Abstract
Microbes in the colon produce compounds, normally excreted by the kidneys, which are potential uremic toxins. Although p-cresol sulfate and indoxyl sulfate are well studied examples, few other compounds are known. Here, we compared plasma from hemodialysis patients with and without colons to identify and further characterize colon-derived uremic solutes. HPLC confirmed the colonic origin of p-cresol sulfate and indoxyl sulfate, but levels of hippurate, methylamine, and dimethylamine were not significantly lower in patients without colons. High-resolution mass spectrometry detected more than 1000 features in predialysis plasma samples. Hierarchical clustering based on these features clearly separated dialysis patients with and without colons. Compared with patients with colons, we identified more than 30 individual features in patients without colons that were either absent or present in lower concentration. Almost all of these features were more prominent in plasma from dialysis patients than normal subjects, suggesting that they represented uremic solutes. We used a panel of indole and phenyl standards to identify five colon-derived uremic solutes: α-phenylacetyl-l-glutamine, 5-hydroxyindole, indoxyl glucuronide, p-cresol sulfate, and indoxyl sulfate. However, compounds with accurate mass values matching most of the colon-derived solutes could not be found in standard metabolomic databases. These results suggest that colonic microbes may produce an important portion of uremic solutes, most of which remain unidentified.
Solutes made by colon microbes may contribute to uremic illness.1,2 Such “putrefaction” products were originally detected in urine and subsequently shown to accumulate in plasma when the kidneys failed.3 Kolff4 emphasized their potential toxicity in his description of the first hemodialysis. Because they are made in an isolated compartment by microbes, their production could prove easier to suppress than that of other uremic solutes. However, only two colon-derived solutes—indoxyl sulfate (IS) and p-cresol sulfate (PCS)—have been extensively studied, and relatively little effort has been made to identify other compounds in this class.
The study presented here used two new strategies to enlarge our knowledge of colon-derived uremic solutes. First, we compared plasma solute concentrations in hemodialysis patients with intact colons with those from hemodialysis patients who had their colons resected. Established assays were used to assess the colon's role in the production of well known uremic solutes. We then used nontargeted high-resolution mass spectrometry (MS) to increase the number of substances detected in uremic plasma. Comparison of results in patients with and without colons using this method revealed the presence of many additional colon-derived uremic solutes.
RESULTS
Characteristics of the patients are summarized in Table 1. Colectomies had been performed in six patients for inflammatory bowel disease (n = 3) and toxic megacolon, ischemia, and cancer (1 each). Patients were all dialyzed three times weekly except for one colectomy patient who was dialyzed twice weekly. Concentrations of uremic solutes measured by HPLC and of urea are summarized in Table 2. Four of these solutes—PCS, IS, hippuric acid (HIPP), and kynurenic acid (KYNA)—were protein-bound organic acids for which the free fractions averaged 0.06 ± 0.02, 0.10 ± 0.05, 0.52 ± 0.07, and 0.14 ± 0.05 of the total concentrations in dialysis patients with colons. As expected, predialysis plasma levels were increased in dialysis patients with intact colons as compared with normal controls. In contrast, PCS and IS levels in dialysis patients without colons were no greater than in normal subjects and were much lower than in dialysis patients with colons. Levels of the two other protein-bound solutes, HIPP and KYNA, and of the aliphatic amines methylamine (MMA) and dimethylamine (DMA) were not significantly different in dialysis patients with and without colons.
Table 1.
Patient characteristics
| Characteristics | Intact Colon (n = 9) | Colectomy (n = 6) |
|---|---|---|
| Women/men | 6/3 | 3/3 |
| Age (years) | 56 ± 7 | 72 ± 14 |
| Body mass index | 26 ± 5 | 26 ± 4 |
| Duration on dialysis (years) | 9 ± 6 | 8 ± 6 |
| Duration colectomy (years) | – | 20 ± 19 |
| Kt/V | 1.77 ± 0.42 | 1.60 ± 0.21 |
| Duration of dialysis session (minutes) | 188 ± 13 | 203 ± 29 |
| Diabetes | 5 of 9 | 3 of 6 |
Values are mean ± SD. Body mass index was not calculated for one colectomy patient who was a double amputee. Values for Kt/V and normalized protein-to-creatinine ratio were calculated from pre- and post-treatment blood urea nitrogen level for the treatment at which samples were collected. Diabetic nephropathy was considered to be the cause of renal failure in four patients with intact colons and in two patients who had undergone colectomy.
Table 2.
