Abstract
Infection of cells by enveloped viruses requires merger of the viral envelope membrane with target cell membranes, resulting in the formation of fusion pores through which the viral genome is released. Since lipid membranes do not mix spontaneously, the fusion process is energy-dependent and mediated by viral envelope glycoprotein complexes. Based on their structural and mechanistic properties, three distinct classes of viral fusion proteins have been identified to date. Despite their diversity, basic principles of viral membrane fusion, simultaneous engagement of both donor and target membrane and refolding into hairpin-like structures, have emerged as universally conserved. This article provides an overview of the basic principles of viral membrane fusion common to all enveloped viruses and discusses the specific structural and functional features of the different fusion protein classes by example of the paramyxovirus, flavivirus and rhabdovirus families.
Introduction
As obligatory intracellular parasites, viruses must transfer their genetic information across cellular membranes to initiate an infectious cycle that ultimately results in the production of progeny virions. For membrane-wrapped enveloped viruses, this requires merger of the viral envelope with target cell membranes and formation of a fusion pore through which the genome of the particle is released. Maintaining membrane integrity is imperative for cellular life, however, and high repulsive energy between approaching membranes prevent spontaneous fusion of distinct lipid vesicles.
Basic principles of viral membrane fusion
Similar to the well-documented vesicular fusion machinery in eukaryotic cells [1], membrane fusion for viral entry is mediated by membrane protein complexes. These envelope glycoproteins are typically observed as characteristic protein “spikes” on the surface of purified virions in electron microscopic images. Significant progress in the structural analysis of spike proteins derived from representatives of a variety of enveloped virus families has resulted in the identification of, thus far, three distinct classes of viral fusogenic membrane glycoprotein (FMG) based on their structural organization and mechanism of fusion (table 1). Independent of this differentiation and the nature of the particular cellular membrane (intracellular versus plasma membrane) that is targeted for viral entry, however, two basic mechanistic principles of membrane fusion have emerged as conserved among enveloped viruses, target membrane engagement and refolding into hairpin-like structures ([2] illustrated in figure 1).
Table 1.
Currently known distinct classes of viral fusiogenic membrane glycoproteins.
| Fusion Protein |
Characteristic Features | Examples |
|---|---|---|
| Class I |
|
Influenza HA [72,73,74] HIV Env [75,76,77,78,79,80] Paramyxovirus F [32,33,34,81] Ebola virus GP [82] |
| Class II |
|
Flavivirus E [48,49,51] Alphavirus E1 [55,56,57,58] |
| Class III |
|
Rhabdovirus G [62,63] Herpes virus gB [59,60] Baculovirus gp64 [61] |
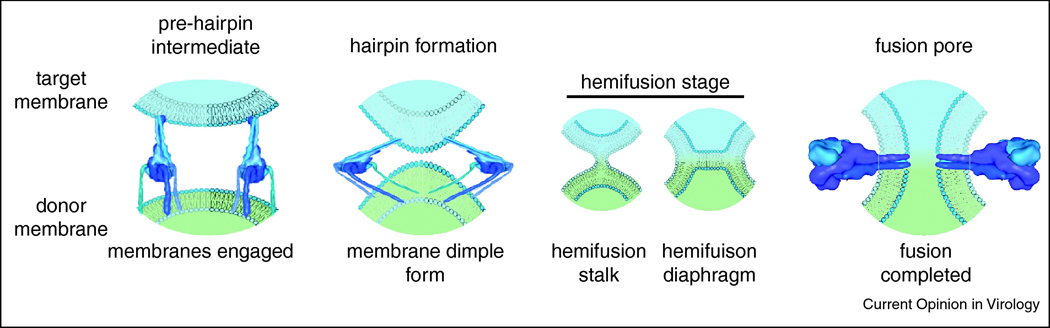
Figure 1.
Schematic of viral membrane merger, exemplified by the paramyxovirus F protein. Upon fusion triggering, conformational changes of the native F glycoprotein result in an extended conformation, in which both viral envelope and target membrane are engaged (left panel). Through refolding of several F trimers, a fusion funnel with high local lipid curvature is thought to form. Merger of the outer monolayers of the approaching membranes creates a lipid stalk, which, according to the common stalk-diaphragm model, expands to hemifusion and formation of a hemifusion diaphragm (center panel). Completion of the F refolding into a thermodynamically stable hairpin structure is considered to advance merger to fusion pore formation and subsequent pore expansion (right panel).
