Abstract
Localization of acetylcholine receptors (AChRs) to neuromuscular synapses is mediated by multiple pathways. Agrin, which is the signal for one pathway, stimulates a redistribution of previously unlocalized AChRs to synaptic sites. The signal for a second pathway is not known, but this signal stimulates selective transcription of AChR genes in myofiber nuclei located near the synaptic site. Neuregulin (NRG) is a good candidate for the extracellular signal that induces synapse-specific gene expression, since NRG is concentrated at synaptic sites and activates AChR gene expression in cultured muscle cells. Previous studies have demonstrated that 181 bp of 5′ flanking DNA from the AChR δ-subunit gene are sufficient to confer synapse-specific transcription in transgenic mice and NRG responsiveness in cultured muscle cells, but the critical sequences within this cis-acting regulatory region have not been identified. We transfected AChR δ-subunit–hGH gene fusions into a muscle cell line, and we show that a potential binding site for Ets proteins is required for NRG-induced gene expression. Furthermore, we produced transgenic mice carrying AChR δ-subunit–hGH gene fusions with a mutation in this NRG-response element (NRE), and we show that this NRE is necessary for synapse-specific transcription in mice. The NRE binds proteins in myotube nuclear extracts, and nucleotides that are important for NRG responsiveness are likewise critical for formation of the protein–DNA complex. This complex contains GABPα, an Ets protein, and GABPβ, a protein that lacks an Ets domain but dimerizes with GABPα, because formation of the protein–DNA complex is inhibited by antibodies to either GABPα or GABPβ. These results demonstrate that synapse-specific and NRG-induced gene expression require an Ets-binding site and suggest that GABPα/GABPβ mediates the transcriptional response of the AChR δ-subunit gene to synaptic signals, including NRG.
Keywords: Ets proteins, GABP, neuregulin, ErbBs, acetylcholine receptor, neuromuscular synapse
Shortly after contact between a growing motor axon and a differentiating myotube is established, signals are exchanged between nerve and muscle that initiate the formation and assembly of a highly differentiated presynaptic nerve terminal and a highly specialized postsynaptic apparatus (Hall and Sanes 1993; Burden 1998). Acetylcholine receptors (AChRs) are among the proteins that become localized to this small patch of the muscle fiber membrane, and their localization to synaptic sites during development is a hallmark of the inductive events of synapse formation.
Current data suggest that postsynaptic differentiation, including clustering of AChRs, is initiated and maintained by two different ligands that stimulate distinct signaling pathways. Agrin, an ∼200-kD protein that is synthesized by motor neurons and deposited into the extracellular matrix at synapses is the signal for one of these pathways (McMahan 1990). Agrin stimulates the reorganization of proteins, including AChRs, associated with the muscle cell membrane and has a critical role in synapse formation, because mice lacking agrin or MuSK, a component of its receptor complex, are unable to form neuromuscular synapses (DeChiara et al. 1996; Gautam et al. 1996).
Studies with transgenic mice that harbor gene fusions between regulatory regions of AChR subunit genes and reporter genes have shown that AChR genes are transcribed at a higher rate in myofiber nuclei positioned near the synaptic site than in nuclei in nonsynaptic regions of the myofiber (Klarsfeld et al. 1991; Sanes et al. 1991; Simon et al. 1992). This second signaling pathway for postsynaptic differentiation leads to the accumulation of AChR mRNAs at synaptic sites (Merlie and Sanes 1985; Fontaine and Changeux 1989; Goldman and Staple 1989), resulting in increased AChR protein synthesis in the synaptic region of the myofiber.
RNAs encoding other synaptic proteins, including AChE, MuSK, rapsyn, s-laminin, N-CAM, utrophin, and the regulatory subunit of protein kinase A are also concentrated at synaptic sites (Jasmin et al. 1993; Valenzuela et al. 1995; Moscoso et al. 1995a; Imaizumi-Scherrer et al. 1996; Gramolini et al. 1997), and these results raise the possibility that these genes are also transcribed preferentially in synaptic nuclei. In support of this idea, a recent study reported that the utrophin gene is transcribed preferentially in synaptic nuclei (Gramolini et al. 1998). Thus, synapse-specific gene expression may be a general and important mechanism for clustering proteins at developing and adult neuromuscular synapses (Chu et al. 1995a; Duclert and Changeux 1995; Burden 1998).
Neuregulin (NRG) is currently the best candidate for the signal that activates synapse-specific transcription. NRG was purified initially on the basis of its activity as a growth factor that stimulates tyrosine phosphorylation of ErbB2 (Neu), a member of the epidermal growth factor (EGF) receptor family, and was termed NDF (Neu differentiation factor) or HRG (heregulin) (Holmes et al. 1992; Peles et al. 1992; Wen et al. 1992). Independent studies, which led to the purification and cloning of GGF (glial growth factor), a series of ligands that regulate Schwann cell survival and proliferation (Marchionni et al. 1993), and ARIA (acetylcholine receptor inducing activity), a factor that stimulates AChR synthesis in muscle cells (Falls et al. 1993), revealed that a single gene encodes NDF, GGF, and ARIA (Peles and Yarden 1993; Carraway and Burden 1995; Fischbach and Rosen 1997).
Four lines of evidence support the idea that NRG may be the signal that activates synapse-specific transcription. First, NRG activates AChR gene expression in cultured muscle cells, and the NRG response element is contained in the same cis-acting region that confers synapse-specific expression in mice (Gundersen et al. 1993; Tang et al. 1994; Chu et al. 1995b; Jo et al. 1995). Second, NRG is concentrated at synaptic sites (Chu et al. 1995b; Goodearl et al. 1995; Jo et al. 1995), and like the signal that activates synapse-specific gene expression, NRG is present in the synaptic basal lamina (Goodearl et al. 1995; Jo et al. 1995). Third, ErbB3 and ErbB4, two members of the EGF receptor family, are receptors for NRG, and both ErbB3 and ErbB4 are concentrated in the postsynaptic membrane at neuromuscular synapses (Altiok et al. 1995; Moscoso et al. 1995b; Zhu et al. 1995). Because mice lacking NRG, ErbB2, or ErbB4 die because of defects in cardiac development at embryonic day 10.5 (E10.5), ∼4 days prior to neuromuscular synapse formation (Gassmann et al. 1995; Lee et al. 1995; Meyer and Birchmeier 1995), it has been difficult to determine whether NRG-mediated signaling is required for synapse-specific gene expression. Nevertheless, adult mice that are heterozygous for the immunoglobulin allele of NRG (NRGIg+/−) have a mild deficiency in synaptic transmission and fewer (50%) AChRs at their neuromuscular synapses (Sandrock et al. 1997), providing further evidence that NRG may have the suspected role in synapse formation.
The sequences in AChR subunit genes that confer synapse-specific gene expression in transgenic mice and NRG responsiveness in cultured muscle cells are contained in <200 bp of 5′ flanking DNA (Gundersen et al. 1993; Tang et al. 1994; Chu et al. 1995b; Jo et al. 1995). A DNA injection assay, in which adult muscle fibers are transfected with gene fusions between an AChR regulatory region and a reporter gene, has been used to further delineate regulatory sequences for synapse-specific expression (Duclert et al. 1993, 1996; Koike et al. 1995). Although this assay allows for rapid analysis of regulatory sequences, there is an imperfect correspondence between results obtained with transgenic mice and with transfected adult muscle fibers, because AChR sequences that confer synapse-specific transcription in transgenic mice direct both synaptic and nonsynaptic expression in transfected muscle fibers (Duclert et al. 1993, 1995; Koike et al. 1995). Thus, although these AChR gene fusions are expressed preferentially in synaptic nuclei of transfected muscle fibers, transfected DNA appears to be subject to less stringent regulation than either endogenous AChR genes or transgenes containing regulatory elements from AChR genes. Nevertheless, DNA injection experiments indicate that a CGGAA sequence, which conforms to a consensus binding site (C/AGGAA/T) for Ets proteins (Koike et al. 1995; Duclert et al. 1996), is required for synaptic expression of the mouse AChR δ and ε subunit genes (Koike et al. 1995; Duclert et al. 1996). We used a cell-culture assay to identify NRG response elements (NRE) in the AChR δ subunit gene, and we produced transgenic mice carrying AChR genes with a mutated NRE to determine whether NREs are required for synapse-specific gene expression. We demonstrate that a CGGAA sequence in the AChR δ subunit gene is an NRE, and we show that this NRE is required for synapse-specific gene expression. Moreover, we show that GABPα, an Ets protein, and GABPβ, a protein that lacks an Ets domain but dimerizes with GABPα (Thompson et al. 1991), bind this NRE. These results, together with similar findings reported by Schaeffer and colleagues (Schaeffer et al. 1998), demonstrate that synapse-specific and NRG-induced gene expression require an Ets-binding site and suggest that GABPα/GABPβ bind the NRE and respond to NRG signaling by stimulating transcription of the AChR δ subunit gene.