Solutes measured by HPLC and urea
| Solute | Dialysis Intact Colon (n = 9) | Dialysis Colectomy (n = 6) | Normal Control (n = 7 to 10) |
|---|---|---|---|
| PCS | |||
| plasma pretreatment mg/dl | 4.1 ± 1.6a,b | 0.06 ± 0.09 | 0.19 ± 0.13 |
| reduction ratio | 30 ± 7 | – | – |
| IS | |||
| plasma pretreatment mg/dl | 2.8 ± 1.3a,b | 0.08 ± 0.06 | 0.06 ± 0.02 |
| reduction ratio | 33 ± 7 | 31 ± 11 | – |
| KYNA | |||
| plasma pretreatment nM | 799 ± 404b | 634 ± 292b | 29 ± 7 |
| reduction ratio | 36 ± 7 | 39 ± 16 | – |
| Hippurate | |||
| plasma pretreatment mg/dl | 7.9 ± 4.5b | 4.6 ± 5.9b | 0.3 ± 0.2 |
| reduction ratio | 68 ± 4 | 72 ± 19 | – |
| DMA | |||
| plasma pretreatment μg/dl | 1032 ± 155b | 890 ± 103b | 218 ± 33 |
| reduction ratio | 38 ± 10 | 43 ± 7 | – |
| MMA | |||
| plasma pretreatment μg/dl | 58 ± 10b | 54 ± 9b | 32 ± 4 |
| reduction ratio | 30 ± 10 | 23 ± 6 | – |
| Urea | |||
| plasma pretreatment mg/dl | 50 ± 8b | 43 ± 16b | 14 ± 3 |
| reduction ratio | 74 ± 4 | 78 ± 7 | – |
Values are mean ± SD. The plasma PCS concentration was below the limits of detection in one normal subject and in three pretreatment samples and four post-treatment samples from dialysis patients without colons; a reduction ratio for PCS in the dialysis colectomy group was therefore not determined. The plasma hippurate was below the limits of detection in one patient without a colon.
aP < 0.05 dialysis intact colon group versus dialysis colectomy group.
bP < 0.05 dialysis intact colon group or dialysis colectomy group versus normal control group.
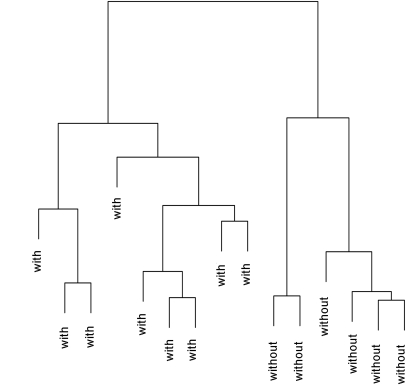
Application of high-resolution mass spectrometry revealed a much broader influence of the colon on uremic solute production. A total of 1055 features representing potential solutes were detected in pretreatment plasma samples from the dialysis patients. Unsupervised hierarchical clustering based on these features revealed a clear separation between dialysis patients with and without colons as depicted in Figure 1. Further analysis detected specific solutes in patients with colons that were absent or present at lower concentration in patients without colons. As summarized in Table 3, statistical analysis identified 35 such solutes, including 19 considered absent in patients without colons and 8 found to have lower levels with significance defined as P < 0.05 and an additional 8 solutes found to have lower levels with significance defined as P < 0.10. These numbers contrasted strongly with the finding of a single solute identified as being at greater concentration in patients without colons using the same criteria. As further summarized in Table 3, almost all of the colon-derived solutes were found to be at higher concentration in dialysis patients than in normal subjects or were undetectable in normal subjects. For several of them the reduction ratio calculated from pre- and post-treatment plasma levels was less than for urea, indicating that they were not effectively removed by conventional hemodialysis.
Figure 1.
Clustering identified the six patients with colectomies and the nine patients with intact colons as belonging to different groups. Unsupervised hierarchical clustering was performed on the distances calculated from the sum of the squared differences between the log-transformed amplitudes of 1055 features detected by MS in predialysis plasma samples from the 15 hemodialysis patients. Distances along the vertical axis provide an index of the aggregate differences in feature amplitudes between individual patients and groups of patients.
Table 3.
