Abstract
The prl1 mutation localized by T-DNA tagging on Arabidopsis chromosome 4-44 confers hypersensitivity to glucose and sucrose. The prl1 mutation results in transcriptional derepression of glucose responsive genes defining a novel suppressor function in glucose signaling. The prl1 mutation also augments the sensitivity of plants to growth hormones including cytokinin, ethylene, abscisic acid, and auxin; stimulates the accumulation of sugars and starch in leaves; and inhibits root elongation. PRL1 encodes a regulatory WD protein that interacts with ATHKAP2, an α-importin nuclear import receptor, and is imported into the nucleus in Arabidopsis. Potential functional conservation of PRL1 homologs found in other eukaryotes is indicated by nuclear localization of PRL1 in monkey COS-1 cells and selective interaction of PRL1 with a nuclear protein kinase C–βII isoenzyme involved in human insulin signaling.
Keywords: Glucose repression, hormone sensitivity, cell elongation, WD-40 protein, α-importin
Coordination of signaling pathways responding to hormonal, metabolic and environmental stress stimuli has a central role in plant growth control. Arabidopsis seedlings developing in the dark undergo fast elongational growth until the depletion of carbon reserves of the cotyledons. For subsequent growth, seedlings require either an external carbon supply or a light signal perceived by the photoreceptors controlling photomorphogenesis and de-etiolation required for autotrophic growth (Chory et al. 1996). In particular, far-red light signaling via the photoreceptor phytochrome A is negatively regulated by sucrose via glucose repression, and this effect is alleviated by the sun mutations (Dijkwel et al. 1997). In addition to glucose and sucrose, photomorphogenesis is antagonized by certain plant hormones, such as brassinosteroids. In contrast, cytokinins synergistically enhance the induction of de-etiolation by light. Brassinosteroid deficiency, as well as cytokinin treatment of wild-type plants, therefore yield a phenocopy of mutations causing de-etiolation (Chory et al. 1994; Li et al. 1996; Szekeres et al. 1996). Mutations of the COP, FUS, and DET genes result in constitutive photomorphogenesis and de-etiolation in the dark (von Arnim and Deng 1996). COP1 encodes a regulatory protein carrying β-transducin-like WD-40 repeats. COP1 is proposed to act as a nuclear repressor of light-regulated genes in concert with the COP9 complex in dark-grown plants (von Arnim and Deng 1994; Chamovitz et al. 1996). Functional analogies between COP1 and the TUP1 WD protein, acting as a general repressor of glucose-regulated genes in yeast (Tzamarias and Struhl 1995), as well as between the COP9 complex and the SWI/SNF modulators of RNA polymerase II (Pol II) have been noted (Chamovitz et al. 1996; Chory et al. 1996; Wilson et al. 1996). Although the role of COP1 in glucose repression is still unknown, its cytoplasmic localization in the light suggests that COP1 is unlikely to function as a TUP1-like repressor in glucose signaling of light-grown plants (von Arnim and Deng 1996).
Carbon partitioning is mediated by sucrose transport in many plant species. Growth control by carbon partitioning is therefore thought to be executed at the cellular level by glucose signaling (Stitt and Sonnewald 1995). In light-grown plants, sucrose feeding and inhibition of sucrose transport, leading to cellular sugar accumulation, result in the inhibition of photosynthesis and chlorophyll biosynthesis, defective root development, as well as induction of stress responses and accumulation of starch and anthocyanins (von Schaewen et al. 1990; Riesmeier et al. 1994; Herbers et al. 1996). As in other eukaryotes, hexose phosphorylation by hexokinases is believed to provide a signal for glucose repression also in plants (Jang et al. 1997). Glucose repression down-regulates the synthesis and stability of mRNAs coding for chlorophyll a/b-binding proteins, enzymes acting in starch degradation, and Calvin and glyoxylate cycles. At the same time, glucose signaling induces the expression of genes encoding storage and defense proteins, and enzymes involved in glycolysis, nitrate assimilation, phosphate mobilization, and anthocyanin biosynthesis (Faure et al. 1994; Smeekens and Rook 1997). In cross-talk with glucose signaling, cytokinins alleviate glucose repression of the photosynthetic genes and synergistically activate the expression of glucose-induced genes. Other plant hormones may have only a secondary role in glucose responses because their synthesis is either directly or indirectly controlled by light-, glucose-, and cytokinin-signaling (for review, see Chory et al. 1996).
In addition to complex cross-talk between hormonal and metabolic regulation, genetic dissection of plant glucose signaling is confronted with the problem that plants themselves produce glucose by CO2 fixation. Because light signaling is modulated by glucose and cytokinin and, vice versa, glucose and cytokinin signaling is controlled by light, mutations affecting glucose regulation may cause either lethality or severe developmental defects. Mutations relieving glucose repression are therefore expected to result in an enhanced expression of glucose responsive genes, as well as in potential defects in cytokinin signaling, root development, general stress responses, and chlorophyll and anthocyanin biosynthesis (Smeekens and Rook 1997). Here we show that such a phenotype is conferred by a recessive mutation in the Pleiotropic regulatory locus 1 (PRL1) encoding a conserved nuclear WD-protein that functions as a pleiotropic regulator of glucose and hormone responses in Arabidopsis.
Results
The prl1 mutation results in altered carbon partitioning and hypersensitivity to glucose and sucrose
A mutant displaying growth arrest in the presence of 175 mm sucrose or glucose (Fig. 1i), but wild-type growth responses to nonmetabolizable sugars and osmolytes (listed in Materials and Methods), was identified in an Arabidopsis T-DNA insertional mutant collection (Koncz et al. 1992). The mutation causing glucose and sucrose hypersensitivity resulted in complex recessive phenotypic defects (Fig. 1) that cosegregated with the hygromycin resistance marker of the T-DNA-tagged locus PRL1 mapped by genetic linkage analysis to chromosome 4-44 (see Materials and Methods). Sugar dose-growth response curves monitoring shoot and root weight, root length, and shoot/root ratio revealed no significant difference between wild-type and prl1 plants grown in the presence of low concentrations [0.1% (3 mm) and 0.5% (15 mm)] of sucrose (Fig. 2a–d). Root elongation of prl1 was reduced two- to threefold in comparison to wild type, independent of the concentration of external carbon and nitrogen sources (Fig. 2c; data not shown). Increasing the sucrose concentration up to 4% (117 mm), however, resulted in severe inhibition of both shoot and root development. Therefore, the shoot/root ratio of prl1 plants growing on higher than 1% sucrose did not change dramatically. On 6% (175 mm) sucrose prl1 barely grew and lost viability within 3 weeks. In comparison with wild type, the onset of growth defects correlated with a two- to fivefold increase of free glucose, fructose, sucrose, and starch content in leaves of prl1 seedlings grown on 2% (59 mm) and 4% sucrose (Fig. 2e–g).
Figure 1.
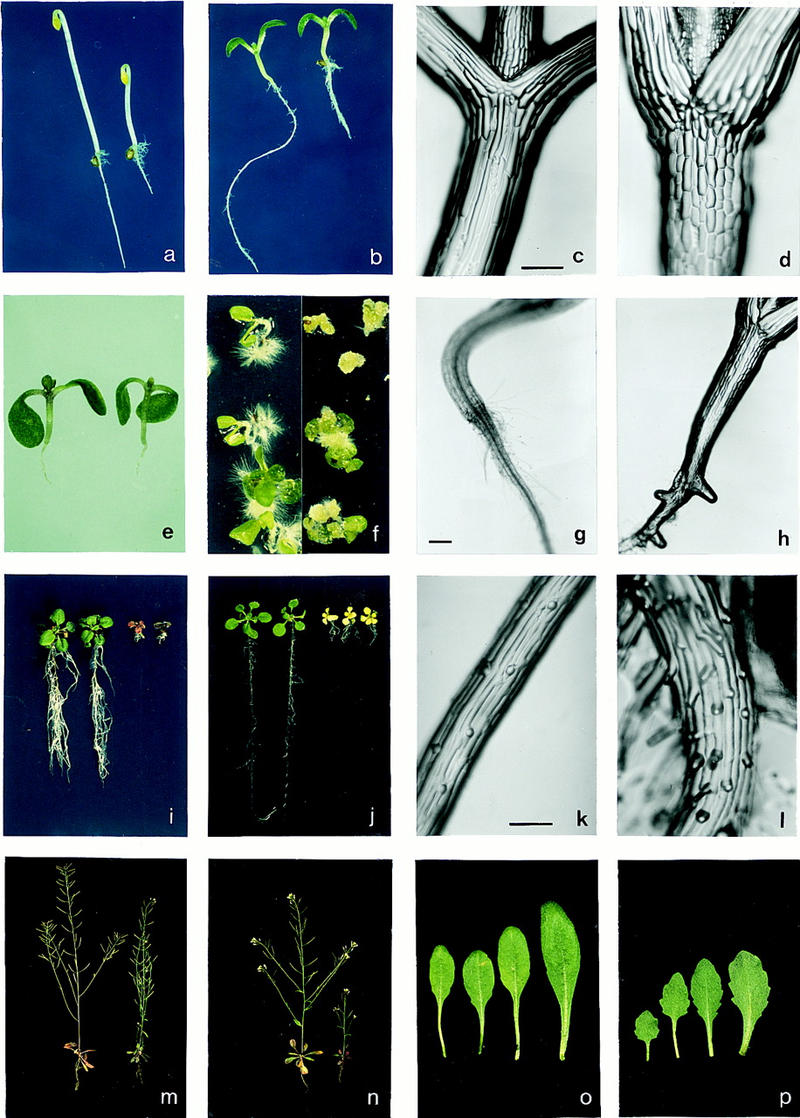
Effects of the prl1 mutation on seedling development and growth responses to glucose, cold stress, and plant hormones. In comparison with wild type (left in a and b), the prl1 mutant (right in a and b) exhibits reduced hypocotyl elongation in the dark (a), and inhibition of root elongation both in the dark (a) and in the light (b). The length of barrel-shaped epidermal cells of the hypocotyl of light-grown prl1 seedlings (d) is about half of that of elongated wild-type epidermal cells (c). When grown on cytokinin and sucrose in the light, the phenotypes of wild-type (left in e) and prl1 (right in e) seedlings are similar. In the presence of auxin, inhibiting the elongation of primary roots, wild-type plants (left in f) develop side roots densely covered by hairs, whereas the roots of prl1 seedlings (right in f) are converted to quickly proliferating, unorganized callus tissues. After 5 days of germination in the light, no side roots are observed on the primary root of wild-type seedlings (g), whereas prl1 develops numerous adventitious root initials (h). In the presence of 175 mm glucose prl1 seedlings (right in i) accumulate anthocyanins and loose viability in contrast to wild type (left in i). Unlike wild-type (left in j), prl1 seedlings (right in j) display bleaching and growth retardation when planted in media containing 0.1 μm ABA. In contrast with alternating files of trichoblasts and atrichoblasts on the wild-type root epidermis (k), adjacent rhizodermal files carry ectopic root hairs in prl1 (l). At 24°C (m) the size of wild type (left in m and n), and prl1 (right in m and n) is comparable, but at 14°C (n) prl1 exhibits a significant growth reduction. In comparison with wild type (o), leaves of the prl1 mutant (p) are smaller and display short petioles and serrated leaf margins. Scale bars in c,g,k, 200 μm.