Results
Identification of a NRG response element
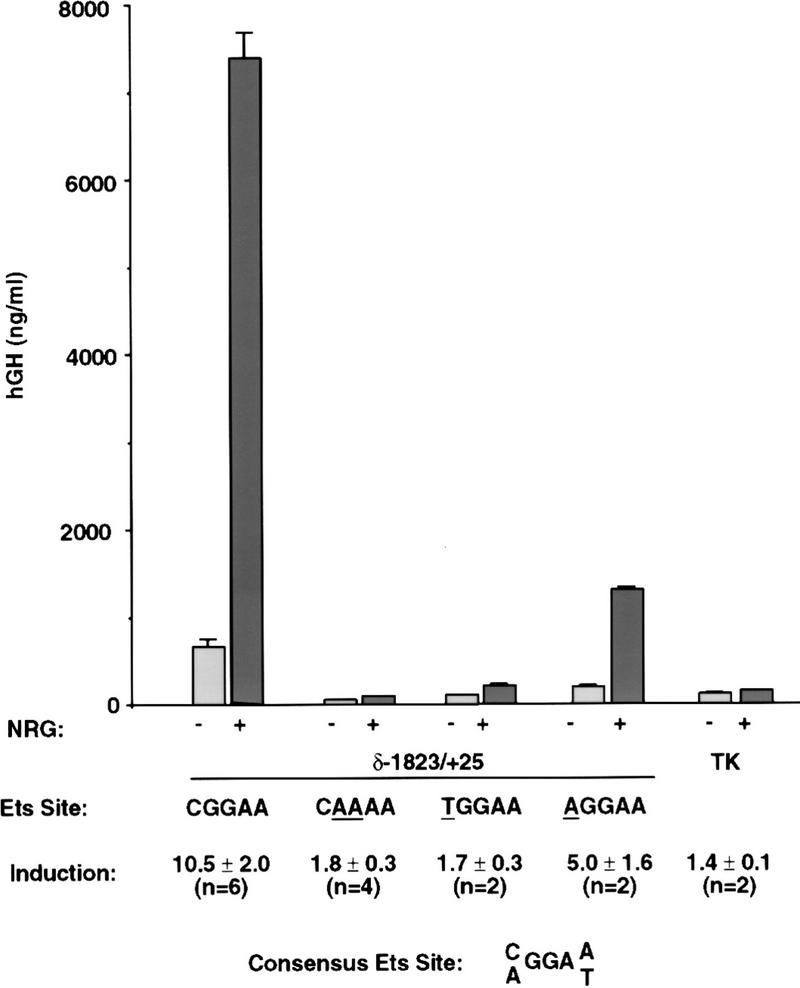
In previous studies we demonstrated that 181 bp of 5′ flanking DNA from the AChR δ subunit gene is sufficient to confer synapse-specific transcription in transgenic mice and NRG-induced transcription in cultured muscle cells (Simon et al. 1992; Tang et al. 1994; Jo et al. 1995). A subsequent study reported that a sequence in the AChR δ subunit gene (CGGAA; nucleotides −54 to −58), which conforms to a consensus Ets-binding site (C/AGGAA/T), is important for synapse-specific expression (Koike et al. 1995). To determine whether this potential binding site for Ets proteins is required for NRG-induced transcription, we transfected Sol8 muscle cells with wild-type or mutant AChR δ subunit–human growth hormone (hGH) gene fusions and measured the amount of hGH expression from untreated or NRG-stimulated myotubes. Figure 1 demonstrates that the potential binding site for Ets proteins is required for NRG-induced transcription. NRG induces an ∼10-fold increase in hGH expression from myotubes transfected with a wild-type AChR δ regulatory region (−1823/+25). In contrast, NRG stimulation causes only a modest (1.8-fold) increase in hGH expression from myotubes transfected with an AChR δ regulatory region containing mutations (CAAAA) in nucleotides that are critical for binding Ets proteins.
Figure 1.
A potential binding site for Ets proteins is required for induction of the AChR δ subunit gene by NRG. Sol8 myotubes were stably transfected with AChR δ subunit–hGH or thymidine kinase (TK)–hGH gene fusions and stimulated with NRG. The amount of hGH secreted into the culture media from NRG-stimulated and untreated myotubes is indicated. NRG induces an ∼10-fold increase in hGH expression from myotubes transfected with an AChR δ (−1823/+25)–hGH gene fusion. In contrast, NRG induces a weak (1.8-fold; 1.7-fold) increase in hGH expression from myotubes transfected with AChR δ subunit–hGH gene fusions containing mutations (underlined) in nucleotides that are critical for binding Ets proteins. A mutated sequence (AGGAA), which still conforms to a consensus binding site (C/AGGAA/T) for Ets proteins, retains a weakened but substantial (fivefold) response to NRG. The mean induction ±s.e.m. is given.
Ets proteins bind sequences containing a C or an A, but not a T (C/AGGAA/T) at the 5′ end of a core Ets-binding site (Wasylyk et al. 1993). To determine whether the sequence requirements for NRG-induced transcription conform to the sequence specificity for binding Ets proteins, we transfected myotubes with gene fusions containing a C, A, or T at the 5′ end of the core Ets-binding site. Figure 1 shows that mutation of the C to an A results in a reduced but substantial (fivefold) response to NRG, whereas mutation of the C to a T results in a weak NRG response (1.7-fold). Thus, these results are consistent with the idea that this NRE binds Ets proteins and that this binding is important for NRG-induced expression of the AChR δ subunit gene.
This NRE is also required for maximal expression from myotubes not treated with NRG, because mutation of the NRE reduces expression from untreated cells by about sevenfold (Fig. 1). Although expression from untreated cells may indeed be NRG-independent, muscle cells synthesize NRG (Moscoso et al. 1995b; Rimer et al. 1998), raising the possibility that AChR gene expression from untreated cells is nevertheless NRG-dependent and regulated by autocrine signaling. In either case, these results indicate that this NRE has a role in stimulating AChR transcription in noninnervated myotubes.
The NRE is required for synapse-specific transcription
In previous studies we analyzed synapse-specific transcription by producing transgenic mice carrying gene fusions between the mouse AChR δ subunit gene and the hGH gene (Simon et al. 1992; Tang et al. 1994). Because hGH is processed in the endoplasmic reticulum and Golgi apparatus and because these organelles are associated closely with nuclei, we were able to infer the nuclear source of hGH transcription by studying the spatial pattern of intracellular hGH using immunohistochemistry. We showed that 181 bp of 5′ flanking DNA from the AChR δ subunit gene is sufficient to confer synapse-specific expression (Tang et al. 1994). To determine whether the NRE is required for synapse-specific transcription, we produced transgenic mice carrying transgenes, containing either 1823 or 181 bp of 5′ flanking DNA from the AChR δ subunit gene, with a mutation in the NRE. Figure 2 shows that transgenes containing a mutation in the NRE, unlike wild-type transgenes, are not expressed selectively from synaptic nuclei (Materials and Methods), demonstrating that the NRE is required for synaptic expression.