Solutes identified by LC/MS
| Solute Name or Mass (D) | Colectomy/with Colon | Hemodialysis/ Normal | Dialytic Reduction Ratio |
|---|---|---|---|
| Colon-derived uremic solutes identified by analysis of chemical standards | |||
| PCSa | 0.01 | 11 | 0.25 ± 0.02 |
| α-N-phenylacetyl-l-glutamineb | 0.07 | 91 | 0.73 ± 0.09 |
| ISb | 0.02 | 23 | 0.31 ± 0.03 |
| indoxyl glucuronidec | 0.02 | e | 0.79 ± 0.06 |
| 5-hydroxyindolec | e | e | 0.83 ± 0.08 |
| Colon-derived uremic solutes not identified by analysis of chemical standards | |||
| 201.0786a | 0.03 | e | 0.10 ± 0.26 |
| 204.0092a | e | e | 0.51 ± 0.15 |
| 229.0043a | e | e | 0.70 ± 0.07 |
| 230.0689a | e | e | 0.07 ± 0.10 |
| 284.0813a | e | e | e |
| 284.0886a | e | e | 0.77 ± 0.06 |
| 301.1058a | e | e | 0.32 ± 0.11 |
| 301.1156a | e | e | 0.83 ± 0.04 |
| 306.0407a | e | e | 0.81 ± 0.07 |
| 306.0709a | e | e | 0.82 ± 0.03 |
| 325.0797a | e | e | 0.81 ± 0.05 |
| 334.1921a | e | 23 | 0.18 ± 0.07 |
| 376.1339a | 0.32 | e | 0.82 ± 0.03 |
| 416.1321a | e | e | 0.80 ± 0.04 |
| 420.1599a | 0.26 | e | 0.82 ± 0.03 |
| 474.1527a | e | 4 | 0.62 ± 0.06 |
| 641.3412a | e | e | 0.73 ± 0.07 |
| 169.0525b | e | e | 0.42 ± 0.09 |
| 229.0045b | e | e | 0.71 ± 0.07 |
| 244.0847b | e | e | 0.26 ± 0.18 |
| 275.0096b | 0.02 | e | 0.70 ± 0.06 |
| 276.0302b | 0.05 | e | 0.79 ± 0.05 |
| 101.0105c | 0.19 | e | 0.67 ± 0.06 |
| 192.0552c | 0.21 | 39 | 0.39 ± 0.25 |
| 195.0529c | 0.02 | 38 | 0.71 ± 0.07 |
| 265.0949c | 0.06 | e | 0.79 ± 0.06 |
| 283.0839c | 0.04 | e | 0.73 ± 0.20 |
| 350.1870c | 0.03 | e | 0.85 ± 0.02 |
| Colon-derived solutes that do not accumulate in uremia | |||
| 3-indolepropionic acida | e | 1 | −0.07 ± 0.05 |
| 129.0578b | 0.04 | 1 | −0.04 ± 0.08 |
| Additional indole and phenyl compounds identified by analysis of chemical standards | |||
| phenyl glucuronided | 0.15 | e | 0.80 ± 0.06 |
| cinnamoylglycined | 0.19 | 14 | 0.22 ± 0.18 |
| HIPPd | 0.28 | 27 | 0.60 ± 0.06 |
| indoleacetic acidd | 0.57 | 5 | 0.48 ± 0.08 |
| 2-hydroxybenzoic acid (salicylic acid)d | 0.68 | 5 | 0.46 ± 0.24 |
| indolelactic acidd | 1.8 | 3 | 0.12 ± 0.09 |
| l-kynurenined | 2.0 | 2 | 0.44 ± 0.07 |
| Found in higher concentration in patients without colons | |||
| 331.3233b | 2.9 | 2 | 0.30 ± 0.35 |
Mass values are for neutral compounds assuming monovalent ion detection by MS. Colectomy/with Colon, estimated ratio of average concentration in hemodialysis patients without colons relative to hemodialysis patients with colons; Hemodialysis/Normal, estimated ratio of average concentration in hemodialysis patients with colons relative to normal subjects; Dialytic Reduction Ratio, mean ± SD fractional reduction in plasma levels achieved by dialysis treatment in patients with colons. Levels were too low to estimate a reduction ratio for one solute.
Difference between patients with and without colons: asolute not detectable in patients without colons;
bP < 0.05;
cP < 0.10;
dP > 0.4.
eConcentrations were too low to be estimated in dialysis patients without colons (for Colectomy/with Colon) or in normal subjects (for Hemodialysis/Normal).