Figure 2.
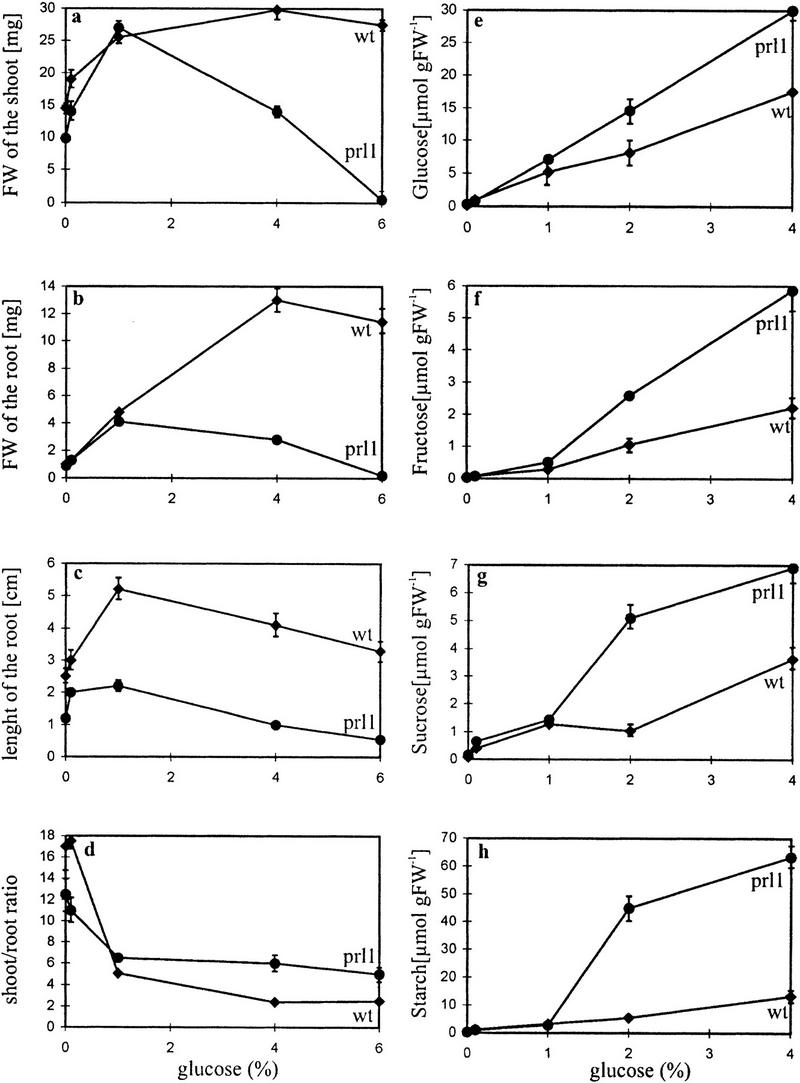
Sugar-dependent growth responses and carbohydrate accumulation in prl1. Comparison of shoot (a) and root (b) weights, root lengths (c), and shoot/root ratios (d) measured in wild-type (♦) and prl1 (•) plants grown in the presence of 0, 1, 4, and 6% glucose. Accumulation of glucose (e), fructose (f), sucrose (g), and starch (h) in the leaves of wild-type (♦) and prl1 (•) plants grown in the presence of 0, 1, 2, and 4% glucose.
Pleiotropic effects of the prl1 mutation on seedling development and hormonal responses
The prl1 mutation resulted in a two- to threefold inhibition of root elongation both in the dark and in the light (Fig. 1a,b). Hypocotyl elongation of prl1 plants was reduced in the dark (Fig. 1a), but was comparable with that of wild-type plants in white (Fig. 1b), red, far-red, and blue light (data not shown). Hypocotyl surface imprints showed a twofold increase in number, contrasting with a twofold decrease in length, of cells in the hypocotyl epidermis of prl1 in comparison with wild type (Fig. 1c,d). Premature initiation of side roots in light-grown prl1 seedlings indicated an enhanced auxin sensitivity (Fig. 1g,h). In the presence of auxins, arresting the elongation of primary roots, wild-type seedlings developed numerous side-roots covered by hairs, whereas primary and adventitious roots of prl1 were converted to undifferentiated callus tissues (Fig. 1f). In contrast with an alternating pattern of root-hair (trichoblast) and non-hair (atrichoblast) cells of wild-type root epidermis (Fig. 1k), adjacent rhizodermal cell files of prl1 carried ectopic root hairs (Fig. 1l), a sign of augmented ethylene sensitivity (Masucci and Schiefelbein 1996). In comparison with wild type, ethylene treatment caused a fivefold reduction of hypocotyl elongation of etiolated prl1 seedlings (Fig. 3g). When grown in soil, prl1 seedlings clearly differed from wild type by their altered leaf morphology and serrated leaf margins (Fig. 1o,p). In contrast, in the presence of cytokinin (4.5 μm isopentenyl adenosine) and 90 mm sucrose the phenotype of light-grown prl1 and wild-type seedlings was nearly identical (Fig. 1e). Unlike wild type plants, however, the prl1 mutant developed short roots and accumulated 20% to 30% more chlorophyll and anthocyanin both in the presence and absence of cytokinin (data not shown).
Figure 3.
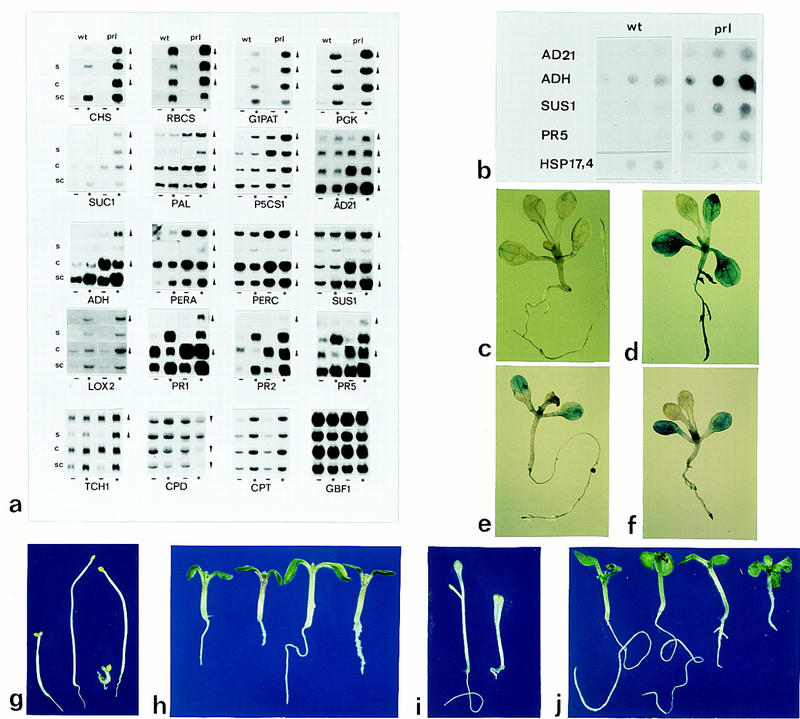
Genetic interactions and effects of the prl1 mutation on the expression of glucose and cytokinin responsive genes. (a) Northern filter quadrates were loaded with 4 × 4 RNA samples of 20 μg of each. In each quadrate, the first upper row is loaded with RNA samples prepared from wild-type (wt) and prl1 (prl) plants grown in the presence of 0.1% (3 mm) sucrose. The second and third rows (s and c) carry RNA samples from wild-type and prl1 plants grown in the presence of 3% (87 mm) sucrose (s), and cytokinin [c; 4.5 μm isopentenyl adenosine (IPAR)], respectively. The fourth row (sc) in each quadrate contains RNA samples from wild-type and prl1 plants subjected to combined sucrose (87 mm) and cytokinin (4.5 μm IPAR) treatments. The first and third columns labeled by − are loaded with RNA samples from dark-grown plants, whereas the second and fourth columns of filter quadrates marked by * carry RNAs from light-grown plants. The filters were hybridized with cDNA probes encoding chalcone synthase (CHS), ribulose-1,5-bisphosphate carboxylase (RBCS), glucose-1-phosphate-adenylate transferase (G1PAT), phosphoglycerate kinase (PGK), sucrose transporter (SUC1), phenylalanine ammonia-lyase (PAL), Δ1-pyrroline-5-carboxylate synthase (P5CS1), late- abundant embryonic protein (AD21), alcohol dehydrogenase (ADH), AP3 anionic peroxidase (PERA), peroxidase C (PERC), sucrose synthase (SUS1), lipoxygenase 2 (LOX2), pathogenesis-related proteins (PR1, PR2, and PR5), calmodulin (TCH1), C23-steroid hydroxylase (CPD), chloroplast triose-phosphate translocator (CPT), and G-box-binding factor (GBF1). (b) Dot-blot hybridization of AD21, ADH, SUS1, PR5, and Hsp17,4 cDNAs (0.4, 2, and 4 μg loaded in each row from left to right) with nuclear run-on RNA samples prepared from isolated wild-type (wt) and prl1 (prl) nuclei. (c–f) Patterns of GUS expression in wild-type (c,e) and prl1 (d,f) plants carrying an uidA reporter gene driven by a wild-type ADH promoter construct, CADH (c,d), and a mutant ADH promoter (ΔG-box2; Dolferus et al. 1994) containing base-pair exchanges in the G-boxII (e,f). (g–j) Phenotypes of prl1 double mutants. Growth response of wild-type, ein2, prl1, and prl1; ein2 (double mutant) seedlings (from left to right in g) to ethylene-treatment for 5 days in the dark. Phenotype of wild-type, prl1, ckr1, and prl1; ckr1 seedlings (from left to right in h) grown in the presence of cytokinin (2 μm N6-benzylaminopurine) for 10 days in the light. Dark-grown amp1 (left in i) and amp1; prl1 (right in i) seedlings 5 days after germination. Phenotypes of wild-type, amp1, prl1, and amp1; prl1 seedlings (from left to right in j) grown for 10 days in the light.
A combination of prl1 with the recessive ein2 mutation and its allele ckr1, conferring cytokinin resistance and ethylene insensitivity (Su and Howell 1992; Ecker 1995), did not suppress the short root prl1 phenotype. Root growth of the homozygous prl1; ckr1 double mutant, as well as wild-type and prl1 seedlings, was inhibited by cytokinin (2 μm 6-benzyl-aminopurine), in contrast to cytokinin resistant root elongation of the ckr1 mutant in the light (Fig. 3h). When treated with ethylene in the dark, the prl1; ein2 double mutant was indistinguishable from ein2, displaying a long hypocotyl and an open apical hook of cotyledons in contrast to short hypocotyls and exaggerated hooks of wild-type and prl1 (Fig. 3g). In addition to the light-dependent reversal of epistasis between prl1 and ein2 (ckr1), an unusual interaction was observed between prl1 and the amp1 mutation, conferring cytokinin overproduction (Chaudhury et al. 1993). The amp1; prl1 double mutant displayed a prl1-like short hypocotyl and root, and amp1-like large, open cotyledons in the dark, indicating additivity (Fig. 3i). amp1 severely aggravated the prl1 phenotype in the light, however, yielding a further size reduction of root, hypocotyl, and leaf (Fig. 3j). Decreasing the temperature from 24 to 14°C also caused a growth inhibition of prl1 (Fig. 1m,n). Cold sensitivity of prl1 correlated with an enhanced sensitivity to abscisic acid (ABA). A treatment of 5-day-old seedlings with 0.1 μm ABA resulted in bleaching and growth reduction of prl1 in contrast to wild type (Fig. 1j). Further assays showed that growth responses to gibberellins, brassinosteroids, methyl jasmonate, salicylic acid, phosphate, NaCl, heavy metals, heat-shock, and drought were unaffected by the prl1 mutation (data not shown).
