Abstract
The human drug metabolizing cytochrome P450 (CYP) 1A2, is one of the major P450 isoforms contributing by about 5–20% to the hepatic P450 pool and catalyzing oxidative biotransformation of up to 10% of clinically relevant drugs including clozapine and caffeine. CYP1A2 activity is interindividually highly variable and although twin studies have suggested a high heritability, underlying genetic factors are still unknown. Here we adopted a pathway-oriented approach using a large human liver bank (n = 150) to elucidate whether variants in candidate genes of constitutive, ligand-inducible, and pathophysiological inhibitory regulatory pathways may explain different hepatic CYP1A2 phenotypes. Samples were phenotyped for phenacetin O-deethylase activity, and the expression of CYP1A2 protein and mRNA was determined. CYP1A2 expression and function was increased in smokers and decreased in patients with inflammation and cholestasis. Of 169 SNPs in 17 candidate genes including the CYP1A locus, 136 non-redundant SNPs with minor allele frequency >5% were analyzed by univariate and multivariate methods. A total of 13 strong significant associations were identified, of which 10 SNPs in the ARNT, AhRR, HNF1α, IL1β, SRC-1, and VDR genes showed consistent changes for at least two phenotypes by univariate analysis. Multivariate linear modeling indicated that the polymorphisms and non-genetic factors together explained 42, 38, and 33% of CYP1A2 variation at activity, protein and mRNA levels, respectively. In conclusion, we identified novel trans-associations between regulatory genes and hepatic CYP1A2 function and expression, but additional genetic factors must be assumed to explain the full extent of CYP1A2 heritability.
Keywords: candidate gene, cytochrome P450, CYP1A2, multivariate analysis, non-genetic factors, pharmacogenetics, pharmacogenomics, SNP
Introduction
The cytochrome P450, CYP1A2, is one of three P450 isoforms of the CYP1 family expressed in humans, the other isoforms being CYP1A1 and CYP1B1. It is predominantly expressed in liver and at lower levels in intestine, pancreas, lung, and brain (Gunes and Dahl, 2008; Zhou et al., 2009). In the liver, CYP1A2 contributes about 5–20% to the total microsomal P450 pool, thus representing one of the major drug metabolizing enzymes and contributing significantly to the oxidative metabolism of 10–15% of clinically relevant drugs. Among these are the antipsychotic clozapine (Bertilsson et al., 1994; Eiermann et al., 1997), the antidepressant duloxetine (Lobo et al., 2008), theophylline (Ha et al., 1995), and caffeine (Butler et al., 1989, which is commonly used for CYP1A2 in vivo phenotyping (e.g., Fuhr et al., 2007). CYP1A2 was also shown to activate procarcinogens, such as aromatic heterocyclic amines, and polycyclic aromatic hydrocarbons (Boobis et al., 1994; Eaton et al., 1995; Kim and Guengerich, 2005).
Like other drug metabolizing enzymes, CYP1A2 activity is highly variable and a number of environmental, non-genetic and genetic as well as epigenetic factors have been shown to play a role (Gunes and Dahl, 2008; Ghotbi et al., 2009).
Drug–drug interactions are one prominent source of clinically relevant variability and several high-affinity and irreversible inhibitors of this enzyme have been identified, including, for example, the potent irreversible inhibitor fluvoxamine (Gunes and Dahl, 2008; Zhou et al., 2009). Furthermore, all three CYP1 genes are coordinately regulated by the Ah-receptor pathway, which leads to induced levels in smokers (Schweikl et al., 1993; Bock et al., 1994; Sesardic et al., 1988; Ghotbi et al., 2007; Pelkonen et al., 2008) and after exposure to other xenobiotics (Pelkonen et al., 2008).
There is also evidence from several studies that CYP1A2 activity is higher in men than in women (Relling et al., 1992; Rasmussen and Brøsen, 1996; Scandlyn et al., 2008).
Additional variability in CYP1A2 expression is observed during inflammation (Vrzal et al., 2004) and several cytokines down-regulate expression in primary human hepatocytes (Abdel-Razzak et al., 1993) or repress inducibility (Muntané-Relat et al., 1995).
Despite these pronounced environmental, sex, and disease-related factors, there is strong evidence for a significant contribution of genetic factors to interindividual variability in CYP1A2 activity. By measuring the caffeine metabolic ratio as a CYP1A2 activity marker in a large cohort (n = 378) of mono- and dizygotic twins selected to exclude the influence of smoking, oral contraceptives, and gender, Brøsen and colleagues found a strong overall heritability of 0.725 (Rasmussen et al., 2002).
To date, 16 defined alleles and additional haplotype variants, comprising more than 30 SNPs have been identified in coding and non-coding regions of the CYP1A2 gene (CYPallele nomenclature website at http://www.cypalleles.ki.se/; inspected August 6, 2010).
The amino acid variants, some of which were shown to be functionally relevant (Chevalier et al., 2001; Murayama et al., 2004; Zhou et al., 2004) are nevertheless of limited clinical use due to their rare occurrence. Among the few SNPs currently considered to be of potential predictive value, are the 5′-upstream variant 3860G > A (CYP1A2*1C) and the intron 1 polymorphism −163C > A (CYP1A2*1F) located downstream of the untranslated first exon. However, attempts to establish phenotype-genotype relationships were overall disappointing and a recent study concluded that no SNP or haplotype in the CYP1A2 gene has a clear predictive value (Jiang et al., 2006). Thus, although some of these variants have been associated with altered drug clearance and response, or with disease susceptibility (Gunes and Dahl, 2008; Zanger et al., 2008), it appears that the basis for inheritable CYP1A2-dependent phenotypes has not been satisfactorily elucidated.
Although novel SNPs within the CYP1A locus may still be discovered, genetic determinants in other genes that participate in the regulation of constitutive and inducible CYP1A2 expression should be expected to contribute to interindividual variability. In particular the liver-enriched transcription factors HNF4α, HNF1α, USF1/2, and the coactivators PGC1α and SRC-1 were shown to be involved in constitutive expression of CYP1A2 (Narvaez et al., 2005; Martínez-Jiménez et al., 2006). Importantly, the aryl hydrocarbon receptor (AhR) pathway coordinately regulates transcription of CYP1 and a battery of additional ADME and other genes that constitute the toxicological response to polycyclic aromatic hydrocarbons and dioxins (Nebert and Dalton, 2006; Pascussi et al., 2008).
While several murine Ah-receptor polymorphisms were shown to affect dose–response curves and toxic effects, the existing polymorphisms in the human Ah-receptor and other genes of the pathway have not been systematically investigated as determinants of downstream transcription (Okey et al., 2005). Further signaling pathways recognized to have an impact on CYP1A2 expression include inflammation and immune response, as it is known that CYP1A expression is downregulated during sepsis or inflammation (Vrzal et al., 2004; Zhou et al., 2008; Tian, 2009).
Here we adopted a pathway-oriented approach to investigate the association of polymorphisms in candidate genes involved in the transcriptional regulation of CYP1A2 gene expression. We included genes that have been previously implicated in CYP1A2 constitutive, inducible, and pathophysiological transcriptional regulation, such as liver-enriched transcription factors and co-regulators, members of the AhR pathway, and mediators of inflammation and immune response, as summarized in Figure 1. In addition, we included several nuclear receptors that function as xenobiotic sensors of other inducible P450s, although direct interaction with CYP1A2 has not been demonstrated. In addition we also re-analyzed numerous SNPs within the CYP1A2 gene that have been previously investigated as determinants of pharmacokinetics or that have other evidence for functional impact. To investigate the possible impact of these genetic variants, we carried out univariate and multivariate association analyses in a large human liver bank phenotyped for CYP1A2 mRNA and protein expression, and enzyme activity. The clinical documentation of the samples furthermore allowed us to include a number of environmental and other non-genetic factors. The results support the view that some polymorphic genes in transcriptional regulatory pathways contribute to hepatic CYP1A2 phenotype, although non-genetic factors appear to be almost as important as genetic ones.
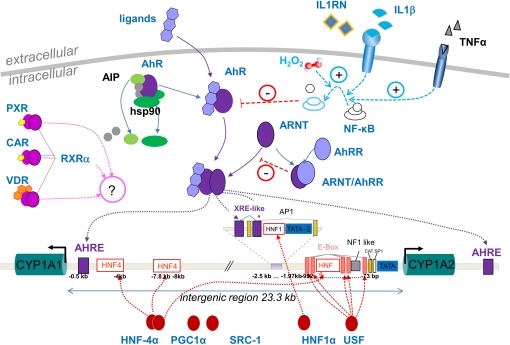
Figure 1.
Overview of known pathways for transcriptional regulation at the CYP1A2 promoter. The scheme shows the 23.3-kb intergenic region between the head-to-head oriented CYP1A1 and 1A2 genes on chromosome 15q24.1. Binding elements located within the common promoter region may thus be shared for transcriptional regulation of both genes. Pathways for constitutive and tissue specific expression are shown by red symbols at the bottom and include the liver-enriched transcription factors HNF4α and HNF1α and their coactivators PGC1α and SRC-1, as well as basic transcriptional activators SP-1 and USF-1 (Quattrochi et al., 1994; Chung and Bresnick, 1997; Narvaez et al., 2005; Martínez-Jiménez et al., 2006). Top-middle: Genes selected for the AhR-pathway include the ligand-binding receptor AhR, the AhR nuclear translocator ARNT, and the AhR regulator AhRR. The activated ligand bound form of AhR is complexed with two heat-shock proteins hsp90 and AIP, and co-chaperone p23, which help to correctly fold and stabilize the AhR and prevent inappropriate trafficking to the nucleus (Petrulis et al., 2000). Upon heterodimerization with activated AhR, the ARNT/AhR complex translocates to the nucleus to activate transcription from AHRE- and XRE-motifs (Okino et al., 2007). AhRR competes with AhR for ARNT binding, resulting in inhibition of AhR-mediated signal transduction. Top-right: Under pathophysiological conditions of inflammation, inflammatory cytokines like IL1ß and TNFα or chronic oxidative stress (H2O2) activate NFκB which in turn inhibits AhR activity thus leading to reduced CYP1A expression. This pathway can be blocked by competing antagonists like IL1RN (Tian et al., 1999; Vrzal et al., 2004; Zhou et al., 2008). Top-left: Exploratory pathways involving possible crosstalk to other known xenobiotic transcriptional regulation networks are indicated by PXR/RXRα, CAR/ RXRα, VDR/ RXRα heterodimers. Proteins designated in black were not included in this study.
Materials and Methods
Patients and liver samples
Liver tissue and corresponding blood samples for genomic DNA extraction were previously collected from patients undergoing liver surgery at the Department of General, Visceral, and Transplantation Surgery (A.K.Nuessler, P.Neuhaus, Campus Virchow, University Medical Center Charité, Humboldt University Berlin, Germany). The study was approved by the ethics committees of the medical faculties of the Charité, Humboldt University, and of the University of Tuebingen and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from each patient. All tissue samples had been examined by a pathologist and only histologically normal liver tissue was collected and stored at −80°C. For each patient, detailed information was available regarding age, sex, smoking status, alcohol consumption, pre-surgery medication (P450 inducers as assigned in Table 1), indication for liver resection, and pre-surgery liver serum parameters (Table 1). Samples from patients with hepatitis, cirrhosis, or chronic alcohol abuse were excluded. A total of 150 liver samples from which high quality RNA and complete documentation could be obtained were finally included.
Table 1.
Population demographics and serum parameters of 150 liver donors.
| Subgroup | Number |
|---|---|
| SEX | |
| Male | 71 |
| Female | 79 |
| AGE | |
| ≤70 | 124 |
| >70 | 26 |
| SMOKING HABITUSa | |
| Non-smoker | 117 |
| Smoker | 29 |
| ALCOHOL CONSUMPTIONa | |
| None | 96 |
| ≥1 times/week | 48 |
| PRE-SURGERY DRUGS | |
| None | 40 |
| P450-inducersb | 40 |
| Other drugs | 70 |
| TBILI (mg/dL)a | |
| Normal (≤1.2) | 125 |
| Elevated (>1.2) | 22 |
| GGT (U/L)a | |
| Normal (f: ≤36; m: ≤64) | 85 |
| Elevated (f: >36; m: >64) | 58 |
| CRP (mg/L)a | |
| Normal (≤8.2) | 140 |
| Elevated (>8.2) | 7 |
| DIAGd | |
| No tumor | 3 |
| Primary liver tumors | 65 |
| Metastases | 82 |
| CHOLESTASISa,c | |
| Non-cholestatic | 121 |
| Cholestatic | 25 |
tBili, serum total bilirubin; GGT, serum gamma glutamyl transferase; CRP, C-reactive protein; DIAG, diagnosis.
aSample number are not summing up to 150 because of missing information.
bP450 inducers: atorvastatin, beclometasondipropionate, budenoside, estradiol, metamizole, nifedipine, omeprazole, pantoprazole, prednisolone, simvastatin, tamoxifen, vitamin D.
cCholestasis was diagnosed according to Nies et al. (2009).
dDiagnosis groups: no tumor; primary benign and malign liver tumors including HCC, cholangiocarcinoma, gallbladder tumor; metastases derived from colon carcinomas (65), or other tumors (17).
High quality total RNA was isolated from liver tissue using Trizol/QiagenRNeasy protocol as described previously (Gomes et al., 2009). Synthesis of cDNA was performed with 1 μg RNA using the TaqMan Reverse Transcription Kit (Applied Biosystems, Darmstadt, Germany) according to the supplier's instructions. Expression of CYP1A2 mRNA in liver tissue was performed using specific primers and a TaqMan-probe as described in Feidt et al. (2010) with a TaqMan 7500 system (Applied Biosystems, Darmstadt, Germany). A standard curve was obtained using CYP1A2 cDNA-containing linearized plasmid DNA (a friendly gift from J. Buters; Munich, Germany). Raw data were normalized to RPLP0 (60S large ribosomal protein P0) expression which was determined in the same samples using the endogenous control assay (4326314E) from Applied Biosystems (Darmstadt, Germany). Normalized values were adjusted to the median value of all samples.
CYP1A2 protein expression was analyzed by Western blot in liver microsomes. Ten micrograms of protein were separated by electrophoresis on a 10% SDS-polyacrylamide gel and blotted onto nitrocellulose membranes. Anti-human CYP1A2 monoclonal antibody (clone 26-7-5 Invitrogen Panvera, Darmstadt, Germany) and IRD800-labeled secondary anti-mouse antibody (Licor, Lincoln, NE, USA) were used for detection with an Odyssey system (Licor, Lincoln, NE, USA). For absolute quantification, a standard curve was generated by coanalyzing 250–4000 fmol of recombinantly expressed CYP1A2 in insect cell membranes (Becton Dickinson Gentest, Heidelberg, Germany) in each experiment. The CYP1A2 enzyme activity was determined by measuring phenacetin-O-deethylation as described in Turpeinen et al. (2009).
SNP selection and genotyping methods
Candidate gene polymorphisms selected for analysis are listed in Table S1 in Supplementary Material. SNPs were selected from public databases1,2,3,4,5 according to available data on functionality, frequency, and linkage based on published research. For each gene, SNP selections were complemented by LD (linkage disequilibrium)-tag SNPs selected using HaploView6 on HapMap-CEU SNP population. Genomic DNA was isolated from EDTA blood samples using the QIAmp DNA Blood Mini Kit System (Qiagen, Hilden, Germany). MALDI-TOF MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) genotyping was performed for 85 SNPs distributed across 12 different assays using the MassARRAY Compact system (Sequenom, San Diego, CA, USA). Primers for MALDI-TOF MS genotyping were ordered from Metabion International (Martinsried, Germany). Data for additional 78 SNPs were obtained from HumanHap300v1.1 chip analysis (Microarray Facility Tübingen Services, Tübingen, Germany). Two polymorphisms were genotyped using predesigned TaqMan allelic discrimination assays (Applied Biosystems, Darmstadt, Germany) using a TaqMan 7900HT (Applied Biosystems, Darmstadt, Germany). Data for four SNPs were kindly provided by E. Schaeffeler (IKP, Stuttgart, Germany). Details of primers and genotyping assays are available upon request. Approximately 10% of samples within each assay were repeated or sequenced in the concerning region to confirm genotyping results as a quality control. Generally, less than 0.1% discordant results were obtained.
Statistical methods
Statistical analyses were performed using software R-2.11.07 with additional packages hwde-0.61 and SNPassoc-1.6-0. Hardy–Weinberg p-values were calculated according to Wigginton et al. (2005). Minor allele frequency (MAF) and missing values frequency of polymorphisms are given in (Table S1 in Supplementary Material). SNPs with MAF < 5% (n = 28) were excluded from further analyses. For polymorphisms having less than three heterozygotes or homozygotes in the remaining data set (n = 14), heterozygotes and homozygotes were combined and compared vs. wild-types. From pairs of 100%-linked polymorphisms (n = 5), the SNP with the fewest missing values, if present, was chosen. Finally, a total of 136 polymorphisms were included in the statistical analyses.
The influence of non-genetic factors on CYP1A2 expression was investigated using Kruskal–Wallis tests for categorical and Spearman correlations for numerical parameters due to the non-normality of the phenotypic and non-genetic data. Associations between polymorphisms and CYP1A2 expression (mRNA, protein, activity) were investigated using the generalized linear model capabilities of the SNPassoc package. In order to satisfy the Gaussian distribution condition, phenotypic data were log-transformed and data distributions were checked using quantile–quantile plots. The SNPassoc package offers four different genetic models: codominant, dominant, recessive, and log-additive. In the codominant, dominant, and recessive models, the genotypic data is treated as a categorical variable. The codominant model compares wild-types vs. heterozygotes vs. homozygotes, while the dominant and recessive model test wildtypes vs. heterozygotes plus homozygotes and wildtypes plus heterozygotes vs. homozygotes, respectively. For the log-additive model, a numeric variable is created with wild-type = 0, heterozygote = 1, and homozygote = 2, assuming a linear allele-dose effect.
Package SNPassoc was also applied to study associations between SNPs and CYP1A2 phenotypes corrected for non-genetic factors. For each phenotype studied, the non-genetic correction factors were chosen based on linear models between phenotypic data and all 10 non-genetic factors considered (age, sex, nicotine and alcohol intake, drug exposure, total bilirubin, GGT, CRP, cholestasis, diagnosis) as well as models derived from these complete models via step-wise model selection using Akaike's information criterion. Non-genetic factors were chosen for correction if the p-value from complete and/or reduced models was <0.15. In all linear models, we used log-transformed phenotypic data as dependent variable. All statistical tests were two-sided and statistical significance was defined as p < 0.05.