Firstly, upon particle attachment to target cells, conformational rearrangements of the fusogenic spike protein complex take place that result in insertion of largely hydrophobic membrane attack domains, termed the fusion peptides or fusion loops, into the target membrane. The resulting pre-hairpin intermediate conformation thus engages both the viral envelope and the host cell membrane.
Secondly, refolding of the trimeric fusogenic complex onto itself into, ultimately, a trimeric hairpin structure is considered to then bring the attack and transmembrane domains, and thus the donor and target membranes, into close proximity. Involving the concomitant engagement of several complexes [3,4,5,6,7,8,9], a current consensus model (however, see [10,11] for alternative hypotheses) proposes progressing of the membranes towards each other through formation of a lipid funnel [12], which, ultimately, sets the stage for merger of the outer leaflets of the approaching monolayers in regions of negative spontaneous curvature [13,14]. This hemifusion step is followed by fusion of the inner leaflets, forming a pore. As alternative pathway to this stalk-diaphragm model, direct transition from stalk to fusion pore was proposed, based on Monte Carlo and coarse-grain ensemble molecular dynamics algorithms [15,16,17].
Once a fusion pore has formed, subsequent pore enlargement appears associated with the most significant energy penalty of the process, resulting in the possibility that newly opened pores regress to the hemifusion stage rather than proceed to expansion for fusion completion and viral infection [7,8,18].
Energy dependence of the process
As depicted schematically in figure 1, membrane fusion requires the introduction of a high degree of local curvature into the lipid bilayer, rendering it an energy-dependent process overall [19]. In the absence of external energy sources, refolding of the FMGs into their thermodynamically stable postfusion conformations is considered to supply the energy that is necessary to overcome the elastic resistance of the lipid bilayers and generate the highly bent intermediate formations. Estimates of the number of individual viral fusion complexes required for productive fusion pore formation vary from a single trimer to over a dozen complexes [6,20]. While the hypothesis that a single HIV Env protein trimer is capable of inducing membrane fusion has been challenged recently [21,22], currently available data do not support a direct calculation of the free energy associated with the refolding of viral fusion complexes. Stoichiometric estimates are therefore based on indirect, functional assays [3,6,23], but typically assume cooperation of several complexes for productive fusion. Definitive estimates are further challenged by the possibility that the stoichiometric requirements for a given viral fusion protein may not be strictly defined, resulting in pore formation and enlargement mediated by different stoichiometric complex assemblies with variable degrees of efficiency. Examining fusion events on a single molecule level as exemplified by the analysis of cellular SNARE proteins [15] will likely be required to address these questions.
Class I fusion proteins
Of all fusogenic viral envelope glycoproteins, representatives of class I proteins were the first to be described and crystallized in both the pre- and postfusion conformations. This class comprises, amongst others, influenza HA, paramyxovirus F, HIV Env and Ebola virus GP proteins. All class I fusion proteins form homo-trimers. They are first synthesized as fusion-inactive precursor molecules that require proteolytic activation to liberate the N-terminus of the fusion peptide and gain fusion competence. Often, cleavage is mediated by the ubiquitous subtilisin-like cellular furin endoproteases in the host secretory system that are most active at acidic pH in the Golgi and/or trans-Golgi network compartments [24], ensuring that proteolytically matured complexes reach the cell surface.
Not surprisingly, the efficiency of fusion protein cleavage can directly affect viral pathogenesis. In the case of influenza virus, for instance, the HA0 precursor proteins of highly pathogenic H5 and H7 viruses are recognized and cleaved by furin and furin-like proteases, enabling systemic infections in the avian host [25]. In contrast, HA proteins of non-avian influenza A and avirulent avian strains are cleaved mostly by trypsin-like proteases such as, amongst others, tryptase Clara, plasmin or miniplasmin [26,27]. Infection by these strains typically remains localized to the respiratory and intestinal tract, lacking a systemic component. While most paramyxovirus fusion (F) proteins are cleaved by furin in transit to the plasma membrane, F proteins of paramyxoviruses of the henipavirus genus are internalized after initial transport to the cell surface for cathepsin L-mediated maturation, followed by re-routing to the plasma membrane [28,29].