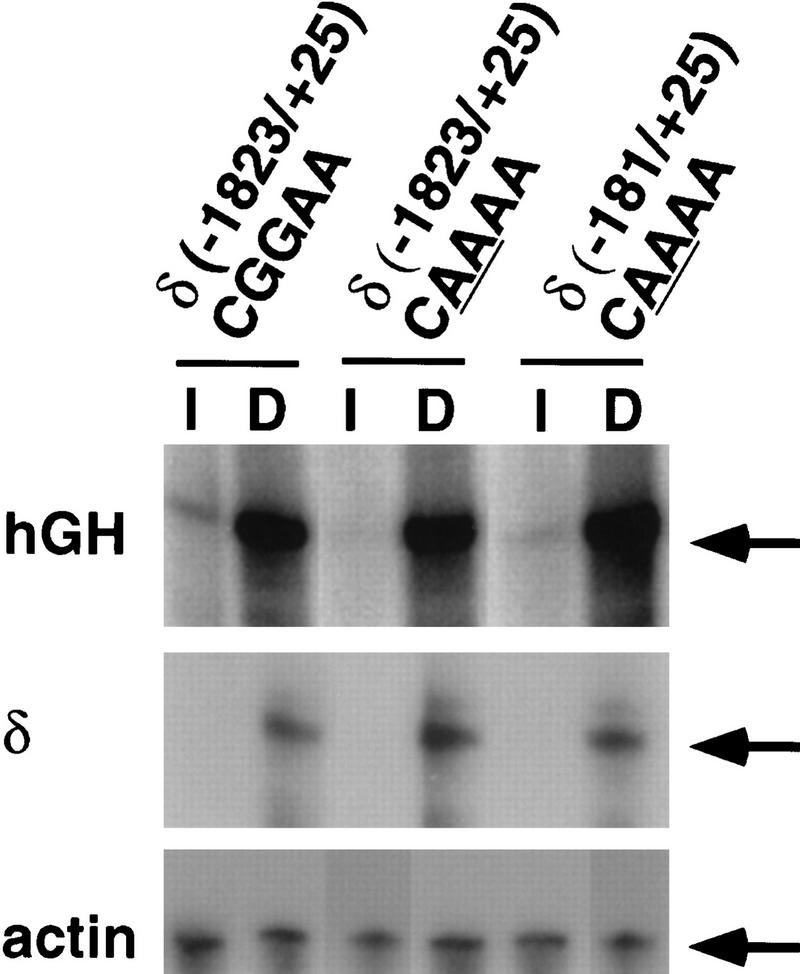
Figure 2.
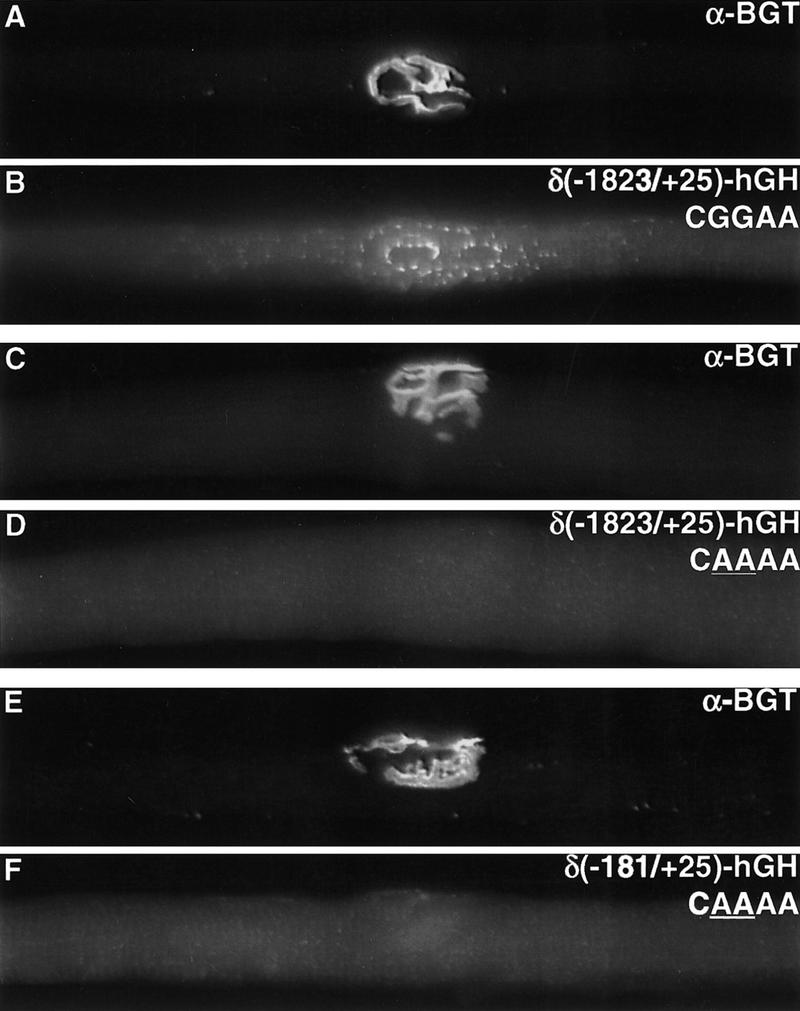
The NRE is required for synapse-specific transcription. hGH expression is restricted to synaptic sites in muscles from mice carrying an AChR δ subunit (−1823/+25)–hGH transgene. In contrast, hGH expression is not detected at synaptic sites in muscle from mice carrying AChR δ–hGH transgenes with a mutation (underlined) in the NRE. Synaptic sites (A,C,E) were identified by staining with Texas Red α-bungarotoxin, and hGH (B,D,F) was detected by indirect immunofluorescence.
An absence of synaptic expression in these transgenic mice could be caused by a specific role for this NRE in synapse-specific transcription. Alternatively, this NRE may be a basal element that is required in vivo for transcription per se. To determine whether transgenes containing a mutant NRE are transcriptionally competent or simply inactive, we asked whether the mutant transgenes could be activated following denervation. AChR gene expression in nonsynaptic nuclei increases substantially following denervation, causing an increase in the abundance of AChR mRNA (Hall and Sanes 1993). Previously, we showed that 181 bp of 5′ flanking DNA from the AChR δ subunit gene is sufficient to confer electrical-activity-dependent regulation, because hGH mRNA levels are ∼25-fold higher in denervated than in innervated muscle from mice carrying an AChR δ (−181/+25)–hGH transgene (Simon et al. 1992; Tang et al. 1994). We denervated muscle from mice carrying transgenes with a mutated NRE, and we measured the level of hGH mRNA in innervated and denervated muscle. Figure 3 shows that the mutant transgenes, like wild-type transgenes are induced following denervation and that the extent of induction following denervation is similar from wild-type and mutant transgenes. Because expression from wild-type and mutant transgenes is identical in denervated muscle, these results demonstrate that the transgenes containing a mutation in this NRE are transcriptionally competent and indicate that the NRE is required specifically for synapse-specific transcription. Furthermore, these results show that the NRE is not required for electrical activity-dependent gene expression.
Figure 3.
The NRE is not required for electrical activity-dependent regulation of the AChR δ subunit gene. The level of AChR δ subunit mRNA is low in innervated muscle and increases ∼25-fold following denervation. This increase in AChR mRNA expression is largely caused by transcriptional mechanisms, because hGH mRNA expression increases ∼25-fold following denervation of muscle from mice carrying a wild-type AChR δ subunit–hGH transgene (Simon et al. 1992). Likewise, hGH expression increases ∼25-fold following denervation of muscle from mice carrying AChR δ subunit–hGH transgenes with a mutation (underlined) in the NRE. The positions of the protected RNA probes are indicated by arrows.
Protein binding to the NRE
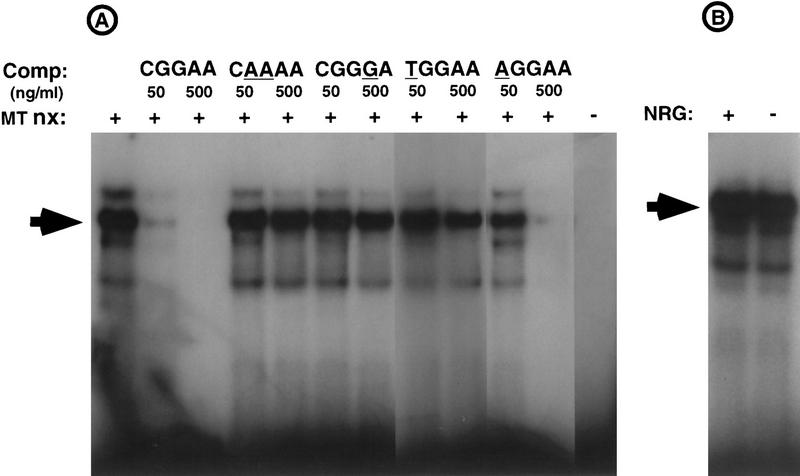
We used an electrophoretic mobility shift assay (EMSA) to identify proteins that might interact with the NRE. Figure 4 demonstrates that protein(s) in myotube nuclear extracts bind an oligonucleotide probe containing the NRE; binding is specific, because an excess of unlabeled, competitor DNA containing a wild-type NRE competes efficiently for binding to the probe. In contrast, competitor DNA containing mutations in nucleotides that are important for NRG responsiveness fail to compete for binding. Because a mutant NRE, in which the C is mutated to an A, retains a weakened, but substantial NRG response, we asked whether this mutant NRE might likewise retain reduced but detectable protein binding. Figure 4 shows that this mutant (AGGAA) NRE indeed has an attenuated but detectable affinity for the DNA-binding protein(s), as a 100-fold but not a 10-fold excess of mutant competitor DNA inhibits binding to the probe. Thus, there is a good correlation between nucleotides required for NRG responsiveness and protein binding.