Results
Population variability of hepatic CYP1A2
CYP1A2 expression in the 150 Caucasian liver samples varied considerably at the different phenotype levels and was unimodally yet not normally distributed (Table 2, Figure 2). Fold-variation was highest (196-fold) for mRNA and lowest (35-fold) for enzyme activity (Table 2). However the coefficient of variation (cv) as a normalized measure of variability was more comparable between the different phenotypes. These values are comparable to previous studies by others (Shimada et al., 1994; Jiang et al., 2006; Gunes and Dahl, 2008), however compared to our studies on other liver drug metabolizing enzymes (e.g., CYPs 2D6, Zanger et al., 2001; 2B6, Hofmann et al., 2008; 3A4, Wolbold et al., 2003) variation was somewhat less pronounced. All three CYP1A2 phenotypes were significantly correlated to each other (Figure 2). As expected due to common regulation, CYP1A2 expression levels were also highly correlated to CYP1A1 (data not shown).
Table 2.
Population variability of hepatic CYP1A2 expression phenotypes (n = 150).
| RNA CYP1A2/ RPLP0 relative units | Protein content pmol/mg | Phenacetin-O-deethylation pmol/min/mg | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Minimum | 0.03 | 3.62 | 130.1 | ||||||
| Median | 1.00 | 64.7 | 1442 | ||||||
| Maximum | 5.88 | 278.7 | 4536 | ||||||
| Ratio maximum/ minimum | 196 | 77.4 | 34.9 | ||||||
| Normal distribution | No | No | No | ||||||
| Coefficient of variation (cv, %) | 77.2 | 69.3 | 56.2 |
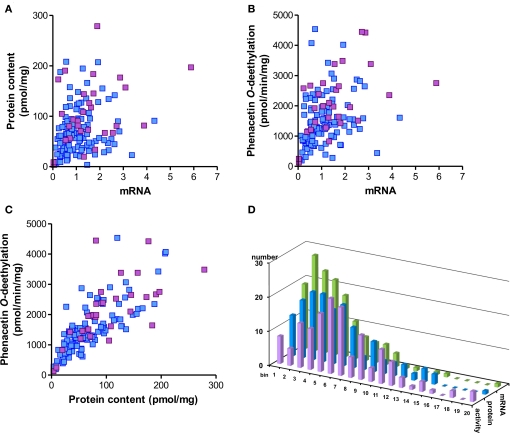
Figure 2.
Spearman correlations and population distribution of hepatic CYP1A2 phenotypes. CYP1A2 mRNA was determined by a specific TaqMan real-time RT-PCR assay, protein was determined by Western blot and enzyme activity was measured by LC-MSMS analysis of phenacetin-O-deethylation in human liver microsomes (n = 150). Results are means of duplicate measurements. (A–C) Spearman rank correlation analysis; blue: non-smokers; red: smokers. The correlation coefficients were (A) rs = 0.36; (B) rs = 0.47; (C) rs = 0.83. The statistical significance for all comparisons was p < 0.0001. (D) Histogram showing population distributions for enzyme activity (red), protein (blue), and mRNA (green) using 20 bins over the entire phenotype range.
Non-genetic factors influencing CYP1A2 expression
Available documentation to the liver donors included demographic, clinical, and self-reported data as shown in Table 1. We used univariate, non-parametric tests to analyze whether any of these parameters had an impact on CYP1A2 phenotype. We identified self-reported cigaret smoking, pre-surgical drug exposure, liver function, inflammation, diagnosis, and cholestasis as factors being significantly associated with microsomal phenacetin O-deethylase activity (Table 3, Figure 3). Regular smoking had the strongest impact on all phenotype levels and increased CYP1A2 activity, protein and mRNA-levels by 1.5-, 1.6-, and 1.5-fold, respectively. Known CYP3A-inducers were associated with decreased CYP1A2 activity and protein levels, whereas patients treated with other drugs had normal levels. As omeprazole had been described as an inducer of caffeine metabolic ratio (Rost et al., 1992), we separately analyzed a subgroup of 12 patients treated with this drug. While mRNA levels were increased by 1.4-fold (not significant), protein and activity were slightly decreased (not significant) compared to controls not exposed to any drug (n = 40). Decreased liver function as indicated by elevated serum gamma glutamyl transferase (GGT) was associated with slightly decreased activity and protein (p < 0.01), but mRNA was unchanged. Pathophysiologically increased levels of C-reactive protein (CRP) were found in seven liver donors who had strongly decreased levels of CYP1A2 activity and protein. Moreover, cholestasis was diagnosed in 25 liver donors who also had decreased activity and protein as well as a trend for decreased mRNA. Finally, we did not find an association between the donor's sex and CYP1A2 at any phenotype level (Figure 3, Table 3), although females had increased protein levels (p 0.05) in the multivariate model (see below). Taken together, these data indicated that environmental and other non-genetic factors have a strong influence on hepatic CYP1A2 phenotype.
Table 3.
Influence of non-genetic factors on CYP1A2 phenotype.
| Non-genetic factora | Correlation testb | mRNAc | Proteinc | Activityc | |
|---|---|---|---|---|---|
| Sex | K | 0.26 | 0.68 | 0.99 | |
| Age | S | 0.27 | 0.045 | 0.24 | |
| NIC | K | 0.018 | 0.001 | 0.0004 | |
| ALC | K | 0.30 | 0.50 | 0.71 | |
| IND | K | 0.38 | 0.025 | 0.03 | |
| BILI | K | 0.001 | 0.0002 | 0.0005 | |
| GGT | K | 0.11 | 0.001 | 0.009 | |
| CRP | K | 0.32 | 0.015 | 0.03 | |
| DIAG | K | 0.08 | 0.09 | 0.009 | |
| Chol | K | 0.06 | 0.008 | 0.004 |
aNIC, nicotine consumption: non smokers vs. smokers (>1 cigarets/day); ALC, alcohol consumption: none vs. >1 times/week; IND, exposure to known inducers of P450; BILI, serum total bilirubin; GGT, serum gamma glutamyl transferase; CRP, C-reactive protein; DIAG, diagnosis; Chol, cholestasis.
bKnuskal-Wallis (K) or Spearman correlation (S) test.
cp-Values for univariate analyses using the given correlation test. Results were not corrected for multiple testing.
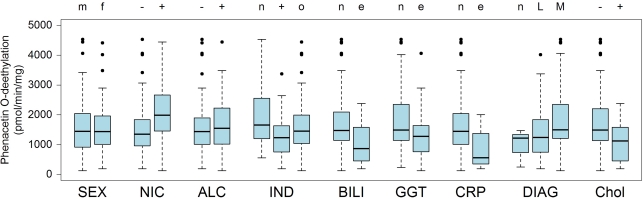
Figure 3.
CYP1A2 enzyme activity in population subgroups. The liver donors (n = 150) were divided into subgroups according to available clinical documentation on environmental exposures and other non-genetic factors: SEX, (m) male, (f) female; NIC, nicotine consumption, (−) non-smokers (+) smokers (>1 cigarets/day); ALC, alcohol consumption (−) none (+) >1 times/week; IND, (n) no drugs, (+) known inducers of P450, (o) other drugs; BILI, serum bilirubin, (n) value in normal range, (e) elevated value; GGT, serum gamma glutamyl transferase, (n) value in normal range, (e) elevated value; CRP, C-reactive protein, (n) value in normal range, (e) elevated value; DIAG, diagnosis leading to liver resection, (n) no tumor, (L) primary liver tumor, (M) metastasis of other primary tumor; Chol, cholestasis, (−): non-cholestatic, (+) cholestatic. For results on mRNA and protein and for statistical information see Table 3. Boxes are defined by the 25 and 75% quantile. The median is displayed as vertical line inside of the box. Whiskers are defined as the lowest/highest data point still within 75%/25% quantile ± 1.5 times the interquantile range (75–25% quantile). Outliers are shown as dots.
SNP-selection and SNP-frequencies in the study population
We analyzed CYP1A locus SNPs for cis-association and polymorphisms in genes of CYP1A2 regulatory pathways for trans-association with CYP1A2 phenotype. Selected SNPs of the CYP1A locus included the most frequent and the commonly studied CYP1A2 alleles8 as well as four tag-SNPs selected to represent other haplotypes not previously investigated (Table S1 in Supplementary Material). Candidate transcriptional pathway genes included the AhR regulatory network (AhR, AhRR, ARNT), liver-enriched transcription factors and coactivators (HNF1α, HNF4α, PPARGC1α, SRC-1, USF1), as well as some key regulators and mediators of inflammation and immune response (NFκB1, IL1β, IL1RN). We also investigated SNPs in the nuclear receptor pathway responsible for induction of CYP2 and CYP3 enzymes (PXR, CAR, RXR, RAR, VDR). These genes were selected for exploratory reasons as direct interaction with CYP1A2 has not been shown.
The selected SNPs were located in intron regions (98), exonic regions (23 non-synonymous, 9 synonymous), promoters (12) and up- or downstream of the genes (27). In total we genotyped 15 SNPs in the CYP1A1/2 locus and 154 SNPs in 16 candidate genes (Table S1 in Supplementary Material). Of these, 4 and 10 SNPs, respectively, were not detected in any of the samples and 5 were in complete linkage to other SNPs and were therefore not included in further analyses. Hardy–Weinberg equilibrium test resulted in p > 0.001 for all tested SNPs (values see Table S1 in Supplementary Material).
The calculated SNP frequencies in our Caucasian liver donor population correlated well (Spearman rS = 0.92) with frequency data compiled from dbSNP database for the HAPMAP-CEU population (n = 153) (Figure S1 in Supplementary Material). Fourteen of the analyzed SNPs were rare (MAF < 5%) and five of these were located in the CYP1A locus. The remaining had MAFs between 5 and 20% (n = 33) and 103 SNPs appeared with frequencies above 20%.
Genotype–phenotype correlation analysis
Univariate analysis
A total of 136 SNPs with MAF > 5% were further investigated for association with phenotypic data by univariate analysis, applying four different genetic models (co-dominant, dominant, recessive, or log-additive, see Materials and Methods). Without correction for non-genetic factors, this resulted in a total of 25, 16, and 16 significant associations at p < 0.05 for activity, protein, and mRNA, respectively. A graphical summary of these results is shown in Figure 4.
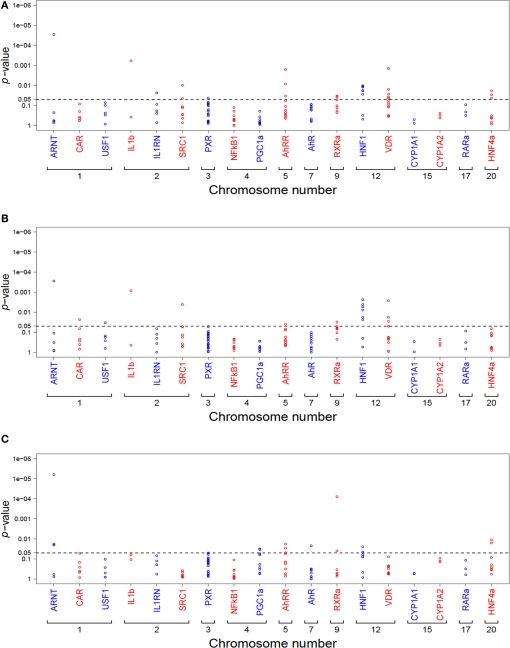
Figure 4.
Summary of univariate analysis of all tested SNPs (n = 136) in relation to CYP1A2 activity (A), protein (B), and mRNA (C). Each dot represents the minimum p-value from all four genetic models (codominant, dominant, recessive, log-additive). Genes are ordered due to chromosomal location. The dotted line marks the 5% significance level p-values are not adjusted for multiple testing.
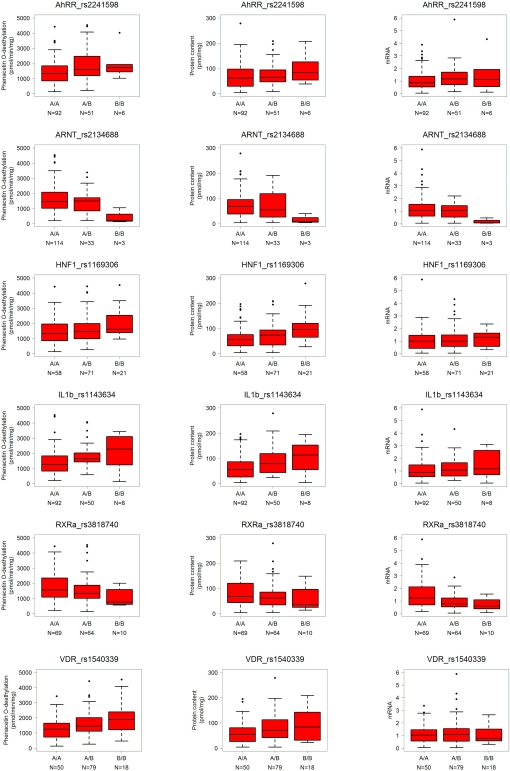
The log-additive model is based on the assumption of a gene-dose effect, such that heterozygotes are phenotypically intermediate between homozygous wild-types and mutants. Limiting the results to this mechanistically most plausible genetic model and restricting to the strongest significant associations with p < 0.01 revealed a total of 11 univariate associations showing a significant linear trend for any one or more phenotypes (Table 4). However, the SNPs in AhRR and HNF1α were pairwise linked with up to 95% (rs2241598/rs10078; rs1169300/rs2464196; rs1169306/rs735396). The SNPs in RXRα and VDR were independent. The SNP rs2134688 in the ARNT gene was consistently associated with reduced levels of all three phenotypes; SNP rs2241598 in the AhRR gene was consistently associated with increased levels as indicated in Table 4 and Figure 5. Seven SNPs in the HNF1α, IL1β, SRC-1, and VDR genes were associated with unidirectional changes in activity and protein, one further AhRR SNP (rs10078) was associated with increased activity. Besides the ARNT SNP mentioned above, only one SNP (rs3818740 in RXRα) was correlated to decreased mRNA.
Table 4.
SNPs with significant association to CYP1A2 phenotype.
| Activity | Protein | mRNA | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Number of homozygous samples | Changea | p-Value (univariate)b | p-Value (multivariate)b | Changea | p-Value (univariate)b | p-value (multivariate)b | Changea | p-Value (univariate)b | p-Value (multivariate)b | |||||||||||||
| rs2241598 | AhRR | 18 | ↑ | 0.0020 | 0.0013 | ↑ | 0.0408 | 0.0344 | ↑ | 0.0349 | 0.0324 | |||||||||||||
| rs10078 | AhRR | 6 | ↑ | 0.0084 | 0.0066 | n.s. | n.s. | n.s. | n.s. | |||||||||||||||
| rs2134688 | ARNT | 3 | ↓ | 0.0018 | 0.0245 | ↓ | 0.0023 | 0.0459 | ↓ | 6.7E-05 | 0.0014 | |||||||||||||
| rs1169300 | HNF1α | 14 | ↑ | 0.0192 | n.s. | ↑ | 0.0080 | 0.0394 | n.s. | n.s. | ||||||||||||||
| rs2464196 | HNF1α | 15 | ↑ | 0.0115 | 0.0295 | ↑ | 0.0055 | 0.0261 | n.s. | n.s. | ||||||||||||||
| rs1169306 | HNF1α | 21 | ↑ | 0.0105 | 0.0304 | ↑ | 0.0024 | 0.0140 | ↑ | n.s. | 0.0461 | |||||||||||||
| rs735396 | HNF1α | 20 | ↑ | 0.0121 | 0.0326 | ↑ | 0.0041 | 0.0217 | n.s. | n.s. | ||||||||||||||
| rs1143634 | IL1β | 8 | ↑ | 0.0022 | 0.0158 | ↑ | 0.0026 | 0.0180 | n.s. | n.s. | ||||||||||||||
| rs3818740 | RXRα | 10 | n.s. | n.s. | ↑ | n.s. | n.s. | ↓ | 1.2E-04 | 0.0026 | ||||||||||||||
| rs3132297 | RXRα | 3 | n.s | n.s. | ↓ | 0.0319 | 0.0082 | n.s. | n.s. | |||||||||||||||
| rs2119115 | SRC-1 | 15 | ↓ | 0.0101 | 0.0411 | ↓ | 0.0041 | 0.0099 | n.s. | n.s. | ||||||||||||||
| rs1540339 | VDR | 18 | ↑ | 0.0015 | 0.0067 | ↑ | 0.0037 | 0.0292 | n.s. | n.s. | ||||||||||||||
| rs10735810 (new: rs2228570) | VDR | 6 | ↑ | 0.0162 | 0.0085 | ↑ | 0.0180 | 0.0143 | n.s. | n.s. | ||||||||||||||
aDirection of expression change: (↑) expression higher in variants, (↓) expression lower in variants.
bOnly the most significant SNPs with p < 0.01 for at least one phenotype in the log-additive model are shown. Multivariate results were corrected for non-genetic factors as described in Materials and Methods; n.s., not significant (p ≥ 0.05 ). p-values are not adjusted for multiple testing.
Figure 5.
Genotype–phenotype correlations for CYP1A2 activity, protein, and mRNA exemplarily for some significant SNPs. Genotypes are indicated by A (major allele) and B (minor allele). N, number of individuals per group; numbers for the three genotypes do not add up to 150 in each case due to missing values. Selected representative results are shown as box-and-whisker plots with outliers for significant SNPs taken from Table 4.
Of note, none of the SNPs of the CYP1A locus were among these most significant relationships when our MAF limit of 5% was applied. However, for the CYP1A2 promoter SNP rs2069522 (MAF = 3.7%), we found a trend to lower protein and mRNA expression (data not shown).
Multivariate analysis
The association between each of the 136 polymorphisms and each phenotype, corrected for chosen non-genetic factors (activity and protein: age, sex, nicotine intake, total bilirubin, CRP, diagnosis; mRNA: sex, nicotine intake, drug treatment, total bilirubin, CRP, diagnosis) was tested by R-package SNPassoc. As described in Section “Statistical Methods,” we selected those non-genetic factors for correction with p < 0.15 in (a) linear models with all 10 non-genetic factors and/or (b) models derived from these models by step-wise model selection.