Depending on the viral family, refolding of matured class I proteins can be initiated through a) acidic pH (i.e. influenza virus HA, b) direct receptor binding, possibly in conjunction with acidic pH (i.e. paramyxoviruses of the Pneumovirinae subfamily [30]), or c) receptor binding by a non-covalently associated attachment envelope glycoprotein that then stimulates refolding of the fusion protein at neutral pH (i.e. most paramyxoviruses of the Paramyxovirinae subfamily). For the latter, receptor binding appears to trigger separation of preformed hetero-oligomers of some Paramyxovirinae such as measles virus and the henipaviruses (fusion triggering through dissociation), whereas receptor binding-induced glycoprotein association was postulated for Newcastle disease virus and the parainfluenzaviruses (fusion triggering through association) [31]. Independent of the mechanism of fusion triggering, characteristic for all class I fusion proteins are two 4-3 heptad repeat (HR) domains, positioned adjacent to the fusion peptide and the transmembrane domain, respectively. Figures 2A and 3A illustrate major structural features of the class I fusion process by example of the paramyxovirus fusion (F) protein.
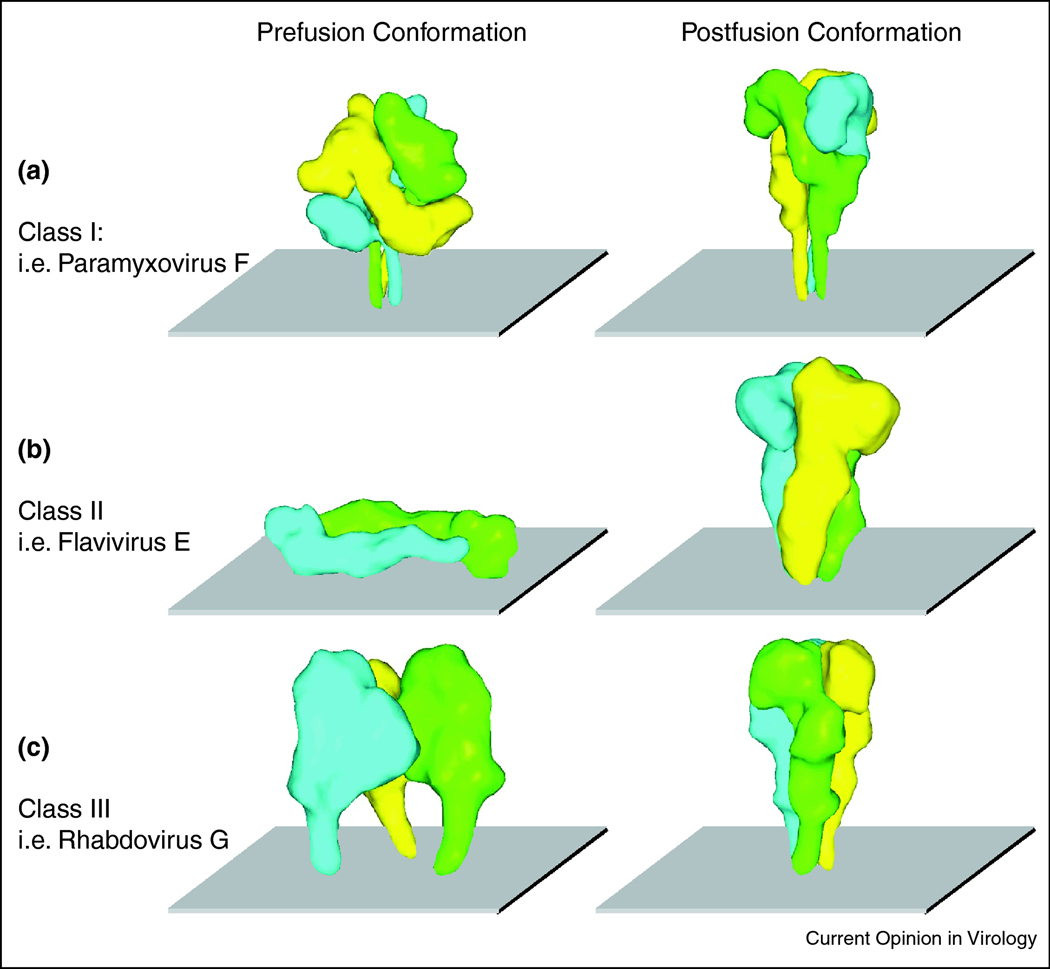
Figure 2.
Renderings of class I (paramyxovirus F (A)), class II (flavivirus E (B)) and class III (rhabdovirus G (C)) viral fusion proteins in the pre- and postfusion conformations. For clarity, high-resolution structural models were colored by monomer and then morphed into lower resolution images using the Sculptor (resolution 12, voxel size 3) software package [83]. Structures are based on PDB coordinates 2B9B and 1ZTM ([32,33] (A)), 1OAN and 1OK8 ([48,49] (B)), and 2J6J and 2CMZ ([62,63] (C)), respectively.