Figure 4.
Myotube nuclear extracts contain a protein(s) that binds specifically to the NRE. (A) EMSA was performed with nuclear extracts prepared from Sol8 myotubes (MT nx) treated with NRG two days. The radiolabeled oligonucleotide probe and unlabeled competitor DNA extend from nucleotides −62 to −47 of the AChR δ subunit gene. The arrow indicates the position of the major protein–DNA complex. Formation of the complex is inhibited by an excess (∼10- or 100-fold) of wild-type competitor DNA. In contrast, unlabeled DNA containing mutations in nucleotides (underlined) that are critical for NRG responsiveness fail to compete for binding. A mutant NRE (AGGAA) that confers a reduced but substantial response to NRG has a reduced but detectable affinity for the DNA-binding protein(s), because a 100-fold but not a 10-fold excess of the mutant competitor inhibits formation of the complex. (B) An indistinguishable protein–DNA complex (arrow) is detected in EMSAs with nuclear extracts from NRG-stimulated or untreated myotubes.
To determine whether NRG stimulates binding of proteins to the NRE we isolated nuclear extracts from NRG-stimulated and untreated myotubes and used an EMSA to measure protein binding. Figure 4 shows that extracts from untreated myotubes contain a protein(s) that binds to the NRE and that NRG stimulation does not appear to alter the capacity of the protein(s) to bind to the NRE. These data favor the idea that NRG stimulation increases the transcriptional activity rather than the DNA-binding activity of the protein(s) that bind to the NRE in the AChR δ subunit gene.
GABPα/GABPβ bind the NRE
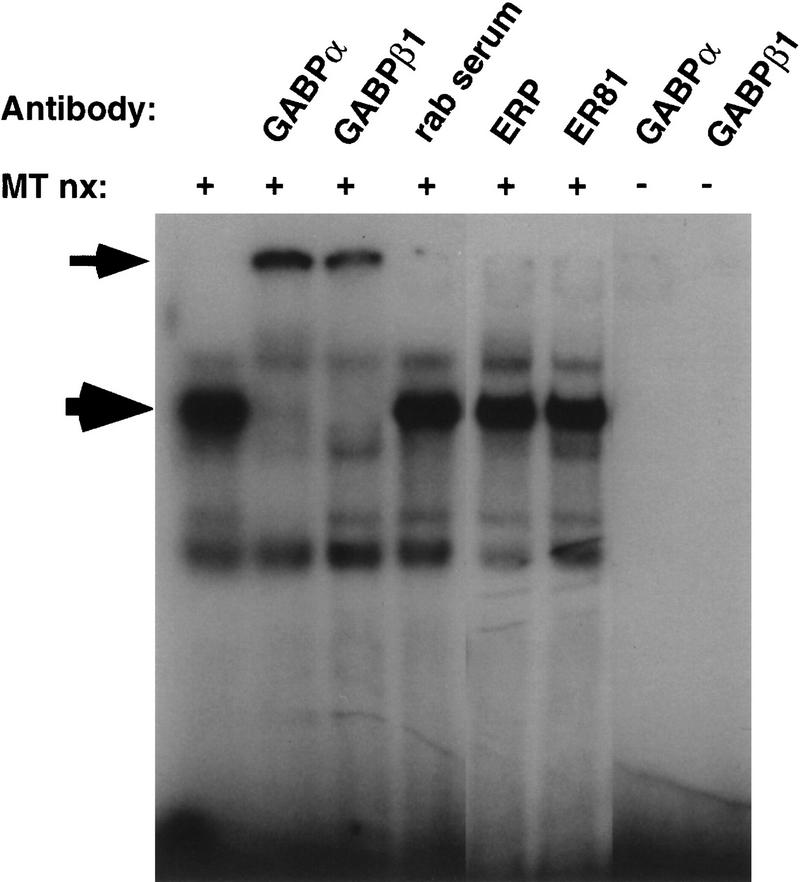
Because the sequence of the NRE conforms to a consensus Ets-binding site, we asked whether known Ets proteins are present in the protein–DNA complex detected in the EMSA. We incubated nuclear extracts from NRG-stimulated myotubes with antibodies to Ets proteins and determined whether these antibodies inhibit and/or super-shift the protein–DNA complex. Antibodies that are specific for GABPα (Brown and McKnight 1992) largely, if not entirely inhibit the formation of the protein–DNA complex and super-shift the complex (Fig. 5). In contrast, antibodies that are selective for other Ets proteins and that can inhibit their ability to bind DNA (Leiden et al. 1992; Monté et al. 1995; Akbarali et al. 1996; Watson et al. 1997; see Materials and Methods), fail to alter the protein–DNA complex.
Figure 5.
GABP binds to the NRE. EMSAs were performed with nuclear extracts from NRG-stimulated Sol8 myotubes (MT nx) and a radiolabeled oligonucleotide containing the NRE. Binding reactions were performed either in the absence of antibodies or after preincubation of nuclear extracts with the indicated antisera or control serum. Antisera directed against either the α or β subunits of GABP inhibit the formation of the major protein–DNA complex (large arrow) and cause the appearance of a super-shifted complex (small arrow). In contrast, antibodies to other Ets proteins, such as ERP and ER81, which are shown here, as well as Elf-1, Erg-1, Ets-1, Ets-2, Fli-1, PEA3, NERF, do not alter the protein–DNA complex.
Importantly, antibodies against GABPβ1, a protein that dimerizes selectively with GABPα but lacks an Ets domain (Thompson et al. 1991; Brown and McKnight 1992), likewise inhibits the formation of the protein–DNA complex (Fig. 5). Because GABPβ1 interacts with GABPα and not with other Ets proteins (Brown and McKnight 1992; Batchelor et al. 1998) and because these antibodies to GABPβ1 are highly selective for GABPβ1 and cross-react only very poorly even with GABPβ2, a GABPβ1-related protein that also interacts selectively with GABPα (de la Brousse et al. 1994), inhibition of the protein–DNA complex by the antibodies to GABPβ1 provides independent evidence that GABPα is the major, if not the only Ets protein in the protein–DNA complex.
Discussion
Our results demonstrate that a binding site for Ets proteins in the AChR δ subunit gene is required for transcriptional induction of the AChR gene by NRG. We show that this NRE is required for synapse-specific gene expression, and these data provide further evidence for the idea that NRG is a signal for synapse-specific transcription. We show that GABPα, an Ets protein, and GABPβ, a protein that lacks an Ets domain but dimerizes with GABPα, bind this NRE, and these results suggest that GABPα/GABPβ respond to NRG signaling by stimulating transcription of the AChR δ subunit gene.
GABP was identified as a multisubunit transcription complex that binds to a cis-acting regulatory element required for activation of herpes simplex virus immediate-early genes by the virion protein VP16 (Triezenberg et al. 1988; LaMarco and McKnight 1989). The structure of the multisubunit complex demonstrates that the Notch-like repeats in GABPβ contact an inhibitory domain in GABPα and suggests that this positioning of the inhibitory domain allows the Ets domain in GABPα to bind DNA (Batchelor et al. 1998). Both GABP subunits are expressed widely in different tissues (LaMarco et al. 1991), but little is known about the cellular genes that are regulated by GABP.