As summarized in Table 4, all SNPs associated to any phenotype in the univariate analysis remained significant (p < 0.05) in the multivariate analysis except rs1169300 in HNF1α which lost association to activity. Both AhRR SNPs and two VDR SNPs remained significantly associated to activity with p < 0.01.
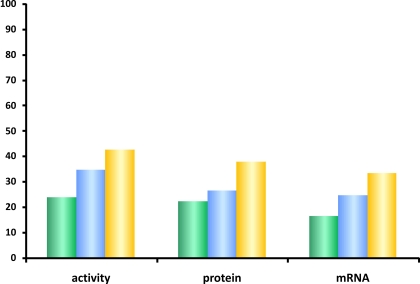
The step-wise model-selection method was furthermore applied to determine the contribution of genetic and non-genetic factors to CYP1A2 variability at each phenotype level (Figure 6). For this purpose, we first defined initial linear models with (a) non-genetic factors only (vide Statistical Methods), (b) all polymorphisms with p < 0.05 in the univariate or multivariate log-additive models described above, and (c) with both, SNPs and non-genetic factors, where the latter were selected as described above. The application of stepwise model-selection using Akaike's information criterion resulted in final linear models containing those SNPs and non-genetic factors which explain each of the phenotypes best. As graphically displayed in Figure 6, the genetic polymorphisms identified in our candidate genes explained a slightly larger fraction of the total variability than the non-genetic factors at each phenotype level. The fraction explained was highest for CYP1A2 activity (42%), however, in total less than half of the hepatic CYP1A2 variability could be explained by the multivariate models.
Figure 6.
Contribution of genetic and non-genetic factors to CYP1A2 expression. Percentage of the total CYP1A2 variation as explained by multivariate linear models containing only non-genetic factors (green), only genetic factors (blue), or both combined (yellow). The bars indicate the coefficient of determination adjusted for the number of factors in the different models. As described in the text, the linear models were obtained by a step-wise model selection procedure using Akaike's information criterion.
Multiple testing correction
To correct for multiple testing we applied the Benjamini–Hochberg adjustment procedure controlling the false discovery rate (FDR) (Benjamini and Hochberg, 1995). With a FDR < 0.05 only the ARNT SNP rs2134688 and the RXRα SNP rs3818740 remained significantly associated with decreased mRNA in the univariate analysis.
Discussion
In this study we searched for genetic polymorphisms that could explain the high heritability that has been proposed for CYP1A2 based on a classical twin-study that compared monozygotic and dizygotic twin-pairs phenotyped for the prototypic CYP1A2 substrate caffeine (Rasmussen et al., 2002). Several previous studies attempted to identify genetic or epigenetic markers for CYP1A2 by concentrating on the CYP1A locus on chromosome 15 (Jiang et al., 2006; Ghotbi et al., 2007; Gunes and Dahl, 2008). However, the results of these former studies have been disappointing in that the few genetic markers identified are unable to explain more than a tiny fraction of the interindividual variability of this enzyme (Jiang et al., 2006) and most of them are still controversial (Zanger et al., 2008). One part of our study dealing with polymorphisms at the CYP1A locus confirmed these findings. We studied 15 well-selected polymorphisms that have been previously associated with changes in phenotype or expression, or represent possible tag-SNPs not studied before. None of these SNPs was significantly associated with CYP1A2 activity or protein expression, and only for one rare promoter SNP a trend to lower protein and mRNA levels was found. A recent genome-wide study that analyzed a total of 782,476 unique SNPs in 466 liver samples also failed to detect any novel markers of CYP1A2 expression (mRNA) or activity within the CYP1A locus (Yang et al., 2010). Future studies by next-generation sequencing and initiatives like the 1000-genomes project9 will reveal novel polymorphisms in the CYP1A locus that can be tested as markers for phenotype. However, given the experience and amount of work done so far, it seems very unlikely that cis-associations make an important contribution to the overall variability of this enzyme.
In our study we have therefore concentrated on other genes and on various non-genetic factors as possible influential factors for expression and function of CYP1A2 in liver. By taking advantage of the detailed documentation available to the samples of our liver bank, we were able to study the influence of diverse environmental and other non-genetic factors at unprecedented detail. It was expected, based on previous work (Sesardic et al., 1988; Bock et al., 1994; Ghotbi et al., 2007), that CYP1A2 will be increased in cigaret smokers, and we confirmed this with about 1.5-fold elevated expression at all levels. Another environmental factor analyzed concerns pre-surgical drug exposure. Interestingly, drugs categorized as typical inducers of CYP2 and CYP3A genes, including nifedipine, atorvastatin, or glucocorticoids, had a repressive effect on CYP1A2. We also confirmed a moderate inducing effect of omeprazole (Rost et al., 1992), although this was not significant at the level of mRNA. We furthermore confirmed that inflammation processes have a decreasing effect on CYP1A2 expression (Congiu et al., 2009). The finding that measurement of CRP and serum bilirubin can predict lower CYP1A2 is new, to our knowledge. Interestingly, a case report on increased serum clozapine levels after bacterial infection indicated a similar effect (Raaska et al., 2002). Furthermore, we also found that other pathophysiological states of the liver are associated with decreased CYP1A2 expression. Thus, cholestasis and/or elevated GGT levels were correlated with decreased CYP1A2 activity, protein and mRNA expression by 1.2-, 1.6-, and 1.3-fold, respectively. To our knowledge this has not been reported before for humans. Reduced activity of CYP1A2 was however found in rats with drug induced cholestasis (Bramow et al., 2001) and following biliary obstruction (Fukushima et al., 2008). A further pathophysiological factor influencing CYP1A2 phenotype was identified by analyzing the reasons leading to the surgical liver resection (diagnosis). The reason for elevated levels of CYP1A2 in livers with metastases of extrahepatic primary tumors, predominantly colon carcinoma, is currently unknown. Finally, we did not find convincing evidence for a sex-related expression difference, as only the multivariate model indicated increased protein in females. Previous in vivo studies have indicated higher activity of CYP1A2 in males vs. females based on in vivo measured probe drugs (Relling et al., 1992; Rasmussen and Brøsen, 1996; Scandlyn et al., 2008). The absence of convincing sex-related differences in our study thus contradicts these earlier findings but is in agreement with the recent study of 466 Caucasian livers, which also failed to detect an unequivocal sex-effect on CYP1A2 activity (Yang et al., 2010). Taken together these data confirm and further extend our knowledge about environmental and pathophysiological factors influencing CYP1A2 activity and expression in human liver.
For the genetic part of our study we applied a pathway-oriented approach and concentrated on transcriptional regulatory routes, because CYP1A2 expression is highly variable and has been shown to be affected by several constitutive and inducible as well as inhibitory pathways (summarized in Figure 1).
From these pathways 136 SNPs in 16 genes and the CYP1A locus were included in our final statistical analysis. The 13 strongest significant associations revealed by univariate and multivariate analysis comprise four SNPs in the ARNT, RXRα, and SRC-1 gene associated with lower expression and/or activity. Only rs2134688 in the ARNT gene consistently showed decreased levels in all phenotypes. This SNP is located in an intron and is member of a large LD block of about 550 kb including SNPs in the promoter region of ARNT and also upstream and downstream neighboring genes. Das et al. (2008) previously described this site in context of acute insulin response in that the insulin secretion in non-diabetic subjects may be altered. The others were intronic SNPs in SRC-1 (rs2119115) and in RXRα (rs3818740, rs3132297), which all were selected as “tag” SNPs representing possible new haplotypes for the genes because of linkage to a set of other polymorphisms in the gene region and throughout the promoter. They have not been analyzed before and may influence their own mRNA expression, thus resulting in lower transcription of CYP1A2. Of note, two of these disabling SNPs (rs2134688 in ARNT, rs3818740 in RXRα) remained significant after multiple testing correction for CYP1A2 mRNA expression.
Interestingly, all other SNPs (n = 9) with strong association to CYP1A2 phenotype caused increased levels for protein and/or activity, except one in AhRR (rs2241598), which showed an additional effect on mRNA level. This SNP is positioned in the 3′ UTR of AhRR, indicating a possible post-transcriptional regulation of the inhibitor AhRR by miRNAs and thus having a positive effect on CYP1A2 expression. The other SNP genes belonged to different regulatory pathways (basal/constitutive, inducible, and cytokine) pointing to the complex network of transcriptional regulation pathways of CYP1A2 expression. The four polymorphisms in the HNF1α gene, consisting of two pairs of strongly linked SNPs, were consistently associated with significantly increased activity and protein levels at least in the univariate analysis. Besides three intronic positions (rs1169300, rs1169306, rs735396), one SNP (rs2464196) affected an amino acid serine (S) at position 487 by change to asparagine (N), which in turn should not be damaging to the HNF1α protein, according to an analysis with PolyPhen10. All four SNPs have been included in previous studies on plasma CRP levels, and S487N was significantly associated with lower CRP and GGT levels (Reiner et al., 2008, 2009; Ridker et al., 2008) indicating a connection to inflammation signaling.
In our study CYP1A2 protein and activity was influenced by an exon SNP in IL1ß (rs1143634) affecting codon 105, but keeping amino acid phenylalanine. This SNP was included in some recent studies and was associated with diseases in Japanese (lung cancer: Kiyohara et al., 2010) and Caucasians (schizophrenia: Xu and He, 2010).
RXRα-participating heterodimers of the xenobiotic translational regulation pathways were included in this study for exploratory reasons. As discussed above, both SNPs in RXRα were associated uniformly with decreased CYP1A2 expression. Of the nuclear receptors included two SNPs in VDR (NR1I1) were significantly associated to higher protein and activity in the univariate model, both remaining significant in the multivariate model. From these the tag SNP rs1540339 is located in an intron and rs10735810 (new rs2228570) is the so-called “FokI” variation often included in pharmacogenomic studies of diseases (Guo et al., 2006; Purdue et al., 2007; Heist et al., 2008). The site is affecting the N-terminus of VDR protein altering the transcriptional start site resulting in a shortened variant with higher transcriptional activity (Uitterlinden et al., 2004). This may explain our finding of increased protein and activity of CYP1A2 associated with that polymorphism, supposing a direct action of VDR in CYP1A2 regulation at unidentified hormone response elements.
When all relevant genetic and non-genetic factors were combined in a multivariate model, they explained 42, 38, and 33% of CYP1A2 variation at activity, protein, and mRNA levels, respectively (Figure 6). While this represents a significant contribution it must be realized that over 50% of the variation in CYP1A2 remains still unexplained and particularly the proposed high heritability requires additional genetic factors for explanation. A different strategy that can be taken is by genome-wide association analysis using high-density microarray technology. This approach is principally hypothesis-free and therefore can detect entirely novel relationships (Daly, 2010; Motsinger-Reif et al., 2010). In a recent genome-wide study of liver gene expression in a liver bank comprising over 400 donors, Schadt et al. (2008) identified 3,210 expression quantitative trait loci, i.e., significant genotype–phenotype relationships. In a follow-up study, Yang et al. (2010) identified a number of novel trans-acting genes with highly significant trans-associations to CYP1A2 activity. None of them was among our selection of candidate genes. The potential role of these genes for CYP1A2 regulation awaits further elucidation.
Conclusion
We performed the first pathway-oriented approach to investigate whether polymorphisms in genes regulating the expression and function of CYP1A2 in human liver in constitutive, ligand-inducible, and pathophysiological manner can explain in part the assumed high heritability of CYP1A2 phenotype. The results are promising in that several polymorphisms in the ARNT, AhRR, HNF1α, IL1β, SRC-1, RXRα, and VDR genes seem to influence hepatic CYP1A2 in a consistent way and that explain more of the phenotypic variability than non-genetic factors including smoking, drug treatment, and inflammation. We think that this represents a significant advance for the pharmacogenomics of this important enzyme.
However, as more than 50% of the variation is still unexplained, additional genetic factors must be assumed. The identified candidate SNPs should now be assessed in larger in vivo studies to evaluate their potential clinical value.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
Comparison of SNP-frequencies to published data (dbSNP). SNP frequencies determined in our study were compared to available SNP data in dbSNP (n = 153). MAF, minor allele frequency.
Included SNPs, genotyping methods and frequencies.
| Gene | SNP rs | Allele/ position | nt | Effect | Type | MAF publisheda | MAFb | HWp | Missing values (%) | Method | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 CHR15; NM_000761 | |||||||||||
| rs2069514 | *1C | G > A | p | 0.081 | 0.007 | 1 | 0 | Tq-AD | Nakajima et al. (1999), Abnet et al. (2007) | ||
| rs2069522 | C > T | p | 0 | 0.037 | 0.173 | 0 | Maldi | Bethke et al. (2007) | |||
| rs12720461 | *1K | C > T | p | 0 | 0.003 | 1 | 0 | Maldi | Aklillu et al. (2003) | ||
| rs762551f | *1F | A > C | p | 0.308 | 0.32 | 0.455 | 0 | Maldi | Han et al. (2002) | ||
| rs72547513 | *11 | C > A | F186L | nc | No data | 0 | 0 | Maldi | Murayama et al. (2004), Pucci et al. (2007) | ||
| rs56276455 | *3 | G > A | D348N | nc | No data | 0.003 | 1 | 0 | Maldi | Zhou et al. (2009) | |
| rs72547516 | *4 | A > T | I386F | nc | No data | 0 | 0 | Maldi | Zhou et al. (2009) | ||
| rs56107638 | *7 | G > A | i | (0.003) | 0 | 0 | Maldi | Allorge et al. (2003) | |||
| rs28399424 | *6 | C > T | R431W | nc | No data | 0 | 0 | Maldi | Zhou et al. (2009) | ||