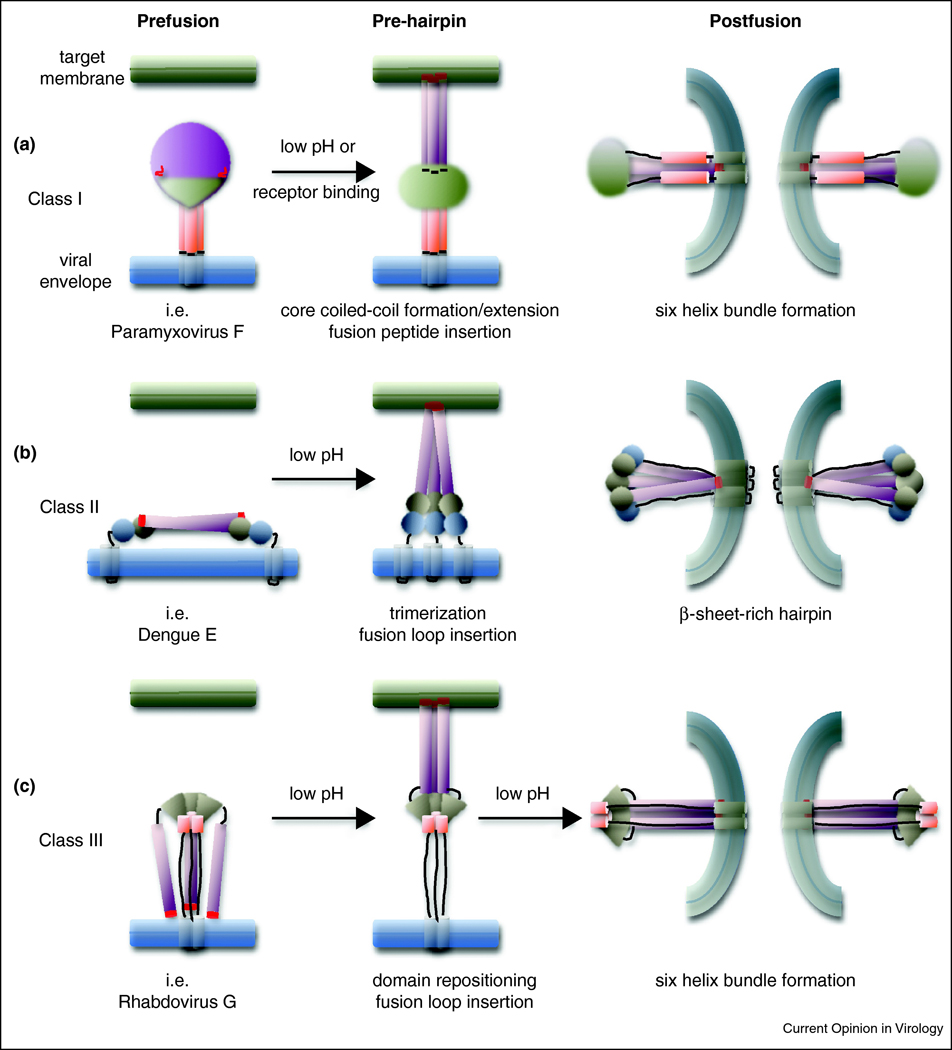
Figure 3.
Schematic representation of the general domain organization and conformational reorganization of the three viral fusion protein classes. Fusion peptides/loops are highlighted in red. A) Paramyxovirus F: fusion peptide-proximal HR domains forming the central trimeric coiled coil (purple) and TMD-proximal HR domains (orange) are highlighted. B) Flavivirus E: the extended domains II harboring the fusion loops are highlighted (purple). C) Rhabdovirus G: the fusion domains (DIV, purple) and the trimerization domains (DII) refolding into the postfusion 6HB (orange) are highlighted.
