ABSTRACT
The use of pneumococcal capsular polysaccharide (PPS)-based vaccines has resulted in a substantial reduction in invasive pneumococcal disease. However, much remains to be learned about vaccine-mediated immunity, as seven-valent PPS-protein conjugate vaccine use in children has been associated with nonvaccine serotype replacement and 23-valent vaccine use in adults has not prevented pneumococcal pneumonia. In this report, we demonstrate that certain PPS-specific monoclonal antibodies (MAbs) enhance the transformation frequency of two different Streptococcus pneumoniae serotypes. This phenomenon was mediated by PPS-specific MAbs that agglutinate but do not promote opsonic effector cell killing of the homologous serotype in vitro. Compared to the autoinducer, competence-stimulating peptide (CSP) alone, transcriptional profiling of pneumococcal gene expression after incubation with CSP and one such MAb to the PPS of serotype 3 revealed changes in the expression of competence (com)-related and bacteriocin-like peptide (blp) genes involved in pneumococcal quorum sensing. This MAb was also found to induce a nearly 2-fold increase in CSP2-mediated bacterial killing or fratricide. These observations reveal a novel, direct effect of PPS-binding MAbs on pneumococcal biology that has important implications for antibody immunity to pneumococcus in the pneumococcal vaccine era. Taken together, our data suggest heretofore unsuspected mechanisms by which PPS-specific antibodies could affect genetic exchange and bacterial viability in the absence of host cells.
IMPORTANCE
Current thought holds that pneumococcal capsular polysaccharide (PPS)-binding antibodies protect against pneumococcus by inducing effector cell opsonic killing of the homologous serotype. While such antibodies are an important part of how pneumococcal vaccines protect against pneumococcal disease, PPS-specific antibodies that do not exhibit this activity but are highly protective against pneumococcus in mice have been identified. This article examines the effect of nonopsonic PPS-specific monoclonal antibodies (MAbs) on the biology of Streptococcus pneumoniae. The results showed that in the presence of a competence-stimulating peptide (CSP), such MAbs increase the frequency of pneumococcal transformation. Further studies with one such MAb showed that it altered the expression of genes involved in quorum sensing and increased competence-induced killing or fratricide. These findings reveal a novel, previously unsuspected mechanism by which certain PPS-specific antibodies exert a direct effect on pneumococcal biology that has broad implications for bacterial clearance, genetic exchange, and antibody immunity to pneumococcus.
Introduction
The introduction of the seven-valent pneumococcal conjugate vaccine (PCV7) in 2000 led to a remarkable decline in invasive pneumococcal disease caused by vaccine-included serotypes (STs) (1). Despite this resounding success, the battle against pneumococcal disease continues to present a formidable challenge in two main areas, namely, that PCV7 use has been associated with ST replacement (2) and the efficacy of the 23-valent PPS vaccine does not extend to the prevention of pneumonia (3, 4). PCV coverage has now been expanded to 13 STs (PCV13), and next-generation pneumococcal vaccines that promise to provide universal (non-ST-specific) coverage are under consideration (5). Nonetheless, the aforementioned challenges underscore the fact that there is more to learn about vaccine-mediated protection against pneumococcal disease.
PPS-based vaccines mediate protection against pneumococcus by inducing PPS-specific antibodies. Given that the Gram-positive cell wall precludes antibody and complement opsonins from exerting a direct effect on pneumococcal viability, the primary mechanism by which such antibodies mediate protection is considered to be that they promote opsonic killing of the homologous ST by host phagocytes (6). ST-specific, antibody-dependent enhancement of phagocyte-mediated (opsonophagocytic) killing of the homologous ST in vitro has emerged as the gold standard for the assessment of PCV immunogenicity (7). Nonetheless, human and mouse PPS-specific monoclonal antibodies (MAbs) that are highly protective against pneumococcal pneumonia and sepsis in mouse models but do not promote opsonic killing in vitro have been identified (8–10). Although the mechanism by which such MAbs induce bacterial clearance remains under investigation, the efficacy of one nonopsonic MAb was shown to depend on its ability to induce bacterial agglutination (8, 11). Agglutination can enhance cell-cell communication and is a characteristic of pneumococcus in its competent state (12). Given that competence is regulated by quorum sensing and certain quorum-sensing molecules have been shown to have immunomodulatory and protective activity in bacterial infection models (13, 14), we investigated the effect of PPS-specific MAbs on pneumococcal quorum sensing. Our data show that certain non-opsonic MAbs increase the frequency of pneumococcal transformation. Further studies with one such MAb showed that it induces a second wave of quorum sensing, an increase in fratricide, and changes in competence- and fratricide-related gene expression compared to competence-stimulating peptide (CSP) alone. These findings demonstrate that certain PPS-specific antibodies can exert a direct effect on pneumococcal biology and viability.
(Some of the data in this article are from a thesis submitted by Masahide Yano in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Science, Albert Einstein College of Medicine, Yeshiva University, Bronx, NY.)
RESULTS
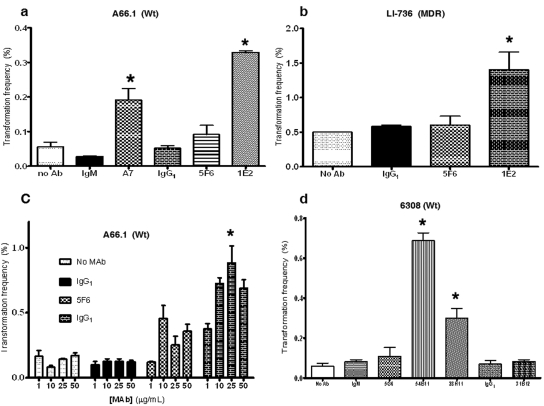
Protective PPS-specific MAbs increase the frequency of pneumococcal transformation and induce bacterial agglutination.
The transformation frequencies of Streptococcus pneumoniae ST3 (A66.1) and ST8 (6308) strains were determined in the presence of pneumococcal CSP with and without PPS-specific and isotype control MAbs. The effect of MAb 1E2, a PPS3-specific mouse IgG1(κ) that protects against ST3 pneumonia and bacteremia in mice but does not promote opsonic killing of ST3 in vitro (9), was investigated by adding it to ST3 (A66.1) or a clinical ST3 strain, LI-736 (15), in the presence of CSP2. Addition of 1E2 resulted in a higher frequency of transformation of A66.1 and the multidrug-resistant (MDR) clinical strain significantly than that observed with CSP2 alone (no MAb) or control MAbs with CSP2 (Fig. 1a). A MAb-induced increase in transformation frequency compared to that with CSP2 alone was also observed with A7, a nonopsonic, human PPS3-specific IgM MAb that is protective against ST3 in mice (11), but not with 5F6, an opsonic mouse PPS3-specific IgG1 that is protective in mice (9); 31B12, a nonopsonic, mouse PPS8-specific IgG1 MAb (see Fig. S1 in the supplemental material) that is protective against another ST (ST8) in mice (16); or an isotype-matched control MAb (Fig. 1a). The 1E2-induced increase in transformation frequency of the MDR ST3 isolate was larger than that of A66.1 (Fig. 1b). The effect of 1E2 (mouse IgG1 to PPS3) on CSP2-mediated transformation exhibited a dose-dependent trend that was statistically significant compared to that obtained with no MAb (Fig. 1c).
FIG 1 .