GABP may be a component of the NRG signaling pathway in skeletal muscle that activates transcription of AChR genes. NRG stimulation results in activation of a Ras/Raf/MAP kinase signaling cascade (Ben-Levy et al. 1994; Marte et al. 1995), and both Ras and MAP kinase kinase are required for induction of AChR genes by NRG (Si et al. 1996; Tansey et al. 1996; Altiok et al. 1997). Ets proteins, including GABPα, are known targets of Ras/MAP kinase (MAPK) signaling (Marais et al. 1993; Brunner et al. 1994; Flory et al. 1996; Ouyang et al. 1996; O’Hagan and Hassell 1998), and in vitro studies have shown that Ets proteins, including GABPα, can be phosphorylated directly by MAPK (Flory et al. 1996). Since GABPβ, which contains the transactivation domain in GABP (Gugneja et al. 1995), is likewise phosphorylated by MAPK (Flory et al. 1996), phosphorylation of GABPα and/or GABPβ may be required for transcriptional activation of GABP. Taken together, these studies raise the possibility that Ras signaling leads to transcriptional activation of GABP and induction of AChR genes. The mechanisms that lead to activation of Ets-mediated transcription have been best studied for Elk-1, and these studies indicate that phosphorylation of Elk-1 stimulates the transcriptional activity of Elk-1 without affecting its capacity to bind DNA (Marais et al. 1993). Our studies suggest that GABP is activated without altering its capacity to bind DNA, and these results suggest that phosphorylation of GABP may be an important step in stimulating its transcriptional activity.
Our results indicate that a complex of GABPα/GABPβ is present in myotube nuclear extracts and binds to the NRE in the AChR δ subunit promoter. Because the major protein–DNA complex is absent following treatment with antibodies to either GABPα or GABPβ, GABPα appears to be the major, if not the only Ets protein in this complex. Because Elf-1, Elf-2, ERP, NERF, ER81, Ets-2, and GABP are known to be expressed in skeletal muscle (LaMarco et al. 1991; Lopez et al. 1994; Monté et al. 1995; Oettgen et al. 1996; Wilkinson et al. 1997; Sapru et al. 1998), it is interesting that GABPα/GABPβ appears to be the major, if not only Ets protein detected by EMSAs with the NRE. In principle, the predominance of GABP in this complex could be caused by a greater abundance of GABPα and GABPβ than other Ets proteins in skeletal muscle. Alternatively, GABPα/GABPβ may bind the NRE with greater affinity than other Ets proteins. Although all known Ets proteins require the same core sequence for DNA binding (C/AGGAA/T) (Wasylyk et al. 1993), there is evidence that the sequence of nucleotides that flank this core sequence can influence the binding affinity for certain Ets proteins. For example, nucleotides that are 3′ to a core Ets binding site have a role in determining whether the site prefers Ets-1 or Elf-1 (Wang et al. 1992). Because the sequence of the NRE conforms perfectly to a 9-bp sequence for GABPα identified in site-selection experiments (Brown and McKnight 1992), the NRE appears to be a particularly favorable binding site for GABP. Comprehensive studies of the binding sites preferred by other Ets proteins expressed in skeletal muscle may reveal whether GABP has the highest affinity for the NRE.
GABP recognizes DNA as a heterotetramer that binds to two directly repeated Ets sites (Thompson et al. 1991), resulting in an increased avidity of GABP for its target site. The 181-bp regulatory region of the AChR δ subunit gene, however, contains only a single Ets site; nevertheless, an increased avidity and specificity could be achieved by interactions between a GABP heterodimer and another transcription factor that binds to an adjacent site in the δ subunit regulatory region.
This is the first study to demonstrate a role for an Ets site in synapse-specific expression using transgenic mice. Changeux and colleagues, using a DNA-injection assay, reported that a potential Ets-binding site, which they termed an N-box, is required for synaptic expression of AChR genes (Koike et al. 1995; Duclert et al. 1996). Because DNA injected into adult myofibers is taken up by a subset of myofiber nuclei in a small number of muscle fibers, the expression pattern of the injected DNA is unintentionally but inevitably restricted to a subset of nuclei. Consequently, interpretation of the expression pattern of the injected DNA requires extensive quantitation and statistical analysis. We believe that transgenic mice provide important advantages for studying synapse-specific gene expression. First, because the transgene is integrated in all myofiber nuclei, selective expression from synaptic nuclei cannot be caused by DNA integration in a subset of nuclei. Second, because wild-type AChR transgenes are expressed in all synaptic nuclei in all myofibers (Sanes et al. 1991; Simon et al. 1992), the significance of mutations that lead to a loss of synaptic expression is readily apparent and requires little quantitation. Third, electrical activity-dependent gene expression can also be studied readily in transgenic mice (Simon et al. 1992; Tang et al. 1994), and we were able to show that transgenes with a mutated Ets-binding site, which are not expressed at synaptic sites, are nevertheless expressed at wild-type levels following denervation. Such control experiments, which would be difficult using a DNA-injection assay, demonstrate that the Ets-site in the AChR δ subunit gene is required selectively for synapse-specific expression and not for gene expression per se.
Our results show that the NRE in the AChR δ subunit gene is a positive regulatory element for NRG-stimulated transcription and for synapse-specific transcription. DNA injection experiments suggest that these Ets-binding sites also function as negative regulatory elements to repress AChR expression in nonsynaptic nuclei, because mutation of an Ets-binding site in the mouse AChR δ subunit gene or rat AChR ε subunit gene results in increased expression in nonsynaptic nuclei (Koike et al. 1995; Sapru et al. 1998). In our experiments with transgenic mice, mutation of the Ets-binding site results in a loss of synaptic expression and no increase in nonsynaptic expression. Consistent with these results in transgenic mice, our cell-culture studies show that mutation of the Ets-binding site results in reduced expression from NRG-treated and untreated myotubes. Thus, our data support the idea that this cis-acting sequence has a selective role in activating AChR gene expression.
An Ets-binding site in the rat AChR ε subunit gene is likewise important for NRG responsiveness (Sapru et al. 1998). Although this study did not attempt to identify the proteins that bind this site, overexpression of Ets-2 increases ε expression (Sapru et al. 1998), raising the possibility that Ets-2 binds this NRE and regulates ε subunit gene expression. It remains unclear, however, whether endogenous Ets-2 binds this site or whether Ets-2 binding and activation occur only after forced overexpression of Ets-2. Sequences in addition to an Ets-binding site may regulate NRG-responsiveness and synapse-specific expression. One study reported that the Ets-site in the ε subunit gene is not required for NRG responsiveness and that a different sequence, termed an ARE, is necessary for a twofold induction by NRG (Si et al. 1997). Other studies, however, have reported that the ARE is not required for NRG responsiveness or synapse-specific expression, using a DNA-injection assay (Duclert et al. 1996; Sapru et al. 1998). The potential role for the ARE in synapse-specific expression may be studied best in transgenic mice.
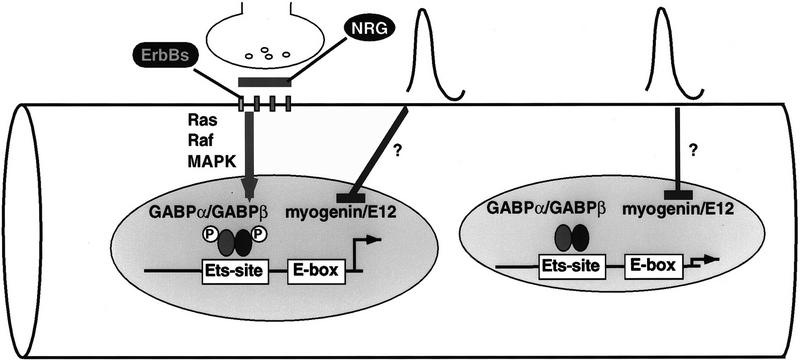
Previous studies demonstrated that a binding site (E-box) for myogenic basic helix–loop–helix (bHLH) proteins is important for electrical activity-dependent but not synapse-specific transcription of AChR genes (Tang et al. 1994; Bessereau et al. 1994). Here, we show that an Ets-binding site is required for synapse-specific but not electrical activity-dependent transcription. Taken together, these studies provide further support for the idea that two separate pathways regulate innervation-dependent transcription: An electrical activity-dependent pathway that is thought to lead to a decrease in the activity and/or expression of myogenic bHLH proteins and a synapse-specific pathway that we propose leads to the activation of GABPα/GABPβ (Fig. 6). Because our experiments that implicate GABP in synapse-specific expression have relied upon in vitro binding studies, the proposed role for GABP in synapse-specific expression clearly needs to be determined by interfering with GABP function in vivo.
Figure 6.
