ABSTRACT
All fully sequenced strains of Streptococcus pneumoniae possess a version of the blp locus, which is responsible for bacteriocin production and immunity. Activation of the blp locus is stimulated by accumulation of the peptide pheromone, BlpC, following its secretion by the ABC transporter, BlpA. The blp locus is characterized by significant diversity in blpC type and in the region of the locus containing putative bacteriocin and immunity genes. In addition, the blpA gene can represent a single large open reading frame or be divided into several smaller fragments due to the presence of frameshift mutations. In this study, we use a collection of strains with blp-dependent inhibition and immunity to define the genetic changes that bring about phenotypic differences in bacteriocin production or immunity. We demonstrate that alterations in blpA, blpC, and bacteriocin/immunity content likely play an important role in competitive interactions between pneumococcal strains. Importantly, strains with a highly conserved frameshift mutation in blpA are unable to secrete bacteriocins or BlpC, but retain the ability to respond to exogenous peptide pheromone produced by cocolonizing strains, stimulating blp-mediated immunity. These “cheater” strains can only coexist with bacteriocin-producing strains that secrete their cognate BlpC and share the same immunity proteins. The variable outcome of these interactions helps to explain the heterogeneity of the blp pheromone, bacteriocin, and immunity protein content.
IMPORTANCE
Streptococcus pneumoniae resides in a polymicrobial environment and competes for limited resources by the elaboration of small antimicrobial peptides called bacteriocins. A conserved cluster of genes in the S. pneumoniae genome is involved in the production of bacteriocins and their associated protective immunity proteins through secretion of a signaling pheromone. In this study, we show that a significant number of strains have lost the ability to secrete bacteriocins and signaling pheromones due to a specific mutation in a dedicated transporter protein. Because the regulatory and immunity portion of the locus is retained, these “cheater” strains can survive in the face of invasion from a bacteriocin-producing strain without the cost of bacteriocin secretion. The outcome of such interactions depends on each strain’s repertoire of pheromone, immunity protein, and bacteriocin genes, such that intrastrain competition drives the diversity in bacteriocin, immunity protein, and pheromone content.
Introduction
Streptococcus pneumoniae (the pneumococcus) is a leading cause of bacteremia, meningitis, pneumonia, and otitis media. The majority of young children are colonized with this organism at some point during their toddler years. Nasopharyngeal colonization is largely asymptomatic but is a prerequisite for the development of invasive disease. In order to survive within the polymicrobial environment of the nasopharynx, the pneumococcus must compete with the endogenous flora, including other pneumococci. The prevalence of colonization in young children is high, and simultaneous carriage of more than a single strain is common. In addition, S. pneumoniae is one of a group of related streptococci colonizing the human upper respiratory tract. It is, therefore, not surprising that there is competition among pneumococci and possibly between pneumococci and other oral streptococcal species.
The blp-encoded bacteriocins have been shown to contribute to intraspecies competition in a murine model of colonization (1). Sequence analysis has demonstrated that the locus is heterogeneous, with the potential to produce a wide array of bacteriocins and associated immunity proteins (1, 2). The locus contains genes that encode a typical three-component regulatory system (blpCRH) and an ABC transporter (blpAB), as well as conserved proteins that may contribute to bacteriocin immunity (BlpYZ and SPO547) (Fig. 1A) (1–3). The peptide pheromone, BlpC, is produced with a typical leader peptide linked to a double-glycine motif. The prepeptide is cleaved and transported out of the cell via BlpAB (4). When levels of BlpC are sufficiently high, the pheromone triggers activation of the histidine kinase BlpH, resulting in activation of the response regulator BlpR (5). BlpR binding has been shown to result in the upregulation of the entire blp locus (3). The region of the locus between blpA and bplY contains genes predicted to encode a variable array of bacteriocin-like peptides and bacteriocin-specific immunity proteins; we refer to this region as the “bacteriocin immunity region” (BIR) (Fig. 1A). The bacteriocin-like peptides in this region are identified by a conserved N-terminal leader sequence followed by a double-glycine motif and are typically cotranscribed with specific immunity proteins. From the sequences available, there are at least 11 distinct putative bacteriocin peptides, and several of these have allelic variability. Specific inhibitory activity has only been experimentally attributed to one subset of the potential bacteriocins, namely, those encoded by specific alleles of blpMN (1). In addition to the variability in the BIR, sequence analysis of the available genomes has demonstrated that there are at least four distinct alleles of the gene encoding the peptide pheromone, blpC (3, 6). Several studies have shown that many strains with apparently intact blp loci lack any appreciable inhibitory activity in vitro, suggesting additional levels of regulation may prevent energetically costly bacteriocin secretion (1, 2, 7). In this work, we demonstrate that interruption of the blpA open reading frame (ORF) by a widely conserved 4-bp insertion results in strains that are unable to secrete peptide pheromone or bacteriocin peptides. These strains, which make up a significant portion of the pneumococcal population, retain the ability to sense exogenous pheromone and thereby initiate production of immunity proteins. The interplay between bacteriocin producers and these “cheater” strains results in selective pressure on the BIR to alter bacteriocin and immunity content and the blpC-blpH pair to alter both the broadcast signal limiting cross talk and the sensor improving detection. These findings explain the enormous diversity of pheromone-functional peptide combinations.