Analysis of a panel of 31 indole and phenyl standards (Supplementary Table 1) established the identity of five potential colon-derived solutes, as further summarized in Table 3. 5-Hydroxyindole and α-N-phenylacetyl-glutamine were found to be not only of colonic origin but present in higher levels in dialysis patients than normal subjects, along with PCS and IS, which had previously been identified as such by the HPLC assays. 3-Indolepropionic acid was also identified as of colonic origin, but it was not more prominent in dialysis patients than in normal subjects and did not appear to be removed by dialysis. However, chemical identities could not be established for most of the features considered to represent uremic solutes of colonic origin. Candidate identities for most of these solutes could not be found in the HMDB,5 METLIN,6 and Metacyc7 databases. When we excluded metabolites of pharmaceuticals that the patients were known not to be taking, the three databases together provided candidates with mass within 3 parts per million (ppm) for only 6 of the 29 unidentified colon-derived solutes. Some of the candidate compounds were known indole or phenyl compounds for which we had not been able to obtain standards. These were p-cresol glucuronide (284.0896 D), indolylacryloylglycine (244.0848 D), and the hydroxyhippuric acid isomers 2-hydroxyhippuric acid and 3-hydroxyhippuric acid (195.0532 D). Review of the uremia literature yielded two additional candidate compounds, the additional hydroxyhippuric acid isomer 4-hydroxyhippuric acid (195.0532 D) and 4-methylcatechol sulfate (204.0092 D).8,9 However, we did not find compounds with mass values corresponding to the other colon-derived solutes detected by MS.
Seven additional substances that we did not categorize as colon-derived were identified by analysis of the panel of indole and phenyl standards, as further shown in Table 3. Two were less prominent in patients without colons than in patients with colons (phenyl glucuronide and cinnamoylglycine), but the differences were NS because of the variable levels in individual patients and the many comparisons made. Consistent with prior HPLC analysis of the same samples, hippurate was also not significantly lower in the patients without colons. We also observed no effect of colectomy on the levels of indoleacetic acid, 2-hydroxy benzoate, indole lactic acid, and l-kynurenine. For the 13 solutes identified by analysis of standards, the average mass error calculated as the true monoisotopic mass subtracted from the value reported by the mass spectrometer was negative 1.4 ± 1.1 ppm.
DISCUSSION
The study presented here compared plasma from dialysis patients with and without colons to enlarge our knowledge of colon-derived solutes. We first assessed the colonic contribution to the production of several well known solutes. HPLC assays revealed that IS and PCS were nearly absent in patients without colons. These findings are consistent with knowledge that IS and PCS are sulfate conjugates of indole and p-cresol produced by colon microbes from tryptophan and phenylalanine/tyrosine, respectively.10,11 The low levels of IS and PCS detected in some colectomy patients could reflect microbial growth in the small intestine or, in one case, solute production in a portion of the left colon that remains in place in a patient with an ileostomy. In contrast, levels of KYNA, which is produced from tryptophan by mammalian cells, were no different in patients with and without colons. We also found no difference in the levels of hippurate, the organic acid that is most abundant in human urine and in dialysis patients' plasma. Hippurate is the glycine conjugate of benzoic acid, which is derived from polyphenols in plant foods and added to processed foods as a preservative. Dietary polyphenols can be converted to benzoate by colon microbes, and hippurate levels are very low in germ-free rats.12–14 However, our findings suggest that in humans a larger portion of hippurate is derived from precursors absorbed in the small intestine as described by Rechner et al.15 It should be noted that diet and species differences may affect the location of hippurate production. Finally, we found no difference in the levels of DMA and MMA between patients with and without colons. Although the metabolic origin of these aliphatic amines is uncertain, gut bacteria have been considered to contribute to the their production, and Simenhoff et al.16 reported that treatment with a nonabsorbable antibiotic reduced DMA levels in dialysis patients.17–19 Of note, these prior studies used different assay methods that yielded much higher values for plasma DMA concentration.
Our use of HPLC to evaluate the colonic production of uremic solutes was limited by the number of assays established in our laboratories. These assays use separate chromatographic runs with fluorescence detection for individual solutes. The number of solutes identified in single chromatographic runs can be greatly increased by MS detection, and it is now possible to perform metabolomic studies in which hundreds of compounds are detected in individual samples.14,20 Niwa21 has reviewed the use of MS to identify uremic solutes, and Kikuchi et al.22 used metabolomic methods to identify solutes for which administration of the oral sorbent AST-120 reduced the plasma levels in rats with renal insufficiency. One approach uses MS to simultaneously assay a large panel of known solutes. Using this approach, Rhee et al.23 found significant differences in the plasma levels of 40 of 353 analytes when hospitalized patients with renal failure were compared with at-risk controls. An alternative “untargeted” approach detects unnamed chemical features characterized by mass-to-charge (m/z) ratio. High-resolution MS detectors can resolve m/z to within a few ppm, and, as seen in the study presented here, such instruments detect hundreds of individual features in plasma samples.