Transcriptional derepression of genes regulated by sucrose and cytokinin in the prl1 mutant
Northern hybridization analysis using RNAs prepared from wild-type and prl1 plants grown in the dark or in white light (excluding UV-A and -B) on either 3 or 90 mm sucrose, with or without 4.5 μm cytokinin, revealed a derepression of glucose- and cytokinin-regulated genes in the prl1 mutant (Fig. 3a). In accordance with an overproduction of anthocyanins, the RNA levels of chalcone synthase (CHS) and phenylalanine ammonia–lyase (PAL) genes were significantly increased in the prl1 mutant as compared with the wild type. In addition, transcript levels of the light-activated and glucose-repressed ribulose-1,5-bisphosphate carboxylase (RBCS), glucose-1-phosphate-adenylate transferase (G1PAT), and phosphoglycerate kinase (PGK) genes were three- to fivefold higher in prl1 than in wild-type plants grown in the absence or presence of either sucrose or cytokinin in the light. The expression of other light-regulated genes encoding, for example, chlorophyll a/b-binding proteins, glucose-6-phosphate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, glutamine/glutamate synthases, superoxide dismutases, malic enzyme, H+/hexose transporters (data not shown), chloroplast triose–phosphate translocator (CPT), and bZIP transcription factors (GBF1, GBF3, TGA1a, and TGA3) showed no difference between wild-type and the prl1 mutant. In accordance with the accumulation of free sugars, one of the sucrose transporter genes (SUC1; Sauer and Stolz 1994) was found to be active in prl1, but not in wild-type plants grown in the absence of cytokinin. The sucrose synthase (SUS1), alcohol dehydrogenase (ADH), anionic peroxidase (PERA), and peroxidase C (PERC) genes showed derepression in the absence and enhanced induction in the presence of cytokinin in prl1, but their activity was sucrose repressible. In contrast, the TCH1 calmodulin gene featured a derepression on sucrose, whereas the steady-state RNA level of LOX2 lipoxygenase was increased by cytokinin in prl1. The abscisic acid-induced genes AD21 and Δ1-pyrroline-5-carboxylate synthase 1 (P5CS) displayed higher expression and inducibility by glucose and cytokinin in prl1, whereas the RNA levels of pathogenesis-related genes PR1, PR2, and PR5 were increased by cytokinin 5- to 10-fold, but their induction by glucose and light was unaltered in prl1. The CPD gene, encoding an essential enzyme in brassinosteroid biosynthesis (Szekeres et al. 1996), proved to be unique among the genes tested because its expression was down-regulated in the prl1 mutant.
Except for AD21, SUS1, PERA, and PERC, the genes affected by the prl1 mutation showed a similar steady-state mRNA level in wild type and prl1 when the seedlings were treated with both cytokinin and sucrose. To determine whether transcription or RNA stability of cytokinin and glucose regulated genes was affected by the prl1 mutation, RNA probes were synthesized in isolated nuclei prepared from wild type and prl1 plants. Hybridization of run-on RNA probes with cDNA dot-blots revealed two- to fivefold higher PR5, SUS1, ADH, and AD21 transcript levels in prl1 as compared with wild type (Fig. 3b), indicating that at least part of the differences detected by Northern hybridization of steady-state RNAs was attributable to transcriptional changes caused by the prl1 mutation. To support this conclusion, a β-glucuronidase (GUS) reporter gene driven by the ADH promoter (Dolferus et al. 1994) was introduced into wild-type and prl1 plants. The ADH–GUS expression was confined to the meristemic junction of rosette leaves in the wild type (Fig. 3c), whereas high ADH–GUS activity was detected in leaves, vascular meristems, and roots of prl1 (Fig. 3d). The difference between ADH–GUS expression in wild-type (Fig. 3e) and prl1 (Fig. 3f) plants was alleviated by a mutation of G-box II sequences within the ADH promoter (Dolferus et al. 1994).
prl1 encodes a conserved WD protein
Southern hybridization mapping of prl1 genomic DNA with probes derived from the T-DNA tagging vector pPCV6NFluxF (Koncz et al. 1994) showed that prl1 contained a tandem repeat of three T-DNAs. Plant DNA fragments linked to the T-DNA ends (LB1 and LB3; Fig. 4a) were isolated by plasmid rescue (Koncz et al. 1990), sequenced and used as probes for the isolation of wild-type genomic and cDNA clones. Sequence comparison of genomic and cDNA clones indicated that the PRL1 gene contained 17 exons. The transcriptional start site was located 38 bp upstream of the ATG codon as determined by primer extension (data not shown). Database searches revealed that the closest neighbor located 3′-downstream of PRL1 was an ABA-induced gene, DI21. Sequence comparison of the wild-type and T-DNA-tagged alleles showed that the T-DNA insertion caused a deletion of sequences between exons 15 and 17, leading to a 3′-truncation of the PRL1-coding sequence (Fig. 4a). In addition to clones carrying the wild-type PRL1 allele, the sequence analysis also identified genomic and cDNA clones encoding a PRL1 homolog, PRL2. Alignment of PRL1- and PRL2-coding sequences, both spanning 1.65 kb, revealed four gaps of 3–12 bp upstream of codons 159 and 153, respectively. Amino-terminal segments of deduced PRL1 and PRL2 protein sequences located upstream of these positions shared only 65% identity, whereas their carboxy-terminal segments showed an amino acid identity of 89% (Fig. 5). With 5′-end-specific cDNA probes, the PRL1 gene was found to hybridize to yeast artificial chromosome (YAC) clones EW22D4, EG23E10, EW14E4, and yUP13C7, and mapped to chromosome 4-44 (Schmidt et al. 1996) confirming the results of the genetic linkage analysis. PRL2 was mapped to YAC clones CIC4H5, CIC11H4, CIC12C2, yUP23E10, and yUP24B8 of contig KG17 located in the vicinity of marker m560B in chromosome 3–24.
Figure 4.
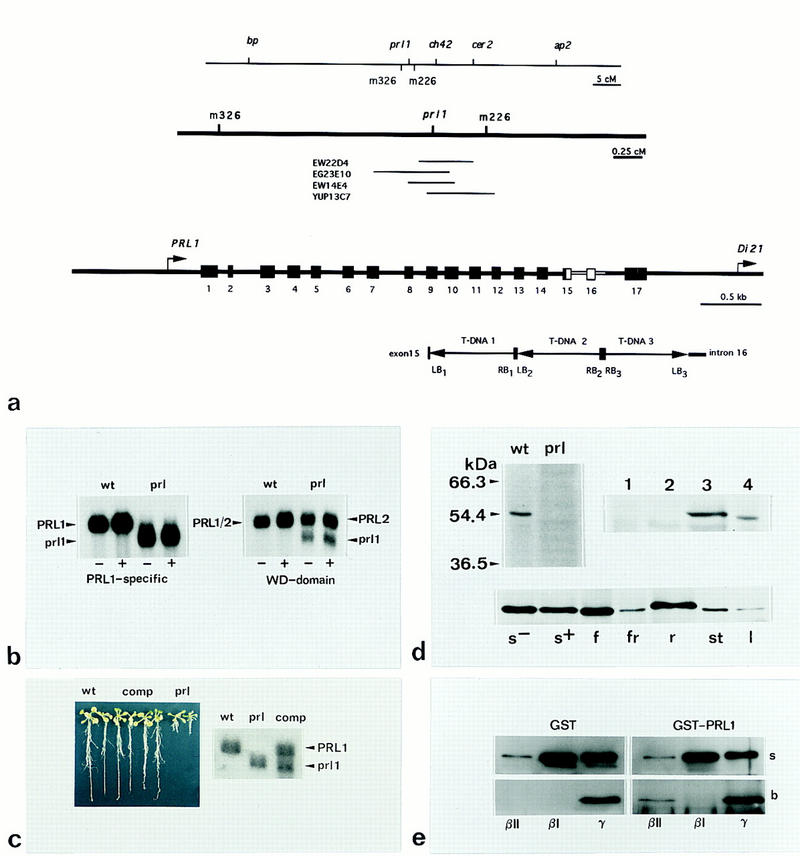
Characterization of wild-type and mutant alleles, genetic complementation of the prl1 mutation, and interaction of PRL1 with human PKC-βII in vitro. (a) The position of prl1 on chromosome 4-44 and YAC clones located between the markers m326 and m226 are shown by the maps in the top two lanes. Structure of the PRL1 gene, position of the neighboring DI21 gene, and location of the trimeric T-DNA insertion replacing sequences between exon 15 and intron 16 in the prl1 mutant are depicted in the bottom section. Left (LB) and right (RB) borders of the T-DNA units within the tandem repeat are numbered. Map distances are indicated by bars. (cM) CentiMorgan. (b) Hybridization of RNA samples prepared from wild-type (wt) and prl1 (prl) plants grown in the absence (−) or presence of 3% sucrose (+) with a PRL1-specific probe derived from the 5′ end of PRL1 cDNA (left section). Hybridization of the same RNA blot with the 3′-end of PRL1 cDNA, encoding the WD-40 repeats conserved between PRL1 and PRL2 (right section). Arrows indicate the position of PRL1, PRL2 and truncated prl1 RNAs. (c) Genetic complementation of the prl1 mutation. Germination test (left) showing normal root elongation of wild-type (wt; first two seedlings) and complemented plants (comp; four seedlings to the right from wild type) in contrast to defective root growth of the prl1 mutant (last two seedlings to the right). Northern hybridization of RNAs (right) prepared from wild-type (wt), prl1 (prl), and complemented (comp) plants with a PRL1-specific probe. Positions of PRL1 and prl1 mRNAs are indicated by arrows. (d) Western blotting with anti-PRL1 antibody (top left) detects the PRL1 protein of 54.4 kD in the wild type (wt), but shows no specific cross-reaction with proteins extracted from the prl1 mutant. Immunoblotting of membrane proteins (top right) obtained from wild-type plants by extraction with (1) 50 mm NaCl, (2) 500 mm NaCl, (3) 0.2 m NaCO3 (pH 11.5), and (4) 2% Triton X-100. Immunoblotting of SDS-solubilized protein extracts (bottom) prepared from wild-type seedlings grown in the absence (s−) or presence (s+) of 3% sucrose, flowers (f), fruits (fr), roots (r), stems (st), and leaves (l). (e) Pull-down PKC assays. PKC-βII (1 μg) and 3 μg of PKC-βI and γ were incubated with GST and GST–PRL1–ΔB proteins immobilized on glutathione–Sepharose. The matrices were extensively washed, then the bound protein fractions were eluted, separated by SDS-PAGE, and immunoblotted with an antibody recognizing all three PKCs. Supernatant fractions in the top lanes (s) show the amount of unbound PKCs. Bound fractions in the bottom lanes (b) indicate binding of PKC-βII to the GST–PRL1–ΔB fusion protein, as well as an unspecific interaction of PKC-γ with the GST bait used as internal control.