In the prefusion conformation, an F protein ectodomain was crystallized in a lollipop-like structure consisting of a globular head domain that is linked to the transmembrane domains (TMDs) and short luminal tails through the TMD-proximal HR sequences [32]. In this metastable prefusion conformation, the fusion peptide-proximal HR domains are broken up into 11 distinct segments forming part of the head structure. Upon fusion activation, major structural rearrangements take place that first result in assembly of head-domain HR segments into a central, trimeric α-helical coiled coil structure, propelling the fusion peptides towards the target membrane [33,34]. Subsequent hairpin-like refolding then positions the individual TMD-proximal HR domains in the grooves of the central triple helix, ultimately resulting in the formation of a thermodynamically extraordinarily stable six-helix bundle (6HB) structure [35,36,37,38]. The final arrangement of a fusion peptide-proximal core coiled-coil structure surrounded by the three TMD-proximal HR helices is a defining feature of class I fusion proteins.
6HB formation appears closely linked to the opening of a fusion pore [39,40] and is considered to provide, possibly augmented by neighboring sequences as in the case of influenza HA [41], a major driving force for pore formation and expansion. Consequently, peptides [36,42] or small molecules [43,44] that prevent final 6HB formation by interfering with refolding of the fusion protein function as potent viral entry inhibitors. Clearly, refolding of the fusion protein trimer as shown schematically in figure 3A cannot proceed symmetrically, as this would wrap the protein into the lipid of the viral envelope. Instead of an umbrella-like folding of the protein “through itself”, an asymmetric process must be assumed in which individual monomers of the trimer progress through the conformational rearrangements at different rates and individual TMD-proximal HR domains possibly engage the grooves in the central triple helix coiled-coil consecutively rather than in a concerted action.
Close confinement of the TMDs and the fusion peptides in the same lipid space after 6HB formation makes conceivable short-range proteinaceous contacts between these membrane-inserted domains in addition to protein-lipid interactions. While still understudied, direct peptidic relations between these domains have been proposed [45,46,47] and may well serve as important driving forces for stabilization and expansion of nascent fusion pores, thus moving beyond fusion flickering through pore closure and reversion to the hemifusion stage. Association studies of isolated TMDs and fusion peptides in artificial lipid environments complemented with mutational analysis of these domains in the context of functional fusion proteins should further test these possibilities.
Class II fusion proteins
Of all fusion protein classes identified thus far, only class II proteins such as flavivirus E show a dimeric stoichiometry in the prefusion conformation. First major structural insight came from the crystallization of Dengue virus E proteins under both neutral (prefusion) and low pH conditions [48,49]. Flavivirus E mediates both receptor binding and membrane fusion, but depends on association with chaperone proteins to stabilize the prefusion fold. Presumably in the endoplasmic reticulum, newly synthesized E forms hetero-dimers with the viral prM glycoprotein. Rearrangement of E into head-to-tail homo-dimers under the low pH conditions of the trans-Golgi network then renders prM accessible for proteolytic cleavage by cellular furin or furin-like endoproteases, and E homo-dimers are present on the surface of mature particles at neutral pH [50,51,52]. Unique to this class, these primed prefusion class II proteins assume an elongated, plate-like shape running parallel to the viral envelope (figure 2B). The E protein ectodomain has a high β-sheet content and is organized into three sections, forming a β-barrel proximal to the TMDs (domain I), generating the rod-like extensions (domain II), and, with an immunoglobulin-like fold, mediating receptor binding (domain III). The internal fusion loops are positioned at the tip of each domain II rod, distal to the TMDs and buried within the dimer interface [53]. Upon triggering through acidification of the endosome, the complex transiently dissociates and domains adjacent to TMDs and fusion loops are rotated into a position perpendicular to the viral envelope and target membrane. Reorganization into homo-trimers in this pre-hairpin intermediate sets the stage for refolding of the complex onto itself through a second rotation step, which again brings TMDs and fusion loops into close proximity (figure 3B and [49,54]. The final E protein structure remains rich in β-sheets in contrast to the high α-helical content that is characteristic for the class I proteins.
Alphavirus E1 proteins present a variation of the theme outlined above. Initially, the E1 protein, resembling the domain architecture of flavivirus E [55,56], is associated with the viral E2 protein, which mediates receptor binding. These hetero-dimer pairs form an organized lattice of 80 spikes on the virion surface [57,58]. Upon low pH triggering, the E1/E2 hetero-dimers dissociate, E1 reorganizes into homo-trimers, and refolds into the class II postfusion hairpin structure.