Wikoff et al.14 recently used untargeted high-resolution liquid chromatography (LC)/MS to identify plasma solutes produced by gut microbes in rats. Of several thousand features detected in plasma, approximately 10% were considered to vary in concentration between conventional and germ-free rats. Most of these features were less prevalent in the germ-free animals and thus considered to be of microbial origin. Using a similar approach, we demonstrated a clear separation between plasma solute profiles in dialysis patients with and without colons. Statistical analysis identified MS features representing more than 20 potential colon-derived solutes. Of note, most of these were more prominent in dialysis patients than in normal subjects, which indicates that they accumulate in the plasma when the kidneys fail. The reduction in plasma concentration during dialysis was less for several of the colon-derived solutes than for urea. Theoretically, this could result from lower clearance by dialysis, as is known to be the case for IS and PCS, and/or a larger volume of distribution.24,25
The features detected by MS represent true circulating compounds and various adducts and fragments formed during the analysis. Many of the excess features can be identified and deleted by software programs or by manual review as was done in the study presented here. However, the correspondence of features detected by MS and compounds circulating in plasma can be established with certainty only when chemical standards are available. Using chemical standards, Wikoff et al.14 identified nine indole and phenyl derivatives and seven other solutes that were present in higher concentration in conventional than in germ-free rats. We identified six indole and phenyl derivatives that were present at higher concentration in patients with colons than in patients without colons. In addition to PCS and IS, these included indoxyl glucuronide, which has previously been identified in uremic plasma by Niwa et al.26 and is presumably formed by hepatic glucuronidation of indoxyl in a manner analogous to the formation of IS by sulfation of indoxyl. We also identified α-N-phenylacetyl-glutamine and 5-hydroxyindole as colon derived. α-N-Phenylacetyl-glutamine has previously been shown to accumulate in uremia, but production by mammalian tissue and microbial decarboxylation of phenylalanine to phenylethylamine with subsequent conjugation have been reported.27–29 The results presented here, like those of Wikoff et al. 14 and Seakins,29 suggest that the microbial pathway is predominant. 5-Hydroxyindole, like indoxyl, is formed by oxidation of indole, which microbes produce from tryptophan. It is excreted largely conjugated with sulfate in rats; the pattern of conjugation in humans has not been studied.30
Chemical identities were not established for most of the colon-derived features we detected in uremic plasma. In four patients we found candidate compounds with closely matching m/z values but were unable to obtain standards. These included p-cresol glucuronide, 2-hydroxyhippuric acid, 4-hydoxyhippuric acid, and 4-methylcatechol sulfate, which have previously been detected in uremic plasma, and 3-hydroxyhippuric acid and indolylacryloylglycine, which have been found in human urine.8,9,31–34 However, for most of the colon-derived solutes, candidate compounds could not be found in the HMDB, METLIN, and Metacyc databases. Mammalian conjugation of microbially derived substances is one important barrier to their identification.14 The problem is twofold. First, databases may include a microbially derived substance but not its conjugates. Second, when the conjugates are known or can be assumed, they are usually not available as standards. Identification of the unidentified colon-derived solutes will thus likely require MS/MS analysis of the product ions produced by their fragmentation and/or synthesis of chemical standards.
In summary, analysis of plasma from hemodialysis patients without colons has allowed us to clarify the origin of some known uremic solutes and to demonstrate the presence of many additional uremic solutes of microbial origin. The major weakness of the study is limited chemical identification of these additional colon-derived solutes. Some of the mass spectrometric features for which magnitude was different in patients with and without colons could represent adducts or fragments that we failed to identify rather than circulating compounds, but the true number of microbially derived compounds is likely to be much larger than described here. First, we used electrospray ionization with a single chromatographic technique (reversed-phase chromatography). The number of compounds identified in biologic samples can generally be increased by using additional chromatographic methods, particularly including those designed to separate highly polar compounds, and using additional ionization methods after LC.14,20 Second, our LC/MS afforded limited sensitivity as evidenced by its failure to detect KYNA. Last and most importantly, we found only six dialysis patients with colectomies, which limited our power to identify significant differences for solutes for which levels varied greatly among individual patients. It is notable that IS and PCS, which are the best known colon-derived solutes, have potential toxic effects.1,2,35 Further studies with additional subjects should provide a fuller profile of microbially derived compounds retained in uremia. This could in turn provide a scientific basis for testing the health benefit of reducing the production of such solutes by nutritional, probiotic, or pharmacologic means.