Figure 5.
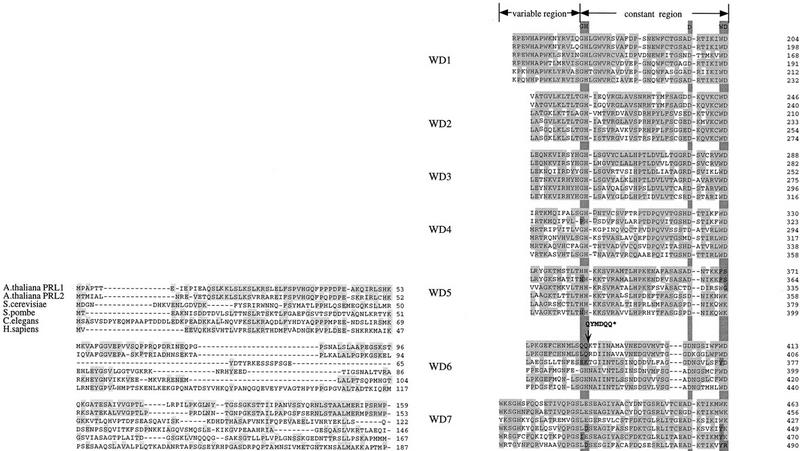
Conservation of PRL1 orthologs in eukaryotes. Alignment of amino acid sequences of PRL1 and PRL2 from Arabidopsis, YPL151c from budding yeast, PRL1 from fission yeast, D1054.15 from Caenorhabditis, and a human PRL1 homolog (GenBank accession no. AF044333). Arrows label the variable and constant regions of WD-40 repeats according to Neer et al. (1994). Conserved amino acids are marked by gray color within the WD repeats and terminal extensions. A putative SV40-type nuclear localization signal is printed in a gray box at the carboxyl terminus. The position of T-DNA insertion in prl1, adding six new amino acid codons to the truncated prl1-coding domain, is indicated by an arrow.
5′ sequences of PRL1 did not hybridize to the PRL2 transcript under stringent conditions and detected only the PRL1 mRNA of 1.75 kb in wild-type plants. The probe hybridized to a mRNA of 1.55 kb in prl1, providing evidence for transcription of the T-DNA-tagged mutant allele (Fig. 4b). Comparable amounts of transcripts were observed in both wild-type and prl1 plants grown in the light in the presence or absence of 90 mm sucrose indicating that transcription of the PRL1 and prl1 alleles was unaffected by sucrose. In addition to the truncated prl1 transcript, probing the same blots with 3′-cDNA sequences conserved between PRL1 and PRL2 detected PRL2 mRNA of 1.75 kb in prl1 plants indicating that transcription of PRL2 was not affected by the prl1 mutation.
The prl1 mutation could be complemented by transformation with the wild-type PRL1 gene carried by the Agrobacterium vector pPCV002 (Koncz and Schell 1986) linked to a kanamycin resistance marker. The prl1 mutation dramatically reduced the frequency of Agrobacterium-mediated transformation. Therefore, Agrobacterium infection of 500,000 prl1 root explants yielded only three kanamycin-resistant transformants that could be regenerated to plants displaying wild-type phenotype concerning all visible and molecular phenotypic traits affected by the prl1 mutation (data not shown). All three complemented lines carried a single copy of wild-type PRL1 gene and their F2 progeny showed a 3:1 segregation ratio of kanamycin resistant wild-type plants with normal root elongation and kanamycin sensitive prl1 plants with short roots (Fig. 4c). Hybridization with the PRL1-specific probe demonstrated that the complemented lines synthesized both wild-type PRL1 and truncated prl1 mRNAs (Fig. 4c).
Analysis of protein sequences deduced from the cDNA indicated that PRL1 is a basically charged protein of 54 kD carrying seven carboxy-terminal β-transducin repeats characteristic for regulatory WD-40 repeat proteins in eukaryotes (Neer et al. 1994). In the database, PRL1 identified a family of WD proteins with unknown function: PRL2 from Arabidopsis shared 83%, PRL1 from fission yeast 69%, YPL151c from budding yeast 63% (Purnelle et al. 1996), and hypothetical gene product D1054.15 from Caenorhabditis 62% sequence identity with PRL1. Expressed sequence tags (ESTs) showing PRL1 homology were also found in Drosophila and mouse. A human ortholog (GenBank accession no. AF044333), showing 59% sequence identity with PRL1, was isolated using expressed sequence tags EST178245 and yw86d09 as probes (Fig. 5; L. Ökrész, unpubl.).
Cellular localization of the PRL1 protein and its interaction with human PKC-βII
Sequence analysis of the mutant prl1 gene showed that the T-DNA insertion interrupted the PRL1 sequence at codon position 392 (after the motif MLSQQ in the sixth WD-40 repeat; Fig. 5) and resulted in the addition of six new carboxy-terminal amino acids. An affinity-purified antibody raised against a unique PRL1 peptide (see Materials and Methods) failed to detect a truncated PRL1 protein with the predicted molecular mass of 43.4 kD in the mutant but recognized a protein of 54 kD in wild-type plants (Fig. 4d). Control experiments, using the synthetic PRL1 peptide as a competitor in immunoblotting with the anti-PRL1 antibody, confirmed that the protein of 54 kD was indeed PRL1. These experiments also demonstrated that the PRL2 protein and its fusion protein derivatives, produced in Escherichia coli and in yeast and lacking the PRL1-specific peptide sequence, were not recognized by the anti-PRL1 antibody (data not shown). PRL1 was detected in microsomal membrane cell fractions prepared from wild-type plants. PRL1 could only be extracted from the membranes with 0.2 m Na2CO3 at pH 11.5, but not by 0.05 or 0.5 m NaCl (Fig. 4d). In SDS-solubilized extracts prepared from different organs, the total amount of PRL1 protein was found to be the highest in roots and flowers, less in stems, and the lowest in leaves (Fig. 4d). Confocal laser microscopy of immunostained prl1 roots detected only a background signal, consistent with a lack of PRL1 protein in the mutant. In wild-type plants, some staining was associated with membrane structures, but the strongest signals overlapped with the DAPI-stained nuclei (Fig. 6a).
Figure 6.
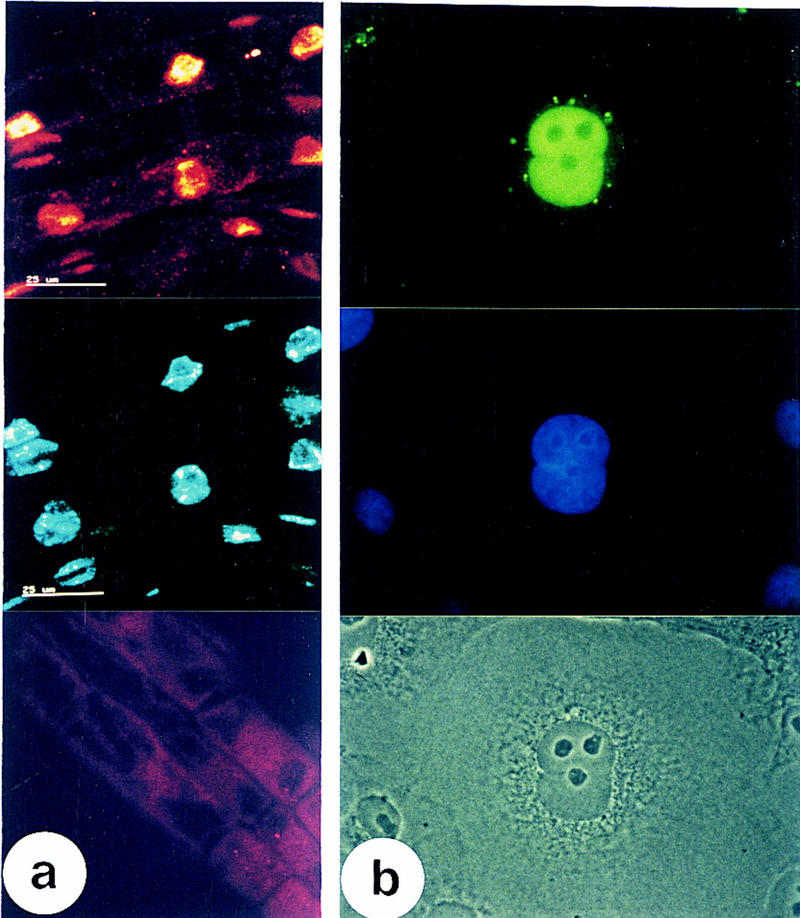
Immunolocalization of the PRL1 protein in Arabidopsis and monkey COS-1 cells. (a) Confocal laser micrographs of wild-type Arabidopsis root cells immunostained with the anti-PRL1 antibody (top) followed by counter-staining the nuclei with DAPI (middle). Immunostaining of root cells in the prl1 mutant (bottom). Scale bars, 25 μm. (b) Detection of MYC-PRL1 fusion protein by indirect immunofluorescence in African green monkey COS-1 cells. Immunostaining of a COS-1 cell with anti-MYC antibody (top), counter-staining the nucleus with DAPI (middle), and light microscopic cell image (bottom).
The remarkable conservation of PRL1 sequences in eukaryotes tempted us to construct and express a MYC-epitope-tagged PRL1 protein in green monkey COS-1 cells. Indirect immunofluorescence microscopy revealed an accumulation of PRL1 in COS-1 cell nuclei counter-stained with DAPI. Some immunostaining was also associated with a filamental halo around the nuclei, whereas MYC–PRL1 was not detected in the nucleoli (Fig. 6b). Nuclear transport of PRL1 in COS-1 cells raised the question about the possible function of mammalian PRL1 orthologs. A similarity to a sequence motif mediating the interaction of RACK1 receptor with activated protein kinase C (PKC) isoenzymes in mammals (Ron et al. 1994) was found within the WD-repeats of PRL1 orthologs. Therefore, a glutathione-S-transferase fusion protein (GST–PRL1–ΔB) carrying an amino-terminal PRL1 segment of 330 amino acids was constructed, purified to homogeneity, immobilized on glutathione-Sepharose, and incubated with activated human PKC-βI, βII, and γ (Stabel et al. 1993). Following stringent washes, immunoblotting of the matrix-bound proteins revealed that GST–PRL1-ΔB retained PKC-βII, but not PKC-βI, whereas PKC-γ displayed a strong binding to the control GST protein providing a suitable internal standard (Fig. 4e). Because PKC-βI and βII sequences only differed by 52 carboxy-terminal amino acids (Kubo et al. 1987), the data also indicated that the carboxyl terminus of PKC-βII was required for PRL1-binding. In addition, nuclear localization of PRL1 in COS1 cells and in vitro interaction of PRL1 with the carboxyl terminus of PKC-βII supported the notion that the carboxyl terminus may be implicated in nuclear import of PKC-βII in mammals (Chalfant et al. 1995; Mochly-Rosen 1995).