Class III fusion proteins
The group of class III fusion proteins currently comprises rhabdovirus G protein, herpesvirus gB and baculovirus gp64. In addition to postfusion conformations of gB [59,60] and gp64 [61], our structural understanding of this class was greatly advanced by crystallization of the vesicular stomatitis virus (VSV) G protein trimer in both the pre- and postfusion conformations (figure 2C and [62,63]). Similar to class II proteins, also G mediates both receptor binding and, triggered by the low pH conditions of the endosome, membrane merger. Major conformational stages, a native (neutral pH) prefusion conformation, an extended conformation that engages both viral envelope and the target membrane, and a hairpin-like postfusion conformation likewise resemble key stages of class I and II fusion protein refolding (figure 3C). In striking contrast to the other fusion protein classes, however, G refolding is reversible, adhering to a pH-driven equilibrium. Consequently, the native prefusion conformation is not metastable [64], enabling the trimer to re-assume its native configuration subsequent to transport through the low-pH environment of the Golgi apparatus [65].
Native G trimers revealed a tripod-like arrangement consisting of four defined sections, a lateral domain (DI) with high β-sheet content, a trimerization domain (DII), a PH domain (DIII) located towards the top of the tripod and, forming the tripod legs, a fusion domain (DIV). The fusion loops are positioned at the tip of each leg, pointing towards the viral envelope [62]. While domains I, III and IV maintain their fold during G transition from pre- to postfusion conformation, the trimerization domain becomes subject to major refolding. Protonation of a cluster of histidine residues located at the intersection between domains at low pH was proposed to have a major effect on destabilizing the native conformation, priming the initiation of refolding [62]. Upon triggering, the tripod legs are proposed to swing upwards, driving the fusion loops towards the target membrane. Repositioning of the domains with conserved tertiary structure relative to each other through secondary structure changes in hinge regions and major reorganization of the trimerization domain then ultimately result in a classic hairpin postfusion conformation, in which fusion loops and TMDs are positioned in proximity in the same lipid space [63].
Despite the absence of heptad repeat domains in VSV G, two helices of each postfusion domain II engage in formation of an antiparallel 6HB in the postfusion trimer in strong reminiscence of the class I protein fusion core structure. A set of acidic amino acids, brought into close proximity in the central 6HB in the postfusion conformation, appear to serve as a second major molecular switch to initiate transition back to the native state upon deprotonation at elevated pH [63].
It was proposed that approximately 15 VSV G trimers are required to mediate membrane fusion [6]. Based on the observation that the prefusion structure of the G trimer supports organization into a hexagonal lattice, the local organization of the G protein spikes on the viral surface may set the stage for concerted conformational changes of multiple G spikes [62], thus optimizing the efficiency of the fusion process.
Conclusions
The basic principles of envelope glycoprotein-mediated membrane fusion, engagement of viral and target membranes followed by hairpin formation that juxtaposes TMDs and fusion peptides/loops in the same lipid environment, have emerged as well conserved in all three classes of enveloped virus fusion proteins identified to date. While X-ray structures of fusion protein ectodomains in pre- and postfusion conformations have resulted in a breakthrough in our general understanding of viral infection, much less is known about the molecular details of lipid merger, the spatial arrangement on the surface of infectious particles, and the nature of short-lived intermediate conformations of the refolding fusion proteins. Addressing these questions will likely require interfacing novel imaging approaches such as cryo-electron tomography reconstructions of emerging fusion pores [12] and refolding complexes arrested in intermediate conformations through addition of peptidic or small-molecule fusion inhibitors [36,43,66,67] with single molecule analyses in analogy to the observation of reconstituted cellular SNAREs [15] and new functional assays [68,69,70,71]. Ultimately, a detailed, high-resolution structural and mechanistic understanding of the viral fusion process has high potential to aid the development of novel virus entry inhibitors through structure-based drug design.