| rs2470890 | C > T | N516 | sc | 0.358 | 0.387 | 0.492 | 0 | Maldi | Gunes and Dahl (2008) | ||
| rs4886406 | A > C | 3′ | 0.292 | 0.323 | 0.34 | 4 | ILM | ||||
| CYP1A1 CHR15; NM_000499 | |||||||||||
| rs2470893 | G > A | 5′ | 0.275 | 0.294 | 0.029 | 1.3 | ILM | ||||
| rs2472299f | G > A | 5′ | 0.308 | 0.319 | 0.571 | 0.7 | ILM | ||||
| rs4646421 | C > T | p | 0.075 | 0.094 | 0.618 | 0.7 | ILM | ||||
| rs1048943 | *2B/C | A > G | I462V | cn | 0.042 | 0.027 | 0.143 | 0.7 | Tq-AD | Hayashi et al. (1991), Zhang et al. (1996) | |
| AHR (ARYL HYDROCARBON RECEPTOR) CHR7; NM_001621 | |||||||||||
| rs10247158 | A > T | 5′ | 0.152 | 0.13 | 1 | 0 | Maldi | Bin et al. (2008) | |||
| rs10250822 | T > C | 5′ | 0.2 | 0.217 | 0.81 | 0 | Maldi | Bin et al. (2008) | |||
| rs4719497 | T > C | 5′ | 0.1 | 0.108 | 1 | 1.3 | ILM | ||||
| rs2282885 | A > G | i | 0.347 | 0.413 | 0.736 | 0 | Maldi | Bin et al. (2008) | |||
| rs1476080 | T > G | i | 0.325 | 0.343 | 0.21 | 0 | Maldi | Bin et al. (2008) | |||
| rs4236290 | T > C | i | 0.125 | 0.122 | 0.697 | 2 | ILM | ||||
| rs6960165 | A > G | i | 0.196 | 0.205 | 1 | 0.7 | Maldi | Bin et al. (2008) | |||
| rs2158041 | C > T | i | 0.183 | 0.203 | 1 | 1.3 | ILM | Chen et al. (2009) | |||
| rs7811989 | G > A | i | 0.233 | 0.233 | 1 | 0 | Maldi | Bin et al. (2008) | |||
| rs2066853 | G > A | R554K | nc | 0.075 | 0.1 | 1 | 0 | Maldi | Long et al. (2006), Abnet et al. (2007), Wong et al. (2001) Cauchi et al. (2001); Merisalu et al. (2007) | ||
| rs4986826 | G > A | V570I | nc | 0 | 0 | 0.7 | Maldi | Wong et al. (2001) | |||
| AHRR(ARYL HYDROCARBON RECEPTOR REGULATOR); CHR5; NM_020731 | |||||||||||
| rs3756712 | T > G | i | 0.342 | 0.361 | 0.723 | 1.3 | ILM | ||||
| rs4957018 | A > G | i | 0.275 | 0.322 | 0.453 | 0.7 | ILM | ||||
| rs3734145 | C > T | i | 0.25 | 0.258 | 0.052 | 0.7 | ILM | ||||
| rs4957028 | T > C | i | 0.424 | 0.463 | 1 | 1.3 | ILM | ||||
| rs908114 | A > G | i | 0.383 | 0.329 | 0.853 | 2.7 | ILM | ||||
| rs2721020 | G > A | i | 0.225 | 0.232 | 1 | 0.7 | ILM | ||||
| rs7731963 | C > A | i | 0.375 | 0.411 | 0.389 | 6.7 | ILM | ||||
| rs2292596 | C > G | P189A | nc | 0.412 | 0.373 | 0.296 | 0 | Maldi | Watanabe et al. (2001) | ||
| rs34453673 | G > C | D641H | nc | 0.31 | 0.403 | 0.612 | 0 | Maldi | Cauchi et al. (2003) | ||
| rs10078 | A > C | 3′u | 0.2 | 0.218 | 0.81 | 0.7 | ILM | ||||
| rs2241598 | C > T | 3′ | 0.183 | 0.211 | 1 | 0.7 | ILM | ||||
| ARNT (HIF1β; ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSLOCATOR) CHR1; NM 001668 | |||||||||||
| rs2292166 | C > T | 5′ | 0.217 | 0.282 | 0.015 | 0.7 | ILM | ||||
| rs7412746 | C > T | 5′ | 0.475 | 0.419 | 0.737 | 1.3 | ILM | ||||
| rs11204735 | C > T | i | 0 | 0.7 | Maldi | Das et al. (2008) | |||||
| rs2134688 | A > G | i | 0.142 | 0.13 | 0.716 | 0 | Maldi | Das et al. (2008) | |||
| rs2228099e | G > C | V189 | sc | 0.35 | 0.307 | 0.255 | 0 | Maldi | Kayano et al. (2004) | ||
| rs1889740e | G > A | i | No data | 0.307 | 0.255 | 0 | Maldi | Das et al. (2008) | |||
| rs10305724 | C > T | i | 0.125 | 0.057 | 0.383 | 0.7 | ILM | ||||
| CAR (NR1I3; CONSTITUTIVE ANDROSTANE RECEPTOR) CHR1; NM_00177469 | |||||||||||
| rs6686001 | C > A | i | 0.13 | 0.143 | 0.739 | 0 | Maldi | ||||
| rs3003596 | T > C | i | 0.49 | 0.363 | 0.289 | 0 | Maldi | ||||
| C > T | i | 0.242 | 0.19 | 0.063 | 0 | Maldi | Oliver et al. (2010) | ||||
| 398T > G | T > G | V133G | nc | No data | 0 | Maldi | Ikeda et al. (2005) | ||||
| rs2307424 | C > T | P180 | sc | 0.471 | 0.311 | 0.252 | 1.3 | ILM | Oliver et al. (2010) | ||
| rs2307418 | A > C | i | 0.183 | 0.153 | 0.529 | 4 | ILM | ||||
| rs4073054 | G > T | i | 0.408 | 0.476 | 0.247 | 2 | ILM | Oliver et al. (2010) | |||
| HNF1α (TCF1; HEPATOCYTE NUCLEAR FACTOR 1 HOMEOBOX A) CHR12; NM_000545 | |||||||||||
| -58 | A > C | p | No data | 0 | 0.7 | Maldi | Gragnoli et al. (1997) | ||||
| rs1169288 | A > G | I27L | nc | 0.283 | 0.393 | 0.864 | 0 | Maldi | Holmkvist et al. (2006), Cauchi et al. (2008), Ryffel (2001) | ||
| rs1800574 | C > T | A98V | nc | 0.029 | 0.023 | 1 | 0 | Maldi | Giuffrida et al. (2009) | ||
| rs1169300 | G > A | i | 0.292 | 0.315 | 0.851 | 0.7 | ILM | Ridker et al. (2008) | |||
| rs2071190 | T > A | i | 0.22 | 0.19 | 1.0 | 0 | Maldi | Weedon et al. (2005) | |||
| rs1169302 | T > G | i | 0.45 | 0.433 | 0.318 | 0.7 | ILM | Ridker et al. (2008) | |||
| rs2464196 | G > A | S487N | nc | 0.292 | 0.317 | 1 | 0 | Maldi | Ridker et al. (2008) | ||
| rs1169306 | C > T | i | 0.375 | 0.377 | 1 | 0 | Maldi | ||||
| rs1169307 | T > C | i | 0.392 | 0.426 | 0.319 | 0.7 | ILM | ||||
| rs735396 | A > G | i | 0.383 | 0.382 | 0.727 | 1.3 | ILM | Reiner et al. (2008), Ridker et al. (2008) | |||
| HNF4α (NR2A1; HEPATOCYTE NUCLEAR FACTOR 4 ALPHA) CHR20; NM_000448 | |||||||||||
| rs4810424 | G > C | p | 0.164 | 0.157 | 0.364 | 0 | Maldi | Bagwell et al. (2005) | |||
| Sookoian et al. (2010) | |||||||||||
| rs2144908 | G > A | p | 0.183 | 0.16 | 0.539 | 0 | Maldi | Lamba et al. (2008), Muller et al. (2005), Holmkvist et al. (2008), Black et al. (2008) | |||
| rs2868094 | T > C | p | 0.317 | 0.28 | 0.316 | 0 | Maldi | ||||
| rs6031544 | C > T | p | 0.283 | 0.289 | 0.842 | 0.7 | ILM | SNP@promoter | |||
| rs3212183 | T > C | i | 0.413 | 0.45 | 0.0029 | 0 | Maldi | Muller et al. (2005) | |||
| rs6073432 | A > C | i | 0.309 | 0.307 | 0.057 | 0 | Maldi | ||||
| rs13041396 | C > G | i | 0.24 | 0.207 | 0.133 | 0 | Maldi | ||||
| c.379 | C > T | R127W | nc | No data | 0 | Maldi | Lausen et al. (2000) | ||||
| rs1800961 | C > T | T139I | nc | No data | 0.02 | 0.05 | 0 | ILM | Sookoian et al. (2010), Lu et al. (2008) | ||
| c.460 | C > T | R154X | nc | No data | 0 | Maldi | Lausen et al. (2000) | ||||
| rs6103731 | G > A | i | 0.484 | 0.423 | 0.404 | 0 | Maldi | ||||
| rs3818247 | G > T | i | 0.39 | 0.367 | 0.86 | 0 | Maldi | Love-Gregory et al. (2004) | |||
| IL1β (INTERLEUKIN 1 BETA) CHR2; NM 000576 | |||||||||||
| rs1143627 | C > T | p | 0.367 | 0.293 | 0.238 | 0 | Maldi | Liu et al. (2010) | |||
| rs1143634 | C > T | F105 | cs | 0.223 | 0.22 | 0.811 | 0 | Maldi | |||
| IL1RN (INTERLEUKIN 1 RECEPTOR ANTAGONIST) CHR2; NM_000577 | |||||||||||
| rs2637988 | G > A | i | 0.392 | 0.399 | 0.733 | 0.7 | ILM | ||||
| rs3213448 | G > A | i | 0.117 | 0.128 | 0.469 | 0.7 | ILM | ||||
| rs419598 | T > C | A57 | Cs | 0.275 | 0.287 | 0.84 | 4.7 | ILM | |||
| rs315952 | T > C | S133 | cs | 0.258 | 0.295 | 0.555 | 0.7 | ILM | |||
| rs397211 | T > C | 3′ | 0.314 | 0.342 | 0.856 | 0.7 | ILM | ||||
| NFKB1 (NUCLEAR FACTOR NF-KAPPA-B P105 SUBUNIT) CHR4; NM_003998 | |||||||||||
| rs28362491 | del > ATT G | 5′ | 0.34 | 0.361 | 0.019 | 2 | Maldi | Karban et al. (2004) | |||
| rs3774937 | T > C | p/i | 0.367 | 0.326 | 0.853 | 0.7 | ILM | ||||
| rs230530 | T > C | i | 0.364 | 0.446 | 0.403 | 2 | ILM | ||||
| rs4647992 | C > T | i | 0.067 | 0.047 | 0.344 | 0.7 | ILM | ||||
| rs230526 | C > T | i | 0.467 | 0.396 | 0.230 | 0.7 | ILM | Liu-Mares et al. (2007) | |||
| rs4648022 | C > T | i | 0.067 | 0.077 | 1 | 0.7 | ILM | Cerhan et al. (2008) | |||
| rs1598859 | T > C | i | 0.343 | 0.379 | 1 | 3.3 | ILM | Liu-Mares et al. (2007) | |||
| rs4648072 | A > G | M507V | nc | 0.075 | 0.013 | 0.049 | 0.7 | Maldi | SIFT-score 0.44 | ||
| rs1609798 | C > T | i | 0.331 | 0.329 | 1 | 0.7 | ILM | ||||
| rs997476 | C > A | 3′ | 0.1 | 0.057 | 0.068 | 1.3 | ILM | ||||
| rs10489113 | A > G | 3′ | 0.225 | 0.182 | 1 | 1.3 | ILM | ||||
| PGC1α (PPARGC1α; PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA COACTIVATOR 1-ALPHA) CHR4; NM_013261 | |||||||||||
| rs4361373 | C > T | i | 0.192 | 0.141 | 1 | 0.7 | ILM | ||||
| rs4550905 | A > G | i | 0.325 | 0.27 | 0.676 | 1.3 | ILM | ||||
| rs6448226 | A > G | i | 0.375 | 0.34 | 0.465 | 2 | ILM | ||||
| rs2970853 | G > A | i | 0.308 | 0.211 | 0.218 | 2 | ILM | Juang et al. (2010) | |||
| rs2932976 | G > A | i | 0.271 | 0.314 | 0.334 | 3.3 | ILM | ||||
| rs2970847 | C > T | T394 | sc | 0.208 | 0.153 | 1 | 0 | Maldi | Bhat et al. (2007), Juang et al. (2010) | ||
| rs8192678 | G > A | G482S | nc | 0.367 | 0.322 | 0.708 | 2.7 | Maldi | Deeb and Brunzell (2009), Wang et al. (2007) | ||
| rs2932965 | G > A | i | 0.25 | 0.171 | 0.771 | 0.7 | ILM | ||||
| rs12650562 | G > A | i | 0.435 | 0.426 | 0.74 | 0.7 | ILM | ||||
| PXR (NR1I2; PREGNANE X RECEPTOR) CHR3; NM_003889 | |||||||||||
| rs1523130 | G > A | 5′ | 0.407 | 0.423 | 0.403 | 0 | Maldi | Lamba et al. (2008) | |||
| rs3814055i | C > T | 5′ | 0.4 | 0.417 | 0.4 | 0 | Maldi* | Zhang et al. (2001) | |||
| rs1523127 | A > C | 5′ | 0.408 | 0.42 | 0.314 | 0 | Maldi* | Zhang et al. (2001), King et al. (2007) | |||
| rs2276706i | G > A | i | 0.39 | 0.417 | 0.4 | 0 | Maldi | Zhang et al. (2001) | |||
| rs4234666 | C > G | i | 0.318 | 0.233 | 1 | 0 | Maldi | ||||
| rs1403527 | T > G | i | 0.11 | 0.226 | 1 | 2.7 | ILM | ||||
| rs1403526 | A > G | i | 0.242 | 0.341 | 0.068 | 1.3 | ILM | ||||
| rs55764158 | C > A | i | 0.293 | 0.29 | 0.078 | 0 | Maldi | Lamba et al. (2008) | |||
| rs16830505 | A > G | i | 0.15 | 0.117 | 0.694 | 0.7 | ILM | ||||
| rs2472677 | C > T | i | 0.5 | 0.427 | 0.096 | 0 | Maldi | Lamba et al. (2008) | |||
| rs4440154 | C > T | i | 0.35 | 0.384 | 0.119 | 2 | ILM | ||||
| rs2461823 | T > C | i | 0.35 | 0.379 | 0.164 | 0.7 | ILM | ||||
| rs13059232 | T > C | i | 0.358 | 0.379 | 0.217 | 3.3 | Maldi | Lamba et al. (2008) | |||
| 69245C > T | C > T | i | No data | 0.077 | 1 | Maldi | Lamba et al. (2008) | ||||
| rs7643645 | A > G | i | 0.43 | 0.363 | 0.598 | 0 | Maldi | Lamba et al. (2008) | |||
| rs12721613 | *2 | C > T | P27S | nc | 0 | 0 | 0.7 | Maldi | Hustert et al. (2001) | ||
| rs12721607 | *3 | G > A | G36R | nc | 0.03 | 0.027 | 1 | 0 | Maldi | Hustert et al. (2001) | |
| rs72551372 | *10 | G > A | nc | No data | 0 | 0.7 | Maldi | Hustert et al. (2001) | |||
| rs72551374 | A > G | i | No data | 0 | 0.7 | Maldi | Hustert et al. (2001), Lim and Huang (2007) | ||||
| rs3732357 | G > A | i | 0.225 | 0.336 | 0.009 | 0.7 | ILM | ||||
| rs6785049 | A > G | i | 0.302 | 0.413 | 0.018 | 0 | Maldi* | Zhang et al. (2001) | |||
| rs2276707 | C > T | i | 0.058 | 0.22 | 0.641 | 0 | Maldi* | Zhang et al. (2001) | |||
| rs3732359 | G > A | 3′u | 0.075 | 0.277 | 0.091 | 6 | Maldi | Oleson et al. (2010) | |||
| rs1054191 | G > A | 3′u | 0.233 | 0.14 | 0.079 | 0 | Maldi | Zhang et al. (2001) | |||
| rs3814057 | T > C | 3′u | 0.188 | 0.218 | 0.81 | 0.7 | Maldi | Zhang et al. (2001) | |||
| RARα (NR1B1; RETINOIC ACID RECEPTOR ALPHA) CHR17; NM_001033603 | |||||||||||
| rs13706g | G > A | V441Id | 5′d | 0.142 | 0.094 | 0.618 | 0.7 | ILM | |||
| rs2715553 | T > C | i | No data | 0.483 | 0.87 | 1.3 | ILM | ||||
| rs9303286g | G > C | i | 0.06 | 0.093 | 0.617 | 0 | Maldi | ||||
| rs482284 | G > A | i | 0.283 | 0.267 | 1 | 0 | Maldi | ||||
| rs4890109 | G > T | i | 0.058 | 0.027 | 0.0016 | 2 | ILM | ||||
| RXRα (NR2B1; RETINOID X RECEPTOR ALPHA) CHR9; NM_002957 | |||||||||||
| rs3818740 | T > C | i | 0.225 | 0.294 | 0.423 | 4.7 | ILM | ||||
| rs3132297 | C > T | i | 0.222 | 0.157 | 1 | 0 | Maldi | ||||
| rs3118529 | T > C | i | 0.227 | 0.29 | 0.557 | 0 | Maldi | ||||
| rs4240705 | A > G | i | 0.4 | 0.369 | 0.597 | 0.7 | ILM | ||||
| rs3118570 | A > C | i | 0.18 | 0.153 | 1 | 0 | Maldi | Cauchi et al. (2003), Kölsch et al. (2009) | |||
| rs1536475 | G > A | i | 0.17 | 0.163 | 1 | 0 | Maldi | Kölsch et al. (2009) | |||
| rs3132293 | G > A | i | 0.22 | 0.209 | 0.451 | 2.7 | Maldi | Cauchi et al. (2003), Kölsch et al. (2009) | |||
| rs1805348 | G > A | A457 | sc | 0.025 | 0.023 | 1 | 0 | Maldi | |||
| SRC-1 (NCOA1; NUCLEAR RECEPTOR COACTIVATOR 1) CHR2; NM_003734 | |||||||||||
| rs995647 | A > G | i | 0.1 | 0.111 | 0.082 | 0.7 | ILM | ||||
| rs11677500 | G > A | i | 0.433 | 0.469 | 1 | 3.3 | ILM | ||||
| rs2119115 | A > C | i | 0.267 | 0.299 | 0.439 | 2 | ILM | ||||
| rs12468225 | A > C | i | 0.136 | 0.137 | 0.481 | 0 | Maldi | ||||
| rs4578807 | A > G | i | 0.133 | 0.145 | 0.512 | 1.3 | ILM | ||||
| rs4665716 | A > G | i | 0.075 | 0.055 | 0.007 | 2.7 | ILM | ||||
| rs11682130 | T > G | i | 0.433 | 0.473 | 0.514 | 1.3 | ILM | ||||
| rs6743362 | A > C | i | 0.058 | 0.034 | 0.205 | 0.7 | ILM | ||||
| rs12617941 | G > A | i | 0.075 | 0.047 | 0.274 | 0.7 | ILM | ||||
| rs6724282 | A > G | 3′ | 0.075 | 0.062 | 0.014 | 2.7 | ILM | ||||
| rs9309308 | T > C | 3′ | 0.225 | 0.297 | 0.242 | 1.3 | ILM | ||||
| USF1 (UPSTREAM STIMULATORY FACTOR 1) CHR1; NM_009053 | |||||||||||
| rs3813609 | C > G | 5′c | 0.3 | 0.373 | 0.601 | 0 | Maldi | Holzapfel et al. (2008) | |||
| rs2774279 | G > A | R873 (ARHG AP30) | 5′c | 0.38 | 0.26 | 0.286 | 0 | Maldi | Auro et al. (2008) | ||
| rs1556259 | T > C | i | 0.083 | 0.127 | 0.47 | 0 | Maldi | Holzapfel et al. (2008) | |||
| rs2774276 | C > G | i | 0.21 | 0.247 | 0.269 | 0 | Maldi | ||||
| rs2073658h | G > A | i | 0.283 | 0.363 | 0.48 | 0 | Maldi | OMIM, Meex et al. (2008) | |||
| rs3737787h | C > T | 3′u | 0.309 | 0.363 | 0.48 | 0 | Maldi | Holzapfel et al. (2008), Collings et al. (2008), Meex et al. (2008), Naukkarinen et al. (2005) | |||
| VDR (NR1I1; VITAMIN D RECEPTOR) CHR12; NM_000376 | |||||||||||
| rs4516035 | T > C | 5′ | 0.458 | 0.493 | 1 | 1.3 | ILM | ||||
| rs3890733 | C > T | i | 0.35 | 0.382 | 0.605 | 1.3 | ILM | ||||
| rs4760648 | C > T | i | 0.457 | 0.433 | 0.183 | 0.7 | ILM | ||||
| rs2853564 | T > C | i | 0.452 | 0.419 | 0.502 | 1.3 | ILM | ||||
| rs10735810(has merged to rs2228570) | T > A | M1T | nc | 0.442 | 0.34 | 0.71 | 4 | Maldi | Guo et al. (2006), | ||
| rs11574090 | C > G | L230V | nc | No data | 0 | 0.7 | Maldi | ||||
| rs2239186 | T > C | i | 0.203 | 0.229 | 0.638 | 2.7 | ILM | ||||
| rs2189480 | C > A | i | 0.398 | 0.413 | 0.731 | 4 | ILM | ||||
| rs2239179 | A > G | i | 0.408 | 0.405 | 0.497 | 1.3 | ILM | ||||
| rs1540339 | G > A | i | 0.398 | 0.391 | 0.165 | 2 | ILM | ||||
| rs7962898 | T > C | i | 0.425 | 0.497 | 0.87 | 0 | Maldi | ||||
| rs1544410 | G > A | i | 0.442 | 0.4 | 0.609 | 0 | Maldi | Purdue et al. (2007) | |||
| rs7975232 | A > C | i | 0.422 | 0.493 | 1 | 0 | Maldi | Purdue et al. (2007) | |||
| rs731236 | T > C | I352 | sc | 0.44 | 0.372 | 1 | 8.7 | Maldi | Purdue et al. (2007) | ||
i, Intronic; nc, non-synonymous coding; sc, synonymous coding; 3′u, 3′ untranslated region; 5′ upstream of mRNA; p, promoter; 3′, downstream of mRNA; Maldi, MALDI-TOF MS (mass assisted light desorption time of flight mass spectrometry) multiplexed genotyping assay; ILM, Illumina 300k Bead Chip; Tq-AD, TaqMan allelic discrimination genotyping assay (Applied Biosystems); calculation of MAF, minor allele frequency; HWp, Hardy–Weinberg p-value and missing values see Materials and Methods section.
aHapMapCEU or alternative population of Caucasian origin (dbSNP-homepage; Build 131).
bMinor allele frequency found in this study.
cOverlap to ARHGAP30 coding region.
dLocated in CDC6 (cn).
e,f,g,h,iSNPs linked pairwise 100% within the one gene.