Separate pathways regulate synapse-specific and electrical activity-dependent transcription. NRG, which is present in the synaptic basal lamina, activates ErbB receptors in the postsynaptic membrane and leads to activation of a Ras signaling pathway. Our experiments sugest that this pathway leads to activation of GABPα/GABPβ and an increase in AChR gene expression selectively in synaptic nuclei. The pathway activated by electrical activity is thought to result in a decrease in the activity and/or abundance of transcriptional activators that bind E-boxes, such as myogenin and E12, leading to a decrease in AChR gene expression in nuclei throughout the muscle fiber.
Utrophin, which is related structurally to dystrophin, is concentrated in the postsynaptic membrane at neuromuscular synapses, and the utrophin gene is expressed selectively in synaptic nuclei of adult muscle fibers (Gramolini et al. 1997). Moreover, synapse-specific expression of the utrophin gene requires an Ets-binding site (Gramolini et al. 1998), and these results raise the possibility that utrophin gene expression is stimulated by NRG activation of GABPα/GABPβ. Duchenne muscular dystrophy, a severe muscle-wasting disease, is caused by a lack of dystrophin, a large intracellular protein associated normally with synaptic and nonsynaptic muscle membranes. Because muscle-wasting is far milder in mice lacking dystrophin and because dystrophin mutant mice express utrophin at high levels at the nonsynaptic membrane, others have proposed that muscle wasting in Duchenne muscular dystrophy may be ameliorated by increasing utrophin expression (Deconinck et al. 1997; Grady et al. 1997). The mechanisms that are responsible for increasing utrophin expression in dystrophin mutant mouse muscle are not known but could require NRG and GABPα/GABPβ signaling. Thus, reagents that stimulate NRG or GABPα/GABPβ signaling may be therapeutic for Duchenne muscular dystrophy.
Materials and methods
Sol 8 myoblasts were transfected, selected, and induced to differentiate into myotubes as described previously (Simon and Burden 1993; Jo et al. 1995). Stably transfected cells were treated with NRG for 48 hr, and the amount of hGH secreted from treated and untreated cells was measured by a radioimmunoassay as described previously (Jo et al. 1995).
Nuclear extracts from Sol8 myotubes were prepared and incubated with a radiolabeled oligonucleotide probe (nucleotides −62 to −47) as described previously (Simon and Burden 1993). The reactions were allowed to proceed for 10 min at room temperature, placed on ice for 5 min, and complexes were resolved by electrophoresis (1.5 hr at 200 V) in a 5% polyacrylamide gel (0.5× Tris-borate-EDTA). The specificity of the binding reaction was determined by the addition of unlabeled competitor DNA (nucleotides −62 to −47) to the reaction mixture. Gels were fixed, dried, and exposed to X-ray film.
Antibodies to Ets proteins were either supplied by colleagues who have characterized these antibodies in previous studies (anti-ERP, Akbarali et al. 1996; anti-ER81, Monté et al. 1995; anti-PEA3, Baert et al. 1997; anti-Elf-1, Leiden et al. 1992; anti-Fli-1, Watson et al. 1997; anti-GABPα and anti-GABPβ, Brown and McKnight 1992; de la Brousse et al. 1994), or were purchased from Santa Cruz Biotechnology (anti-Ets-1; anti-Ets-2; anti-PEA3; anti-Erg-1; anti-Fli-1). In addition, we found that the antibodies to Ets-1, NERF, ERP, and Fli-1 inhibit the gel shift detected between the NRE and the appropriate recombinant Ets protein expressed either in reticulocyte lysates (NERF, ERP, Fli-1) or as recombinant protein in Escherichia coli (Ets-1) (data not presented). Antibodies (0.1–0.5 μl of serum or 0.6–4 μg of purified antibodies) were incubated with nuclear extracts for 2 hr on ice prior to the addition of the labeled probe (Simon and Burden 1993).
Founder mice carrying AChR δ subunit–hGH transgenes with a mutation in the NRE were produced using methods similar to those described previously (Simon et al. 1992; Tang et al. 1994). We studied two lines carrying AChR δ subunit–hGH transgenes with a mutation (CAAAA) in the NRE in detail; one line carries an AChR δ (−1823/mutant NRE/+25)–hGH transgene and the other line carries an AChR δ (−181/mutantNRE/+25)–hGH transgene. These two lines were chosen for study, because hGH expression in denervated muscle from these lines is similar to that from lines carrying wild-type AChR transgenes (see above); thus, a lack of synaptic expression in these lines could not be caused by a low level of transgene expression per se. Two other AChR δ (−1823/mutant NRE/+25)–hGH lines expressed hGH and responded to denervation, but the absolute level of hGH expression in these lines was lower than the wild-type lines and the two NRE-mutant lines studied here in detail. We excluded these lines from further study because an absence of synaptic expression in these mice could be caused by a low level of transgene expression. We stained diaphragm muscles for hGH expression as described previously (Simon et al. 1992; Tang et al. 1994). The wild-type transgene was expressed at 100% of synaptic sites (>300 synaptic sites in three muscles from three mice); in contrast, synaptic expression was not detected in muscles from mice carrying either the −1823/+25 mutant transgene (>300 synaptic sites in two muscles from two mice) or the −181/+25 mutant transgene (>300 synaptic sites in three muscles from three mice). The levels of hGH, AChR δ subunit, and actin mRNA expression in innervated and five-day denervated leg muscles were measured by RNase protection and quantitated with a PhosphorImager (Simon et al. 1992; Tang et al. 1994).
Acknowledgments
We thank Towia Libermann (anti-ERP), Yvan de Launoit (anti-PEA3 and anti-ER81), Jeff Leiden (anti-Elf-1), Dennis Watson (anti-Fli-1), Sylvia Arbor and Tom Jessell (anti-PEA3), and Fabienne de la Brousse (anti-GABP) for providing us with antibodies to Ets proteins. We thank Elizabeth Prescott and Erika Kinetz for excellent technical assistance and Ruth Herbst, Xia Yang, Moses Chao, Ed Skolnik, and Ruth Lehmann for comments on the manuscript. This work was supported by research grants from the National Institutes of Health (NIH) (NS27963) and the Muscular Dystrophy Association and by a postdoctoral fellowship from the NIH to L.F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL burden@saturn.med.nyu.edu; FAX (212) 263-8214.