CONCISE METHODS
Studies were carried out in 15 stable hemodialysis patients: 9 had intact colons and 6 had undergone colectomy. All of the patients had ileostomies. They were selected for study because they were reported to have total colectomies, but in one patient further record review revealed that a portion of the left colon remained in place. Patients were maintained on their routine diets and were not asked to fast before the study. They had no antibiotic treatment for at least 1 month and no active gastrointestinal disease. Residual urea clearance was recorded at 0.3 and 1.0 ml/min in two colectomy patients; other subjects described negligible urine output. Plasma samples were obtained pre- and postdialysis with the postsample obtained in a manner appropriate for estimation of urea kinetics.36 Concentrations of PCS, IS, HIPP, KYNA, and MMA were measured by HPLC as previously described,37,38 and DMA was measured by a modification of the MMA method. Free levels of PCS, IS, HIPP, and KYNA were measured in plasma ultrafiltrate prepared with Nanosep 30K Omega separators (Pall, Ann Arbor, MI), and the free fraction was calculated as the level in the ultrafiltrate divided by the level in the plasma. Plasma samples were obtained from ten normal subjects for assay of MMA and DMA and from another group of seven normal subjects for other HPLC assays and for mass spectrometry. Urea was measured in the clinical laboratory.
Plasma samples for LC/MS analysis were deproteinized by the addition of four parts methanol, dried by vacuum centrifugation, resuspended in 95:5 water:acetonitrile, and clarified by centrifugation. Chromatography was performed on an ACQUITY UPLC system (Waters, Milford, MA); 5 μl of extracted plasma was loaded to a Kinetex 150 mm × 2.1 mm × 1.7 μm particle-size fused core C18 column (Phenomenex, Torrance, CA). The column was held at 40°C. Buffer flow was 0.25 ml/min using 0.1% (v) formic acid in water (A) and 0.1% (v) formic acid in acetonitrile (B) with a gradient from 5% B to 10% B over 14 minutes and then from 10% B to 100% B to 22 minutes. MS was performed on an Exactive orbitrap mass spectrometer (ThermoFisher, San Jose, CA) with data collected over the range from 70 to 800 m/z with 50,000 FWHM resolution using electrospray ionization. Samples from patients with and without colons were first run in negative ionization and then in positive ionization mode with the order mixed to minimize any effects of instrument drift. Pre- and postdialysis plasma samples from patients with colons and predialysis plasma samples from seven of the nine patients with colons and normal subjects were subsequently analyzed in separate LC/MS runs using the same techniques. Manufacturer's software (Sieve 1.3, ThermoFisher, San Jose, CA) was used to identify features characterized by retention time and m/z values in data files from each LC/MS run and to assign amplitudes to these features in individual samples on the basis of integration of the ion current values over ranges of time and m/z. The software identified 819 negative ion features and 900 positive ion features in the predialysis samples from patients with and without colons. These totals were reduced to 519 negative ion and 536 positive ion features by inspection with elimination of those considered to represent isotopic peaks and adducts or not to represent defined chromatographic peaks. An unsupervised hierarchical cluster analysis was performed on the log-transformed feature amplitudes derived by the Sieve software for the total of 1055 negative and positive features. The clustering was performed using complete linkage with the hclust command in R software.39 Comparison of individual features in dialysis patients with and without colons was accomplished by first removing features that did not have a median signal above a noise threshold in at least one of the two groups. The noise threshold was set based on examination of a QQplot comparing the feature amplitudes in the two groups. Features were then considered to represent uremic solutes absent in the colectomy group but present in the patients with colons if the amplitudes were below the noise threshold in all of the colectomy patients but greater than the noise threshold in at least five of the nine patients with colons. Conventional significance values cannot be calculated for this comparison because the variance of amplitudes for features below the noise threshold in all patients in a group is artificially low, leading to inflation of t statistics. However, there were no features characterized by the opposite pattern of having amplitudes below the noise threshold in the patients with colons and amplitudes above the noise threshold in at least three of the six colectomy patients. The significance of differences between the remaining features in the two groups was then assessed using a nonparametric t test with P values adjusted for multiple testing. Features were chemically identified by comparing m/z and retention time to a panel of standards for 31 indole and phenyl compounds identified by literature review as possible uremic and/or colon-derived solutes (described in Supplementary Table 1).
Amplitudes for features identified as having different amplitude in patients with and without colons and of seven additional features identified by analysis of standards were re-integrated manually using LCquant software (ThermoFisher, San Jose, CA). These amplitudes were used to estimate relative concentrations of individual solutes among different patients but could not be used to estimate relative concentrations of different solutes because ionization of different solutes by electrospray is highly variable. Estimates of the ratios of solute concentrations in patients without colons relative to patients with colons were calculated as the inverse logarithms of the difference in mean logarithms for the feature amplitudes as assessed by manual re-integration in the two groups. These calculations were performed only when peaks were measurable in at least four of the six subjects without colons; missing values were replaced with the average of the three lowest measureable values. Estimates of ratios of solute concentration in dialysis patients with colons relative to normal subjects were again obtained using manually re-integrated feature amplitudes. Solute reduction ratios were calculated by comparison of manually re-integrated feature amplitudes in pre- and postdialysis samples in the patients with colons. Values for the reduction ratio were not calculated in the small number of cases in which feature amplitudes could not be assigned in pre- and postdialysis samples, and the mean ratio was calculated with these cases omitted.
