763–772. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- Zyskind JW, Smith DW. DNA replication, the bacterial cell cycle, and cell growth. Cell. 1992;69:5–8. doi: 10.1016/0092-8674(92)90112-p. [DOI] [PubMed] [Google Scholar]