Acknowledgements
I thank A. L. Hammond and G. B. Melikyan for comments on the manuscript, and M. A. Brindley and A. Prussia for computational help with figure preparation. The work of the author was supported, in part, by U.S. Public Health Service grants AI071002 and AI083402 from the NIH/NIAID and a seed grant from the Children’s Healthcare of Atlanta Vaccines & Immunology Center. The funders had no role in the design of this review article, data analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danieli T, Pelletier SL, Henis YI, White JM. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbons DL, Erk I, Reilly B, Navaza J, Kielian M, et al. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell. 2003;114:573–583. doi: 10.1016/s0092-8674(03)00683-4. [DOI] [PubMed] [Google Scholar]
- 5.Markovic I, Leikina E, Zhukovsky M, Zimmerberg J, Chernomordik LV. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J Cell Biol. 2001;155:833–844. doi: 10.1083/jcb.200103005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roche S, Gaudin Y. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 2002;297:128–135. doi: 10.1006/viro.2002.1429. [DOI] [PubMed] [Google Scholar]
- 7.Markosyan RM, Cohen FS, Melikyan GB. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol Biol Cell. 2000;11:1143–1152. doi: 10.1091/mbc.11.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chernomordik LV, Frolov VA, Leikina E, Bronk P, Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heldwein EE, Krummenacher C. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci. 2008;65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bentz J. Membrane fusion mediated by coiled coils: a hypothesis. Biophys J. 2000;78:886–900. doi: 10.1016/S0006-3495(00)76646-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozlov MM, Chernomordik LV. A mechanism of protein-mediated fusion: coupling between refolding of the influenza hemagglutinin and lipid rearrangements. Biophys J. 1998;75:1384–1396. doi: 10.1016/S0006-3495(98)74056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee KK. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–1311. doi: 10.1038/emboj.2010.13. **Important study to better elucidate the mechanism of membrane fusion through merger of X-ray information with native structural insight derived from cryo-electron tomography. Reconstructions show early fusion intermediates, progressing from local lipid nipples to the formation of a liposomal funnel structure, while the viral matrix protein layer preserves the virion shape.
- 13.Kozlovsky Y, Chernomordik LV, Kozlov MM. Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys J. 2002;83:2634–2651. doi: 10.1016/S0006-3495(02)75274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlovsky Y, Kozlov MM. Stalk model of membrane fusion: solution of energy crisis. Biophys J. 2002;82:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brunger AT, Weninger K, Bowen M, Chu S. Single-molecule studies of the neuronal SNARE fusion machinery. Annu Rev Biochem. 2009;78:903–928. doi: 10.1146/annurev.biochem.77.070306.103621. *Excellent discussion of recent advances towards single-molecule approaches to study membrane fusion by reconstituted SNARE complexes.
- 16.Muller M, Katsov K, Schick M. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys J. 2003;85:1611–1623. doi: 10.1016/S0006-3495(03)74592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasson PM, Kelley NW, Singhal N, Vrljic M, Brunger AT, et al. Ensemble molecular dynamics yields submillisecond kinetics and intermediates of membrane fusion. Proc Natl Acad Sci U S A. 2006;103:11916–11921. doi: 10.1073/pnas.0601597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borrego-Diaz E, Peeples ME, Markosyan RM, Melikyan GB, Cohen FS. Completion of trimeric hairpin formation of influenza virus hemagglutinin promotes fusion pore opening and enlargement. Virology. 2003;316:234–244. doi: 10.1016/j.virol.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 19.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Kurteva S, Ren X, Lee S, Sodroski J. Stoichiometry of envelope glycoprotein trimers in the entry of human immunodeficiency virus type 1. J Virol. 2005;79:12132–12147. doi: 10.1128/JVI.79.19.12132-12147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369:245–262. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magnus C, Rusert P, Bonhoeffer S, Trkola A, Regoes RR. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83:1523–1531. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunther-Ausborn S, Schoen P, Bartoldus I, Wilschut J, Stegmann T. Role of hemagglutinin surface density in the initial stages of influenza virus fusion: lack of evidence for cooperativity. J Virol. 2000;74:2714–2720. doi: 10.1128/jvi.74.6.2714-2720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenk HD, Garten W, Rott R. Inhibition of proteolytic cleavage of the hemagglutinin of influenza virus by the calcium-specific ionophore A23187. EMBO J. 1984;3:2911–2915. doi: 10.1002/j.1460-2075.1984.tb02231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kido H, Murakami M, Oba K, Chen Y, Towatari T. Cellular proteinases trigger the infectivity of the influenza A and Sendai viruses. Mol Cells. 1999;9:235–244. [PubMed] [Google Scholar]
- 27.Kido H, Yokogoshi Y, Sakai K, Tashiro M, Kishino Y, et al. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem. 1992;267:13573–13579. [PubMed] [Google Scholar]
- 28.Pager CT, Craft WW, Jr, Patch J, Dutch RE. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79:12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J Virol. 2009;83:1511–1522. doi: 10.1128/JVI.01381-08. *First demonstration of a bona fide pH-dependent entry pathway for a member of the paramyxovirus family.