Disclosures
None.
Acknowledgments
F.J-G.L was supported by a National Institutes of Health (NIH) Training Grant (DK7357) and subsequently by a National Kidney Foundation Fellowship. Other support was provided by NIH (R21 DK077326 to T.H.H., R21 DK084439 to T.W.M., and R01 GM086884 to S.H.). The authors are grateful to nephrologists who collaborated in identifying hemodialysis patients who had undergone colectomy.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1. Evenepoel P, Meijers BK, Bammens BR, Verbeke K: Uremic toxins originating from colonic microbial metabolism. Kidney Int Suppl 114: S12–S19, 2009 [DOI] [PubMed] [Google Scholar]
- 2. Schepers E, Glorieux G, Vanholder R: The gut: The forgotten organ in uremia? Blood Purif 29: 130–136, 2010 [DOI] [PubMed] [Google Scholar]
- 3. Schreiner G, Maher J: Biochemistry of uremia. In: Uremia, Springfield, MA, Charles Thomas, 1960, pp 55–85 [Google Scholar]
- 4. Kolff WJ: The Artificial Kidney, Kampen, The Netherlands, J H. Kok, 1946 [Google Scholar]
- 5. Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I: HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res 37: D603–D610, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G: METLIN: A metabolite mass spectral database. Ther Drug Monit 27: 747–751, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD: The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 38: D473–D479, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Niwa T, Maeda K, Ohki T, Saito A, Kobayashi K: A gas chromatographic-mass spectrometric analysis for phenols in uremic serum. Clin Chim Acta 110: 51–57, 1981 [DOI] [PubMed] [Google Scholar]
- 9. Schoots AC, De Vries PM, Thiemann R, Hazejager WA, Visser SL, Oe PL: Biochemical and neurophysiological parameters in hemodialyzed patients with chronic renal failure. Clin Chim Acta 185: 91–107, 1989 [DOI] [PubMed] [Google Scholar]
- 10. Niwa T: Organic acids and the uremic syndrome: Protein metabolite hypothesis in the progression of chronic renal failure. Semin Nephrol 16: 167–182, 1996 [PubMed] [Google Scholar]
- 11. Bone E, Tamm A, Hill M: The production of urinary phenols by gut bacteria and their possible role in the causation of large bowel cancer. Am J Clin Nutr 29: 1448–1454, 1976 [DOI] [PubMed] [Google Scholar]
- 12. Mulder TP, Rietveld AG, van Amelsvoort JM: Consumption of both black tea and green tea results in an increase in the excretion of hippuric acid into urine. Am J Clin Nutr 81: 256S–260S, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Gonthier MP, Remesy C, Scalbert A, Cheynier V, Souquet JM, Poutanen K, Aura AM: Microbial metabolism of caffeic acid and its esters chlorogenic and caftaric acids by human faecal microbiota in vitro. Biomed Pharmacother 60: 536–540, 2006 [DOI] [PubMed] [Google Scholar]
- 14. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G: Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106: 3698–3703, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rechner AR, Kuhnle G, Hu H, Roedig-Penman A, van den Braak MH, Moore KP, Rice-Evans CA: The metabolism of dietary polyphenols and the relevance to circulating levels of conjugated metabolites. Free Radic Res 36: 1229–1241, 2002 [DOI] [PubMed] [Google Scholar]
- 16. Simenhoff ML, Burke JF, Saukkonen JJ, Ordinario AT, Doty R: Biochemical profile of uremic breath. N Engl J Med 297: 132–135, 1977 [DOI] [PubMed] [Google Scholar]
- 17. Smith JL, Wishnok JS, Deen WM: Metabolism and excretion of methylamines in rats. Toxicol Appl Pharmacol 125: 296–308, 1994 [DOI] [PubMed] [Google Scholar]
- 18. Zeisel SH, Wishnok JS, Blusztajn JK: Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther 225: 320–324, 1983 [PubMed] [Google Scholar]
- 19. Zhang AQ, Mitchell SC, Ayesh R, Smith RL: Dimethylamine formation in man. Biochem Pharmacol 45: 2185–2188, 1993 [DOI] [PubMed] [Google Scholar]