References
- Akbarali Y, Oettgen P, Boltax J, Libermann TA. ELF-1 interacts with and transactivates the IgH enhancer π site. J Biol Chem. 1996;271:26007–26012. doi: 10.1074/jbc.271.42.26007. [DOI] [PubMed] [Google Scholar]
- Altiok N, Bessereau JL, Changeux JP. ErbB3 and ErbB2/neu mediate the effect of heregulin on acetylcholine receptor gene expression in muscle: Differential expression at the endplate. EMBO J. 1995;14:4258–4566. doi: 10.1002/j.1460-2075.1995.tb00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altiok N, Altiok X, Changeux JP. Heregulin-stimulated acetylcholine receptor gene expression in muscle: Requirement for MAP kinase and evidence for a parallel inhibitory pathway independent of electrical activity. EMBO J. 1997;16:717–725. doi: 10.1093/emboj/16.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baert JL, Monté D, Musgrove EA, Albagli O, Sutherland RL, de Launoit Y. Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int J Cancer. 1997;70:590–597. doi: 10.1002/(sici)1097-0215(19970304)70:5<590::aid-ijc17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPα/β: An Ets domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Ben-Levy R, Paterson HF, Marshall CJ, Yarden Y. A single autophosphorylation site confers oncogenicity to the neu/ErbB-2 receptor and enables coupling to the MAP kinase pathway. EMBO J. 1994;13:3302–3311. doi: 10.1002/j.1460-2075.1994.tb06632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessereau JL, Stratfored-Perricaudet LD, Piette J, LePoupon C, Changeux JP. In vivo and in vitro analysis of electrical activity-dependent expression of muscle acetylcholine receptor genes using adenovirus. Proc Natl Acad Sci. 1994;91:1304–1308. doi: 10.1073/pnas.91.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, McKnight SL. Specificities of protein–protein and protein–DNA interaction of GABPα and two newly defined ets-related proteins. Genes & Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinasein the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Burden SJ. The formation of neuromuscular synapses. Genes & Dev. 1998;12:133–148. doi: 10.1101/gad.12.2.133. [DOI] [PubMed] [Google Scholar]
- Carraway KL, III, Burden SJ. Neuregulins and their receptors. Curr Opin Neurobiol. 1995;5:606–612. doi: 10.1016/0959-4388(95)80065-4. [DOI] [PubMed] [Google Scholar]
- Chu GC, Moscoso LM, Sliwkowski MX, Merlie JP. Regulation of the acetylcholine receptor e subunit gene by recombinant ARIA: An in vitro model for transynaptic gene regulation. Neuron. 1995b;14:329–339. doi: 10.1016/0896-6273(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Chu GC, Velleca MA, Merlie JP. Synapse-specific gene expression. Sem Dev Biol. 1995a;6:175–183. [Google Scholar]
- DeChiara TM, Bowen DC, Valenzuela DM, Simmons MV, Poueymirou WT, Thomas S, Kinetz E, Compton DL, Park JS, Smith C, DiStefano PS, Glass DJ, Burden SJ, Yancopoulos GD. The receptor tyrosine kinase, MuSK, is required for neuromuscular junction formation in vivo. Cell. 1996;85:501–512. doi: 10.1016/s0092-8674(00)81251-9. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JJA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson G, Tinsley JM, Davies KE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- de la Brousse FC, Birkenmeier EH, King DS, Rowe LB, McKnight SL. Molecular and genetic characterization of GABPβ. Genes & Dev. 1994;8:1853–1865. doi: 10.1101/gad.8.15.1853. [DOI] [PubMed] [Google Scholar]
- Duclert A, Changeux JP. Acetylcholine receptor gene expression at the developing neuromuscular junction. Physiol Rev. 1995;75:339–368. doi: 10.1152/physrev.1995.75.2.339. [DOI] [PubMed] [Google Scholar]
- Duclert A, Savatier N, Changeux JP. An 83-nucleotide promoter of the acetylcholine receptor ε-subunit gene confers preferential synaptic expression in mouse muscle. Proc Natl Acad Sci. 1993;90:3043–3047. doi: 10.1073/pnas.90.7.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclert A, Savatier N, Schaeffer L, Changeux JP. Identification of an element crucial for the sub-synaptic expression of the acetylcholine receptor epsilon-subunit gene. J Biol Chem. 1996;271:17433–17438. doi: 10.1074/jbc.271.29.17433. [DOI] [PubMed] [Google Scholar]
- Falls DL, Rosen KM, Corfas G, Lane WS, Fischbach GD. ARIA, a protein that stimulates acetylcholine receptor synthesis, is a member of the neu ligand family. Cell. 1993;72:801–815. doi: 10.1016/0092-8674(93)90407-h. [DOI] [PubMed] [Google Scholar]
- Fischbach GD, Rosen KM. A neuromuscular junction neuregulin. Annu Rev Neurosci. 1997;20:429–458. doi: 10.1146/annurev.neuro.20.1.429. [DOI] [PubMed] [Google Scholar]
- Flory E, Hoffmeyer A, Smola U, Rapp UR, Bruder JT. Raf-1 kinse targets GA-binding protein in transcriptional regulation of the human immunodeficiency virus type 1 promoter. J Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine B, Changeux JP. Localization of nicotinic acetylcholine receptor α-subunit transcripts during myogenesis and motor endplate development in the chick. J Cell Biol. 1989;108:1025–1037. doi: 10.1083/jcb.108.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- Gautam M, Noakes PG, Moscoso L, Rupp F, Scheller RH, Merlie JP, Sanes JR. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell. 1996;85:525–535. doi: 10.1016/s0092-8674(00)81253-2. [DOI] [PubMed] [Google Scholar]
- Goldman D, Staple J. Spatial and temporal expression of acetylcholine receptor mRNAs in innervated and denervated rat soleus muscle. Neuron. 1989;3:219–228. doi: 10.1016/0896-6273(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Goodearl AD, Yee AG, Sandrock AW, Jr, Corfas G, Fischbach GD. ARIA is concentrated in the synaptic basal lamina of the developing chick neuromuscular junction. J Cell Biol. 1995;130:1423–1434. doi: 10.1083/jcb.130.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: A model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- Gramolini AO, Dennis CL, Tinsley JM, Robertson GS, Cartaud J, Davies KE, Jasmin BJ. Local transcriptional control of utrophin expression at the neuromuscular synapse. J Biol Chem. 1997;272:8117–8120. doi: 10.1074/jbc.272.13.8117. [DOI] [PubMed] [Google Scholar]
- Gramolini AO, Burton EA, Tinsley JM, Ferns MJ, Cartaud A, Cartaud J, Davies KE, Lunde JA, Jasmin BJ. Muscle and neural isoforms of agrin increase utrophin expression in cultured myotubes via a transcriptional regulatory mechanism. J Biol Chem. 1998;273:736–743. doi: 10.1074/jbc.273.2.736. [DOI] [PubMed] [Google Scholar]
- Gugneja S, Virbasius JV, Scarpulla RC. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol Cell Biol. 1995;15:102–111. doi: 10.1128/mcb.15.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Sanes JR, Merlie JP. Neural regulation of muscle acetylcholine receptor epsilon- and alpha-subunit gene promoters in transgenic mice. J Cell Biol. 1993;123:1535–1544. doi: 10.1083/jcb.123.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall ZW, Sanes JR. Synaptic structure and development: The neuromuscular junction. Cell/Neuron (Suppl.) 1993;72/10:99–121. doi: 10.1016/s0092-8674(05)80031-5. [DOI] [PubMed] [Google Scholar]
- Holmes WE, Sliwkowski MX, Akita RW, Henzel WJ, Lee J, Park JW, Yansura D, Abadi N, Raab H, Lewis GD, et al. Identification of heregulin, a specific activator of p185erbB2. Science. 1992;256:1205–1210. doi: 10.1126/science.256.5060.1205. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Scherrer T, Faust DM, Benichou JC, Hellio R, Weiss MC. Accumulation in fetal muscle and localization to the neuromuscular junction of cAMP dependent protein kinase A regulatory and catalytic subunits RI alpha and C alpha. J Cell Biol. 1996;134:1241–1254. doi: 10.1083/jcb.134.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin BJ, Lee RK, Rotundo RL. Compartmentalization of acetylcholinesterase mRNA and enzyme at the vertebrate neuromuscular junction. Neuron. 1993;11:467–477. doi: 10.1016/0896-6273(93)90151-g. [DOI] [PubMed] [Google Scholar]
- Jo SA, Zhu X, Marchionni MA, Burden SJ. NRGs are concentrated at nerve-muscle synapses and activate Ach-receptor gene expression. Nature. 1995;373:158–161. doi: 10.1038/373158a0. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Bessereau JL, Salmon AM, Triller A, Babinet C, Changeux JP. An acetylcholine receptor α-subunit promoter conferring preferential synaptic expression in muscle of transgenic mice. EMBO J. 1991;10:625–632. doi: 10.1002/j.1460-2075.1991.tb07990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Schaeffer L, Changeux JP. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor delta-subunit gene. Proc Natl Acad Sci. 1995;92:10624–10628. doi: 10.1073/pnas.92.23.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarco KL, McKnight SL. Purification of a set of cellular factors that bind to the purine-rich cis-regulatory element of herpes simplex virus early genes. Genes & Dev. 1989;3:1372–1383. doi: 10.1101/gad.3.9.1372. [DOI] [PubMed] [Google Scholar]