PRL1 interacts with α-importin ATHKAP2, a novel Arabidopsis nuclear import receptor
To screen for Arabidopsis cDNAs encoding PRL1-interacting proteins (PIPs), the full-length PRL1 protein and an amino-terminal PRL1 segment of 321 amino acids were expressed as baits carrying the Gal4p DNA-binding domain in yeast using the two-hybrid vector pAS2 (Durfee et al. 1993). From 18.4 × 107 transformants obtained with a cDNA expression library prepared from an Arabidopsis cell suspension in pACT2 (Durfee et al. 1993), 342 clones showed His+ and LacZ+ phenotype indicating an interaction between the PRL1 baits and cDNA encoded proteins fused to the activation domain in pACT2 (Fig. 7a). Classification of PIP clones identified a family of 62 cDNAs coding for carboxy-terminal segments of PIP-B corresponding to a novel class of Arabidopsis α-importins, ATHKAP2. The amino acid sequence of ATHKAP2 deduced from a full-length cDNA of 2 kb (EMBL accession no. Y09511) showed a remarkably high sequence identity with Arabidopsis ATHKAP1, human HSRP1, Xenopus IMP1, yeast YSRP1, and other α-importins involved in the nuclear import of proteins and RNAs. ATHKAP2 carried all amino-terminal sequence motifs required for nuclear localization and interaction with β-importins followed by eight highly conserved internal armadillo repeats (Merkle and Nagy 1997), but its carboxyl terminus was shorter than that of ATHKAP1 (Hicks et al. 1996; Fig. 7d). Using a 3′-end specific cDNA probe, the ATHKAP2 gene was mapped to YAC clones CIC9F6 and CIC10H3 in chromosome 4-10.8. Northern analyses showed that the ATHKAP2 mRNA levels were high in stems and flowers, but lower in leaves and roots. Similarly to PRL1, ATHKAP2 mRNA levels were comparable in wild-type and prl1 plants grown in the presence or absence of sucrose (data not shown).
Figure 7.
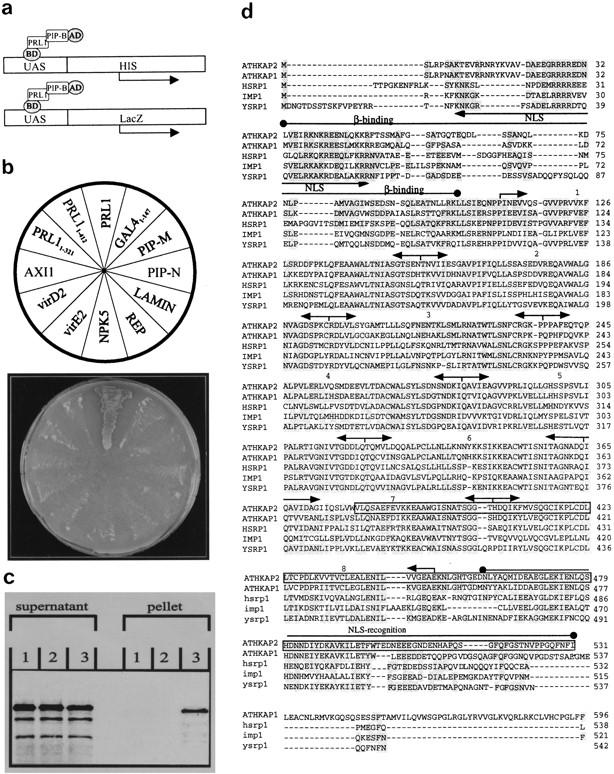
Identification of α-importin ATHKAP2 by screening for PIPs in the yeast two-hybrid system. (a) PRL1 in fusion with the Gal4p DNA-binding domain (BD) was used as a bait to screen for PRL1-binding proteins (e.g., PIP-B) carrying an activation domain (AD) in a yeast two-hybrid system by monitoring the activation of His3 and lacZ reporter genes as described (Durfee et al. 1993). (b) A pACT2 construct, expressing a carboxy-terminal ATHKAP2 domain of 152 amino acids, was combined with different pAS2-baits encoding a full-length PRL1 protein (PRL1), amino-terminal PRL1 segments of 321 and 412 amino acids (PRL11–321 and PRL11–412), DNA-binding domain of Gal4p in fusion with SV40 NLS (GAL41–147), cystatin protease-inhibitor (PIP-M), unknown PRL1-binding protein (PIP-N), lamin, replicator protein of wheat-dwarf geminivirus (REP), tobacco SNF1 kinase (NPK5), tobacco AXI1, and Agrobacterium virulence proteins VirE2 and VirD2. Specific interaction between the carboxyl terminus of ATHKAP2 and PRL1 was confirmed by growing the yeast strains on histidinefree medium. (c) A GST–fusion protein carrying the carboxy-terminal domain of ATHKAP2 and a control glutathione–S–transferase (GST) protein were immobilized on glutathione–Sepharose. Equal aliquots of the [35S]methionine PRL1 protein were incubated with the glutathione–Sepharose (1); GST (2); and GST–ATHKAP2 (3) matrices followed by extensive washing and elution of bound proteins. The labeled PRL1 protein in the supernatant and bound protein fractions was resolved by SDS-PAGE and detected by autoradiography. (d) Amino acid sequence alignment of ATHKAP2 with Arabidopsis ATHKAP1 (Hicks et al. 1996), human HSRP1 (GenBank accession no. U28386), Xenopus IMP1 (accession no. L36339), and yeast YSRP1 (accession no. M75849) α-importins. Identical amino acids are labeled by gray color. Nuclear localization sequences (NLS), β-importin-binding, and NLS-recognition domains are marked by lines above the sequence. ARMADILLO repeats are numbered and labeled by arrow-headed separating lines. The carboxy-terminal domain of ATHKAP2 used in the yeast two-hybrid tests is boxed.
The His− phenotype of yeast strains carrying an ATHKAP2-fused activation domain in combination with the Gal4p DNA-binding domain either alone or in fusion with unrelated proteins (NPK5, PIP-M, and PIP-N; Fig. 7b) showed that the PRL1–ATHKAP2 interaction was specific. To support these data, [35S]methionine-labeled PRL1 was synthesized by coupled transcription and translation using the cDNA template, and equal aliquots of PRL1 protein were incubated with GST–ATHKAP2 and GST proteins immobilized on glutathione–S–Sepharose, as well as with the empty Sepharose matrix (see Materials and Methods). PRL1 was quantitatively removed from the control Sepharose and GST matrices by stringent washes, but remained tightly-bound to GST–ATHKAP2, confirming an interaction of ATHKAP2 with PRL1 in vitro (Fig. 7c). A carboxy-terminal segment of ATHKAP2 of 152 amino acids interacted only with the full-length PRL1 bait in yeast, but not with truncated PRL1 proteins carrying either 321 or 412 amino-terminal amino acids. Binding of ATHKAP2 was therefore mapped to a carboxy-terminal PRL1 domain of 74 amino acids, carrying the last WD-40 repeat followed by a putative SV40-type nuclear localization signal (NLS; Figs. 5, 7b). The carboxy-terminal segment of ATHKAP2, however, interacted neither with SV40–NLS in fusion with the Gal4 DNA-binding domain nor with monopartite and bipartite NLS sequences carried by the VirD2, VirE2, lamin and AXI1 baits, and the replication protein (REP) of wheat-dwarf gemini virus (Fig. 7b; see Materials and Methods). The data therefore indicated that either the putative carboxy-terminal PRL1–NLS represented a specific ligand for ATHKAP2, or the interaction was not confined to recognition of PRL1–NLS by the carboxy-terminal NLS-recognition domain of ATHKAP2 α-importin.
Discussion
Implication of PRL1 in glucose regulation
Glucose repression has a major role in the regulation of carbon metabolism in higher plants as in other organisms (Sheen 1990; Stitt and Sonnewald 1995). Feeding of plants with glucose or sucrose (which is converted to glucose and fructose) results in either transcriptional or post-transcriptional down-regulation (or both) of genes involved in chlorophyll biosynthesis, Calvin cycle, gluconeogenesis, starch degradation and glyoxylate cycle, but leads to the activation of genes in glycolysis, defense responses, nitrate and phosphate metabolism, and biosynthesis of anthocyanin pigments and storage proteins (for review, see Koch 1996; Jang and Sheen 1997). Arabidopsis plants can tolerate as high as 300–400 mm glucose or sucrose present in growth media (Smeekens and Rook 1997). Therefore, mutations affecting potential regulatory functions in glucose signaling can simply be isolated by screening for plants showing hypersensitive or insensitive growth response to glucose or sucrose. Using this strategy, we have isolated a mutation, prl1, from a T-DNA-tagged Arabidopsis collection (Koncz et al. 1992) that displays a hypersensitive growth arrest and ultimate lethality in the presence of 175 mm glucose or sucrose. Remarkable accumulation of glucose, fructose, starch, chlorophyll, and anthocyanin pigments in the leaves suggested that the prl1 mutation may relieve glucose repression of metabolic pathways and simultaneously enhance the activation of other pathways by glucose. Therefore, we tested the steady-state transcript levels of numerous genes in the prl1 mutant that were reported to be either repressed or induced by glucose in plants. It was found that many genes that are repressed or induced by glucose showed higher steady-state mRNA levels in prl1 in comparison with the wild type. For example, among the glucose-repressible genes, the light-induced ribulose 1,5-bisphosphate carboxylase, glucose-1-phosphate-adenylate transferase, and phosphoglycerate kinase genes acting in photosynthesis, the light- and cytokinin-inducible PAL1 gene required for flavonoid biosynthesis, the cytokinin-inducible alcohol dehydrogenase, sucrose synthase and peroxidase (PERA and PERC) genes, the LOX2 gene involved in jasmonate synthesis, the abscisic acid and salt-regulated P5CS1 gene controlling proline biosynthesis, the TCH1 calmodulin gene implicated in Ca2+ signaling and the sucrose transporter SUC1 gene showed de-repressed expression in the prl1 mutant. Similarly, several glucose-inducible genes, such as the chalcone synthase gene in anthocyanin biosynthesis, the AD21 gene coding for an embryo-specific late-abundant protein, and the cytokinin-inducible pathogenesis-related PR genes displayed higher transcript levels in the prl1 mutant as compared with the wild type. These data suggested that PRL1 may act as a negative regulator of glucose responsive genes. To support this conclusion, a hybridization analysis with nuclear run-on RNAs was performed that confirmed at least for four different genes (ADH, AD21, SUS1, and PR5) that derepression of gene expression was indeed attributable to transcriptional changes caused by the prl1 mutation. Furthermore, a mutation in the G-boxII bZIP-binding site within the ADH promoter (Schindler et al. 1992; Dolferus et al. 1994) was demonstrated to alleviate the differences observed in ADH gene expression between prl1 and wild-type plants by defining a common promoter upstream element required for negative regulation of transcription by PRL1.