- 20. Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S: Mass-spectrometry-based metabolomics: Limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 5: 435–458, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niwa T: Recent progress in the analysis of uremic toxins by mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 877: 2600–2606, 2009 [DOI] [PubMed] [Google Scholar]
- 22. Kikuchi K, Itoh Y, Tateoka R, Ezawa A, Murakami K, Niwa T: Metabolomic search for uremic toxins as indicators of the effect of an oral sorbent AST-120 by liquid chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 878: 2997–3002, 2010 [DOI] [PubMed] [Google Scholar]
- 23. Rhee EP, Souza A, Farrell L, Pollak MR, Lewis GD, Steele DJ, Thadhani R, Clish CB, Greka A, Gerszten RE: Metabolite profiling identifies markers of uremia. J Am Soc Nephrol 21: 1041–1051, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lesaffer G, De Smet R, Lameire N, Dhondt A, Duym P, Vanholder R: Intradialytic removal of protein-bound uraemic toxins: Role of solute characteristics and of dialyser membrane. Nephrol Dial Transplant 15: 50–57, 2000 [DOI] [PubMed] [Google Scholar]
- 25. Martinez AW, Recht NS, Hostetter TH, Meyer TW: Removal of p-cresol sulfate by hemodialysis. J Am Soc Nephrol 16: 3430–3436, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Niwa T, Miyazaki T, Tsukushi S, Maeda K, Tsubakihara Y, Owada A, Shiigai T: Accumulation of indoxyl-beta-d-glucuronide in uremic serum: Suppression of its production by oral sorbent and efficient removal by hemodialysis. Nephron 74: 72–78, 1996 [DOI] [PubMed] [Google Scholar]
- 27. Zimmerman L, Jornvall H, Bergstrom J: Phenylacetylglutamine and hippuric acid in uremic and healthy subjects. Nephron 55: 265–271, 1990 [DOI] [PubMed] [Google Scholar]
- 28. Moldave K, Meister A: Synthesis of phenylacetylglutamine by human tissue. J Biol Chem 229: 463–476, 1957 [PubMed] [Google Scholar]
- 29. Seakins JW: The determination of urinary phenylacetylglutamine as phenylacetic acid. Studies on its origin in normal subjects and children with cystic fibrosis. Clin Chim Acta 35: 121–131, 1971 [DOI] [PubMed] [Google Scholar]
- 30. King LJ, Parke DV, Williams RT: The metabolism of [2–14C] indole in the rat. Biochem J 98: 266–277, 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Loor H, Bammens B, Evenepoel P, De Preter V, Verbeke K: Gas chromatographic-mass spectrometric analysis for measurement of p-cresol and its conjugated metabolites in uremic and normal serum. Clin Chem 51: 1535–1538, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Armstrong MD, Shaw KN, Gortatowski MJ, Singer H: The indole acids of human urine; paper chromatography of indole acids. J Biol Chem 232: 17–30, 1958 [PubMed] [Google Scholar]
- 33. Carr K, Whiteley P, Shattock P: Development and reproducibility of a novel high-performance liquid-chromatography monolithic column method for the detection and quantification of trans-indolyl-3-acryloylglycine in human urine. Biomed Chromatogr 23: 1108–1115, 2009 [DOI] [PubMed] [Google Scholar]
- 34. Armstrong MD, Wall PE, Parker VJ: The excretion of m-hydroxyhippuric acid by humans. J Biol Chem 218: 921–927, 1956 [PubMed] [Google Scholar]
- 35. Niwa T: Indoxyl sulfate is a nephro-vascular toxin. J Ren Nutr 20: S2–S6, 2010 [DOI] [PubMed] [Google Scholar]
- 36. Depner TA, Daugirdas JT, Goldstein S, Ing TS, Kumar V, Meyer KB, Norris K: Hemodialysis Adequacy 2006: Guideline 3. Methods for post dialysis blood sampling. Am J Kidney Dis 48: S24–S27, 2006 [Google Scholar]
- 37. Luo FJ, Patel KP, Marquez IO, Plummer NS, Hostetter TH, Meyer TW: Effect of increasing dialyzer mass transfer area coefficient and dialysate flow on clearance of protein-bound solutes: A pilot crossover trial. Am J Kidney Dis 53: 1042–1049, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ponda MP, Quan Z, Melamed ML, Raff A, Meyer TW, Hostetter TH: Methylamine clearance by haemodialysis is low. Nephrol Dial Transplant 25: 1608–1613, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. R DCT: A Language and Environment for Statistical Computing, Vienna, Austria, R Foundation for Statistical Computing, 2009. Available at: http://www.R-project.org [Google Scholar]