MAbs 1E2 and A7 increase the transformation frequency of ST3 pneumococcal cells. (a) 1E2 (nonopsonic mouse IgG1 to PPS3) increases the CSP2-mediated transformation frequency of ST3 (A66.1) pneumococcal cells, in contrast to opsonic ST3 MAb 5F6 (mouse IgG1 to PPS3) (P < 0.05 by one-way ANOVA, P < 0.05 by Tukey’s multiple-comparison test) and isotype-matched control MAb 31B12 (P < 0.05 by Tukey’s multiple-comparison test). Human IgM MAb A7 (IgM to PPS3) also led to a statistically significant increase in the frequency of transformation, in contrast to control MAb (CT antibody [Ab]) G19 (P < 0.05 by Tukey’s multiple-comparison test). G19 (human IgM) binds to C. neoformans capsular polysaccharide and was used as a matched isotype-matched control. (b) 1E2 increases the transformation frequency of an MDR ST3 clinical strain (LI-736), in contrast to 31B12 (opsonic mouse IgG1 to PPS8; P < 0.05 by one-way ANOVA, P < 0.05 by Tukey’s multiple-comparison test) and 5F6 (mouse IgG1 to PPS3; P < 0.05 by Tukey’s multiple-comparison test). (c) 1E2 enhances the transformation frequency of A66.1 in a dose-dependent manner (P < 0.05 by two-way ANOVA, P < 0.050 comparing untreated versus 1 µg and 1 µg versus 10 µg by Tukey’s multiple-comparison test). Experiments were performed as described in Materials and Methods (see supplemental material). (d) Two IgM MAbs, 54B11 (human IgM to PPS8) and 28H11 (nonopsonic mouse IgM to PPS8), increase the CSP1-mediated transformation frequency of ST8 pneumococcal cells (6308), in contrast to the isotype- and species-matched control MAb (P < 0.05 by one-way ANOVA, P < 0.05 by Tukey’s multiple-comparison test). 31B12 (opsonic mouse IgG1 to PPS8) does not change the transformation frequency of 6308 cells. Wt, wild type.
Experiments with the ST8 strain (6308) were performed with 54B11, a human IgM PPS8-specific MAb that is protective in mice (unpublished data); 28H11, a mouse IgM PPS8-specific MAb that is protective in mice (16) and nonopsonic in vitro (see Fig. S1 in the supplemental material); 31B12, a mouse IgG1 PPS8-specific MAb that induces opsonic killing of ST8 in vitro (see Fig. S1 in the supplemental material); and 5G6, an MAb to another pneumococcal determinant, phosphorylcholine (PC), that is not protective in mice (16). MAbs 54B11 (human IgM to PPS8) and 28H11 (mouse IgM to PPS8) each increased the transformation frequency of 6308 more than CSP alone, but 31B12 (mouse IgG1 to PPS8) and an isotype-matched control did not (Fig. 1d).
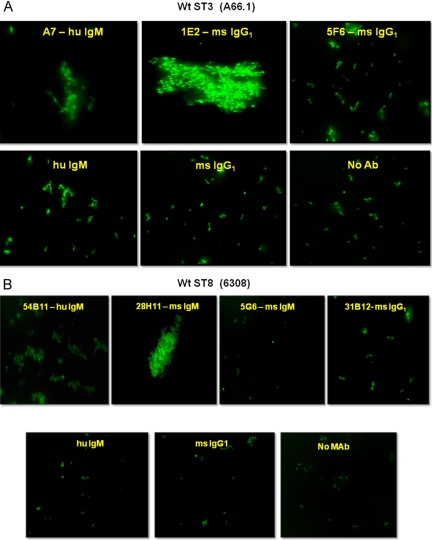
To investigate the association between MAb-mediated increases in transformation frequency and bacterial agglutination, we mixed 10 µg PPS-specific MAb with 106 fluorescently labeled CFU of the homologous ST and examined the reaction product for agglutination by fluorescence microscopy. A7 (nonopsonic human IgM to PPS3), 1E2 (nonopsonic mouse IgG1 to PPS3) (Fig. 2a), 54B11 (human IgM to PPS8), and 28H11(nonopsonic mouse IgM to PPS8) (Fig. 2b) agglutinated the respective ST, whereas 5F6 (opsonic mouse IgG1 to PPS3), 31B12 (opsonic mouse IgG1 to PPS8), and isotype-matched controls did not (Fig. 2a and b).
FIG 2 .
PPS-specific MAbs induce agglutination of pneumococcus. (a) MAbs A7 (nonopsonic human [hu] IgM to PPS3) and 1E2 (nonopsonic mouse [ms] IgG1 to PPS3) induce agglutination of ST3 (A66.1); MAb 5F6 (opsonic mouse IgG1 to PPS3) and human and mouse isotype-matched controls do not. (b) 54B11 (human IgM to PPS8) and 28H11 (nonopsonic mouse IgM to PPS8) induce agglutination of ST8 (6308); 5G6 (nonopsonic mouse IgM to PC), 31B12 (opsonic mouse IgG1 to PPS8), and human and mouse isotype-matched controls do not. Wt, wild type; Ab antibody.
To further determine the relationship between the increase in the frequency of transformation and the expression of the com two-component regulatory system (TCS), we constructed an A66.1::comE mutant strain that is unable to induce comX-mediated competence (see Fig. S2 in the supplemental material). No transformants were recovered when A66.1::comE was treated with 1E2 and/or CSP2 (data not shown).
MAb 1E2 induces late pneumococcal competence and alters gene expression.
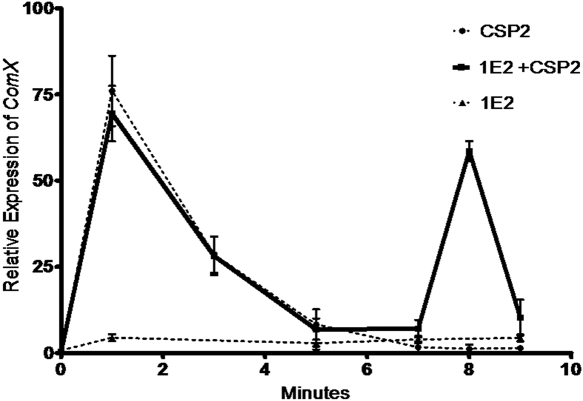
Induction of pneumococcal competence is controlled by comX, an alternative sigma factor that acts on the promoter sites of downstream pneumococcal competence genes (17). The gene com X is quiescent in the noncompetent state but rapidly activated by CSP (17). We determined the effect of 1E2 (nonopsonic mouse IgG1 to PPS3) on the induction of comX expression by quantitative reverse transcription (qRT)-PCR. Two minutes after the incubation of A66.1 with CSP2 or CSP2 plus 1E2, there were similar degrees of comX expression (Fig. 3). However, at 8 min after incubation, there was a new second peak of comX expression with CSP2 plus 1E2 that was not observed with CSP2 alone (Fig. 3) or with a control MAb (data not shown). Samples collected up to 30 min after CSP2 stimulation did not exhibit the increase in comX expression that was observed with CSP2 plus 1E2 at 8 min after incubation.
FIG 3 .
Expression of comX mRNA in the presence of CSP and 1E2. The expression of comX mRNA was determined by qRT-PCR. Expression was maximal approximately 2 min after CSP2 stimulation. When 1E2 (nonopsonic mouse IgG1 to PPS3) and CSP2 were combined, a second peak of comX gene expression was observed 8 min after initiation of the reaction. This experiment was repeated multiple times with comparable results.
MAb 1E2 alters global pneumococcal gene expression.