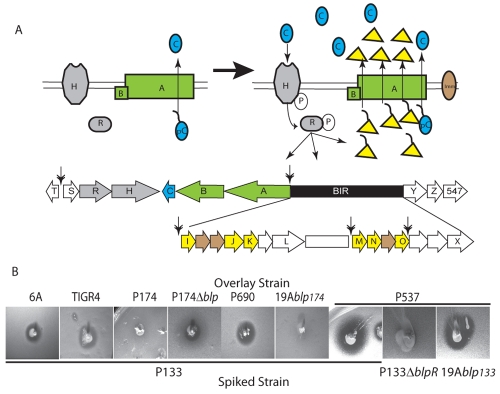
FIG 1 .
Model for blp locus activation and demonstration of the inhibitory phenotype of isolate P133. (A) Diagrammatic representation of the blp locus showing the location of the BIR between the conserved genes blpA and blpY. The genomic organization of the P133/TIGR4 BIR is shown as an example. Under low-density conditions, BlpC is secreted at a low basal levels by the BlpAB complex. When local BlpC concentrations are sufficiently high, BlpC binding to BlpH results in activation of the response regulator BlpR. Activated BlpR upregulates the production of blp transcripts at the sites marked by double arrows. Activation of the locus results in production of immunity proteins and accumulation of prebacteriocin, which is processed and secreted by BlpAB. Putative bacteriocins are shown in yellow, putative bacteriocin-specific immunity proteins are shown in brown, the ABC transporter is shown in green, the regulatory proteins are shown in gray, and BlpC is shown in blue. Genes of unknown significance are shown in white. Genes with existing blp annotation are labeled. (B) Isolates were tested for inhibition (spiked strain) or immunity (overlay strain) using overlay assays. Zones of clearing surrounding the spiked isolate signify inhibition.
RESULTS
Characterization of the blp locus from an inhibitory strain, P133.
In an attempt to identify novel bacteriocin activity, we screened a large number of pneumococcal isolates for their ability to inhibit the prototypic BlpMN producer, strain 6A, in plate overlay assays. We identified P133 as an inhibitory strain that produced a clear zone of inhibition when tested against strain 6A and the fully sequenced strain TIGR4 (Fig. 1B). Deletion of blpRH in P133 resulted in loss of inhibition, suggesting that the inhibitory activity requires a functional blp locus (Fig. 1B). To further clarify the dependence of inhibition on the blp locus, the locus of P133 was moved into a heterologous type 19A background. This strain gained inhibitory activity, demonstrating that the locus is both necessary and sufficient for the inhibitory activity of P133 (Fig. 1B). Sequence analysis of the BIR of P133 demonstrated significant homology to the BIR of the fully sequenced TIGR4 strain, containing the putative bacteriocin gene clusters blpIJK and blpMNO (Fig. 1A).
Characterization of the blp locus of a strain with immunity to P133.
We identified a strain, P174, that had immunity to the inhibitory activity of P133 but lacked any blp-dependent inhibition (Fig. 1B) (data not shown). We used an approach similar to that used with the P133 locus to confirm that P174 immunity was blp dependent by examining the immunity of both a blp deletion in P174 (P174∆blp) and a 19A strain containing the P174 locus (19Ablp174). P174∆blp lost immunity to P133, while 19Ablp174 gained immunity to P133 in overlay assays compared with the original 19A strain, P690 (Fig. 1B), demonstrating that immunity to the P133 elaborated bacteriocins requires the P174 blp locus. The content of the BIR was examined by comparing restriction profiles of P174 with those of P133 and TIGR4. The restriction profiles of the three strains were identical, suggesting similar bacteriocin/immunity protein contents (see Fig. S1 in the supplemental material). The presence of a similar BIR to P133 combined with the lack of blp-dependent inhibition suggested that P174 lacks a functional bacteriocin/pheromone secretion system. Analysis of the blpA and blpC genes from P133 and P174 demonstrated that the pair had identical BlpC types, but their blpA genes differed with respect to a 4-bp repeat insertion in the transporter gene, blpA, found only in P174. This insertion results in a frameshift mutation that disrupts the single open reading frame of blpA (Fig. 2A). The repeat has been identified in the blpA sequence of a number of fully sequenced strains, including the TIGR4 strain, but its significance was unknown.