So far, most known regulatory functions required for glucose repression have been identified by genetic dissection of glucose signaling in yeast and molecular studies of glucose-controlled insulin production in pancreatic β-cells. In addition to important functions of different hexose transporters, hexokinases (HXK2 in yeast and glucokinase in pancreas) were found to be essential for monitoring the rate of hexose phosphorylation and thus generating a signal for glucose repression. As in yeast and pancreatic β-cells, inhibition of the hexokinase was demonstrated recently to relieve glucose repression causing glucose insensitivity, whereas overexpression of hexokinase was found to augment glucose repression resulting in glucose hypersensitivity in Arabidopsis (Jang et al. 1997). In addition to HXK2, many other signaling functions, such as GRR1, RTG1, GLC7, REG1, MIG1, TUP1, and SSN6, were demonstrated to mediate glucose repression in yeast (Johnston and Carlson 1992; Özcan and Johnston 1995; Ronne 1995). The deficiency of these functions in yeast leads to derepression of glucose responsive genes resulting in glucose insensitivity. Therefore, PRL1 clearly differs from these regulators of glucose repression, because the prl1 mutation causes glucose hypersensitivity by simultaneous derepression of glucose-regulated genes.
To explain how a repressor mutation, such as prl1, can increase glucose sensitivity, we follow a model suggesting that PRL1 acts as a negative regulator of a function that counteracts the activity of factors that mediate glucose repression. The serine/threonine kinase SNF1 and its activator subunit SNF4 are known to perform such a function in yeast. By controlling its phosphorylation and nuclear import, SNF1 inactivates MIG1, which acts as a negative regulator of glucose responsive genes by binding to their promoters and recruiting the TUP1/SSN6 general repressors (Trietel and Carlson 1995). The fact that a similar regulatory mechanism exists in plants is indicated by the involvement of SNF1-like kinases in the control of key metabolic enzymes (Huber et al. 1994), as well as by the characterization of plant protein kinases that can functionally complement the snf1 mutation and interact with several regulatory subunits of SNF1 in yeast (Jiang and Carlson 1997).
Pleiotropic effects of the prl1 mutation
Characterization of the prl1 mutation indicated that tight cross talk exists between glucose, cytokinin, and light signaling. Thus, simultaneous cytokinin and sucrose treatment of wild-type plants resulted in a prl1-like mutant phenocopy and abolished the differences seen in gene expression between wild-type and prl1 plants. Furthermore, most developmental, hormonal and molecular alterations caused by the prl1 mutation were detectable only in light-grown plants, suggesting a possible light-dependence of the PRL1 regulatory function. Nonetheless, PRL1 probably acts downstream and independently of the photoreceptor–mediated light signaling pathways because the prl1 mutant shows normal light responses in hypocotyl elongation assays, and because mutations causing constitutive photomorphogenesis and de-etiolation, including cop1 and det1, are epistatic to prl1 (C. Koncz, unpubl.).
Because cytokinin is known to counteract, rather than enhance, glucose repression of the light-regulated genes, it is unlikely that PRL1 acts as a cytokinin-dependent repressor. It is more likely that cytokinin signaling converges on a function that can alleviate glucose repression, and that PRL1 is a light-dependent negative regulator of this function. Genetic data derived from preliminary analyses of prl1 double mutants seem to support this model because in light-grown plants prl1 is epistatic to the ein2 (ckr1) mutation that confers cytokinin resistance, whereas the amp1 mutation, which activates cytokinin signaling by stimulation of the synthesis of this hormone (Chaudhury et al. 1993), severely aggravates the phenotype of the prl1 mutant. A light dependence of these genetic interactions is indicated by the observations that the prl1; ein2 double mutant shows ein2 phenotype in the dark, but prl1 phenotype in the light, whereas the amp1; prl1 double mutant displays an additivity of phenotypic traits in the dark in contrast to the ‘super-prl1’ phenotype in the light. Cross talk mediated by EIN2 between ethylene and cytokinin signaling (Ecker 1995) may therefore provide a possible explanation for the ethylene hypersensitive phenotype and ectopic root hair development caused by the prl1 mutation. The fact that prl1 also augments the sensitivity of plants to ABA and auxin indicates cross talk with other hormonal signaling pathways. Auxin-mediated induction of lateral root development observed in wild-type Arabidopsis plants thus occurs in the absence of auxin stimulus in the prl1 mutant correlating with its increased auxin-sensitivity. Down-regulation of the CPD gene encoding a steroid C23-hydroxylase suggests that PRL1 may even modulate (in this case positively) the biosynthesis of brassinosteroids that have an essential role in skotomorphogenic development and antagonize de-etiolation (Szekeres et al. 1996). The observation that prl1 affects the transcription of genes such as CHS and PAL implicated in UV light and fungal elicitor-induced signaling, TCH1 in touch-signaling, PR genes in salicylic acid signaling, ADH in cold-stress signaling, and P5CS1 and AD21 in abscisic acid and salt signaling (for review, see Meyerowitz and Somerville 1994) also illustrates a role of PRL1 in modulating genes controlled by other regulatory pathways. Sensitivity of the prl1 mutant to low-temperature stress therefore not only correlates with the ABA hypersensitive phenotype, but also suggests that PRL1 may pleiotropically affect the regulation of cell elongation. In fact, the prl1 mutation results in the inhibition of root elongation, a phenotype that is not glucose-dependent and cannot be compensated by known plant hormones. It is not surprising that mutations in glucose signaling affect the regulation of cell shape and elongation pleiotropically, as GRR1 and REG1 in yeast are known to control cell size and polarity independently of their function in glucose repression (Ronne 1995). A possible conservation of PRL1-like functions is not only indicated by the identification of PRL1 orthologs in yeast, but also by the recent observations demonstrating that overexpression of truncated PRL1 proteins or antisense transcripts in fission yeast result in large, barrel-shape budding cells displaying a loss of polarity (Xia et al. 1996; unpubl.).
Nuclear transport and interacting partners of PRL1
PRL1 encodes a novel protein carrying seven WD-40 repeats that share homology with the β-subunits of trimeric GTP-binding proteins and many other WD-proteins that perform different regulatory functions in eukaryotes (Neer et al. 1994). WD-40 repeats and terminal extensions of PRL1 are distinct from those of so far characterized WD-proteins indicating that PRL1 represents a novel class of regulatory factors. Nonetheless, connections between PRL1 and regulation of transcription suggest some functional analogies with the Arabidopsis COP1 and yeast TUP1 WD proteins. TUP1, together with SSN6, forms a repressor complex with the MIG1 transcription factor (Trietel and Carlson 1995; Tzamarias and Struhl 1995), whereas COP1, a repressor of photomorphogenic development, directly interacts with and negatively regulates the function of the HY5 bZIP transcription factor that binds to G-box sequences within the Arabidopsis chalcone synthase promoter (Ang et al. 1998). Because transcriptional regulation by PRL1 also converges on a G-box sequence within the ADH promoter (Dolferus et al. 1994), it would be interesting to determine whether PRL1 can interact with bZIP-like transcription factors. In particular, possible interaction of PRL1 with HY5 needs to be tested because both prl1 and hy5 mutations induce the initiation of lateral roots in Arabidopsis.
As expected for a regulatory protein functioning as a potential repressor, PRL1 is a basically charged protein that is imported into the nucleus, but also detected in association with membrane fractions consisting of fragments of nuclear envelope and endoplasmic reticulum. PRL1 interacts with ATHKAP2, a novel class of Arabidopsis α-importins in the yeast two-hybrid system and in vitro. Although ATHKAP2 shares >90% sequence identity with other eukaryotic α-importins, it cannot recognize proteins carrying prototypes of monopartite and bipartite nuclear localization signals in the yeast two-hybrid system. Albeit PRL1 contains a putative SV40-type NLS within its carboxyl terminus, the binding of PRL1 to ATHKAP2 probably reflects a more intricate, possibly regulatory, interaction suggesting a potential role for PRL1 in regulation of nuclear import. PRL1 orthologs, sharing >55% sequence identity are not only found in budding and fission yeasts, but also in Caenorhabditis, Drosophila, mouse, and man. Therefore, it may be relevant that PRL1 serves as heterologous receptor in vitro for a nuclear protein kinase C-βII isoenzyme. In addition to nuclear import of PRL1 in COS-1 cells, selective interaction of PRL1 with human PKC-βII, but not with PKC-βI, is intriguing because PKC-βI and βII differ only by 52 carboxy-terminal amino acids (Kubo et al. 1987) required for nuclear import of PKC-βII during insulin signaling (Chalfant et al. 1995; Mochly-Rosen 1995). Carboxy-terminal sequences of Arabidopsis PRL1 bind a highly conserved α-importin nuclear receptor, ATHKAP2, and share a high sequence identity with a human PRL1 ortholog. Therefore, binding of the carboxyl terminus of PKC-βII to the amino-terminal domain of PRL1 in a complex with α-importin may mediate nuclear targeting of PKC-βII. Although protein interactions in heterologous systems have to be interpreted with caution, further functional study of PRL1-homologs in eukaryotes certainly deserves attention.
Materials and methods
Mutant selection, physiological assays, genetic analysis, and physical mapping
Seeds from 1200 M2 families of T-DNA-tagged Arabidopsis lines were germinated in MS medium (Koncz et al. 1994) containing either glucose or sucrose (0.1, 0.5, 2, 4, 6, 8, or 10%). Mutants showing growth retardation on glucose and sucrose were further tested by germination in the presence of fructose, raffinose, mannose, galactose, lactose, maltose, xylose, ribose, mannitol, sorbitol (each used at concentrations 1, 5, 10, 50, 100 and 200 mm), 3-O-methylglucose (0.1, 1. 5, 10, 50, 100 mm), 6-deoxyglucose (0.001, 0.01, 0.05 mm), and polyethylene-glycol [0.1, 0.5, 1, and 2% (weight/volume)]. Other growth responses were assayed by supplementing the media with auxins (2,4-dichlorophenoxyacetic acid or 1-naphthaleneacetic acid), cytokinins [N6-(2-isopentenyl)adenosine riboside or N6-benzyladenine], ABA, salicylic acid, methyl-jasmonate, brassinosteroids (each used at concentrations 0.01, 0.1, 0.5, 1.0, 2.0, and 4.5 μm), gibberellins (0.5, 1.0, or 5.0 μm GA3, GA4, or GA7), ethephone (25 or 50 mg/l), NaCl, KCl, KNO3, K2HPO4/KH2PO4, LiCl, and CsCl (50 to 500 mm). Physiological parameters, including shoot and root weight, root length, and sugar and starch content were determined by growing plants in either MS or Hoagland media containing different concentrations of glucose at 24°C under 200 to 400 μEinstein m−2s−1 irradiance for 2, 3, or 4 weeks using 16-hr light and 8-hr dark cycle as described (Krapp et al. 1993). Data in dose response curves indicate the mean ± standard deviation of four independent metabolite assays or six to 16 biomass measurements. Chlorophyll and anthocyanin concentrations were determined according to Chory (1992). Histological analysis and preparation of contact surface imprints were as described (Szekeres et al. 1996).