We used a pneumococcal DNA microarray (Roche NimbleGen, Inc., Madison, WI) to determine if 1E2 had an effect on A66.1 gene expression 2 and 8 min after incubation with CSP2 or CSP2 plus 1E2. These times were chosen based on the times at which increased comX expression was observed with 1E2 plus CSP2 (Fig. 3). Heat maps showing the effects of CSP and 1E2 on global gene expression of A66.1 are depicted in Figure S3. Tables 1 to 3 (see also Table S2 in the supplemental material) show the genes that were differentially expressed by 2.5-fold or more by CSP2-treated versus untreated A66.1 cells (Table S2a), CSP2-treated versus CSP2-plus-1E2-treated cells at 2 (Table 1) and 8 (Table 2) min, 1E2-treated versus untreated cells (Table 3), and/or controls. CSP2 alone markedly upregulated competence-related genes but downregulated very few genes (see Table S2a and b). At 2 min, the gene expression of CSP2- and CSP2-plus-1E2-treated cells was not significantly different, although there was more expression of some competence-related genes in the presence of 1E2 (Table 1). However, at 8 min, there was a marked increase in DNA processing-, competence induction-, and fratricide-related genes in the presence of CSP2 plus 1E2 (Table 2) but very few downregulated genes (see Table S2d). Hence, the second wave of CSP2-plus-1E2-induced comX expression (see Fig. 3) was associated with increased expression of competence and fratricide genes. Treatment of A66.1 with 1E2 alone also induced the expression of bacteriocin genes, including blpN, blpO, blpM, blpI, and blpC (Table 2), which was not observed with a control (PPS8-specific) MAb or with CSP2 alone (see Table S2a, b, and f).
TABLE 1 .
Genes differentially expressed by CSP2- and CSP2-plus-1E2 (mouse IgG1 to PPS3)-treated A66.1 cells at 2 min after incubationa
| Seq_id | Function | Fold change |
|---|---|---|
| SP_0978080700000908 | Competence protein CoiA | 4.9 |
| SP_2207080700002076 | Competence protein ComF, putative | 4.7 |
| SP_1017080700000942 | 4-Oxalocrotonate tautomerase | 3.8 |
| SP_2206080700002075 | Ribosomal subunit interface protein | 3.6 |
| SP_0395080700000368 | Transcriptional regulator, putative | 3.4 |
| SP_0047080700000043 | Phosphoribosylaminoimidazole synthetase | 3.2 |
| SP_0957080700000887 | ABC transporter, ATP-binding protein | 3.2 |
| SP_2035080700001917 | 3-Keto-l-gulonate-6-phosphate decarboxylase | 3.1 |
| SP_0515080700000482 | Heat-inducible transcription repressor | 2.9 |
| SP_0021080700000020 | Deoxyuridine 5′-triphosphate nucleotidohydrolase | 2.9 |
| SP_2208080700002077 | Helicase, putative | 2.8 |
| SP_2186080700002055 | Glycerol kinase | 2.8 |
| SP_1113080700001034 | DNA-binding protein HU | 2.7 |
| SP_1856080700001743 | MerR family transcriptional regulator | 2.7 |
| SP_1809080700001697 | Transcriptional regulator | 2.6 |
| SP_1689080700001582 | ABC transporter, permease protein | 2.6 |
| SP_0516080700000483 | Heat shock protein GrpE | 2.6 |
| SP_0321080700000304 | Phosphotransferase system, IIA component | 2.6 |
| SP_2051080700001932 | Competence protein CglC | 2.6 |
| SP_1898080700001784 | α-Galactosidase | 2.6 |
| SP_0090080700000084 | ABC transporter, permease protein | 2.6 |
| SP_2031080700001913 | Putative l-ascorbate 6-phosphate lactonase | 2.5 |
| SP_2036080700001918 | Phosphotransferase system, IIA component | 2.5 |
| SP_1813080700001701 | N-(5′-Phosphoribosyl)anthranilate isomerase | 2.5 |
Genes that exhibited a >2.5-fold increase are shown. For the hypothetical genes not included here, see Table S3 in the supplemental material.
TABLE 2 .
Genes differentially expressed by CSP2- and CSP2-plus-1E2 (mouse IgG1 to PPS3)-treated cells 8 min after incubationa
| Seq_id | Function | Fold change |
|---|---|---|
| SP_1266080700001180 | DNA processing protein DprA, putative | 64.1 |
| SP_2207080700002076 | Competence protein ComF, putative | 49.3 |
| SP_0955080700000885 | Competence protein CelB | 46.8 |
| SP_2208080700002077 | Helicase, putative | 39.7 |
| SP_0954080700000884 | Competence protein CelA | 38.4 |
| SP_0957080700000887 | ABC transporter, ATP-binding protein | 31.0 |
| SP_1808080700001696 | Type IV prepilin peptidase, putative | 29.7 |
| SP_2201080700002070 | Choline-binding protein D | 29.0 |
| SP_1908080700001793 | Single-stranded DNA-binding protein | 27.7 |
| SP_2053080700001934 | Competence protein CglA | 26.4 |
| SP_2050080700001931 | Competence protein CglD | 26.4 |
| SP_0042080700000038 | Competence factor transporting ATP-binding/permease protein ComA | 24.7 |
| SP_2052080700001933 | Competence protein CglB | 21.5 |
| SP_0978080700000908 | Competence protein CoiA | 21.4 |
| SP_2236080700002101 | Putative sensor histidine kinase ComD | 17.5 |
| SP_1988080700001872 | Immunity protein, putative | 17.0 |
| SP_1941080700001825 | Competence damage-inducible protein A | 16.6 |
| SP_1717080700001609 | ABC transporter, ATP-binding protein | 16.1 |
| SP_0544080700000510 | Immunity protein BlpX | 15.1 |
| SP_0014080700000014 | Transcriptional regulator ComX1 | 14.7 |
| SP_1088080700001010 | DNA repair protein RadC | 14.5 |
| SP_0043080700000039 | Competence factor transport protein ComB | 13.6 |
| SP_2235080700002100 | Response regulator ComE | 12.9 |
| SP_0030080700000027 | Competence-induced protein Ccs16 | 9.4 |
| SP_1987080700001871 | ABC transporter, ATP-binding protein | 9.0 |
| SP_1809080700001697 | Transcriptional regulator | 8.4 |
| SP_0529080700000496 | BlpC ABC transporter | 8.0 |
| SP_0545080700000511 | Immunity protein BlpY | 7.4 |
| SP_1110080700001031 | Bifunctional riboflavin kinase/flavin mononucleotide adenylyltransferase | 6.3 |
| SP_2051080700001932 | Competence protein CglC | 5.6 |
| SP_0524080700000491 | BlpT protein, fusion | 5.1 |
| SP_0021080700000020 | Deoxyuridine 5′-triphosphate nucleotidohydrolase | 5.1 |
| SP_0528080700000495 | Peptide pheromone BlpC | 4.7 |
| SP_1714080700001606 | GntR family transcriptional regulator | 4.5 |
| SP_1089080700001011 | Glutamine amidotransferase, class I | 4.3 |
| SP_1812080700001700 | Tryptophan synthase subunit beta | 3.8 |
| SP_0942080700000874 | IS1381 transposase protein A | 3.7 |
| SP_1894080700001780 | Sucrose phosphorylase | 3.7 |
| SP_2206080700002075 | Ribosomal subunit interface protein | 3.5 |
| SP_1310080700001221 | IS1381 transposase protein A | 3.1 |
| SP_2239080700002104 | Serine protease | 3.1 |
| SP_1939080700001823 | MATE efflux family protein DinF | 3.0 |
| SP_1940080700001824 | Recombinase A | 3.0 |
| SP_1549080700001446 | Peptide deformylase | 3.0 |
| SP_1811080700001699 | Tryptophan synthase subunit alpha | 3.0 |
| SP_1072080700000995 | DNA primase | 3.0 |
| SP_0048080700000044 | Phosphoribosylglycinamide formyltransferase | 2.8 |
| SP_2240080700002105 | spspoJ protein | 2.7 |
| SP_0050080700000046 | Bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase | 2.7 |
| SP_0647080700000602 | Phosphotransferase system, IIC component, putative | 2.7 |
| SP_0062080700000058 | Phosphotransferase system, IIC component | 2.7 |
| SP_0049080700000045 | VanZ protein, putative | 2.7 |
Genes that exhibited a >2.5-fold increase are shown. For the hypothetical genes not included here, see Table S3 in the supplemental material.