FIG 2 .
Correction of the AAGC repeat in P174 results in active transcription of the blp locus and recovery of inhibitory activity. (A) Demonstration of the effect of the 4-bp repeat on the blpA ORF. The sequence starts at nt +433 from the blpA start codon for the predicted translated product. The 4-bp repeat sequence is underlined. Amino acids highlighted in light gray are found in the intact BlpA product; dark gray amino acids are found only in the disrupted BlpA protein. Predicted new stop and start codons resulting from the frameshift mutation are marked by boxes. (B) Diagramatic representation of construction of the on and off reporter strains in P174. P174 transformed with pE65 was selected on kanamycin to recover strains with plasmid integration. pE65 has the 5′ region of the non-repeat-containing blpA gene from P133 followed by divergent promoters driving blpA (PA) and blpI transcription (PI). PI drives lacZ expression. Integration at point 1 results in repair of the disrupted blpA ORF in P174, while integration at point 2 results in retention of the disrupted blpA gene. Promoters are shown with solid arrows signifying the direction of transcription. (C) Phenotypic confirmation of P174on-repaired and P174off demonstrating transcriptional activity on plates containing X-Gal with and without 100 ng/ml of BlpCR6 in strains with the full-length blpA ORF and dependence on exogenous BlpC in the strains with a disrupted blpA ORF. (D) P133 induces blp transcription in the P174off strain when X-Gal is included in the overlay. (E) BlpC6A secretion confirmed in strain 19Ablp133blpC6A using reporter strain P1802 and X-Gal in the overlay. 19Ablp133CR6 was unable to inhibit 19Ablp174 (F), while 19Ablp133C6A showed clear inhibition against 19Ablp174 in the overlay (G). (H) Inhibitory activity was noted against P537 in P174onΔlanC but not in P174offΔlanC.
Characterization of the role of the 4-bp repeat in regulation of the activity of the blp locus.
To further examine the correlation between the presence of an intact blpA gene and the ability to secrete Blp bacteriocins, we constructed an integration plasmid containing 700 bp of P133 blp sequence that was designed to both repair the 4-bp insertion in the P174 blpA gene and function as a reporter for blp transcriptional activity by taking advantage of the proximity of the divergent promoters driving blpI and blpA to the 4-bp repeat region (Fig. 2B). The resultant P174 transformants fell into one of two phenotypes when plated on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-containing media, depending on the site of integration of the plasmid. When plasmid integration occurred upstream of the repeat, resulting in an intact blpA ORF, the strains had endogenous responsiveness to their own BlpC secretion (P174on-repaired). Plasmid integration downstream of the repeat (P174off) resulted in strains that were only transcriptionally active when BlpCR6 peptide was added exogenously to the media (Fig. 2C). P133 was able to induce transcription of the blp locus in the P174off strain when tested in overlay media containing X-Gal, demonstrating transcriptional response to P133-secreted pheromone (Fig. 2D). To further demonstrate the dependence of P174 immunity on BlpCR6 secretion, a chimeric strain, 19Ablp133C6A, was created in which P133 BIR transcription was driven by a Blp6A regulatory region, which is of a distinct pherotype (Fig. 2E). Unlike the original 19Ablp133 strain (Fig. 2F), 19Ablp133C6A was able to inhibit the growth of 19Ablp174 (Fig. 2G), verifying that P174 immunity is activated by BlpCR6 that is secreted by P133. As further confirmation of the importance of an intact blpA gene, repair of the repeat in P174 (P174on) resulted in a strain with inhibitory activity when tested in overlay assays (Fig. 2H). These findings confirm that the presence of the frameshift mutation results in a strain that can only turn on the blp locus in response to exogenous peptide pheromone. Remote activation provides immunity but not inhibitory activity because the defect in the ABC transporter prevents secretion of both BlpC pheremone and bacteriocin.
Demonstration of the cheater phenotype in vivo.