Cosegregation analysis of phenotypic traits and T-DNA-encoded antibiotic resistance markers was performed as described (Koncz et al. 1990). Seeds from 199 F2 families (18464) obtained by crossing of prl1 with wild type yielded 4603 hygromycin sensitive and 9239 hygromycin-resistant wild type, and 4622 hygromycin resistant prl1 segregants (1:2:1 ratio, P = 0.999). A cross between ch42(a) and prl1(b) in repulsion resulted in 4 AABB, 163 AaBB, 1553 aaBB, 165 AABb, 3008 AaBb, 165 aaBb, 1514 AAbb, 161 Aabb, and 5 aabb F3 progeny, resulting in a map distance D = 5.13 ± 0.2 cM (P = 0.985, χ2 = 1.443, d.f. 7). A four-point test cross between a bp1/prl1/cer2/ap2 line and F1 yielded the following parental (P) and recombinant (R) classes within the intervals: bp/prl1 (P = 1304, R = 536), prl1/cer2 (P = 1651, R = 189), cer2/ap2 (P = 1537, R = 303), prl1/ap2 (P = 1380, R = 460), bp1/cer2 (P = 1167, R = 673), and bp1/ap2 (P = 1068, R = 772). Classes of double crossover were between bp1–prl1–ap2 (P = 1820; R = 20), prl1–cer2–ap2 (P = 1825, R = 15), and bp1–prl1/cer2–ap2 (P = 1752, R = 88), whereas a triple crossover class was bp1–prl1–cer2–ap2 (P = 1838, R = 2). Calculation of recombination frequencies and derived map distances, as well as determination of physical map positions of PRL1, PRL2, and ATHKAP2 genes using colony-filter hybridization of YAC libraries with cDNA probes were as described (Szekeres et al. 1996). Double mutants were constructed by crossing prl1 with ein2 (ckr1; Ecker 1995) and amp1 (Chaudhury et al. 1993), followed by isolation of homozygous ein2 and amp1 lines carrying the hygromycin resistance marker of the prl1 locus. The prl1 double mutants were germinated in either dark or light in seed medium or in media containing either cytokinin (4.5 μm N6-benzyladenine) or ethephone (25 mg/l) as described (Su and Howell 1992; Szekeres et al. 1996).
Characterization of PRL1 alleles and genetic complementation of the prl1 mutation
The prl1 locus was mapped by Southern hybridization using fragments of the T-DNA vector pPCV6NFLuxF as probes (Koncz et al. 1994). Plant DNA fragments flanking the T-DNA ends were isolated and used as probes for isolation of genomic and cDNA clones from Arabidopsis λEMBL3 and λgt10 libraries, respectively, as described (Koncz et al. 1990). The screening resulted in 16 cDNA clones that all but one showed perfect sequence identity with plant DNA segments of the T-DNA tagged prl1 locus. The longest PRL1 cDNA of 1742 bp (EMBL accession no. X82825) carried 21 bp corresponding to the 5′-untranslated leader of mRNA. One cDNA clone carried a full-length coding sequence of PRL2 (EMBL accession no. X82826). Genomic clones (85) hybridizing with the PRL1 and PRL2 cDNAs were isolated and fingerprinted. PRL1 genomic clones (6) were subjected to physical mapping followed by sequencing of overlapping DNA fragments covering a region of ∼10 kb, including the PRL1 gene of 5455 bp (EMBL accession no. X82824) and a neighboring gene, DI21 (EMBL accession no. Z97339). Sequence comparison of wild-type and mutant loci showed that the T-DNA insertion in prl1 deleted a segment of 344 bp from the PRL1 gene. Breakpoints of this deletion (marked by the T-DNA ends LB1 and LB3; Fig. 4a) were located, respectively, 10 bp 3′-downstream from the 5′-end of exon 15 and 96 bp downstream from the 3′ end of exon 16 of PRL1. PRL1 homologs were identified in the database using the BLASTN and BLASTX programs. A cDNA encoding a human PRL1 ortholog was isolated using the ESTs EST178245 and yw86d09 as probes and sequenced (GenBank accession no. AF044333).
To complement the prl1 mutation genetically, a SpeI–XbaI fragment of 7.9 kb spanning the entire PRL1 gene was isolated from the genomic clone pgcPRL16, cloned into the XbaI site of pPCV002 (Koncz and Schell 1986), transferred to Agrobacterium GV3101 (pMP90RK), and used for transformation of root explants of the homozygous hygromycin-resistant prl1 mutant as described (Koncz et al. 1994). The copy number of pPCV002–PRL1 T-DNA construct was determined as described (Koncz et al. 1990). All three complemented lines carried a single copy of wild-type PRL1 gene in linkage with a selectable kanamycin resistance marker of pPCV002 T-DNA. By selfing of these lines, 36 F2 families were obtained that showed a 3:1 segregation of 2354 kanamycin-resistant wild-type and 793 kanamycin-sensitive prl1 F3 progeny.
Analysis of gene expression in the prl1 mutant and immunolocalization of the PRL1 protein
RNA isolation from Arabidopsis and Northern filter hybridizations with CHS (GenBank accession no. M20308), RBCS (accession no. X13611), G1PAT (accession no. T46127), PGK (accession no. T04348), SUC1 (Sauer and Stolz 1994), PAL (accession no. L33677), P5CS1 (accession no. X86778), AD21 (AtDi21; EMBL accession no. X78585), ADH (GenBank accession no. M12196), PERA (accession no. M58380), PERC (accession no. T03969), SUS1 (Martin et al. 1993), LOX2 (Bell and Mullett 1993), PR1, PR2, PR5 (Uknes et al. 1992), TCH1 (Braam 1992), CPD (Szekeres et al. 1996), CPT (accession no. T04248), GBF1 (Schindler et al. 1992) and other cDNA probes were as described (Szekeres et al. 1996). Nuclei were isolated from wild-type and prl1 plants and run-on transcripts were labeled with 32P[UTP] in vitro (Somssich 1994). Nuclear run-on RNA probes, showing equal specific activity, were hybridized to dot-blots loaded with aliquots (0.4, 2, and 4 μg) of PR5, SUS1, ADH, AD21, and Hsp17,4 (EMBL accession no. X17293) cDNAs (Fig. 3b). The activity of ADH promoter-driven GUS reporter constructs (CADH and Δ G-box-2; Dolferus et al. 1994) was assayed by histochemical staining of Arabidopsis seedlings for 6 hr with X-gluc [1 mg/liter in 50 mm Na-phosphate buffer (pH 7.0) containing 0.5 mm K-ferricyanide and 0.5 mm K-ferrocyanide] as described (Mathur et al. 1998).
From a polyclonal serum raised against a PRL1-specific peptide (VVSQPPRQPDRINEQPGPS located between amino acid positions 64 and 83; Fig. 5) in rabbit, an IgG fraction was purified by (NH4)2SO4-fractionation, protein A–Sepharose binding, and affinity chromatography on the PRL1 peptide (15 mg) coupled to Affigel 10 (Bio-Rad) as described (Harlow and Lane 1988). A BamHI fragment of PRL1 cDNA, carrying 5′-coding sequences of 1 kb, was cloned in pGEX-2T (Pharmacia) and transformed into E. coli BL21DE3 (Novagen, UK) to purify a GST–PRL1–ΔB fusion protein on glutathione–S–Sepharose (Pharmacia; Ausubel et al. 1989). Peptide competition assays were performed with immunoblotted GST–PRL1–ΔB protein using anti-PRL1 IgG incubated with various amounts of PRL1 peptide, as well as with control peptides from PRL2 and other unrelated proteins. SDS-solubilized protein extracts were prepared from plant organs using a glass homogenizer and extraction buffer [100 mm Tris-HCl (pH 6.8) 0.6 m dithiothreitol, 1% SDS, 0.01% bromphenol blue], boiled for 10 min, pelletted by centrifugation (20,000g for 15 min) and separated by SDS-PAGE after loading 30 μg protein in each lane (Ausubel et al. 1989). Crude membrane fractions were prepared from Arabidopsis cells maintained in a suspension culture by disruption of cells in extraction buffer [20 mm Tris-HCl (pH 8.0), 50 mm NaCl, 2 mm EDTA, and 1 mm PMSF], and separation of soluble and membrane fractions by centrifugation (100,000g, for 1 hr at 2°C). The membrane fraction was extracted with 500 mm NaCl (at 0°C for 1 hr), pelletted, and re-extracted with 0.2 m Na2CO3 (pH 11.5). The final membrane pellet was solubilized with extraction buffer containing 2% Triton X-100, and together with the extracted protein fractions subjected to SDS-PAGE separation followed by immunoblotting with anti-PRL1 IgG. Microsomal and plasma membranes were purified as described (Larsson et al. 1987).
For immunolocalization, 4-day-old wild-type and prl1 seedlings were fixed in MTBS [50 mm PIPES (pH 7.0), 5 mm EGTA, 5 mm MgSO4) containing 4% paraformaldehyde for 1 hr, washed with 0.5 ml MTBS, incubated with 2% Driselase in MTBS for 15 min to digest the cell walls, treated with MTBS containing 10% DMSO and 3% NP-40 to permeabilize the membranes, and washed four times with MTBS. The specimens were incubated for 1 hr with a 1:300 dilution of anti-PRL1 IgG, then treated for 1 hr with an anti-rabbit Cy3 antibody (1:300 dilution) in the dark, washed four times with MTBS, stained with DAPI (1 μg/ml in H2O), and inspected by confocal laser microscopy.
The PRL1 cDNA was cloned into the BamHI site of the MYC-tag expression vector pEFmPLINK (Marais et al. 1995), then 15 μg of DNA was transfected by Lipofectin (Life Technologies, Grand Island, NY) into 2 × 105 COS-1 cells on cover slips. After 48 hr, the cells were fixed in 3.5% paraformaldehyde in PBS, and permeabilized with 0.1% Triton X-100 in PBS for 20 min. The specimens were incubated with anti-MYC antibody 9E10 (Evan et al. 1985), followed by treatment with a fluorescein-labeled anti-mouse IgG (Amersham) and staining of the nuclei with DAPI (Puls et al. 1997).
Analysis of PRL1 protein interactions in vitro and in the yeast two-hybrid system
Human PKC-βI and βII enzymes were expressed and purified as described (Stabel et al. 1993). Purified GST and GST–PRL1ΔB proteins were bound to glutathione–S–Sepharose and incubated with PKC-βI (3 μg) and βII (1 μg) activated with phosphatidylserine and phorbol-12–myristate-13–acetate in binding buffer [20 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm CaCl2, 0.1% Triton X-100, and 1 mg/ml of bovine serum albumin) for 1 hr at 4°C as described (Puls et al. 1997). The beads were washed extensively with TENNS [2.5 mm Tris-HCl (pH 7.4), 2.5 mm EDTA, 250 mm NaCl, 1% NP-40, and 2.5% sucrose], then the bound proteins were eluted with 6× SDS-sample buffer, resolved by SDS-PAGE (Ausubel et al. 1989) and immunoblotted using an anti-PKC-β antibody (Stabel et al. 1993) and ECL detection kit (Amersham).