TABLE 3 .
Genes differentially expressed by 1E2 (mouse IgG1 to PPS3)-treated and untreated A66.1 bacterial cells grown to an OD405 of 0.032a
| Seq_id | Function | Fold change |
|---|---|---|
| SP_0540080700000506 | BlpN protein | 10.2 |
| SP_0541080700000507 | Bacteriocin BlpO | 5.6 |
| SP_1330080700001238 | N-Acetylmannosamine-6-phosphate 2-epimerase | 4.8 |
| SP_0464080700000433 | Cell wall surface anchor family protein | 4.6 |
| SP_0539080700000505 | Bacteriocin BlpM | 3.8 |
| SP_0468080700000437 | Sortase, putative | 3.5 |
| SP_0528080700000495 | Peptide pheromone BlpC | 3.3 |
| SP_1336080700001244 | Type II DNA modification methyltransferase Spn5252IP | 3.2 |
| SP_0053080700000049 | Phosphoribosylaminoimidazole carboxylase catalytic subunit | 3.2 |
| SP_2092080700001969 | UTP-glucose-1-phosphate uridylyltransferase | 3.1 |
| SP_0074080700000070 | Acetyltransferase | 2.9 |
| SP_0978080700000908 | Competence protein CoiA | 2.8 |
| SP_0046080700000042 | Amidophosphoribosyltransferase | 2.7 |
| SP_0044080700000040 | Phosphoribosylaminoimidazolesuccinocarboxamide synthase | 2.7 |
| SP_0531080700000497 | Bacteriocin BlpI | 2.7 |
| SP_1343080700001251 | Prolyl oligopeptidase family protein | 2.7 |
| SP_0048080700000044 | Phosphoribosylglycinamide formyltransferase | 2.7 |
| SP_0047080700000043 | Phosphoribosylaminoimidazole synthetase | 2.7 |
| SP_0466080700000435 | Sortase, putative | 2.6 |
| SP_2142080700002017 | ROK family protein | 2.6 |
Genes that exhibited a >2.5-fold increase are shown. For the hypothetical genes not included here, see Table S3 in the supplemental material.
MAb 1E2 increases CSP2-mediated agglutination and fratricide.
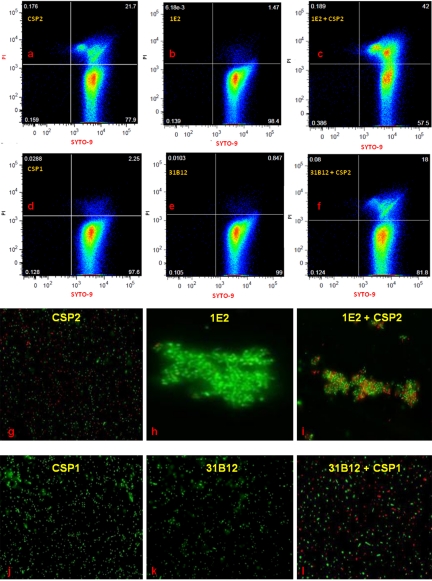
The effect of CSP2 with or without 1E2 (nonopsonic mouse IgG1 to PPS3) on A66.1 agglutination and fratricide was investigated by flow cytometry and fluorescence microscopy. A66.1 cells were stained with SYTO-9 (to detect all bacterial cells) and propidium iodide (PI; to detect dead cells), and cell viability was examined by flow cytometry (Fig. 4a to f). Incubation of A66.1 with CSP2 alone resulted in 21.7% death (Fig. 4a), while incubation with CSP2 plus 1E2 resulted in 42% death. Although this nearly 2-fold increase did not reach statistical significance (P = 0.11, Student’s t test, Fig. 4c), an increase was not observed with CSP2 plus a control MAb, CSP1, or 1E2 alone. Less than 1% killing was observed after the incubation of A66.1::comE with either CSP2 or CSP2 plus 1E2 (mouse IgG1 to PPS3) (Fig. S4). The ability of 1E2 to increase CSP2-mediated agglutination and death of A66.1 was confirmed by live-dead staining and fluorescence microscopy (Fig. 4g to l).
FIG 4 .
(a to f) Quantification of pneumococcal fratricide by flow cytometry. Pneumococcal viability studies were performed by live-dead staining as described in Materials and Methods. Dead ST3 (A66.1) cells appear in the upper right quadrant as doubly positive for green fluorescence (SYTO-9) and red fluorescence (PI). Live cells appear in the lower right quadrant as singly positive for SYTO-9. Bacterial killing by CSP2 (a), 1E2 (nonopsonic mouse IgG1 to PPS3) (b), CSP2 plus 1E2 (c), CSP1 (control CSP) (d), 31B12 (opsonic mouse IgG1 to PPS8) (e), and CSP1 plus 31B12 (f) is shown. (g to l) Immunofluorescence imaging of pneumococci treated with CSP2 and 1E2. Live bacteria fluoresce green, and dead bacteria fluoresce red. Bacterial killing by CSP2 (g), 1E2 (nonopsonic mouse IgG1 to PPS3) (h), CSP2 plus 1E2 (I), CSP1 (j), 31B12 (opsonic mouse IgG1 to PPS8) (k), and CSP1 plus 31B12 (l) is shown.
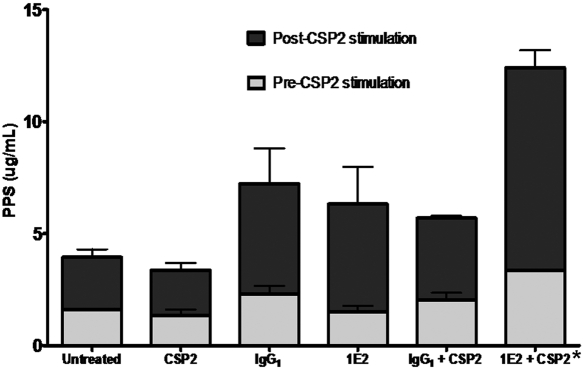
MAb 1E2 increases CSP2-mediated PPS secretion.
The amount of PPS3 secreted by A66.1 cells after incubation with MAbs was quantified by enzyme-linked immunosorbent assay (ELISA). There was no significant difference in the amount of PPS3 secreted by MAb-treated cells prior to the addition of CSP2 in any of the groups tested in this study. The addition of CSP2 to 1E2 (nonopsonic mouse IgG1 to PPS3)-treated A66.1 cells resulted in a statistically significant increase in PPS3 secretion compared to that of CSP2-treated A66.1 cells treated with CSP2 alone or with control MAb 31B12 (opsonic mouse IgG1 to PPS8; P < 0.05 by one-way analysis of variance [ANOVA] and P < 0.05 by Tukey’s multiple-comparison test comparing CSP2 to 1E2 plus CSP2) (Fig. 5).