- LaMarco K, Thompson CC, Byers BP, Walton EM, McKnight SL. Identification of Ets- and Notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- Leiden JM, Wang C-Y, Petryniak B, Markovitz DM, Nabel GJ, Thompson CB. A novel Ets-related transcription factor, Elf-1, binds to human immunodeficiency virus type 2 regulatory elements that are required for inducible trans activation in T cells. J Virol. 1992;66:5890–5897. doi: 10.1128/jvi.66.10.5890-5897.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M, Oettgen P, Akbarali Y, Dendorfer U, Libermann TA. ERP, a new member of the Ets transcription factor/oncoprotein family: Cloning, characterization, and differential expression during B-lymphocyte development. Mol Cell Biol. 1994;14:3292–3309. doi: 10.1128/mcb.14.5.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- Marchionni MA, Goodearl AD, Chen MS, Bermingham-McDonogh O, Kirk C, Hendricks M, Danehy F, Misumi D, Sudhalter J, Kobayashi K, et al. Glial growth factors are alternatively spliced erbB2 ligands expressed in the nervous system. Nature. 1993;362:312–318. doi: 10.1038/362312a0. [DOI] [PubMed] [Google Scholar]
- Marte BM, Graus-Porta D, Jeschke M, Doriano F, Nynes NE, Taverna D. NDF/heregulin activates MAP kinase and p70/p85 S6 kinase during proliferation or differentiation of mammary epithelial cells. Oncogene. 1995;10:167–175. [PubMed] [Google Scholar]
- McMahan UJ. The agrin hypothesis. Cold Spring Harb Symp Quant Biol. 1990;55:407–418. doi: 10.1101/sqb.1990.055.01.041. [DOI] [PubMed] [Google Scholar]
- Merlie JP, Sanes JR. Concentration of acetylcholine receptor mRNA in synaptic regions of adult muscle fibers. Nature. 1985;317:66–68. doi: 10.1038/317066a0. [DOI] [PubMed] [Google Scholar]
- Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- Monté D, Coutte L, Baert J-L, Angeli I, Stéhelin D, de Launoit Y. Molecular characterization of the ets-related human transcription factor ER81. Oncogene. 1995;11:771–779. [PubMed] [Google Scholar]
- Moscoso LM, Chu GC, Gautam M, Noakes PG, Merlie JP, Sanes JR. Synapse-associated expression of an acetylcholine receptor-inducing protein, ARIA/heregulin, and its putative receptors, ErbB2 and ErbB3, in developing mammalian muscle. Dev Biol. 1995b;172:158–169. doi: 10.1006/dbio.1995.0012. [DOI] [PubMed] [Google Scholar]
- Moscoso LM, Merlie JP, Sanes JR. N-CAM, 43k-rapsyn, and s-laminin mRNAs are concentrated at synaptic sites in muscle fibers. Mol Cell Neurosci. 1995a;6:80–89. doi: 10.1006/mcne.1995.1008. [DOI] [PubMed] [Google Scholar]
- Oettgen P, Akbarali Y, Boltax J, Best J, Kunsch C, Libermann TA. Characterization of NERF, a novel transcription factor related to the Ets factor ELF-1. Mol Cell Biol. 1996;16:5091–5106. doi: 10.1128/mcb.16.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan RC, Hassell JA. The PEA3 Ets transcription factor is a downstream target of the HER2/Neu receptor tyrosine kinase. Oncogene. 1998;16:301–310. doi: 10.1038/sj.onc.1201547. [DOI] [PubMed] [Google Scholar]
- Ouyang L, Jacob KK, Stanley FM. GABP mediates insulin-increased prolactin gene transcription. J Biol Chem. 1996;271:10425–10428. doi: 10.1074/jbc.271.18.10425. [DOI] [PubMed] [Google Scholar]
- Peles E, Yarden Y. Neu and its ligands: From an oncogene to neural factors. BioEssays. 1993;15:815–824. doi: 10.1002/bies.950151207. [DOI] [PubMed] [Google Scholar]
- Peles E, Bacus SS, Koski RA, Lu HS, Wen D, Ogden SC, Ben-Levy R, Yarden Y. Isolation of the Neu/HER-2 stimulatory ligand: A 44 kd glycoprotein that induces differentiation of mammary tumor cells. Cell. 1992;69:205–216. doi: 10.1016/0092-8674(92)90131-u. [DOI] [PubMed] [Google Scholar]
- Rimer M, Cohen I, Lømo T, Burden SJ, McMahan UJ. Neuregulins and erbB receptors at neuromuscular junctions and at agrin-induced postsynaptic-like apparatus in skeletal muscle. Mol Cell Neurosci. 1998;12:1–15. doi: 10.1006/mcne.1998.0695. [DOI] [PubMed] [Google Scholar]
- Sandrock AW, Dryer SE, Rosen KM, Gozani SN, Kramer R, Theil LE, Fischbach GD. Maintenance of acetylcholine receptor number by neuregulins at the neuromuscular junction in vivo. Science. 1997;276:599–603. doi: 10.1126/science.276.5312.599. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Johnson YR, Kotzbauer PT, Mudd J, Hanley T, Martinou JC, Merlie JP. Selective expression of an acetylcholine receptor-lacZ transgene in synaptic nuclei of adult muscle fibers. Development. 1991;113:1181–1191. doi: 10.1242/dev.113.4.1181. [DOI] [PubMed] [Google Scholar]
- Sapru MK, Florance SK, Kirk C, Goldman D. Identification of a neuregulin and protein-tyrosine phosphatase response element in the nicotinic acetylcholine receptor ε subunit gene: Regulatory role of an Ets transcription factor. Proc Natl Acad Sci. 1998;95:1289–1294. doi: 10.1073/pnas.95.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S, Duclert N, Huchet-Dymanus M, Changeux J-P. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 1998;17:3078–3090. doi: 10.1093/emboj/17.11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si J, Luo Z, Mei L. Induction of acetylcholine receptor gene expression by ARIA requires activation of mitogen-activated protein kinase. J Biol Chem. 1996;271:19752–19759. doi: 10.1074/jbc.271.33.19752. [DOI] [PubMed] [Google Scholar]
- Si J, Miller DS, Mei L. Identification of an element required for acetylcholine receptor-inducing activity (ARIA)-induced expression of the acetylcholine receptor epsilon subunit gene. J Biol Chem. 1997;272:10367–10371. doi: 10.1074/jbc.272.16.10367. [DOI] [PubMed] [Google Scholar]
- Simon AM, Burden SJ. An E box mediates activation and repression of the acetylcholine receptor δ-subunit gene during myogenesis. Mol Cell Biol. 1993;13:5133–5150. doi: 10.1128/mcb.13.9.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AM, Hoppe P, Burden SJ. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 1992;114:545–553. doi: 10.1242/dev.114.3.545. [DOI] [PubMed] [Google Scholar]
- Tang J, Jo SA, Burden SJ. Separate pathways for synapse-specific and electrical activity-dependent gene expression in skeletal muscle. Development. 1994;120:1799–1804. doi: 10.1242/dev.120.7.1799. [DOI] [PubMed] [Google Scholar]
- Tansey MG, Chu GC, Merlie JP. ARIA/HRG regulates AChR ε subunit gene expression at the neuromuscular synapse via activation of phosphatidylinositol 3-kinase and Ras/MAPK pathway. J Cell Biol. 1996;134:465–476. doi: 10.1083/jcb.134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Brown TA, McKnight SL. Convergence of Ets- and Notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ, LaMarco KL, McKnight SL. Evidence of DNA: Protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes & Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Stitt TN, DiStefano PS, Rojas E, Mattsson K, Compton DL, Lunez L, Park JS, Stark JL, Gies DR, Thomas S, Copeland NG, Jenkins NA, Burden SJ, Glass DJ, Yancopoulos GD. Identification of a receptor tyrosine kinase specific for the skeletal muscle lineage: Expression in embryonic muscle, at the neuromuscular junction and after injury. Neuron. 1995;15:573–584. doi: 10.1016/0896-6273(95)90146-9. [DOI] [PubMed] [Google Scholar]
- Wang CY, Petryniak B, Ho IC, Thompson CB, Leiden JM. Evolutionary conserved Ets family members display distinct DNA binding specificites. J Exp Med. 1992;175:1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B, Hahn SL, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Watson DK, Robinson L, Hodge DR, Kola I, Papas TS, Seth A. FLI1 and EWS-FLI1 function as ternary complex factors and ELK1 and SAP1a function as ternary and quaternary complex factors on the Egr1 promoter serum response elements. Oncogene. 1997;14:213–221. doi: 10.1038/sj.onc.1200839. [DOI] [PubMed] [Google Scholar]
- Wen D, Peles E, Cupples R, Suggs SV, Bacus SS, Luo Y, Trail G, Hu S, Silbiger SM, Levy RB, et al. Neu differentiation factor: A transmembrane glycoprotein containing an EGF domain and an immunoglobulin homology unit. Cell. 1992;69:559–572. doi: 10.1016/0092-8674(92)90456-m. [DOI] [PubMed] [Google Scholar]
- Wilkinson DA, Neale G, Mao S, Naeve CW, Goorha RM. Elf-2, a rhombotin-2 binding Ets transcription factor: Discovery and potential role in T cell leukemia. Leukemia. 1997;11:86–96. doi: 10.1038/sj.leu.2400516. [DOI] [PubMed] [Google Scholar]
- Zhu X, Lai C, Thomas S, Burden SJ. Neuregulin receptors, erbB3 and erbB4, are localized at neuromuscular synapses. EMBO J. 1995;14:5842–5848. doi: 10.1002/j.1460-2075.1995.tb00272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]