The PRL1-coding sequence was PCR amplified as a BamHI–XhoI fragment, sequenced, and cloned into the yeast vector pAS2 by generating a fusion between PRL1 and the DNA-binding (DB) domain of Gal4p (Durfee et al. 1993). The pAS2–PRL1 bait was transformed into the yeast strain Y190, and the Gal4p–DB–PRL1 expression was confirmed by immunoblotting with an anti-Gal4p–DB antibody (Clontech). An oligo(dT) primed cDNA library was prepared in plasmid pACT2 using mRNA from an Arabidopsis cell suspension and a cDNA synthesis kit (BRL). Yeast host Y190 carrying the pAS2–PRL1 bait was transformed with 0.3 mg of DNA from the pACT2 cDNA library, then the cells were plated on SD-medium containing 50 mm 3-aminotriazole (3-AT) and lacking leucine, tryptophan, and histidine (Durfee et al. 1993). Transformants were inoculated on nylon filters and grown on SD-plates with 25 mm 3-AT to verify their LacZ+ phenotype by β-galactosidase assays. pACT clones coding for PIPs were isolated and transformed into yeast strains Y187 and Y190 (Durfee et al. 1993) carrying either no bait, pAS2–PRL1, or different control baits coding for amino-terminal PRL1 segments of 321 and 412 amino acids, replicator protein of wheat dwarf geminivirus (GenBank accession no. S49387), lamin (Matchmaker System, Clontech), VirD2 (accession no. P18592), VirE2 (accession no. S11844), NPK5 (accession no. D26602), AXI1 (accession no. X80301), cystatin proteinase inhibitor PIP-M and PIP-N proteins (K. Salchert, unpubl.). cDNAs coding for carboxy-terminal segments of α-importin ATHKAP2 were cloned as BamHI–XhoI fragments in pGEX-5X-1 (Pharmacia) to isolate GST–ATHKAP2 fusion proteins from E. coli by purification on glutathione–S–Sepharose (Ausubel et al 1989). PRL1 cDNA was PCR-amplified using a 5′-primer carrying a T7 promoter, sequenced, and used as template to synthesize [35S]methionine PRL1 protein using an in vitro coupled transcription and translation kit (Promega). GST and GST–ATHKAP2 proteins were immobilized on glutathione–S–Sepharose, then equal aliquots from the 35S-labeled PRL1 protein were incubated with these matrices, as well as with the empty Sepharose matrix, in a binding buffer [20 mm Tris-HCl (pH 7.0), 150 mm NaCl, 2 mm EDTA, and 0.1% NP-40) for 1 hr at 4°C. The matrices were washed four times with binding buffer, then the bound proteins were eluted with 4× SDS-sample buffer (Ausubel et al. 1989), and together with the supernatant fractions were size-fractionated on an SDS-PAGE to visualize the labeled PRL1 protein by autoradiography.
Acknowledgments
We thank Drs. Peter Huijser and Gerd Jürgens for their help in the immunolocalization experiments, as well as the Ohio and Nottingham Arabidopsis Biological Resource Centers and the research community for kindly providing cDNA and seed stocks. This work was supported as part of a joint project between the Max-Planck Institut (Köln) and the Biological Research Center (Szeged) by the Deutsche Forschungsgemeinschaft (DFG) and the Hungarian Academy of Sciences and by grants from the European Commission Project of Technological Priority (PL 920401.22), DFG Arabidopsis Schwerpunkt (II B1-1438/1-1), and OTKA T13182.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL koncz@mpiz-koeln.mpg.de; FAX 49-221-5062-213.
References
- Ang L-H, Chattopadhyay S, Wie N, Oyama T, Okada K, Batschauer A, Deng X-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: Greene/Wiley; 1989. [Google Scholar]
- Bell E, Mullet JE. Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 1993;103:1133–1137. doi: 10.1104/pp.103.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J. Regulated expression of the calmodulin-related TCH gene in cultured Arabidopsis cells: Induction by calcium and heat shock. Proc Natl Acad Sci. 1992;89:3213–3216. doi: 10.1073/pnas.89.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfant CE, Mischak H, Watson JE, Winkler BC, Goodnight J, Farese RV, Cooper DR. Regulation of alternative splicing of protein kinase C-beta by insulin. J Biol Chem. 1995;270:13326–13332. doi: 10.1074/jbc.270.22.13326. [DOI] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub MT, Matsui M, Deng X-W. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–121. doi: 10.1016/s0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES. amp1—a mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 1993;4:907–916. [Google Scholar]
- Chory J. A genetic model for light-regulated seedling development in Arabidopsis. Development. 1992;115:337–354. [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M. A role for cytokinins in de-etiolation in Arabidopsis. Plant Physiol. 1994;104:339–347. doi: 10.1104/pp.104.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Chatterjee M, Cook RK, Elich T, Frankhauser C, Li J, Nagpal P, Neff M, Pepper A, Poole D, Reed J, Vitart V. From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc Natl Acad Sci. 1996;93:12066–12071. doi: 10.1073/pnas.93.22.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel PP, Huijser C, Weisbeek PJ, Chua N-H, Smeekens SCM. Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell. 1997;9:583–595. doi: 10.1105/tpc.9.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 1994;105:1075–1087. doi: 10.1104/pp.105.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn A, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Ecker J. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop MJ. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure J-D, Jullien M, Caboche M. Zea3: A pleiotropic mutation affecting cotyledon development, cytokinin resistance and carbon-nitrogen metabolism. Plant J. 1994;5:481–491. doi: 10.1046/j.1365-313x.1994.5040481.x. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Herbers K, Meuwly P, Frommer WB, Métraux J-P, Sonnewald U. Systemic acquired resistance mediated by the ectopic expression of invertase: Possible hexose sensing in the secretory pathway. Plant Cell. 1996;8:793–803. doi: 10.1105/tpc.8.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Smith HMS, Lobreaux S, Raikhel NV. Nuclear import in permeabilized protoplasts from higher plants has unique features. Plant Cell. 1996;8:1337–1352. doi: 10.1105/tpc.8.8.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Huber JL, McMichael RW. Control of plant enzyme activity by reversible phosphorylation. Int Rev Cytol. 1994;149:47–98. [Google Scholar]
- Jang J-C, Sheen J. Sugar sensing in higher plants. Trends Plant Sci. 1997;2:208–214. [Google Scholar]
- Jang J-C, León P, Zhou L, Sheen J. Hexokinase as sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Carlson M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol Cell Biol. 1997;17:2099–2106. doi: 10.1128/mcb.17.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 193–231. [Google Scholar]
- Koch KE. Carbonhydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue specific expression of foreign genes carried by a novel type of Agrobacterium binary vector. Mol & Gen Genet. 1986;204:383–396. [Google Scholar]
- Koncz C, Mayerhofer R, Koncz-Kálmán Z, Nawrath C, Reiss B, Rédei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990;9:1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Németh K, Rédei GP, Schell J. T-DNA insertional mutagenesis. Plant Mol Biol. 1992;20:963–976. doi: 10.1007/BF00027166. [DOI] [PubMed] [Google Scholar]
- Koncz C, Martini N, Szabados L, Hrouda M, Bachmair A, Schell J. Specialized vectors for gene tagging and expression studies. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. B2. Dordrecht, The Netherlands: Kluwer Academic; 1994. pp. 1–22. [Google Scholar]
- Krapp A, Hofmann B, Schäfer C, Stitt M. Regulation of the expression of rbcS and other photosynthetic genes by carbonhydrates: A mechanism for the ‘sink regulation’ of photosynthesis? Plant J. 1993;3:817–828. [Google Scholar]
- Kubo K, Ohno S, Suzuki K. Primary structures of human protein kinase CβI and βII differ only in their C-terminal sequences. FEBS Lett. 1987;223:138–142. doi: 10.1016/0014-5793(87)80524-0. [DOI] [PubMed] [Google Scholar]
- Larsson C, Widell S, Kjellbom P. Preparation of high-purity plasma membranes. Methods Enzymol. 1987;148:558–568. [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T, Frommer WB, Salanoubat M, Willmitzer L. Expression of an Arabidopsis sucrose synthase gene indicates the role in metabolization of sucrose both during phloem loading and in sink organs. Plant J. 1993;4:367–377. doi: 10.1046/j.1365-313x.1993.04020367.x. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Szabados L, Schaefer S, Grunenberg B, Lossow A, Jonas-Straube E, Schell J, Koncz C, Koncz-Kálmán Z. Gene identification with sequenced T-DNA tags generated by transformation of Arabidopsis cell suspensions. Plant J. 1998;13:707–716. doi: 10.1046/j.1365-313x.1998.00059.x. [DOI] [PubMed] [Google Scholar]
- Merkle T, Nagy F. Nuclear import of proteins: Putative import factors and development of in vitro import systems in higher plants. Trends Plant Sci. 1997;2:458–464. [Google Scholar]
- Meyerowitz EM, Somerville CR. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Mochly-Rosen D. Localization of protein kinases by anchoring proteins: A theme in signal transduction. Science. 1995;268:247–251. doi: 10.1126/science.7716516. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C ζ with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnelle B, Coster F, Goffeau A. The sequence of 55 kb on the left arm of yeast chromosome XVI identifies a small nuclear RNA, a new putative protein kinase and two new putative regulators. Yeast. 1996;12:1483–1492. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1483::AID-YEA34%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB. Evidence for an essential role of the sucrose transporter in phloem loading and assimilate partitioning. EMBO J. 1994;13:1–7. doi: 10.1002/j.1460-2075.1994.tb06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Chen C-H, Caldwell J, Jamieson L, Orr E, Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: A homolog of the β subunit of G proteins. Proc Natl Acad Sci. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronne H. Glucose repression in yeast. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J. SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana: Expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- Schindler U, Beckmann H, Cashmore AR. TGA1 and G-box binding factors: Two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell. 1992;4:1309–1319. doi: 10.1105/tpc.4.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, West J, Cnops G, Love K, Balestrazzi A, Dean C. Detailed description of four YAC contigs representing 17 Mb of chromosome 4 of Arabidopsis thaliana ecotype Columbia. Plant J. 1996;9:755–765. doi: 10.1046/j.1365-313x.1996.9050755.x. [DOI] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell. 1990;2:1027–1038. doi: 10.1105/tpc.2.10.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S, Rook F. Sugar sensing and sugar-mediated signal transduction in plants. Plant Physiol. 1997;115:7–13. doi: 10.1104/pp.115.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich IE. Assay for gene expression using run-on transcription in isolated nuclei. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. E1. Dordrect, The Netherlands: Kluwer Academic; 1994. pp. 1–11. [Google Scholar]
- Stabel S, Liyanage M, Frith D. Expression of protein kinase C isoenzymes in insect cells and isolation of recombinant proteins. Methods Neurosci. 1993;18:154–173. [Google Scholar]
- Stitt M, Sonnewald U. Regulation of metabolism in transgenic plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:341–368. [Google Scholar]
- Su W, Howell SH. A single locus, CKR1, defines Arabidopsis mutants in which root growth is resistant to low concentrations of cytokinin. Plant Physiol. 1992;99:1569–1574. doi: 10.1104/pp.99.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Trietel MA, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor activator protein. Proc Natl Acad Sci. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes & Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng X-D. Light activation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- ————— Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- von Schaewen A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. Expression of a yeast-derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbonhydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO J. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Xia G, Ramachandran S, Hong Y, Chan Y-S, Simanis V, Chua N-H. Identification of plant cytoskeletal, cell cycle-related and polarity-related proteins using Schizosaccharomyces pombe. Plant J. 1996;10:761–769. doi: 10.1046/j.1365-313x.1996.10040761.x. [DOI] [PubMed] [Google Scholar]