To demonstrate that the in vitro interactions that we have observed between bacteriocin-sensitive, producer and immune strains are relevant on mucosal surfaces, we examined the interactions between otherwise isogenic strains with these three phenotypes in a mouse model of colonization. The type 19A strain with the P133 blp locus (19Ablp133; bacteriocin producer) was competed against a 19A derivative with a deletion in its BIR (19AΔBIR; bacteriocin sensitive) or the 19A strain containing the entire P174 blp locus (bacteriocin immune). Each strain was inoculated singly or in combination into the mouse nasopharynx, and colonization was assessed by nasal wash. All strains colonized at similar levels when given alone (Fig. 3A). The 19Ablp133 bacteriocin producer strain outcompeted 19AΔBIR but not 19Ablp174 (Fig. 3B), suggesting that the blp-mediated competition and induced immunity observed in plate overlay assays also occur on mucosal surfaces.
FIG 3 .
The 19A strain expressing the P133 blp locus outcompetes 19AΔBIR but not the 19A strain expressing the P174 blp locus during mouse nasopharyngeal colonization. (A) Mice were singly colonized with 19AΔBIR, 19Ablp133CR6, or 19Ablp174, and colonization density was determined at day 4 postinoculation. Colonization levels were not significantly different, as determined by Mann-Whitney analysis. A dotted line indicates the limit of detection. (B) Competitive index of 19AΔBIR or 19Ablp174 when cocolonized with 19Ablp133CR6. The horizontal line represents the mean for each group. The competitive index (CI) was calculated for each mouse, and P values comparing CI values in each group were determined by Mann-Whitney analysis. The dotted line denotes equivalent colonization at a competitive index of 1.
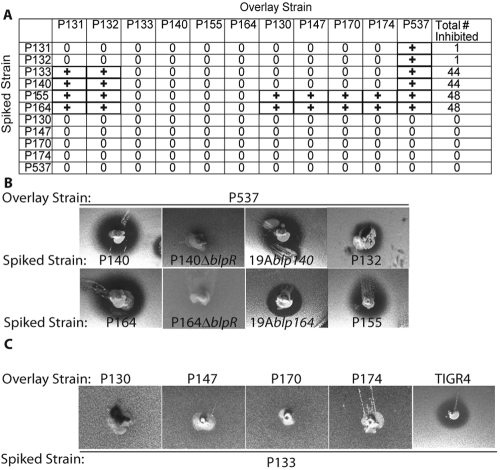
Characterization of the inhibitory profile of 51 colonizing and invasive isolates from South Africa.
To verify the correlation of an intact blpA with endogenous activity of the locus, we expanded our study to include the remainder of the 51-member strain collection to which P133 and P174 belong. This collection was derived from strains compiled from ongoing surveillance studies in South Africa (SA isolates) and includes colonizing and invasive isolates of 15 distinct serotypes (see Tables S1 and S2 in the supplemental material). Using agar overlay assays, we tested each isolate against itself and the remaining isolates (Fig. 4A). Three isolates (P140, P155, and P164) inhibited the majority of other strains with a spectrum similar to that of P133 (Fig. 4B). Two isolates, P131 and P132, had inhibition limited to the blp deletion strain P537 (Fig. 4B) (data not shown). The BIR of these strains was sequenced, and putative bacteriocins were identified. P140 had only the blpIJK cluster, while P155 and P164 had identical BIR sequences and contained genes encoding BlpIJ followed by two putative bacteriocin-like proteins not found in the existing sequence database. The BIR sequence of P132 contained only blpK. Deletion and heterologous expression strains were created using the blp loci from P140 and P164 to verify the dependence of the blp locus on inhibitory activity in both isolates (Fig. 4B). The spectra of inhibition of isolates P133 and P140 were identical and included all but each other, P164, P155, and four “immune-only” isolates, P130, P147, P170, and P174 (Fig. 4A and 4C). The fact that P133 did not inhibit P140 or P164/P155 suggests that the inhibitory activity of P133 is largely attributed to the shared blpIJK cluster rather than the blpMNO cluster found only in P133. Unlike P133 and P140, P164 and P155 were able to inhibit the growth of the immune-only isolates (Fig. 4A).
FIG 4 .
Summary of inhibitory and immunity profiles of the South African isolates. (A) Table summarizing the results of overlays of each of the 51 isolates and P537 against each other. 0 denotes no inhibition, and + denotes inhibition of the overlay. (B) Overlay assays demonstrating the inhibitory activity of selected South African isolates and their derivatives demonstrating that the locus is necessary and sufficient for inhibitory activity. (C) P133 spiked into a plate fails to inhibit the growth of P130, P147, P170, and P174 but not TIGR4.