*MALDI-TOF genotype data provided by E. Schaeffeler, IKP, Stuttgart
Acknowledgments
We gratefully acknowledge the expert technical assistance of Igor Liebermann and Britta Klumpp. We also thank Dr. Elke Schaeffeler for providing selected PXR genotype data and Alice Klär for help with data normalization. This study was supported by the German Federal Ministry of Education and Research (Virtual Liver grant 0315755 and grant 03 IS 2061C) and the Robert Bosch Foundation, Stuttgart, Germany.
Footnotes
Abbreviations
AhR, aryl hydrocarbon receptor; AhRR, aryl hydrocarbon receptor regulator; AHRE, aryl hydrocarbon response element; AIP, aryl hydrocarbon interacting protein; AP1, activator protein 1; ARNT, aryl hydrocarbon receptor nuclear translocator; CAR, constitutive androstene receptor alpha; CAT, CCAAT box; E-box, enhancer element where factors of the basic helix-loop-helix family bind; HNF1α, hepatocyte nuclear factor 1 alpha; HNF4α, hepatocyte nuclear factor 4 alpha; hsp90, heat shock protein 90; IL1β, interleukin 1 beta; IL1RN, interleukin 1 receptor antagonist; NF1-like, nuclear factor 1-like binding motif; NFkB1, nuclear factor kappa-light-chain-enhancer of activated B-cells; PGC1α, peroxisomal proliferator activated receptor coactivator 1 alpha; PXR, pregnane X receptor; RXRα, retinoid X receptor alpha; SP1, specificity protein 1 (member of the sp/KLF family of transcription factors); SRC-1, steroid receptor coactivator 1; TATA, TATA box binding motif; TNFα, tumor necrosis factor alpha; USF, upstream stimulating factor; VDR, vitamin D receptor; XRE-like, xenobiotic response element
References
- Abdel-Razzak Z., Loyer P., Fautrel A., Gautier J. C., Corcos L., Turlin B., Beaune P., Guillouzo A. (1993). Cytokines down-regulate expression of major cytochrome P-450 enzymes in adult human hepatocytes in primary culture. Mol. Pharmacol. 44, 707–715 [PubMed] [Google Scholar]
- Bahlo M., Stankovich J., Danoy P., Hickey P. F., Taylor B. V., Browning S. R., Brown M. A., Rubio J. P. (2010). Saliva-derived DNA performs well in large-scale, high-density single-nucleotide polymorphism microarray studies. Cancer Epidemiol. Biomarkers Prev. 19, 794–798 10.1158/1055-9965.EPI-09-0812 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol. 57, 289–300 [Google Scholar]
- Bertilsson L., Carrillo J. A., Dahl M. L., Llerena A., Alm C., Bondesson U., Lindström L., Rodriguez de la Rubia I., Ramos S., Benitez J. (1994). Clozapine disposition covaries with CYP1A2 activity determined by a caffeine test. Br. J. Clin. Pharmacol. 38, 471–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock K. W., Schrenk D., Forster A., Griese E. U., Mörike K., Brockmeier D., Eichelbaum M. (1994). The influence of environmental and genetic factors on CYP2D6, CYP1A2 and UDP-glucuronosyltransferases in man using sparteine, caffeine, and paracetamol as probes. Pharmacogenetics 4, 209–218 10.1097/00008571-199408000-00005 [DOI] [PubMed] [Google Scholar]
- Boobis A. R., Lynch A. M., Murray S., de la Torre R., Solans A., Farré M., Segura J., Gooderham N. J., Davies D. S. (1994). CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 54, 89–94 [PubMed] [Google Scholar]
- Bramow S., Ott P., Thomsen Nielsen F., Bangert K., Tygstrup N., Dalhoff K. (2001). Cholestasis and regulation of genes related to drug metabolism and biliary transport in rat liver following treatment with cyclosporine A and sirolimus (Rapamycin). Pharmacol. Toxicol. 89, 133–139 10.1034/j.1600-0773.2001.d01-147.x [DOI] [PubMed] [Google Scholar]
- Butler M. A., Iwasaki M., Guengerich F. P., Kadlubar F. F. (1989). Human cytochrome P-450PA (P-450IA2), the phenacetin O-deethylase, is primarily responsible for the hepatic 3-demethylation of caffeine and N-oxidation of carcinogenic arylamines. Proc. Natl. Acad. Sci. U.S.A. 86, 7696–7700 10.1073/pnas.86.20.7696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D., Cauffiez C., Allorge D., Lo-Guidice J. M., Lhermitte M., Lafitte J. J., Broly F. (2001). Five novel natural allelic variants-951A>C, 1042G>A (D348N), 1156A>T (I386F), 1217G>A (C406Y) and 1291C>T (C431Y)-of the human CYP1A2 gene in a French Caucasian population. Hum. Mutat. 17, 355–356 [PubMed] [Google Scholar]
- Chung I., Bresnick E. (1997). Identification of positive and negative regulatory elements of the human cytochrome P4501A2 (CYP1A2) gene. Arch. Biochem. Biophys. 338, 220–226 10.1006/abbi.1996.9815 [DOI] [PubMed] [Google Scholar]
- Congiu M., Mashford M. L., Slavin J. L., Desmond P. V. (2009). Coordinate regulation of metabolic enzymes and transporters by nuclear transcription factors in human liver disease. J. Gastroenterol. Hepatol. 24, 1038–1044 10.1111/j.1440-1746.2009.05800.x [DOI] [PubMed] [Google Scholar]
- Daly A. K. (2010). Genome-wide association studies in pharmacogenomics. Nat. Rev. Genet. 11, 241–246 10.1038/nrg2751 [DOI] [PubMed] [Google Scholar]
- Das S. K., Sharma N. K., Chu W. S., Wang H., Elbein S. C. (2008). Aryl hydrocarbon receptor nuclear translocator (ARNT) gene as a positional and functional candidate for type 2 diabetes and prediabetic intermediate traits: mutation detection, case-control studies, and gene expression analysis. BMC Med. Genet. 9, 16. 10.1186/1471-2350-9-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton D. L., Gallagher E. P., Bammler T. K., Kunze K. L. (1995). Role of cytochrome P4501A2 in chemical carcinogenesis: implications for human variability in expression and enzyme activity. Pharmacogenetics 5, 259–274 10.1097/00008571-199510000-00001 [DOI] [PubMed] [Google Scholar]
- Eiermann B., Engel G., Johansson I., Zanger U. M., Bertilsson L. (1997). The involvement of CYP1A2 and CYP3A4 in the metabolism of clozapine. Br. J. Clin. Pharmacol. 44, 439–446 10.1046/j.1365-2125.1997.t01-1-00605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feidt D. M., Klein K., Hofmann U., Riedmaier S., Knobeloch D., Thasler W. E., Weiss T. S., Schwab M., Zanger U. M. (2010). Profiling induction of cytochrome P450 enzyme activity by statins using a new LC-MS/MS cocktail assay in human hepatocytes. Drug Metab. Dispos. 38, 1589–1597 10.1124/dmd.110.033886 [DOI] [PubMed] [Google Scholar]
- Fuhr U., Jetter A., Kirchheiner J. (2007). Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the “cocktail” approach. Clin. Pharmacol. Ther. 81, 270–283 10.1038/sj.clpt.6100050 [DOI] [PubMed] [Google Scholar]
- Fukushima S., Okuno H., Shibatani N., Nakahashi Y., Seki T., Okazaki K. (2008). Effect of biliary obstruction and internal biliary drainage on hepatic cytochrome P450 isozymes in rats. World J. Gastroenterol. 14, 2556–2560 10.3748/wjg.14.2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghotbi R., Christensen M., Roh H., Ingelman-Sundberg M., Aklillu E., Bertilsson L. (2007). Comparisons of CYP1A2 genetic polymorphisms, enzyme activity and the genotype-phenotype relationship in Swedes and Koreans. Eur. J. Clin. Pharmacol. 63, 537–546 10.1007/s00228-007-0288-2 [DOI] [PubMed] [Google Scholar]
- Ghotbi R., Gomez A., Milani L., Tybring G., Syvänen A., Bertilsson L., Ingelman-Sundberg M., Aklillu E. (2009). Allele-specific expression and gene methylation in the control of CYP1A2 mRNA level in human livers. Pharmacogenomics J. 9, 208–217 10.1038/tpj.2009.4 [DOI] [PubMed] [Google Scholar]
- Gomes A. M., Winter S., Klein K., Turpeinen M., Schaeffeler E., Schwab M., Zanger U. M. (2009). Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10, 579–599 10.2217/pgs.09.7 [DOI] [PubMed] [Google Scholar]
- Gunes A., Dahl M. (2008). Variation in CYP1A2 activity and its clinical implications: influence of environmental factors and genetic polymorphisms. Pharmacogenomics 9, 625–637 10.2217/14622416.9.5.625 [DOI] [PubMed] [Google Scholar]
- Guo S., Magnuson V. L., Schiller J. J., Wang X., Wu Y., Ghosh S. (2006). Meta-analysis of vitamin D receptor polymorphisms and type 1 diabetes: a HuGE review of genetic association studies. Am. J. Epidemiol. 164, 711–724 10.1093/aje/kwj278 [DOI] [PubMed] [Google Scholar]
- Ha H. R., Chen J., Freiburghaus A. U., Follath F. (1995). Metabolism of theophylline by cDNA-expressed human cytochromes P-450. Br. J. Clin. Pharmacol. 39, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heist R. S., Zhou W., Wang Z., Liu G., Neuberg D., Su L., Asomaning K., Hollis B. W., Lynch T. J., Wain J. C., Giovannucci E., Christiani D. C. (2008). Circulating 25-hydroxyvitamin D, VDR polymorphisms, and survival in advanced non-small-cell lung cancer. J. Clin. Oncol. 26, 5596–5602 10.1200/JCO.2008.18.0406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Blievernicht J., Klein K., Saussele T., Schaeffeler E., Schwab M., Zanger U. (2008). Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J. Pharmacol. Exp. Ther. 325, 284–292 10.1124/jpet.107.133306 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Dragin N., Jorge-Nebert L. F., Martin M. V., Guengerich F. P., Aklillu E., Ingelman-Sundberg M., Hammons G. J., Lyn-Cook B. D., Kadlubar F. F., Saldana S. N., Sorter M., Vinks A. A., Nassr N., von Richter O., Jin L., Nebert D. W. (2006). Search for an association between the human CYP1A2 genotype and CYP1A2 metabolic phenotype. Pharmacogenet. Genomics 16, 359–367 10.1097/01.fpc.0000204994.99429.46 [DOI] [PubMed] [Google Scholar]
- Kim D., Guengerich F. P. (2005). Cytochrome P450 activation of arylamines and heterocyclic amines. Annu. Rev. Pharmacol. Toxicol. 45, 27–49 10.1146/annurev.pharmtox.45.120403.100010 [DOI] [PubMed] [Google Scholar]
- Kiyohara C., Horiuchi T., Takayama K., Nakanishi Y. (2010). IL1B rs1143634 polymorphism, cigarette smoking, alcohol use, and lung cancer risk in a Japanese population. J. Thorac. Oncol. 5, 299–304 10.1097/JTO.0b013e3181c8cae3 [DOI] [PubMed] [Google Scholar]
- Lobo E. D., Bergstrom R. F., Reddy S., Quinlan T., Chappell J., Hong Q., Ring B., Knadler M. P. (2008). In vitro and in vivo evaluations of cytochrome P450 1A2 interactions with duloxetine. Clin. Pharmacokinet. 47, 191–202 10.2165/00003088-200847030-00005 [DOI] [PubMed] [Google Scholar]
- Martínez-Jiménez C. P., Castell J. V., Gómez-Lechón M. J., Jover R. (2006). Transcriptional activation of CYP2C9, CYP1A1, and CYP1A2 by hepatocyte nuclear factor 4alpha requires coactivators peroxisomal proliferator activated receptor-gamma coactivator 1alpha and steroid receptor coactivator 1. Mol. Pharmacol. 70, 1681–1692 10.1124/mol.106.025403 [DOI] [PubMed] [Google Scholar]
- Motsinger-Reif A. A., Jorgenson E., Relling M. V., Kroetz D. L., Weinshilboum R., Cox N. J., Roden D. M. (2010). Genome-wide association studies in pharmacogenomics: successes and lessons. Pharmacogenet. Genomics 1, 10.1097/FPC.0b013e32833d7b45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntané-Relat J., Ourlin J. C., Domergue J., Maurel P. (1995). Differential effects of cytokines on the inducible expression of CYP1A1, CYP1A2, and CYP3A4 in human hepatocytes in primary culture. Hepatology 22, 1143–1153 [PubMed] [Google Scholar]
- Murayama N., Soyama A., Saito Y., Nakajima Y., Komamura K., Ueno K., Kamakura S., Kitakaze M., Kimura H., Goto Y., Saitoh O., Katoh M., Ohnuma T., Kawai M., Sugai K., Ohtsuki T., Suzuki C., Minami N., Ozawa S., Sawada J. (2004). Six novel nonsynonymous CYP1A2 gene polymorphisms: catalytic activities of the naturally occurring variant enzymes. J. Pharmacol. Exp. Ther. 308, 300–306 10.1124/jpet.103.055798 [DOI] [PubMed] [Google Scholar]
- Narvaez M. J., Anderson G. R., Pickwell G. V., Quattrochi L. C. (2005). Characterization of adjacent E-box and nuclear factor 1-like DNA binding sequence in the human CYP1A2 promoter. J. Biochem. Mol. Toxicol. 19, 78–86 10.1002/jbt.20063 [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Dalton T. P. (2006). The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer 6, 947–960 10.1038/nrc2015 [DOI] [PubMed] [Google Scholar]
- Nies A. T., Koepsell H., Winter S., Burk O., Klein K., Kerb R., Zanger U. M., Keppler D., Schwab M., Schaeffeler E. (2009). Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology 50, 1227–1240 10.1002/hep.23103 [DOI] [PubMed] [Google Scholar]
- Okey A. B., Franc M. A., Moffat I. D., Tijet N., Boutros P. C., Korkalainen M., Tuomisto J., Pohjanvirta R. (2005). Toxicological implications of polymorphisms in receptors for xenobiotic chemicals: the case of the aryl hydrocarbon receptor. Toxicol. Appl. Pharmacol. 207, 43–51 10.1016/j.taap.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Okino S. T., Quattrochi L. C., Pookot D., Iwahashi M., Dahiya R. (2007). A dioxin-responsive enhancer 3′ of the human CYP1A2 gene. Mol. Pharmacol. 72, 1457–1465 10.1124/mol.107.039826 [DOI] [PubMed] [Google Scholar]
- Pascussi J., Gerbal-Chaloin S., Duret C., Daujat-Chavanieu M., Vilarem M., Maurel P. (2008). The tangle of nuclear receptors that controls xenobiotic metabolism and transport: crosstalk and consequences. Annu. Rev. Pharmacol. Toxicol. 48, 1–32 10.1146/annurev.pharmtox.47.120505.105349 [DOI] [PubMed] [Google Scholar]
- Pelkonen O., Turpeinen M., Hakkola J., Honkakoski P., Hukkanen J., Raunio H. (2008). Inhibition and induction of human cytochrome P450 enzymes: current status. Arch. Toxicol. 82, 667–715 10.1007/s00204-008-0332-8 [DOI] [PubMed] [Google Scholar]
- Petrulis J. R., Hord N. G., Perdew G. H. (2000). Subcellular localization of the aryl hydrocarbon receptor is modulated by the immunophilin homolog hepatitis B virus X-associated protein 2. J. Biol. Chem. 275, 37448–37453 10.1074/jbc.M006873200 [DOI] [PubMed] [Google Scholar]
- Purdue M. P., Lan Q., Kricker A., Vajdic C. M., Rothman N., Armstrong B. K. (2007). Vitamin D receptor gene polymorphisms and risk of non-Hodgkin's lymphoma. Haematologica 92, 1145–1146 10.3324/haematol.11053 [DOI] [PubMed] [Google Scholar]
- Quattrochi L. C., Vu T., Tukey R. H. (1994). The human CYP1A2 gene and induction by 3-methylcholanthrene. A region of DNA that supports AH-receptor binding and promoter-specific induction. J. Biol. Chem. 269, 6949–6954 [PubMed] [Google Scholar]
- Raaska K., Raitasuo V., Arstila M., Neuvonen P. J. (2002). Bacterial pneumonia can increase serum concentration of clozapine. Eur. J. Clin. Pharmacol. 58, 321–322 10.1007/s00228-002-0486-x [DOI] [PubMed] [Google Scholar]
- Rasmussen B. B., Brix T. H., Kyvik K. O., Brøsen K. (2002). The interindividual differences in the 3-demethylation of caffeine alias CYP1A2 is determined by both genetic and environmental factors. Pharmacogenetics 12, 473–478 10.1097/00008571-200208000-00008 [DOI] [PubMed] [Google Scholar]