FIG 5 .
Quantification of secreted pneumococcal capsules treated with CSP2 and 1E2.The amount of PPS3 secreted by A66.1 cells left unstimulated or stimulated with CSP2, as indicated, is shown on the y axis for the MAbs indicated on the x axis. Treatment of CSP2-stimulated cells with 1E2 (mouse IgG1 to PPS3), but not the other MAbs, resulted in increased PPS3 secretion, in contrast to CSP2 and control MAb (*, P < 0.05 by one-way ANOVA and P < 0.05 by Tukey’s multiple-comparison test).
DISCUSSION
In this article, we report that certain PPS-specific MAbs that protect against ST3 and ST8 pneumonia and/or sepsis in mice (8–10) increase the transformation frequency of the homologous ST in the presence of CSP. For ST3 (A66.1), this phenomenon was mediated by protective, nonopsonic MAbs to PPS3, including a human IgM (A7) and a mouse IgG1 (1E2) that induced the agglutination of ST3, but not a protective, opsonic mouse IgG1 (5F6) that did not induce agglutination. Notably, 1E2-mediated enhancement of pneumococcal transformation frequency was even greater for an MDR ST3 clinical strain (LI-736). Further work is required to identify an explanation for this phenomenon; however, as the fold increases in 1E2-mediated enhancement of A66.1 and LI-736 transformation were similar, our result could reflect the transformability and plasticity of drug-resistant pneumococci (2). For ST8 (6308), an increase in transformation frequency was mediated by two protective IgM MAbs to PPS8 (54B11 and 28H11) that induced agglutination of ST8 but not by a protective opsonic IgG1 (31B12) that did not induce agglutination. Neither a nonprotective MAb to PC (5G6) nor a human IgM to another capsular polysaccharide (G19) increased the transformation frequency of or agglutinated ST8 or ST3, respectively. Hence, our data reveal a relationship between MAb-induced agglutination and the bacterial transformation frequency.
The PPS3-specific MAb 1E2 is highly protective and induces bacterial clearance in mice, despite being unable to induce opsonophagocytic killing in vitro (9). Our data show that 1E2 (nonopsonic mouse IgG1 to PPS3) promotes ST3 agglutination. To investigate the effect of such binding on pneumococcal biology, we performed gene expression profiling with a ST3 strain (A66.1). These experiments revealed two main findings. First, incubation of A66.1 with CSP2 plus 1E2 induced a new, second wave of competence induction 8 min after incubation manifested by an increase in comX expression that was not observed with CSP2 alone. This second wave of comX expression was associated with a marked increase in the expression of pneumococcal competence-related and bacteriocin, including immunity, genes such as blpX and blpY. The mechanism by which this second wave of comX expression was induced is unknown. However, given that 1E2 induces bacterial agglutination, this state could enhance or prolong cell-cell communication, thereby triggering quorum sensing and fratricide (12, 18). We found that the addition of 1E2 to CSP2 resulted in an almost 2-fold increase in fratricide and cell death by flow cytometry and fluorescence microscopy, respectively. Although this increase was not statistically significant, we note that a change of this magnitude could be biologically significant.
The second main finding in our microarray experiments was that 1E2 (mouse IgG1 to PPS3) alone increased the expression of blp TCS bacteriocin genes, irrespective of the presence of CSP2. Hence, 1E2 (mouse IgG1 to PPS3) had a distinct effect on A66.1 that was independent of CSP2. Expression of blp family genes occurs during stationary-phase growth (19). Certain bacteriocins have been implicated in driving interspecies competition among pneumococci (20). Hence, although the direct effect of 1E2 (mouse IgG1 to PPS3) on bacteriocins and its possible effect on pneumococcal fratricide, fitness, and biology require further study, our data suggest that in the presence of CSP2, 1E2 (mouse IgG1 to PPS3)-induced changes in A66.1 gene expression could translate into reduced bacterial viability via fratricide of noncompetent cells, while increasing competence for genetic exchange.
Antibiotics have been shown to exert direct effects on pneumococcal quorum sensing and gene expression (21, 22). Other groups have shown that while penicillin upregulates the ciaR-ciaH TCS and reduces com TCS (22), mitomycin C, aminoglycosides, and fluoroquinolones increase com TCS (21) expression. In contrast to the foregoing effect of penicillin, 1E2 (mouse IgG1 to PPS3) plus CSP2 increased transformation frequency and competence-related gene expression of A66.1 and 1E2 alone increased fratricide-related gene expression. These findings establish that PPS-specific antibody can affect pneumococcal biology, revealing an effect that is different from that of penicillin. Thus, our data suggest that penicillin and antibody could exert different effects on the host response. Consistent with this idea, penicillin and the protective, nonopsonic MAb A7 (human IgM to PPS3) affected the splenic cytokine response differently, even though each mediated bacterial clearance in a mouse model (8). The ability of a MAb to affect microbial biology has been demonstrated previously for a MAb to the capsular polysaccharide of Cryptococcus neoformans, which altered fungal gene expression and antifungal susceptibility (23).
More work is required to unravel the mechanism by which 1E2 binding to the A66.1 capsule affects gene expression. Given that the blp locus is a bacterial TCS (24), one possibility is that 1E2 (nonopsonic mouse IgG1 to PPS3) binding evokes signal transduction pathways that occur under conditions of stress. A mechanism by which this could occur is via 1E2-induced changes to capsular morphology and/or induction of capsular shedding by CSP2-treated cells. The latter was observed in this study. In other words, 1E2 could enhance quorum sensing by enhancing the ability of CSP2 to bind COMD, the CSP-binding protein on the cell membrane, by making it more accessible. There is a precedent for this phenomenon; capsular binding of MAbs to cryptococcal polysaccharide has been shown to expose otherwise cryptic or hidden fungal determinants (25, 26).
The findings in this study raise the possibility that certain PPS-specific MAbs could reduce bacterial viability in vivo by enhancing fratricide. We realize that the idea that an antibody to a Gram-positive organism could have a direct effect on bacterial viability is unconventional. Nonetheless, new mechanisms of antibody action, including direct antimicrobial effects (27), suggest that this idea warrants further consideration. Our data show that PPS-specific MAbs can increase the frequency of pneumococcal transformation and that such MAbs induce bacterial agglutination, suggesting that PPS-specific antibodies that bring pneumococci in close proximity to one another could trigger and/or enhance quorum-sensing-related phenomena such as competence induction and fratricide. Fratricide could translate into a bacterial burden reduction. Another way in which PPS-specific MAbs that affect quorum sensing could influence bacterial biology in the host is via immunomodulation. Immunomodulation has been associated with the efficacy of nonopsonic MAbs to PPS3 (8, 9, 11). Along these lines, a bacterial quorum-sensing peptide was recently shown to modulate the inflammatory response to Aeromonas in mice (14), and CSP treatment prolonged the survival of mice in a pneumococcal infection model (13). On the other hand, our data raise questions regarding the influence of PPS-specific antibody on ST replacement and antibiotic resistance. Do certain antibodies promote these phenomena by increasing pneumococcal competence for transformation and genetic exchange? More work is needed to answer this question and understand the myriad effects that antibody-induced and/or enhanced quorum sensing have on pneumococcal biology and host defense. Nonetheless, our findings reveal a novel, heretofore unknown, mechanism by which certain PPS-specific antibodies interact with pneumococci and suggest new avenues of exploration to enhance our understanding of host-pneumococcus interaction and antibody-mediated immunity to pneumococci.
MATERIALS AND METHODS
Strains and growth conditions.