RFLP mapping of the BIR in the SA isolate collection demonstrating heterogeneity.
In order to study the heterogeneity of the BIR in the SA isolate collection, we analyzed this region from each isolate by restriction fragment length polymorphism (RFLP) analysis. The BIR from each isolate was amplified by PCR using conserved sequence primers within blpA and blpY. PCR products were obtained in 49/51 isolates. Attempts to amplify the BIR of the two remaining isolates using different primers in blpA and blpY were unsuccessful. The 49 BIR products were then purified and digested separately with AseI or MfeI to create a restriction map of the region (Table 1; see Fig. S1 in the supplemental material) (data not shown). The resultant banding patterns were compared with predicted patterns from the 15 known distinct BIR regions in the sequence database and the newly sequenced broadly active isolates (see Table S3 in the supplemental material) (2, 8–13). If an isolate had different restriction patterns from any other isolate in one of the two digests, it was assigned to a separate group. Using this strategy, we were able to divide the 51 isolates into 17 groups. Ten of these could be assigned to known BIR sequences in the database, based on their restriction patterns (Table 1). P131 and P132 had identical BIR sequences by RFLP analysis. The four immune-only isolates had identical RFLP patterns to P133, suggesting that their bacteriocin and immunity contents were identical (see Fig. S1 in the supplemental material).
TABLE 1 .
BIR group assignment of the South African isolates.
| Group no. (pattern type)a | No. of isolates | RFLP match to GSSb or known BIR sequence | Serotype(s) represented | blpC type(s) |
|---|---|---|---|---|
| 1 | 12 | 2306 | 6B, 9N, 15B, 18A, 23A, 23F | P164, R6, 6A |
| 2* | 6 | TIGR4, P133 | 6A, 6B, 23F | R6, 6A |
| 3 | 5 | 1031 | 6A, 11A, 22 | P164, R6, T4 |
| 4 | 4 | 19F | 6A | |
| 5* | 3 | P132 | 23F, 29 | 6A |
| 6 | 4 | TCH8431 | 19F, 45 | P164, 6A |
| 7 | 2 | ATCC 700669 | 6A, 14 | T4 |
| 8 | 2 | 22 | 6A | |
| 9* | 2 | P164, P155 | 6B, 45 | P164,P155 |
| 10 | 2 | CDC 1873 | 19A, 23F | 6A, T4 |
| 11 | 1 | SP3BS71 | 3 | R6 |
| 12* | 1 | P140 | 35B | R6 |
| 13 | 1 | 6A | R6 | |
| 14 | 1 | 14BS69 | 14 | 6A |
| 15 | 1 | 3 | 6A | |
| 16 | 1 | 19F | 6A | |
| 17 | 1 | 35 | 6A | |
| NAc | 2 | 15B, 19F | 6A |
Groups with an asterisk have members with inhibitory activity.
GSS, genome sequenced strains represented by GenBank designations. Isolate numbers preceded by “P” represent strains described in this study with available BIR sequence information.
NA, not amplified.
Determination of the blpC type in the SA isolate collection.
The blpC gene was sequenced in all isolates (Table 1; see Table S2 in the supplemental material) and was found to be largely unlinked to BIR group. The largest BIR group had representatives from three of the four known pherotypes. The variability of the blpC type suggests that the pherotype is under selective pressure with new pherotypes acquired by horizontal gene transfer, similar to what has been observed with the homologous competence-stimulating peptide locus (14). P155 had a novel blpC sequence (see Table S4 in the supplemental material) that was clearly distinct from the four known pherotypes and notably from the blpC164 type produced by P164 which carries an identical BIR sequence. P133 and P140 share the same blpCR6 pheromone as the four immune-only isolates, suggesting that, like P174, the immune-only strains may have disrupted blpA genes. P164 and P155, which retain the ability to inhibit the immune-only isolates, express non-blpCR6 pheromones. These observations, in combination with the distinct inhibitory profile of pherotype switch strains, confirm that pherotype is critical in dictating the outcome of producer-cheater interactions. Matching pheromone response systems, if combined with appropriate immunity proteins, will result in protection from bacteriocin-mediated inhibition even in transporter-defective strains.
Construction of type 1, 2, and 3 reporter strains.