- Rasmussen B. B., Brøsen K. (1996). Determination of urinary metabolites of caffeine for the assessment of cytochrome P4501A2, xanthine oxidase, and N-acetyltransferase activity in humans. Ther. Drug. Monit. 18, 254–262 10.1097/00007691-199606000-00006 [DOI] [PubMed] [Google Scholar]
- Reiner A. P., Barber M. J., Guan Y., Ridker P. M., Lange L. A., Chasman D. I., Walston J. D., Cooper G. M., Jenny N. S., Rieder M. J., Durda J. P., Smith J. D., Novembre J., Tracy R. P., Rotter J. I., Stephens M., Nickerson D. A., Krauss R. M. (2008). Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 82, 1193–1201 10.1016/j.ajhg.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A. P., Gross M. D., Carlson C. S., Bielinski S. J., Lange L. A., Fornage M., Jenny N. S., Walston J., Tracy R. P., Williams O. D., Jacobs D. R., Nickerson D. A. (2009). Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older European-American adults: the Coronary Artery Risk Development in Young Adults Study and The Cardiovascular Health Study. Circ. Cardiovasc. Genet. 2, 244–254 10.1161/CIRCGENETICS.108.839506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling M. V., Lin J. S., Ayers G. D., Evans W. E. (1992). Racial and gender differences in N-acetyltransferase, xanthine oxidase, and CYP1A2 activities. Clin. Pharmacol. Ther 52, 643–658 10.1038/clpt.1992.203 [DOI] [PubMed] [Google Scholar]
- Ridker P. M., Pare G., Parker A., Zee R. Y. L., Danik J. S., Buring J. E., Kwiatkowski D., Cook N. R., Miletich J. P., Chasman D. I. (2008). Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am. J. Hum. Genet. 82, 1185–1192 10.1016/j.ajhg.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost K. L., Brösicke H., Brockmöller J., Scheffler M., Helge H., Roots I. (1992). Increase of cytochrome P450IA2 activity by omeprazole: evidence by the 13C-[N-3-methyl]-caffeine breath test in poor and extensive metabolizers of S-mephenytoin. Clin. Pharmacol. Ther. 52, 170–180 10.1038/clpt.1992.126 [DOI] [PubMed] [Google Scholar]
- Scandlyn M. J., Stuart E. C., Rosengren R. J. (2008). Sex-specific differences in CYP450 isoforms in humans. Expert Opin. Drug Metab. Toxicol. 4, 413–424 10.1517/17425255.4.4.413 [DOI] [PubMed] [Google Scholar]
- Schadt E. E., Molony C., Chudin E., Hao K., Yang X., Lum P. Y., Kasarskis A., Zhang B., Wang S., Suver C., Zhu J., Millstein J., Sieberts S., Lamb J., GuhaThakurta D., Derry J., Storey J. D., Avila-Campillo I., Kruger M. J., Johnson J. M., Rohl C. A., van Nas A., Mehrabian M., Drake T. A., Lusis A. J., Smith R. C., Guengerich F. P., Strom S. C., Schuetz E., Rushmore T. H., Ulrich R. (2008). Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 6, e107. 10.1371/journal.pbio.0060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweikl H., Taylor J. A., Kitareewan S., Linko P., Nagorney D., Goldstein J. A. (1993). Expression of CYP1A1 and CYP1A2 genes in human liver. Pharmacogenetics 3, 239–249 10.1097/00008571-199310000-00003 [DOI] [PubMed] [Google Scholar]
- Sesardic D., Boobis A. R., Edwards R. J., Davies D. S. (1988). A form of cytochrome P450 in man, orthologous to form d in the rat, catalyses the O-deethylation of phenacetin and is inducible by cigarette smoking. Br. J. Clin. Pharmacol. 26, 363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Huang J., Fitch K. R., Truong V. B., Kirby A., Chen W., Zhang J., Liu G., McCarroll S. A., Jones K. W., Shapero M. H. (2008). Improved detection of global copy number variation using high density, non-polymorphic oligonucleotide probes. BMC Genet. 9, 27. 10.1186/1471-2156-9-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Yamazaki H., Mimura M., Inui Y., Guengerich F. P. (1994). Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 270, 414–423 [PubMed] [Google Scholar]
- Tian Y. (2009). Ah receptor and NF-kappaB interplay on the stage of epigenome. Biochem. Pharmacol. 77, 670–680 10.1016/j.bcp.2008.10.023 [DOI] [PubMed] [Google Scholar]
- Tian Y., Ke S., Denison M. S., Rabson A. B., Gallo M. A. (1999). Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J. Biol. Chem. 274, 510–515 10.1074/jbc.274.1.510 [DOI] [PubMed] [Google Scholar]
- Turpeinen M., Hofmann U., Klein K., Mürdter T., Schwab M., Zanger U. M. (2009). A predominate role of CYP1A2 for the metabolism of nabumetone to the active metabolite, 6-methoxy-2-naphthylacetic acid, in human liver microsomes. Drug Metab. Dispos. 37, 1017–1024 10.1124/dmd.108.025700 [DOI] [PubMed] [Google Scholar]
- Uitterlinden A. G., Fang Y., Van Meurs J. B. J., Pols H. A. P., Van Leeuwen J. P. T. M. (2004). Genetics and biology of vitamin D receptor polymorphisms. Gene 338, 143–156 [DOI] [PubMed] [Google Scholar]
- Vrzal R., Ulrichová J., Dvorák Z. (2004). Aromatic hydrocarbon receptor status in the metabolism of xenobiotics under normal and pathophysiological conditions. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 148, 3–10 [PubMed] [Google Scholar]
- Wigginton J. E., Cutler D. J., Abecasis G. R. (2005). A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 76, 887–893 10.1086/429864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbold R., Klein K., Burk O., Nussler A., Neuhaus P., Eichelbaum M., Schwab M., Zanger U. (2003). Sex is a major determinant of CYP3A4 expression in human liver. Hepatology 38, 978–988 [DOI] [PubMed] [Google Scholar]
- Xu M., He L. (2010). Convergent evidence shows a positive association of interleukin-1 gene complex locus with susceptibility to schizophrenia in the Caucasian population. Schizophr. Res. 120, 131–142 10.1016/j.schres.2010.02.1031 [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang B., Molony C., Chudin E., Hao K., Zhu J., Gaedigk A., Suver C., Zhong H., Leeder J. S., Guengerich F. P., Strom S. C., Schuetz E., Rushmore T. H., Ulrich R. G., Slatter J. G., Schadt E. E., Kasarskis A., Lum P. Y. (2010). Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 20, 1020–1036 10.1101/gr.103341.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger U., Turpeinen M., Klein K., Schwab M. (2008). Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 392, 1093–1108 10.1007/s00216-008-2291-6 [DOI] [PubMed] [Google Scholar]
- Zanger U. M., Fischer J., Raimundo S., Stüven T., Evert B. O., Schwab M., Eichelbaum M. (2001). Comprehensive analysis of the genetic factors determining expression and function of hepatic CYP2D6. Pharmacogenetics 11, 573–585 10.1097/00008571-200110000-00004 [DOI] [PubMed] [Google Scholar]
- Zhou H., Josephy P. D., Kim D., Guengerich F. P. (2004). Functional characterization of four allelic variants of human cytochrome P450 1A2. Arch. Biochem. Biophys. 422, 23–30 10.1016/j.abb.2003.11.019 [DOI] [PubMed] [Google Scholar]
- Zhou M., Maitra S. R., Wang P. (2008). The potential role of transcription factor aryl hydrocarbon receptor in downregulation of hepatic cytochrome P-450 during sepsis. Int. J. Mol. Med. 21, 423–428 [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Chan E., Zhou Z., Xue C. C., Lai X., Duan W. (2009). Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr. Drug Metab. 10, 713–729 10.2174/138920009789895552 [DOI] [PubMed] [Google Scholar]
References
- Abnet C. C., Fagundes R. B., Strickland P. T., Kamangar F., Roth M. J., Taylor P. R., Dawsey S. M. (2007). The influence of genetic polymorphisms in Ahr, CYP1A1, CYP1A2, CYP1B1, GST M1, GST T1 and UGT1A1 on urine 1-hydroxypyrene glucuronide concentrations in healthy subjects from Rio Grande do Sul, Brazil. Carcinogenesis 28, 112–117 10.1093/carcin/bgl131 [DOI] [PubMed] [Google Scholar]
- Aklillu E., Carrillo J. A., Makonnen E., Hellman K., Pitarque M., Bertilsson L., Ingelman-Sundberg M. (2003). Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1. Mol. Pharmacol. 64, 659–669 10.1124/mol.64.3.659 [DOI] [PubMed] [Google Scholar]
- Allorge D., Chevalier D., Lo-Guidice J., Cauffiez C., Suard F., Baumann P., Eap C. B., Broly F. (2003). Identification of a novel splice-site mutation in the CYP1A2 gene. Br. J. Clin. Pharmacol. 56, 341–344 10.1046/j.1365-2125.2003.01858.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auro K., Kristiansson K., Zethelius B., Berne C., Lannfelt L., Taskinen M., Jauhiainen M., Perola M., Peltonen L., Syvänen A. (2008). USF1 gene variants contribute to metabolic traits in men in a longitudinal 32-year follow-up study. Diabetologia 51, 464–472 10.1007/s00125-007-0892-9 [DOI] [PubMed] [Google Scholar]
- Bagwell A. M., Bento J. L., Mychaleckyj J. C., Freedman B. I., Langefeld C. D., Bowden D. W. (2005). Genetic analysis of HNF4A polymorphisms in Caucasian-American type 2 diabetes. Diabetes 54, 1185–1190 10.2337/diabetes.54.4.1185 [DOI] [PubMed] [Google Scholar]
- Bethke L., Webb E., Sellick G., Rudd M., Penegar S., Withey L., Qureshi M., Houlston R. (2007). Polymorphisms in the cytochrome P450 genes CYP1A2, CYP1B1, CYP3A4, CYP3A5, CYP11A1, CYP17A1, CYP19A1 and colorectal cancer risk. BMC Cancer 7, 123. 10.1186/1471-2407-7-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat A., Koul A., Rai E., Sharma S., Dhar M. K., Bamezai R. N. K. (2007). PGC-1alpha Thr394Thr and Gly482Ser variants are significantly associated with T2DM in two North Indian populations: a replicate case-control study. Hum. Genet. 121, 609–614 10.1007/s00439-007-0352-0 [DOI] [PubMed] [Google Scholar]
- Bin P., Leng S., Cheng J., Dai Y., Huang C., Pan Z., Niu Y., Duan H., Li H., Liu Q., Chen W., Zheng Y. (2008). Association of aryl hydrocarbon receptor gene polymorphisms and urinary 1-hydroxypyrene in polycyclic aromatic hydrocarbon-exposed workers. Cancer Epidemiol. Biomarkers Prev. 17, 1702–1708 10.1158/1055-9965.EPI-07-2812 [DOI] [PubMed] [Google Scholar]
- Black M. H., Fingerlin T. E., Allayee H., Zhang W., Xiang A. H., Trigo E., Hartiala J., Lehtinen A. B., Haffner S. M., Bergman R. N., McEachin R. C., Kjos S. L., Lawrence J. M., Buchanan T. A., Watanabe R. M. (2008). Evidence of interaction between PPARG2 and HNF4A contributing to variation in insulin sensitivity in Mexican Americans. Diabetes 57, 1048–1056 10.2337/db07-0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchi S., Nead K. T., Choquet H., Horber F., Potoczna N., Balkau B., Marre M., Charpentier G., Froguel P., Meyre D. (2008). The genetic susceptibility to type 2 diabetes may be modulated by obesity status: implications for association studies. BMC Med. Genet. 9, 45. 10.1186/1471-2350-9-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchi S., Stücker I., Cénée S., Kremers P., Beaune P., Massaad-Massade L. (2003). Structure and polymorphisms of human aryl hydrocarbon receptor repressor (AhRR) gene in a French population: relationship with CYP1A1 inducibility and lung cancer. Pharmacogenetics 13, 339–347 10.1097/00008571-200306000-00005 [DOI] [PubMed] [Google Scholar]
- Cauchi S., Stücker I., Solas C., Laurent-Puig P., Cénée S., Hémon D., Jacquet M., Kremers P., Beaune P., Massaad-Massade L. (2001). Polymorphisms of human aryl hydrocarbon receptor (AhR) gene in a French population: relationship with CYP1A1 inducibility and lung cancer. Carcinogenesis 22, 1819–1824 10.1093/carcin/22.11.1819 [DOI] [PubMed] [Google Scholar]
- Cerhan J. R., Liu-Mares W., Fredericksen Z. S., Novak A. J., Cunningham J. M., Kay N. E., Dogan A., Liebow M., Wang A. H., Call T. G., Habermann T. M., Ansell S. M., Slager S. L. (2008). Genetic variation in tumor necrosis factor and the nuclear factor-kappaB canonical pathway and risk of non-Hodgkin's lymphoma. Cancer Epidemiol. Biomarkers Prev. 17, 3161–3169 10.1158/1055-9965.EPI-08-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Tian T., Wang H., Liu H., Hu Z., Wang Y., Liu Y., Ma H., Fan W., Miao R., Sun W., Wang Y., Qian J., Jin L., Wei Q., Shen H., Huang W., Lu D. (2009). Association of human aryl hydrocarbon receptor gene polymorphisms with risk of lung cancer among cigarette smokers in a Chinese population. Pharmacogenet. Genomics 19, 25–34 10.1097/FPC.0b013e328316d8d8 [DOI] [PubMed] [Google Scholar]
- Collings A., Höyssä S., Fan M., Kähönen M., Hutri-Kähönen N., Marniemi J., Juonala M., Viikari J. S. A., Raitakari O. T., Lehtimäki T. J. (2008). Allelic variants of upstream transcription factor 1 associate with carotid artery intima-media thickness: the Cardiovascular Risk in Young Finns study. Circ. J. 72, 1158–1164 10.1253/circj.72.1158 [DOI] [PubMed] [Google Scholar]
- Das S. K., Sharma N. K., Chu W. S., Wang H., Elbein S. C. (2008). Aryl hydrocarbon receptor nuclear translocator (ARNT) gene as a positional and functional candidate for type 2 diabetes and prediabetic intermediate traits: mutation detection, case-control studies, and gene expression analysis. BMC Med. Genet. 9, 16. 10.1186/1471-2350-9-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb S. S., Brunzell J. D. (2009). The role of the PGC1alpha Gly482Ser polymorphism in weight gain due to intensive diabetes therapy. PPAR Res. 2009, 649286. 10.1155/2009/649286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrida F. M. A., Furuzawa G. K., Kasamatsu T. S., Oliveira M. M., Reis A. F., Dib S. A. (2009). HNF1A gene polymorphisms and cardiovascular risk factors in individuals with late-onset autosomal dominant diabetes: a cross-sectional study. Cardiovasc. Diabetol. 8, 28. 10.1186/1475-2840-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragnoli C., Lindner T., Cockburn B. N., Kaisaki P. J., Gragnoli F., Marozzi G., Bell G. I. (1997). Maturity-onset diabetes of the young due to a mutation in the hepatocyte nuclear factor-4 alpha binding site in the promoter of the hepatocyte nuclear factor-1 alpha gene. Diabetes 46, 1648–1651 10.2337/diabetes.46.10.1648 [DOI] [PubMed] [Google Scholar]
- Gunes A., Dahl M. (2008). Variation in CYP1A2 activity and its clinical implications: influence of environmental factors and genetic polymorphisms. Pharmacogenomics 9, 625–637 10.2217/14622416.9.5.625 [DOI] [PubMed] [Google Scholar]
- Guo S., Magnuson V. L., Schiller J. J., Wang X., Wu Y., Ghosh S. (2006). Meta-analysis of vitamin D receptor polymorphisms and type 1 diabetes: a HuGE review of genetic association studies. Am. J. Epidemiol. 164, 711–724 10.1093/aje/kwj278 [DOI] [PubMed] [Google Scholar]