The pneumococcal strains used in this study were A66.1 (28) (originally provided by D. Briles, University of Alabama), LI-736 (15) (provided Nurith Porat, Ben-Gurion University of the Negev), 6308 (ATCC, Manassas, VA), and A66.1::comE (constructed for this study). All strains were propagated in Todd-Hewitt broth (Difco Laboratories, Detroit, MI) supplemented with 0.5% yeast extract (Difco Laboratories, Detroit, MI) in liquid phase or in solid phase on agar plates containing 5% sheep blood (BD, Franklin Lakes, NJ), unless otherwise indicated. Recombinant strains were propagated in medium containing 200 µg/ml kanamycin. The strains were grown in 5% CO2 at 37°C (unless otherwise indicated) and frozen in Todd-Hewitt broth with 0.5% yeast extract medium in 10% glycerol.
MAbs.
The MAbs used in this study are listed in Table 4.
TABLE 4 .
MAbs used in this study
| MAb | Specificitya | Species/isotype | Efficacy in mice | Reference(s) |
|---|---|---|---|---|
| 1E2 | ST3 | Mouse IgG1 | Protective | 9 |
| A7 | ST3 | Human IgM | Protective | 8, 33 |
| 5F6 | ST3 | Mouse IgG1 | Protective | 9 |
| 31B12 | ST8 | Mouse IgG1 | Protective | 16 |
| 54B11 | ST8 | Human IgM | Protectivec | 11 |
| 28H11 | ST8 | Mouse IgM | Protective | 16 |
| G19 | GXMb | Mouse IgM | NTd | 34 |
| 5G6 | PC | Mouse IgM | Nonprotective | 16 |
ST of capsular polysaccharide recognized by MAb.
GXM, C. neoformans capsular polysaccharide glucuronoxylomannan.
Unpublished.
NT, not tested against pneumococci.
Opsonophagocytosis killing assays.
Opsonophagocytosis killing assays were performed with PPS-specific MAbs 31B12 and 28H11 to evaluate their abilities to enhance the phagocytosis and killing of bacteria by the murine macrophage-like cell line J774.16 as previously described (9). MAb concentrations of 0.1 µg/ml, 1 µg/ml, and 10 µg/ml were added to 103 CFU of ST8 strain 6308 (ATCC), and the volume was adjusted to 30 µl with Veronal buffer (Lonza, Walkersville, MD). After incubation for 15 min at 37°C, 10% (by volume) mouse serum was added as a complement source (Sigma-Aldrich, St. Louis, MO) and then 4 × 105 J774.16 cells/well were added to achieve an effector-to-target cell (E/T) ratio of 400:1. This E/T ratio has been used previously in opsonophagocytosis killing assays with ST8 (29). After incubation for 45 min at 37°C, the mixture was diluted with Veronal buffer (Lonza, Walkersville, MD) and plated onto blood agar plates (BD, Franklin Lakes, NJ) in duplicate. The plates were incubated overnight at 37°C with 5% CO2, and colonies were counted. The number of colonies for each condition was determined as a percentage of the number of colonies with no MAb, e.g., for bacteria and MAb, for bacteria only; for bacteria, MAb, and J774.1 cells, for bacteria and J774.1 cells, etc.
Competence induction and determination of pneumococcal transformation frequencies.
Pneumococcal cultures were initiated by growth of a 1:100 dilution of an overnight culture to early log-phase: an optical density at 550 nm (OD550) of approximately 0.02 (measured with a Thermo Scientific Biomate 3) with the culture pH adjusted to 7.8 with 0.1 M NaOH. Subsequently, 1-ml aliquots of bacteria were made and 10 µg of antibody was added, except where noted otherwise. Cells were then grown to an OD550 of approximately 0.032. CSP2 (100 ng/ml) for the ST3 strain, CSP1 (100 ng/ml) for the ST8 strain (AnaSpec), and 100 ng of transforming plasmid pPM2-Kan DNA (30) were added to cultures supplemented with 0.1 mM CaCl2, and then the cultures were incubated for 30 min at 30°C and additionally for 30 min at 37°C. After incubation, the cultures were serially diluted and plated on kanamycin (200 µg/ml)-containing plates. Positive colonies were identified by drug resistance (growth) the next day. The transformation frequency was calculated as a percentage of the input number of CFU by dividing the number of CFU after the reaction by the number of CFU at the time competence induction was started.
Fluorescence microscopy to determine agglutination.
A suspension of approximately 106 CFU of bacteria was labeled with Oregon Green 488-carboxylic acid diacetate succinimidyl ester mixed isomers (600:1; Invitrogen, Carlsbad, CA) overnight at 4°C. On the following day, labeled bacteria were incubated with the PPS3-specific, the PPS8-specific, or the PC-reactive MAb in tryptic soy broth for 1 h at 37°C. After three washes (centrifugation at 8,000 × g for 20 min), cells were resuspended in 5 to 10 µl of ProLong gold antifade reagent (Invitrogen, Carlsbad, CA). All fluorescent images were taken on an Axio Imager microscope (Carl Zeiss MicroImaging) at final magnifications of ×630 and ×1,000. Photomicrographs were captured using an AxioCam MRm camera (Carl Zeiss MicroImaging). Captured photomicrographs were adjusted by Axio vision 4.7.2 (Carl Zeiss MicroImaging) for contrast, balance, and color saturation. All resizing of images was done by taking care to keep dimensions and size ratios intact.
Construction of A66.1::comE strain.
Mini prep (Qiagen), Clone Jet PCR cloning (Fermentas), and Rapid DNA ligation (Fermentas) kits were used for strain construction according to the manufacturers’ instructions. To construct the A66.1::comE strain, genomic DNA of CPM17 containing a kanamycin resistance cassette in the ComE-encoding region (provided by D. Morrison, University of Illinois, Chicago) was amplified with primers ComEup5 (CTC ATA AAG TTC TGG AGA AAG AGA TTG CTT TGA AGC A) and ComEdown3 (GGC GCG CCA ATA ACA TCT TCT AAA ATT AAA ACT TTC AT) and then cloned into vector pJET1.3 (see Fig. S2 in the supplemental material). The vector containing the comE mutant gene (kanamycin cassette) was subsequently transformed into wild-type strain A66.1 as described above.
qRT-PCR.
Aliquots from cultures in which competence was induced as described above were taken every 30 s for 20 min after CSP stimulation, immediately spun down at 4°C for 15 min at 17,000 × g, and resuspended in RNAprotect Bacteria Reagent (Invitrogen), and pellets were stored at −20°C until use. To prepare cells for PCR, glass beads were used to mechanically disrupt the cells and mRNA was isolated using the MICROBExpress bacterial mRNA enrichment kit (Ambion) according to the manufacturer’s instructions. Isolated mRNA was reverse transcribed to cDNA with a Maxima Universal first-strand cDNA synthesis kit (Fermentas), and qRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer’s instructions. For the primers used in this experiment, see Table S1 in the supplemental material. The 16S rRNA housekeeping gene was used as an internal control.
Flow cytometry.
Upon competence induction as described above, bacteria were pipetted, vortexed, and sonicated to disrupt clump formation. Approximately 106 bacteria were stained with a 1.5:1 ratio of green fluorescent SYTO-9, which stains all cells with intact membranes and red fluorescent PI, which penetrates only bacteria with damaged cell membranes (LIVE/DEAD BacLight bacterial viability kit; Invitrogen). Cells were incubated at 4°C for 20 min, centrifuged, washed, and resuspended in 2% paraformaldehyde. Data were acquired on a FACSCanto II (BD) flow cytometer interfaced to BD FACSDiva software with the forward scatter threshold adjusted to 300 and single-color controls for compensation. Positive controls with various concentrations of live and dead (heat-killed) bacteria were included, as well as single-color controls for each sample to ensure proper gating. As many events were acquired as possible, with a minimum of 100,000 events per sample, each of which was run in duplicate. All analyses were performed using FlowJo software version 9.2 (Treestar). Pneumococcal viability was confirmed by fluorescence microscopy to evaluate live (green) and dead (red) bacteria.