- 31.Plemper RK, Brindley MA, Iorio RM. Structural and mechanistic studies of measles illuminate paramyxovirus entry. PLoS Pathog. doi: 10.1371/journal.ppat.1002058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Gorman JJ, McKimm-Breschkin J, Lawrence LJ, Tulloch PA, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Camb) 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 35.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirusmediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 36.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, Singh M, Malashkevich VN, Kim PS. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci U S A. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 39.Markosyan RM, Cohen FS, Melikyan GB. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol Biol Cell. 2003;14:926–938. doi: 10.1091/mbc.E02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park HE, Gruenke JA, White JM. Leash in the groove mechanism of membrane fusion. Nat Struct Biol. 2003;10:1048–1053. doi: 10.1038/nsb1012. [DOI] [PubMed] [Google Scholar]
- 42.Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle J, Prussia A, White LK, Sun A, Liotta DC, et al. Two domains that control prefusion stability and transport competence of the measles virus fusion protein. J Virol. 2006;80:1524–1536. doi: 10.1128/JVI.80.3.1524-1536.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roymans D, De Bondt HL, Arnoult E, Geluykens P, Gevers T, et al. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci U S A. 2010;107:308–313. doi: 10.1073/pnas.0910108106. *Detailed mechanism of action study of a small-molecule respiratory syncytial virus entry inhibitor through co-crystallization, showing that potent inhibiton is achievable through disturbance of the 6HB conformation.
- 45.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong RT, Kushnir AS, White JM. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamm LK. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim Biophys Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- 48.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 203;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-San Martin C, Liu CY, Kielian M. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 2009;17:514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 52.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 53.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:2941–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 54.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, et al. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, et al. The Fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 56.Roussel A, Lescar J, Vaney MC, Wengler G, Rey FA. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure. 2006;14:75–86. doi: 10.1016/j.str.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 57. Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, et al. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. **Together with reference 58, groundbreaking insight into the regulation of alphavirus fusion proteins. Molecular structure of the viral E1/E2 protein complex and the regulation of E1 activity through E2 during for particle assembly and entry.
- 58. Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. **Together with reference 57, groundbreaking insight into the regulation of alphavirus fusion proteins. Molecular structure of the viral E1/E2 protein complex and the regulation of E1 activity through E2 during for particle assembly and entry.
- 59.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 60. Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106:2880–2885. doi: 10.1073/pnas.0810530106. *Report of the X-ray structure of EBV gB in the postfusion conformation. A comparison with known structures of HSV-1 gB and VSV G identifies distinct features of the EBV gB structure, adding to the better mechanistic understanding of class III fusion proteins and highlighting potential targets for strucutre-based drug design.
- 61.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 62.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 63.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 64.Gaudin Y. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell Biochem. 2000;34:379–408. doi: 10.1007/0-306-46824-7_10. [DOI] [PubMed] [Google Scholar]
- 65.Gaudin Y, Tuffereau C, Durrer P, Flamand A, Ruigrok RW. Biological function of the low-pH, fusion-inactive conformation of rabies virus glycoprotein (G): G is transported in a fusion-inactive state-like conformation. J Virol. 1995;69:5528–5534. doi: 10.1128/jvi.69.9.5528-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J Cell Biol. 2005;171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000851. e1000851. *Structure-guided mechanistic characterization of a peptidic dengue virus fusion inhibitor, revealing a two-stage process of nonspecific binding to the viral envelope followed by specific docking to refolding E in the endosome. This antiviral strategy should conceptually be transferrable to other viral pathogens that enter target cells pH-dependent through the endosomal route.
- 68.Brindley MA, Plemper RK. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J Virol. 2010;84:12174–12184. doi: 10.1128/JVI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, et al. Bimolecular complementation defines functional regions of Herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol. 2010;84:3825–3834. doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol. 2009;83:10857–10868. doi: 10.1128/JVI.01191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salzwedel K, Berger EA. Complementation of diverse HIV-1 Env defects through cooperative subunit interactions: a general property of the functional trimer. Retrovirology. 2009;6:75. doi: 10.1186/1742-4690-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 73.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 74.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 80. White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, et al. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001249. e1001249. *Using cryo-electrontomography, this study provides a first demonstration that virus strain-specific variations can lead to distinct glycoprotein conformations on the virion surface, providing a structural explanation for phenotypic differences between these strains.
- 81.Swanson K, Wen X, Leser GP, Paterson RG, Lamb RA, et al. Structure of the Newcastle disease virus F protein in the post-fusion conformation. Virology. 2010;402:372–379. doi: 10.1016/j.virol.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Birmanns S, Rusu M, Wriggers W. Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J Struct Biol. 2011;173:428–435. doi: 10.1016/j.jsb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]