- Han X., Ouyang D., Chen X., Shu Y., Jiang C., Tan Z., Zhou H. (2002). Inducibility of CYP1A2 by omeprazole in vivo related to the genetic polymorphism of CYP1A2. Br. J. Clin. Pharmacol. 54, 540–543 10.1046/j.1365-2125.2002.01686.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S. I., Watanabe J., Nakachi K., Kawajiri K. (1991). PCR detection of an A/G polymorphism within exon 7 of the CYP1A1 gene. Nucleic Acids Res. 19, 4797. 10.1093/nar/19.17.4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmkvist J., Almgren P., Lyssenko V., Lindgren C. M., Eriksson K., Isomaa B., Tuomi T., Nilsson P., Groop L. (2008). Common variants in maturity-onset diabetes of the young genes and future risk of type 2 diabetes. Diabetes 57, 1738–1744 10.2337/db06-1464 [DOI] [PubMed] [Google Scholar]
- Holmkvist J., Cervin C., Lyssenko V., Winckler W., Anevski D., Cilio C., Almgren P., Berglund G., Nilsson P., Tuomi T., Lindgren C. M., Altshuler D., Groop L. (2006). Common variants in HNF-1 alpha and risk of type 2 diabetes. Diabetologia 49, 2882–2891 10.1007/s00125-006-0450-x [DOI] [PubMed] [Google Scholar]
- Holzapfel C., Baumert J., Grallert H., Müller A. M., Thorand B., Khuseyinova N., Herder C., Meisinger C., Hauner H., Wichmann H. E., Koenig W., Illig T., Klopp N. (2008). Genetic variants in the USF1 gene are associated with low-density lipoprotein cholesterol levels and incident type 2 diabetes mellitus in women: results from the MONICA/KORA Augsburg case-cohort study, 1984–2002. Eur. J. Endocrinol. 159, 407–416 10.1530/EJE-08-0356 [DOI] [PubMed] [Google Scholar]
- Hustert E., Zibat A., Presecan-Siedel E., Eiselt R., Mueller R., Fuss C., Brehm I., Brinkmann U., Eichelbaum M., Wojnowski L., Burk O. (2001). Natural protein variants of pregnane X receptor with altered transactivation activity toward CYP3A4. Drug Metab. Dispos. 29, 1454–1459 [PubMed] [Google Scholar]
- Ikeda S., Kurose K., Jinno H., Sai K., Ozawa S., Hasegawa R., Komamura K., Kotake T., Morishita H., Kamakura S., Kitakaze M., Tomoike H., Tamura T., Yamamoto N., Kunitoh H., Yamada Y., Ohe Y., Shimada Y., Shirao K., Kubota K., Minami H., Ohtsu A., Yoshida T., Saijo N., Saito Y., Sawada J. (2005). Functional analysis of four naturally occurring variants of human constitutive androstane receptor. Mol. Genet. Metab. 86, 314–319 10.1016/j.ymgme.2005.05.011 [DOI] [PubMed] [Google Scholar]
- Juang J. J., de Las Fuentes L., Waggoner A. D., Gu C. C., Dávila-Román V. G. (2010). Association and interaction of PPAR-complex gene variants with latent traits of left ventricular diastolic function. BMC Med. Genet. 11, 65. 10.1186/1471-2350-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban A. S., Okazaki T., Panhuysen C. I. M., Gallegos T., Potter J. J., Bailey-Wilson J. E., Silverberg M. S., Duerr R. H., Cho J. H., Gregersen P. K., Wu Y., Achkar J., Dassopoulos T., Mezey E., Bayless T. M., Nouvet F. J., Brant S. R. (2004). Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum. Mol. Genet. 13, 35–45 10.1093/hmg/ddh008 [DOI] [PubMed] [Google Scholar]
- Kayano S., Suzuki Y., Kanno K., Aoki Y., Kure S., Yamada A., Matsubara Y. (2004). Significant association between nonsyndromic oral clefts and arylhydrocarbon receptor nuclear translocator (ARNT). Am. J. Med. Genet. A 130A, 40–44 10.1002/ajmg.a.30023 [DOI] [PubMed] [Google Scholar]
- King C. R., Xiao M., Yu J., Minton M. R., Addleman N. J., Van Booven D. J., Kwok P., McLeod H. L., Marsh S. (2007). Identification of NR1I2 genetic variation using resequencing. Eur. J. Clin. Pharmacol. 63, 547–554 10.1007/s00228-007-0295-3 [DOI] [PubMed] [Google Scholar]
- Kölsch H., Lütjohann D., Jessen F., Popp J., Hentschel F., Kelemen P., Friedrichs S., Maier W., Heun R. (2009). RXRA gene variations influence Alzheimer's disease risk and cholesterol metabolism. J. Cell. Mol. Med. 13, 589–598 10.1111/j.1582-4934.2009.00383.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba J., Lamba V., Strom S., Venkataramanan R., Schuetz E. (2008). Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab. Dispos. 36, 169–181 10.1124/dmd.107.016600 [DOI] [PubMed] [Google Scholar]
- Lausen J., Thomas H., Lemm I., Bulman M., Borgschulze M., Lingott A., Hattersley A. T., Ryffel G. U. (2000). Naturally occurring mutations in the human HNF4alpha gene impair the function of the transcription factor to a varying degree. Nucleic Acids Res. 28, 430–437 10.1093/nar/28.2.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Huang J. (2007). Pregnane X receptor polymorphism affects CYP3A4 induction via a ligand-dependent interaction with steroid receptor coactivator-1. Pharmacogenet. Genomics 17, 369–382 10.1097/FPC.0b013e32803e40d7 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang Z., Yu J., Lei G., Wang S. (2010). Three polymorphisms in interleukin-1beta gene and risk for breast cancer: a meta-analysis. Breast Cancer Res. Treat. 10.1007/s10549-010-0910-3 [DOI] [PubMed] [Google Scholar]
- Liu-Mares W., Sun Z., Bamlet W. R., Atkinson E. J., Fridley B. L., Slager S. L., de Andrade M., Goode E. L. (2007). Analysis of variation in NF-kappaB genes and expression levels of NF-kappaB-regulated molecules. BMC Proc. 1(Suppl. 1), S126. 10.1186/1753-6561-1-s1-s126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Egan K. M., Dunning L., Shu X., Cai Q., Cai H., Dai Q., Holtzman J., Gao Y., Zheng W. (2006). Population-based case-control study of AhR (aryl hydrocarbon receptor) and CYP1A2 polymorphisms and breast cancer risk. Pharmacogenet. Genomics 16, 237–243 10.1097/01.fpc.0000189803.34339.ed [DOI] [PubMed] [Google Scholar]
- Love-Gregory L. D., Wasson J., Ma J., Jin C. H., Glaser B., Suarez B. K., Permutt M. A. (2004). A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 alpha gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an ashkenazi jewish population. Diabetes 53, 1134–1140 10.2337/diabetes.53.4.1134 [DOI] [PubMed] [Google Scholar]
- Lu P., Liu J., Melikishvili M., Fried M. G., Chi Y. (2008). Crystallization of hepatocyte nuclear factor 4 alpha (HNF4 alpha) in complex with the HNF1 alpha promoter element. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 313–317 10.1107/S1744309108007136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meex S. J. R., van Vliet-Ostaptchouk J. V., van der Kallen C. J. H., van Greevenbroek M. M. J., Schalkwijk C. G., Feskens E. J. M., Blaak E. E., Wijmenga C., Hofker M. H., Stehouwer C. D. A., de Bruin T. W. A. (2008). Upstream transcription factor 1 (USF1) in risk of type 2 diabetes: association study in 2000 Dutch Caucasians. Mol. Genet. Metab. 94, 352–355 10.1016/j.ymgme.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Merisalu A., Punab M., Altmäe S., Haller K., Tiido T., Peters M., Salumets A. (2007). The contribution of genetic variations of aryl hydrocarbon receptor pathway genes to male factor infertility. Fertil. Steril. 88, 854–859 10.1016/j.fertnstert.2006.12.041 [DOI] [PubMed] [Google Scholar]
- Muller Y. L., Infante A. M., Hanson R. L., Love-Gregory L., Knowler W., Bogardus C., Baier L. J. (2005). Variants in hepatocyte nuclear factor 4alpha are modestly associated with type 2 diabetes in Pima Indians. Diabetes 54, 3035–3039 10.2337/diabetes.54.10.3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama N., Soyama A., Saito Y., Nakajima Y., Komamura K., Ueno K., Kamakura S., Kitakaze M., Kimura H., Goto Y., Saitoh O., Katoh M., Ohnuma T., Kawai M., Sugai K., Ohtsuki T., Suzuki C., Minami N., Ozawa S., Sawada J. (2004). Six novel nonsynonymous CYP1A2 gene polymorphisms: catalytic activities of the naturally occurring variant enzymes. J. Pharmacol. Exp. Ther. 308, 300–306 10.1124/jpet.103.055798 [DOI] [PubMed] [Google Scholar]
- Nakajima M., Yokoi T., Mizutani M., Kinoshita M., Funayama M., Kamataki T. (1999). Genetic polymorphism in the 5′-flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humans. J. Biochem. 125, 803–808 [DOI] [PubMed] [Google Scholar]
- Naukkarinen J., Gentile M., Soro-Paavonen A., Saarela J., Koistinen H. A., Pajukanta P., Taskinen M., Peltonen L. (2005). USF1 and dyslipidemias: converging evidence for a functional intronic variant. Hum. Mol. Genet. 14, 2595–2605 10.1093/hmg/ddi294 [DOI] [PubMed] [Google Scholar]
- Oleson L., von Moltke L. L., Greenblatt D. J., Court M. H. (2010). Identification of polymorphisms in the 3′-untranslated region of the human pregnane X receptor (PXR) gene associated with variability in cytochrome P450 3A (CYP3A) metabolism. Xenobiotica 40, 146–162 10.3109/00498250903420243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P., Lubomirov R., Carcas A. (2010). Genetic polymorphisms of CYP1A2, CYP3A4, CYP3A5, pregnane/steroid X receptor and constitutive androstane receptor in 207 healthy Spanish volunteers. Clin. Chem. Lab. Med. 48, 635–639 10.1515/CCLM.2010.130 [DOI] [PubMed] [Google Scholar]
- Pucci L., Geppetti A., Maggini V., Lucchesi D., Maria Rossi A., Longo V. (2007). CYP1A2 F21L and F186L polymorphisms in an Italian population sample. Drug Metab. Pharmacokinet. 22, 220–222 10.2133/dmpk.22.220 [DOI] [PubMed] [Google Scholar]
- Purdue M. P., Lan Q., Kricker A., Vajdic C. M., Rothman N., Armstrong B. K. (2007). Vitamin D receptor gene polymorphisms and risk of non-Hodgkin's lymphoma. Haematologica 92, 1145–1146 10.3324/haematol.11053 [DOI] [PubMed] [Google Scholar]
- Reiner A. P., Barber M. J., Guan Y., Ridker P. M., Lange L. A., Chasman D. I., Walston J. D., Cooper G. M., Jenny N. S., Rieder M. J., Durda J. P., Smith J. D., Novembre J., Tracy R. P., Rotter J. I., Stephens M., Nickerson D. A., Krauss R. M. (2008). Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 82, 1193–1201 10.1016/j.ajhg.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker P. M., Pare G., Parker A., Zee R. Y. L., Danik J. S., Buring J. E., Kwiatkowski D., Cook N. R., Miletich J. P., Chasman D. I. (2008). Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am. J. Hum. Genet. 82, 1185–1192 10.1016/j.ajhg.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryffel G. U. (2001). Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 27, 11–29 10.1677/jme.0.0270011 [DOI] [PubMed] [Google Scholar]
- Sookoian S., Gemma C., Pirola C. J. (2010). Influence of hepatocyte nuclear factor 4alpha (HNF4alpha) gene variants on the risk of type 2 diabetes: a meta-analysis in 49,577 individuals. Mol. Genet. Metab. 99, 80–89 10.1016/j.ymgme.2009.08.004 [DOI] [PubMed] [Google Scholar]
- Wang S., Fu C., Wang H., Shi Y., Xu X., Chen J., Song X., Sun K., Wang J., Fan X., Wang H., Yang X., Huan T., Hui R. (2007). Polymorphisms of the peroxisome proliferator-activated receptor-gamma coactivator-1alpha gene are associated with hypertrophic cardiomyopathy and not with hypertension hypertrophy. Clin. Chem. Lab. Med. 45, 962–967 10.1515/CCLM.2007.189 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Imoto I., Kosugi Y., Fukuda Y., Mimura J., Fujii Y., Isaka K., Takayama M., Sato A., Inazawa J. (2001). Human arylhydrocarbon receptor repressor (AHRR) gene: genomic structure and analysis of polymorphism in endometriosis. J. Hum. Genet. 46, 342–346 10.1007/s100380170070 [DOI] [PubMed] [Google Scholar]
- Weedon M. N., Owen K. R., Shields B., Hitman G., Walker M., McCarthy M. I., Hattersley A. T., Frayling T. M. (2005). A large-scale association analysis of common variation of the HNF1alpha gene with type 2 diabetes in the U.K. Caucasian population. Diabetes 54, 2487–2491 10.2337/diabetes.54.8.2487 [DOI] [PubMed] [Google Scholar]
- Wong J. M., Okey A. B., Harper P. A. (2001). Human aryl hydrocarbon receptor polymorphisms that result in loss of CYP1A1 induction. Biochem. Biophys. Res. Commun. 288, 990–996 10.1006/bbrc.2001.5861 [DOI] [PubMed] [Google Scholar]
- Zhang J., Kuehl P., Green E. D., Touchman J. W., Watkins P. B., Daly A., Hall S. D., Maurel P., Relling M., Brimer C., Yasuda K., Wrighton S. A., Hancock M., Kim R. B., Strom S., Thummel K., Russell C. G., Hudson J. R., Schuetz E. G., Boguski M. S. (2001). The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics 11, 555–572 10.1097/00008571-200110000-00003 [DOI] [PubMed] [Google Scholar]
- Zhang Z. Y., Fasco M. J., Huang L., Guengerich F. P., Kaminsky L. S. (1996). Characterization of purified human recombinant cytochrome P4501A1-Ile462 and -Val462: assessment of a role for the rare allele in carcinogenesis. Cancer Res. 56, 3926–3933 [PubMed] [Google Scholar]
- Zhou S., Chan E., Zhou Z., Xue C. C., Lai X., Duan W. (2009). Insights into the structure, function, and regulation of human cytochrome P450 1A2. Curr. Drug Metab. 10, 713–729 10.2174/138920009789895552 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of SNP-frequencies to published data (dbSNP). SNP frequencies determined in our study were compared to available SNP data in dbSNP (n = 153). MAF, minor allele frequency.
Included SNPs, genotyping methods and frequencies.
| Gene | SNP rs | Allele/ position | nt | Effect | Type | MAF publisheda | MAFb | HWp | Missing values (%) | Method | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 CHR15; NM_000761 | |||||||||||
| rs2069514 | *1C | G > A | p | 0.081 | 0.007 | 1 | 0 | Tq-AD | Nakajima et al. (1999), Abnet et al. (2007) | ||
| rs2069522 | C > T | p | 0 | 0.037 | 0.173 | 0 | Maldi | Bethke et al. (2007) | |||
| rs12720461 | *1K | C > T | p | 0 | 0.003 | 1 | 0 | Maldi | Aklillu et al. (2003) | ||
| rs762551f | *1F | A > C | p | 0.308 | 0.32 | 0.455 | 0 | Maldi | Han et al. (2002) | ||
| rs72547513 | *11 | C > A | F186L | nc | No data | 0 | 0 | Maldi | Murayama et al. (2004), Pucci et al. (2007) | ||
| rs56276455 | *3 | G > A | D348N | nc | No data | 0.003 | 1 | 0 | Maldi | Zhou et al. (2009) | |
| rs72547516 | *4 | A > T | I386F | nc | No data | 0 | 0 | Maldi | Zhou et al. (2009) | ||
| rs56107638 | *7 | G > A | i | (0.003) | 0 | 0 | Maldi | Allorge et al. (2003) | |||
| rs28399424 | *6 | C > T | R431W | nc | No data | 0 | 0 | Maldi | Zhou et al. (2009) | ||
| rs2470890 | C > T | N516 | sc | 0.358 | 0.387 | 0.492 | 0 | Maldi | Gunes and Dahl (2008) | ||
| rs4886406 | A > C | 3′ | 0.292 | 0.323 | 0.34 | 4 | ILM | ||||
| CYP1A1 CHR15; NM_000499 | |||||||||||
| rs2470893 | G > A | 5′ | 0.275 | 0.294 | 0.029 | 1.3 | ILM | ||||
| rs2472299f | G > A | 5′ | 0.308 | 0.319 | 0.571 | 0.7 | ILM | ||||
| rs4646421 | C > T | p | 0.075 | 0.094 | 0.618 | 0.7 | ILM | ||||
| rs1048943 | *2B/C | A > G | I462V | cn | 0.042 | 0.027 | 0.143 | 0.7 | Tq-AD | Hayashi et al. (1991), Zhang et al. (1996) | |
| AHR (ARYL HYDROCARBON RECEPTOR) CHR7; NM_001621 | |||||||||||