Microarray experiments.
Microarrays were prepared with the S. pneumoniae TIGR4 385K 4-plex array (Roche NimbleGen). This microarray has been used extensively for studies of pneumococcal gene expression (31, 32). Microarray analysis was performed for the following conditions: (i) untreated cells; (ii) CSP2 alone at 2 min, (iii) CSP2 alone at 8 min, (iv) CSP plus 1E2 (nonopsonic mouse IgG1 to PPS3) at 2 min, (v) CSP2 plus 1E2 at 8 min, (vi) 1E2 alone, and (vii) 31B12 (opsonic mouse IgG1 to PPS8). All mRNAs were prepared as described above. Subsequently, they were amplified using the MICROBExpress bacterial mRNA enrichment kit (Ambion). All of the samples were duplicated by separate preparation. Labeling, hybridization, scanning, and data analysis were done at Roche NimbleGen, Inc. (Madison, WI). This array detects 2,094 S. pneumoniae open reading frames. Prepared slides were hybridized with Cy-3-labeled samples and scanned with a NimbleGen MS200 microarray scanner. Data analysis was performed using raw fluorescence intensity values with microarray suite version 5.0. Gene functions were annotated from the National Center for Biotechnology Information (NCBI) database, and further data analysis was done with ArrayStar (DNASTAR, Inc., Madison, WI).
Quantification of capsular polysaccharide secretion by cultured cells.
Pneumococcal cultures were initiated by growing a 1:100 dilution of an overnight culture to early log phase, i.e., an OD550 of approximately 0.02, under the culture conditions described above. Subsequently, 1.0-ml aliquots of bacteria were prepared and 10 µg of MAb was added to the cells, which were then grown to an OD550 of approximately 0.032. CSP2 (100 ng/ml) was then added, and the reaction mixtures were incubated for 30 min at 30°C and then for 30 min at 37°C. Pre- and post-CSP stimulation samples were collected immediately prior to the addition of CSP2 and 30 min after incubation at 37°C, respectively. The supernatants were collected by centrifugation, heat killed at 65°C for 15 min, and stored at 4°C until use. Secreted PPS was harvested by centrifugation for 12 min at 14,000 × g, and culture supernatant fluids were filtered and saved. PPS was quantified by capture ELISA as follows. Wells of polystyrene ELISA plates (Corning Glass Works, Corning, NY) were coated with 1.0 µg/ml PPS3-specific MAb A7 (human IgM) (8, 11, 33) or PPS8-specific MAb 28H11 (mouse IgM) (16) and incubated with serially diluted supernatants from the cell cultures. After being washed, the plates were incubated at 37°C for 1 h with secondary MAbs to PPS3 (1E2, mouse IgG1 to PPS3) and PPS8 (31B12, mouse IgG1 to PPS8). Alkaline phosphatase-conjugated goat anti-human secondary antibody (Southern Biotechnology, Birmingham, AL) was then added. After the plates were washed, bound antibody was detected by developing the plates with p-nitrophenylphosphate substrate (Sigma-Aldrich, St. Louis, MO). A Sunrise absorbance reader (Tecan, Durham, NC) was used to record ODs. Relative quantities of capsular polysaccharides were determined by normalizing the OD values obtained to standard curves that were generated by quantified capsular polysaccharide (ATCC, Manassas, VA).
Statistical evaluation.
Differences in the transformation frequency of and PPS secretion by A66.1 cells were analyzed using Student’s t test or Turkey’s multiple-comparison test after ascertaining the normality distribution of the data by one-way ANOVA. A two-tailed P value of ≤0.05 was considered significant. All statistical analyses were performed using Prism (version 5.02 for Windows; GraphPad Software, San Diego, CA).
Microarray data accession number.
The NCBI Gene Expression Omnibus accession number for the microarray data in this report is GSE27258.
SUPPLEMENTAL MATERIAL
Opsonic activities of ST8-binding MAbs. Opsonophagocytic killing was determined as described in the text with the MAbs indicated in the title of each panel. The percent CFU for each reaction was determined by dividing the number of CFU for the condition containing the indicated MAb by the number of CFU obtained with the same reactants without the MAb. The bars on the x axis represent (i) the MAb alone, (ii) the MAb with serum (mouse) complement (SC), (iii) the MAb with J774 cells, and (iv) the MAb with SC and J774 cells. (A) 28H11 (nonopsonic mouse IgM to PPS8). (B) 31B12 (opsonic mouse IgG1 to PPS8). (C and D) Species- and isotype-matched control MAbs. Download Figure S1, TIF file, 0.7 MB.
Construction of comE mutant strain. A66.1::comE was constructed by inserting a kanamycin resistance marker into the comE gene. Lane 1 shows the wild-type comE gene (774 bp), and lane 2 shows A66.1::comE (1,500 bp). Download Figure S2, TIF file, 1.2 MB.
Heat maps showing the effect of CSP2 and MAbs. Global gene expression profiling of early-log-phase strain A66.1 was done after incubation with CSP2 or with CSP2 plus 1E2 (opsonic mouse IgG1 to PPS3) for different times in the presence (a) or absence (b) of CSP2. Untreated cells were not treated with any additional reagents. Genes that were increased in expression by >2.5 fold are shown in Table S3. Download Figure S3, TIF file, 4.1 MB.
MAb has no effect on comE mutant pneumococcal strain A66.1::comE. This experiment was performed as described in the legend to Fig. 2, except that the comE-deficient mutant A66.1::comE was used. Less than 1% killing was observed after incubation with CSP2 alone or with CSP2 plus 1E2 (nonopsonic mouse IgG1 to PPS3). Download Figure S4, TIF file, 4 MB.
Primers used in this study.
Gene expression of A66.1 cells treated with CSP2, MAbs, or CSP2 plus MAbs.
Top 25 hypothetical genes increased or decreased by >2.5 fold by 1E2, CSP2, and/or controls.
ACKNOWLEDGMENTS
We thank D. Morrison for discussion, reagents, and critical reading of the manuscript and M. Feldmesser for critical reading of the manuscript and helpful suggestions.
This work was supported by the National Institutes of Health (NIH) (R01AI045459 and R01AI044374 to L.P), the NIAID training grant Geographic Medicine and Emerging Infections (5T32AI070117 to S.G), and the NIH Ruth L. Kirschstein Molecular Pathogenesis Training Grant (5T32AI007506 to J.R.C).
Footnotes
Citation Yano M, Gohil S, Coleman JR, Manix C, Pirofski L. 2011. Antibodies to Streptococcus pneumoniae capsular polysaccharide enhance pneumococcal quorum sensing. mBio 2(5):e00176-11. doi:10.1128/mBio.00176-11.