| rs10247158 | A > T | 5′ | 0.152 | 0.13 | 1 | 0 | Maldi | Bin et al. (2008) | |||
| rs10250822 | T > C | 5′ | 0.2 | 0.217 | 0.81 | 0 | Maldi | Bin et al. (2008) | |||
| rs4719497 | T > C | 5′ | 0.1 | 0.108 | 1 | 1.3 | ILM | ||||
| rs2282885 | A > G | i | 0.347 | 0.413 | 0.736 | 0 | Maldi | Bin et al. (2008) | |||
| rs1476080 | T > G | i | 0.325 | 0.343 | 0.21 | 0 | Maldi | Bin et al. (2008) | |||
| rs4236290 | T > C | i | 0.125 | 0.122 | 0.697 | 2 | ILM | ||||
| rs6960165 | A > G | i | 0.196 | 0.205 | 1 | 0.7 | Maldi | Bin et al. (2008) | |||
| rs2158041 | C > T | i | 0.183 | 0.203 | 1 | 1.3 | ILM | Chen et al. (2009) | |||
| rs7811989 | G > A | i | 0.233 | 0.233 | 1 | 0 | Maldi | Bin et al. (2008) | |||
| rs2066853 | G > A | R554K | nc | 0.075 | 0.1 | 1 | 0 | Maldi | Long et al. (2006), Abnet et al. (2007), Wong et al. (2001) Cauchi et al. (2001); Merisalu et al. (2007) | ||
| rs4986826 | G > A | V570I | nc | 0 | 0 | 0.7 | Maldi | Wong et al. (2001) | |||
| AHRR(ARYL HYDROCARBON RECEPTOR REGULATOR); CHR5; NM_020731 | |||||||||||
| rs3756712 | T > G | i | 0.342 | 0.361 | 0.723 | 1.3 | ILM | ||||
| rs4957018 | A > G | i | 0.275 | 0.322 | 0.453 | 0.7 | ILM | ||||
| rs3734145 | C > T | i | 0.25 | 0.258 | 0.052 | 0.7 | ILM | ||||
| rs4957028 | T > C | i | 0.424 | 0.463 | 1 | 1.3 | ILM | ||||
| rs908114 | A > G | i | 0.383 | 0.329 | 0.853 | 2.7 | ILM | ||||
| rs2721020 | G > A | i | 0.225 | 0.232 | 1 | 0.7 | ILM | ||||
| rs7731963 | C > A | i | 0.375 | 0.411 | 0.389 | 6.7 | ILM | ||||
| rs2292596 | C > G | P189A | nc | 0.412 | 0.373 | 0.296 | 0 | Maldi | Watanabe et al. (2001) | ||
| rs34453673 | G > C | D641H | nc | 0.31 | 0.403 | 0.612 | 0 | Maldi | Cauchi et al. (2003) | ||
| rs10078 | A > C | 3′u | 0.2 | 0.218 | 0.81 | 0.7 | ILM | ||||
| rs2241598 | C > T | 3′ | 0.183 | 0.211 | 1 | 0.7 | ILM | ||||
| ARNT (HIF1β; ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSLOCATOR) CHR1; NM 001668 | |||||||||||
| rs2292166 | C > T | 5′ | 0.217 | 0.282 | 0.015 | 0.7 | ILM | ||||
| rs7412746 | C > T | 5′ | 0.475 | 0.419 | 0.737 | 1.3 | ILM | ||||
| rs11204735 | C > T | i | 0 | 0.7 | Maldi | Das et al. (2008) | |||||
| rs2134688 | A > G | i | 0.142 | 0.13 | 0.716 | 0 | Maldi | Das et al. (2008) | |||
| rs2228099e | G > C | V189 | sc | 0.35 | 0.307 | 0.255 | 0 | Maldi | Kayano et al. (2004) | ||
| rs1889740e | G > A | i | No data | 0.307 | 0.255 | 0 | Maldi | Das et al. (2008) | |||
| rs10305724 | C > T | i | 0.125 | 0.057 | 0.383 | 0.7 | ILM | ||||
| CAR (NR1I3; CONSTITUTIVE ANDROSTANE RECEPTOR) CHR1; NM_00177469 | |||||||||||
| rs6686001 | C > A | i | 0.13 | 0.143 | 0.739 | 0 | Maldi | ||||
| rs3003596 | T > C | i | 0.49 | 0.363 | 0.289 | 0 | Maldi | ||||
| C > T | i | 0.242 | 0.19 | 0.063 | 0 | Maldi | Oliver et al. (2010) | ||||
| 398T > G | T > G | V133G | nc | No data | 0 | Maldi | Ikeda et al. (2005) | ||||
| rs2307424 | C > T | P180 | sc | 0.471 | 0.311 | 0.252 | 1.3 | ILM | Oliver et al. (2010) | ||
| rs2307418 | A > C | i | 0.183 | 0.153 | 0.529 | 4 | ILM | ||||
| rs4073054 | G > T | i | 0.408 | 0.476 | 0.247 | 2 | ILM | Oliver et al. (2010) | |||
| HNF1α (TCF1; HEPATOCYTE NUCLEAR FACTOR 1 HOMEOBOX A) CHR12; NM_000545 | |||||||||||
| -58 | A > C | p | No data | 0 | 0.7 | Maldi | Gragnoli et al. (1997) | ||||
| rs1169288 | A > G | I27L | nc | 0.283 | 0.393 | 0.864 | 0 | Maldi | Holmkvist et al. (2006), Cauchi et al. (2008), Ryffel (2001) | ||
| rs1800574 | C > T | A98V | nc | 0.029 | 0.023 | 1 | 0 | Maldi | Giuffrida et al. (2009) | ||
| rs1169300 | G > A | i | 0.292 | 0.315 | 0.851 | 0.7 | ILM | Ridker et al. (2008) | |||
| rs2071190 | T > A | i | 0.22 | 0.19 | 1.0 | 0 | Maldi | Weedon et al. (2005) | |||
| rs1169302 | T > G | i | 0.45 | 0.433 | 0.318 | 0.7 | ILM | Ridker et al. (2008) | |||
| rs2464196 | G > A | S487N | nc | 0.292 | 0.317 | 1 | 0 | Maldi | Ridker et al. (2008) | ||
| rs1169306 | C > T | i | 0.375 | 0.377 | 1 | 0 | Maldi | ||||
| rs1169307 | T > C | i | 0.392 | 0.426 | 0.319 | 0.7 | ILM | ||||
| rs735396 | A > G | i | 0.383 | 0.382 | 0.727 | 1.3 | ILM | Reiner et al. (2008), Ridker et al. (2008) | |||
| HNF4α (NR2A1; HEPATOCYTE NUCLEAR FACTOR 4 ALPHA) CHR20; NM_000448 | |||||||||||
| rs4810424 | G > C | p | 0.164 | 0.157 | 0.364 | 0 | Maldi | Bagwell et al. (2005) | |||
| Sookoian et al. (2010) | |||||||||||
| rs2144908 | G > A | p | 0.183 | 0.16 | 0.539 | 0 | Maldi | Lamba et al. (2008), Muller et al. (2005), Holmkvist et al. (2008), Black et al. (2008) | |||
| rs2868094 | T > C | p | 0.317 | 0.28 | 0.316 | 0 | Maldi | ||||
| rs6031544 | C > T | p | 0.283 | 0.289 | 0.842 | 0.7 | ILM | SNP@promoter | |||
| rs3212183 | T > C | i | 0.413 | 0.45 | 0.0029 | 0 | Maldi | Muller et al. (2005) | |||
| rs6073432 | A > C | i | 0.309 | 0.307 | 0.057 | 0 | Maldi | ||||
| rs13041396 | C > G | i | 0.24 | 0.207 | 0.133 | 0 | Maldi | ||||
| c.379 | C > T | R127W | nc | No data | 0 | Maldi | Lausen et al. (2000) | ||||
| rs1800961 | C > T | T139I | nc | No data | 0.02 | 0.05 | 0 | ILM | Sookoian et al. (2010), Lu et al. (2008) | ||
| c.460 | C > T | R154X | nc | No data | 0 | Maldi | Lausen et al. (2000) | ||||
| rs6103731 | G > A | i | 0.484 | 0.423 | 0.404 | 0 | Maldi | ||||
| rs3818247 | G > T | i | 0.39 | 0.367 | 0.86 | 0 | Maldi | Love-Gregory et al. (2004) | |||
| IL1β (INTERLEUKIN 1 BETA) CHR2; NM 000576 | |||||||||||
| rs1143627 | C > T | p | 0.367 | 0.293 | 0.238 | 0 | Maldi | Liu et al. (2010) | |||
| rs1143634 | C > T | F105 | cs | 0.223 | 0.22 | 0.811 | 0 | Maldi | |||
| IL1RN (INTERLEUKIN 1 RECEPTOR ANTAGONIST) CHR2; NM_000577 | |||||||||||
| rs2637988 | G > A | i | 0.392 | 0.399 | 0.733 | 0.7 | ILM | ||||
| rs3213448 | G > A | i | 0.117 | 0.128 | 0.469 | 0.7 | ILM | ||||
| rs419598 | T > C | A57 | Cs | 0.275 | 0.287 | 0.84 | 4.7 | ILM | |||
| rs315952 | T > C | S133 | cs | 0.258 | 0.295 | 0.555 | 0.7 | ILM | |||
| rs397211 | T > C | 3′ | 0.314 | 0.342 | 0.856 | 0.7 | ILM | ||||
| NFKB1 (NUCLEAR FACTOR NF-KAPPA-B P105 SUBUNIT) CHR4; NM_003998 | |||||||||||
| rs28362491 | del > ATT G | 5′ | 0.34 | 0.361 | 0.019 | 2 | Maldi | Karban et al. (2004) | |||
| rs3774937 | T > C | p/i | 0.367 | 0.326 | 0.853 | 0.7 | ILM | ||||
| rs230530 | T > C | i | 0.364 | 0.446 | 0.403 | 2 | ILM | ||||
| rs4647992 | C > T | i | 0.067 | 0.047 | 0.344 | 0.7 | ILM | ||||
| rs230526 | C > T | i | 0.467 | 0.396 | 0.230 | 0.7 | ILM | Liu-Mares et al. (2007) | |||
| rs4648022 | C > T | i | 0.067 | 0.077 | 1 | 0.7 | ILM | Cerhan et al. (2008) | |||
| rs1598859 | T > C | i | 0.343 | 0.379 | 1 | 3.3 | ILM | Liu-Mares et al. (2007) | |||
| rs4648072 | A > G | M507V | nc | 0.075 | 0.013 | 0.049 | 0.7 | Maldi | SIFT-score 0.44 | ||
| rs1609798 | C > T | i | 0.331 | 0.329 | 1 | 0.7 | ILM | ||||
| rs997476 | C > A | 3′ | 0.1 | 0.057 | 0.068 | 1.3 | ILM | ||||
| rs10489113 | A > G | 3′ | 0.225 | 0.182 | 1 | 1.3 | ILM | ||||
| PGC1α (PPARGC1α; PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA COACTIVATOR 1-ALPHA) CHR4; NM_013261 | |||||||||||
| rs4361373 | C > T | i | 0.192 | 0.141 | 1 | 0.7 | ILM | ||||
| rs4550905 | A > G | i | 0.325 | 0.27 | 0.676 | 1.3 | ILM | ||||
| rs6448226 | A > G | i | 0.375 | 0.34 | 0.465 | 2 | ILM | ||||
| rs2970853 | G > A | i | 0.308 | 0.211 | 0.218 | 2 | ILM | Juang et al. (2010) | |||
| rs2932976 | G > A | i | 0.271 | 0.314 | 0.334 | 3.3 | ILM | ||||
| rs2970847 | C > T | T394 | sc | 0.208 | 0.153 | 1 | 0 | Maldi | Bhat et al. (2007), Juang et al. (2010) | ||
| rs8192678 | G > A | G482S | nc | 0.367 | 0.322 | 0.708 | 2.7 | Maldi | Deeb and Brunzell (2009), Wang et al. (2007) | ||
| rs2932965 | G > A | i | 0.25 | 0.171 | 0.771 | 0.7 | ILM | ||||
| rs12650562 | G > A | i | 0.435 | 0.426 | 0.74 | 0.7 | ILM | ||||
| PXR (NR1I2; PREGNANE X RECEPTOR) CHR3; NM_003889 | |||||||||||
| rs1523130 | G > A | 5′ | 0.407 | 0.423 | 0.403 | 0 | Maldi | Lamba et al. (2008) | |||
| rs3814055i | C > T | 5′ | 0.4 | 0.417 | 0.4 | 0 | Maldi* | Zhang et al. (2001) | |||
| rs1523127 | A > C | 5′ | 0.408 | 0.42 | 0.314 | 0 | Maldi* | Zhang et al. (2001), King et al. (2007) | |||
| rs2276706i | G > A | i | 0.39 | 0.417 | 0.4 | 0 | Maldi | Zhang et al. (2001) | |||
| rs4234666 | C > G | i | 0.318 | 0.233 | 1 | 0 | Maldi | ||||
| rs1403527 | T > G | i | 0.11 | 0.226 | 1 | 2.7 | ILM | ||||
| rs1403526 | A > G | i | 0.242 | 0.341 | 0.068 | 1.3 | ILM | ||||
| rs55764158 | C > A | i | 0.293 | 0.29 | 0.078 | 0 | Maldi | Lamba et al. (2008) | |||
| rs16830505 | A > G | i | 0.15 | 0.117 | 0.694 | 0.7 | ILM | ||||
| rs2472677 | C > T | i | 0.5 | 0.427 | 0.096 | 0 | Maldi | Lamba et al. (2008) | |||
| rs4440154 | C > T | i | 0.35 | 0.384 | 0.119 | 2 | ILM | ||||
| rs2461823 | T > C | i | 0.35 | 0.379 | 0.164 | 0.7 | ILM | ||||
| rs13059232 | T > C | i | 0.358 | 0.379 | 0.217 | 3.3 | Maldi | Lamba et al. (2008) | |||
| 69245C > T | C > T | i | No data | 0.077 | 1 | Maldi | Lamba et al. (2008) | ||||
| rs7643645 | A > G | i | 0.43 | 0.363 | 0.598 | 0 | Maldi | Lamba et al. (2008) | |||
| rs12721613 | *2 | C > T | P27S | nc | 0 | 0 | 0.7 | Maldi | Hustert et al. (2001) | ||
| rs12721607 | *3 | G > A | G36R | nc | 0.03 | 0.027 | 1 | 0 | Maldi | Hustert et al. (2001) | |
| rs72551372 | *10 | G > A | nc | No data | 0 | 0.7 | Maldi | Hustert et al. (2001) | |||
| rs72551374 | A > G | i | No data | 0 | 0.7 | Maldi | Hustert et al. (2001), Lim and Huang (2007) | ||||
| rs3732357 | G > A | i | 0.225 | 0.336 | 0.009 | 0.7 | ILM | ||||
| rs6785049 | A > G | i | 0.302 | 0.413 | 0.018 | 0 | Maldi* | Zhang et al. (2001) | |||
| rs2276707 | C > T | i | 0.058 | 0.22 | 0.641 | 0 | Maldi* | Zhang et al. (2001) | |||
| rs3732359 | G > A | 3′u | 0.075 | 0.277 | 0.091 | 6 | Maldi | Oleson et al. (2010) | |||
| rs1054191 | G > A | 3′u | 0.233 | 0.14 | 0.079 | 0 | Maldi | Zhang et al. (2001) | |||
| rs3814057 | T > C | 3′u | 0.188 | 0.218 | 0.81 | 0.7 | Maldi | Zhang et al. (2001) | |||
| RARα (NR1B1; RETINOIC ACID RECEPTOR ALPHA) CHR17; NM_001033603 | |||||||||||
| rs13706g | G > A | V441Id | 5′d | 0.142 | 0.094 | 0.618 | 0.7 | ILM | |||
| rs2715553 | T > C | i | No data | 0.483 | 0.87 | 1.3 | ILM | ||||
| rs9303286g | G > C | i | 0.06 | 0.093 | 0.617 | 0 | Maldi | ||||
| rs482284 | G > A | i | 0.283 | 0.267 | 1 | 0 | Maldi | ||||
| rs4890109 | G > T | i | 0.058 | 0.027 | 0.0016 | 2 | ILM | ||||
| RXRα (NR2B1; RETINOID X RECEPTOR ALPHA) CHR9; NM_002957 | |||||||||||
| rs3818740 | T > C | i | 0.225 | 0.294 | 0.423 | 4.7 | ILM | ||||
| rs3132297 | C > T | i | 0.222 | 0.157 | 1 | 0 | Maldi | ||||
| rs3118529 | T > C | i | 0.227 | 0.29 | 0.557 | 0 | Maldi | ||||
| rs4240705 | A > G | i | 0.4 | 0.369 | 0.597 | 0.7 | ILM | ||||
| rs3118570 | A > C | i | 0.18 | 0.153 | 1 | 0 | Maldi | Cauchi et al. (2003), Kölsch et al. (2009) | |||
| rs1536475 | G > A | i | 0.17 | 0.163 | 1 | 0 | Maldi | Kölsch et al. (2009) | |||
| rs3132293 | G > A | i | 0.22 | 0.209 | 0.451 | 2.7 | Maldi | Cauchi et al. (2003), Kölsch et al. (2009) | |||
| rs1805348 | G > A | A457 | sc | 0.025 | 0.023 | 1 | 0 | Maldi | |||
| SRC-1 (NCOA1; NUCLEAR RECEPTOR COACTIVATOR 1) CHR2; NM_003734 | |||||||||||
| rs995647 | A > G | i | 0.1 | 0.111 | 0.082 | 0.7 | ILM | ||||
| rs11677500 | G > A | i | 0.433 | 0.469 | 1 | 3.3 | ILM | ||||
| rs2119115 | A > C | i | 0.267 | 0.299 | 0.439 | 2 | ILM | ||||
| rs12468225 | A > C | i | 0.136 | 0.137 | 0.481 | 0 | Maldi | ||||
| rs4578807 | A > G | i | 0.133 | 0.145 | 0.512 | 1.3 | ILM | ||||
| rs4665716 | A > G | i | 0.075 | 0.055 | 0.007 | 2.7 | ILM | ||||
| rs11682130 | T > G | i | 0.433 | 0.473 | 0.514 | 1.3 | ILM | ||||
| rs6743362 | A > C | i | 0.058 | 0.034 | 0.205 | 0.7 | ILM | ||||
| rs12617941 | G > A | i | 0.075 | 0.047 | 0.274 | 0.7 | ILM | ||||
| rs6724282 | A > G | 3′ | 0.075 | 0.062 | 0.014 | 2.7 | ILM | ||||
| rs9309308 | T > C | 3′ | 0.225 | 0.297 | 0.242 | 1.3 | ILM | ||||
| USF1 (UPSTREAM STIMULATORY FACTOR 1) CHR1; NM_009053 | |||||||||||
| rs3813609 | C > G | 5′c | 0.3 | 0.373 | 0.601 | 0 | Maldi | Holzapfel et al. (2008) | |||
| rs2774279 | G > A | R873 (ARHG AP30) | 5′c | 0.38 | 0.26 | 0.286 | 0 | Maldi | Auro et al. (2008) | ||
| rs1556259 | T > C | i | 0.083 | 0.127 | 0.47 | 0 | Maldi | Holzapfel et al. (2008) | |||
| rs2774276 | C > G | i | 0.21 | 0.247 | 0.269 | 0 | Maldi | ||||
| rs2073658h | G > A | i | 0.283 | 0.363 | 0.48 | 0 | Maldi | OMIM, Meex et al. (2008) | |||
| rs3737787h | C > T | 3′u | 0.309 | 0.363 | 0.48 | 0 | Maldi | Holzapfel et al. (2008), Collings et al. (2008), Meex et al. (2008), Naukkarinen et al. (2005) | |||
| VDR (NR1I1; VITAMIN D RECEPTOR) CHR12; NM_000376 | |||||||||||
| rs4516035 | T > C | 5′ | 0.458 | 0.493 | 1 | 1.3 | ILM | ||||
| rs3890733 | C > T | i | 0.35 | 0.382 | 0.605 | 1.3 | ILM | ||||
| rs4760648 | C > T | i | 0.457 | 0.433 | 0.183 | 0.7 | ILM | ||||
| rs2853564 | T > C | i | 0.452 | 0.419 | 0.502 | 1.3 | ILM | ||||
| rs10735810(has merged to rs2228570) | T > A | M1T | nc | 0.442 | 0.34 | 0.71 | 4 | Maldi | Guo et al. (2006), | ||
| rs11574090 | C > G | L230V | nc | No data | 0 | 0.7 | Maldi | ||||
| rs2239186 | T > C | i | 0.203 | 0.229 | 0.638 | 2.7 | ILM | ||||
| rs2189480 | C > A | i | 0.398 | 0.413 | 0.731 | 4 | ILM | ||||
| rs2239179 | A > G | i | 0.408 | 0.405 | 0.497 | 1.3 | ILM | ||||
| rs1540339 | G > A | i | 0.398 | 0.391 | 0.165 | 2 | ILM | ||||
| rs7962898 | T > C | i | 0.425 | 0.497 | 0.87 | 0 | Maldi | ||||
| rs1544410 | G > A | i | 0.442 | 0.4 | 0.609 | 0 | Maldi | Purdue et al. (2007) | |||
| rs7975232 | A > C | i | 0.422 | 0.493 | 1 | 0 | Maldi | Purdue et al. (2007) | |||
| rs731236 | T > C | I352 | sc | 0.44 | 0.372 | 1 | 8.7 | Maldi | Purdue et al. (2007) | ||
i, Intronic; nc, non-synonymous coding; sc, synonymous coding; 3′u, 3′ untranslated region; 5′ upstream of mRNA; p, promoter; 3′, downstream of mRNA; Maldi, MALDI-TOF MS (mass assisted light desorption time of flight mass spectrometry) multiplexed genotyping assay; ILM, Illumina 300k Bead Chip; Tq-AD, TaqMan allelic discrimination genotyping assay (Applied Biosystems); calculation of MAF, minor allele frequency; HWp, Hardy–Weinberg p-value and missing values see Materials and Methods section.
aHapMapCEU or alternative population of Caucasian origin (dbSNP-homepage; Build 131).
bMinor allele frequency found in this study.
cOverlap to ARHGAP30 coding region.
dLocated in CDC6 (cn).
e,f,g,h,iSNPs linked pairwise 100% within the one gene.
*MALDI-TOF genotype data provided by E. Schaeffeler, IKP, Stuttgart