REFERENCES
- 1. Whitney CG, et al. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737–1746 [DOI] [PubMed] [Google Scholar]
- 2. Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jackson LA, et al. 2003. Effectiveness of pneumococcal polysaccharide vaccine in older adults. N. Engl. J. Med. 348:1747–1755 [DOI] [PubMed] [Google Scholar]
- 4. Weinberger DM, et al. 2010. Association of serotype with risk of death due to pneumococcal pneumonia: a meta-analysis. Clin. Infect. Dis. 51:692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson LA, Janoff EN. 2008. Pneumococcal vaccination of elderly adults: new paradigms for protection. Clin. Infect. Dis. 47:1328–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robbins JB, Schneerson R, Szu SC. 1995. Perspective: hypothesis: serum IgG antibody is sufficient to confer protection against infectious diseases by inactivating the inoculum. J. Infect. Dis. 171:1387–1398 [DOI] [PubMed] [Google Scholar]
- 7. Romero-Steiner S, et al. 1997. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin. Diagn. Lab. Immunol. 4:415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabrizio K, Groner A, Boes M, Pirofski LA. 2007. A human monoclonal immunoglobulin M reduces bacteremia and inflammation in a mouse model of systemic pneumococcal infection. Clin. Vaccine Immunol. 14:382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian H, Weber S, Thorkildson P, Kozel TR, Pirofski LA. 2009. Efficacy of opsonic and nonopsonic serotype 3 pneumococcal capsular polysaccharide-specific monoclonal antibodies against intranasal challenge with Streptococcus pneumoniae in mice. Infect. Immun. 77:1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burns T, Zhong Z, Steinitz M, Pirofski LA. 2003. Modulation of polymorphonuclear cell interleukin-8 secretion by human monoclonal antibodies to type 8 pneumococcal capsular polysaccharide. Infect. Immun. 71:6775–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fabrizio K, Manix C, Guimaraes AJ, Nosanchuk JD, Pirofski LA. 2010. Aggregation of Streptococcus pneumoniae by a pneumococcal capsular polysaccharide-specific human monoclonal IgM correlates with antibody efficacy in vivo. Clin. Vaccine Immunol. 17:713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Håvarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. 2006. New insights into the pneumococcal fratricide: relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 59:1297–1307 [DOI] [PubMed] [Google Scholar]
- 13. Oggioni MR, et al. 2004. Antibacterial activity of a competence-stimulating peptide in experimental sepsis caused by Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:4725–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khajanchi BK, Kirtley ML, Brackman SM, Chopra AK. 2011. Immunomodulatory and protective roles of quorum-sensing signaling molecules N-acyl homoserine lactones during infection of mice with Aeromonas hydrophila. Infect. Immun. 79:2646–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Porat N, et al. 2008. An international serotype 3 clone causing pediatric noninvasive infections in Israel, Costa Rica, and Lithuania. Pediatr. Infect. Dis. J. 27:709–712 [DOI] [PubMed] [Google Scholar]
- 16. Yano M, Pirofski LA. 2011. Characterization of gene use and efficacy of mouse monoclonal antibodies to Streptococcus pneumoniae serotype 8. Clin. Vaccine Immunol. 18:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo P, Morrison DA. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451–475 [DOI] [PubMed] [Google Scholar]
- 19. Reichmann P, Hakenbeck R. 2000. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol. Lett. 190:231–236 [DOI] [PubMed] [Google Scholar]
- 20. Dawid S, Sebert ME, Weiser JN. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92 [DOI] [PubMed] [Google Scholar]
- 22. Rogers PD, et al. 2007. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 59:616–626 [DOI] [PubMed] [Google Scholar]
- 23. McClelland EE, Nicola AM, Prados-Rosales R, Casadevall A. 2010. Ab binding alters gene expression in Cryptococcus neoformans and directly modulates fungal metabolism. J. Clin. Invest. 120:1355–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin B, Quentin Y, Fichant G, Claverys JP. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14:339–345 [DOI] [PubMed] [Google Scholar]
- 25. Kmetzsch L, et al. 2011. Role for Golgi reassembly and stacking protein (GRASP) in polysaccharide secretion and fungal virulence. Mol. Microbiol. 81:206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taborda CP, Casadevall A. 2002. CR3 (CD11b/CD18) and CR4 (CD11c/CD18) are involved in complement-independent antibody-mediated phagocytosis of Cryptococcus neoformans. Immunity 16:791–802 [DOI] [PubMed] [Google Scholar]
- 27. Torosantucci A, et al. 2005. A novel glyco-conjugate vaccine against fungal pathogens. J. Exp. Med. 202:597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McDaniel LS, Scott G, Kearney JF, Briles DE. 1984. Monoclonal antibodies against protease-sensitive pneumococcal antigens can protect mice from fatal infection with Streptococcus pneumoniae. J. Exp. Med. 160:386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burns T, Abadi M, Pirofski LA. 2005. Modulation of the lung inflammatory response to serotype 8 pneumococcal infection by a human immunoglobulin M monoclonal antibody to serotype 8 capsular polysaccharide. Infect. Immun. 73:4530–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coleman JR, Papamichail D, Yano M, Garcia-Suarez MD, Pirofski LA. 2011. Designed reduction of Streptococcus pneumoniae pathogenicity via synthetic changes in virulence factor codon-pair bias. J. Infect. Dis. 203:1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar R, et al. 2010. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics 11:350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feng J, et al. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res. 19:1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang Q, Zhong Z, Lees A, Pekna M, Pirofski L. 2002. Structure-function relationships for human antibodies to pneumococcal capsular polysaccharide from transgenic mice with human immunoglobulin loci. Infect. Immun. 70:4977–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maitta RW, et al. 2004. Protective and nonprotective human immunoglobulin M monoclonal antibodies to Cryptococcus neoformans glucuronoxylomannan manifest different specificities and gene use profiles. Infect. Immun. 72:4810–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Opsonic activities of ST8-binding MAbs. Opsonophagocytic killing was determined as described in the text with the MAbs indicated in the title of each panel. The percent CFU for each reaction was determined by dividing the number of CFU for the condition containing the indicated MAb by the number of CFU obtained with the same reactants without the MAb. The bars on the x axis represent (i) the MAb alone, (ii) the MAb with serum (mouse) complement (SC), (iii) the MAb with J774 cells, and (iv) the MAb with SC and J774 cells. (A) 28H11 (nonopsonic mouse IgM to PPS8). (B) 31B12 (opsonic mouse IgG1 to PPS8). (C and D) Species- and isotype-matched control MAbs. Download Figure S1, TIF file, 0.7 MB.
Construction of comE mutant strain. A66.1::comE was constructed by inserting a kanamycin resistance marker into the comE gene. Lane 1 shows the wild-type comE gene (774 bp), and lane 2 shows A66.1::comE (1,500 bp). Download Figure S2, TIF file, 1.2 MB.
Heat maps showing the effect of CSP2 and MAbs. Global gene expression profiling of early-log-phase strain A66.1 was done after incubation with CSP2 or with CSP2 plus 1E2 (opsonic mouse IgG1 to PPS3) for different times in the presence (a) or absence (b) of CSP2. Untreated cells were not treated with any additional reagents. Genes that were increased in expression by >2.5 fold are shown in Table S3. Download Figure S3, TIF file, 4.1 MB.
MAb has no effect on comE mutant pneumococcal strain A66.1::comE. This experiment was performed as described in the legend to Fig. 2, except that the comE-deficient mutant A66.1::comE was used. Less than 1% killing was observed after incubation with CSP2 alone or with CSP2 plus 1E2 (nonopsonic mouse IgG1 to PPS3). Download Figure S4, TIF file, 4 MB.
Primers used in this study.
Gene expression of A66.1 cells treated with CSP2, MAbs, or CSP2 plus MAbs.
Top 25 hypothetical genes increased or decreased by >2.5 fold by 1E2, CSP2, and/or controls.