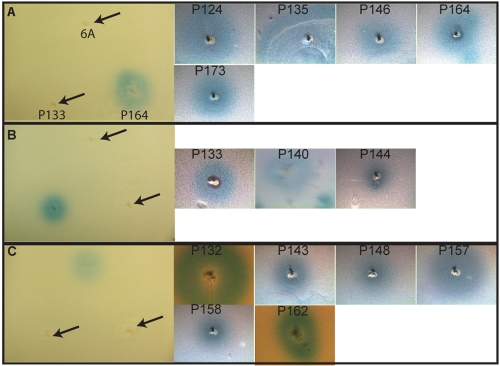
To address the possibility that more isolates had active blp loci but lacked inhibition in overlay assays, we designed a series of reporter strains to evaluate for pheromone secretion. We theorized that pheromone secretion could serve as a correlate for bacteriocin secretion because the same machinery is required for the secretion of both peptides, and detection of pheromone secretion does not require knowledge of the target organisms. To assess whether the isolate collection contained isolates that were capable of secreting peptide pheromone, we constructed three strains with lacZ reporter genes fused to the BIR promoter in strains responsive to BlpC types P164, R6, and 6A. No reporter could be constructed against BlpCT4 because no active isolates with this type could be identified. The blpA gene was disrupted in each reporter so that strains would respond only to exogenously added BlpC. The absence of cross stimulation of the reporter constructs by noncognate pherotypes was verified using known pheromone secretors (Fig. 5A to C). The SA isolate collection was tested in reporter overlay assays with each of the three reporter strains. Isolates secreting pheromone were identified by a zone of blue surrounding the spiked growth. Using this strategy, we were able to identify 13 additional noninhibitory isolates that were actively secreting peptide pheromone (Fig. 5).
FIG 5 .
Representative isolates with pheromone secretion detected in overlays with reporter strains specific for BlpC164 (A), BlpCR6 (B), or BlpC6A (C). Isolates were inoculated into TSA plates 6 h before a soft agar overlay was applied containing the BlpC-specific reporter overlay strain and X-Gal. The specificity of the reporters is demonstrated in the first panel of each row. Arrows demonstrate where secreting strains of the different pherotype were spiked. In the case of P133, a zone of inhibition can be seen inside the larger zone of pheromone secretion.
blpA sequence variation in active and inactive isolates.
To verify the correlation of pheromone secretion with an intact blpA ORF, we sequenced the region surrounding the repeats in all isolates. Twenty-five of 51 isolates lacked the 4-bp repeat consistent with an intact blpA ORF. These 25 isolates included all 6 inhibitory isolates and each of the 13 pheromone-secreting isolates. Eighteen of 51 isolates had a blpA gene with the 4-bp repeat, including P174 and the three additional immune-only isolates. Like P174, the remaining immune isolates lacked the ability to secrete bacteriocin or pheromone but could respond to exogenous BlpCR6, resulting in bacteriocin-specific immunity. The blpA gene could not be amplified in 8 isolates, suggesting the presence of either significant sequence variation in the primer binding site or a large deletion.
blpA sequence alignment and phylogenetic analysis.
To address the question of whether the repeat insertion or deletion of the 4-bp sequence is likely to occur within a clonal population, we performed phylogenetic analysis of the 500 bp of the blpA sequence which flank the repeat region and compared this with the 3′-most 500 bp of the gene. To remove the variation in repeat sequence from the analysis, the 4-bp repeat was removed from all repeat-containing genes. All available full-length blpA sequences from our data and the database were used for this analysis (2, 8–13). A neighbor-joining phylogenetic tree was created for both regions (Fig. 6A to C). The tree derived from the region flanking the repeats demonstrated clustering of repeat-containing strains. This clustering became much less apparent when the 3′ regions were analyzed and when the blpC type was considered. This finding suggests that acquisition of repeats is a result of homologous recombination with DNA from a repeat-containing strain rather than as a result of variation within a single clonal population.
FIG 6 .
Phylogenetic tree of blpA sequences in genome sequenced strains and South African isolates. (A) Phylogenetic tree of 500 bp of blpA sequence flanking the repeat region determined by the neighbor-joining method (25) after removal of the repeat sequence. Partitions reproduced in less than 25% of bootstrap replicates are collapsed. Percentages of replicate trees in which taxa cluster together in the bootstrap test are shown next to branches. Repeat-containing strains are designated by circles; isolates without the repeat are designated by squares. Pherotypes are designated by colors, as follows: P164, blue; R6, red; 6A, green; TIGR4, black; and P155, yellow. (B) Phylogenetic tree of 500 bp at the 3′ end of the blpA open reading frame analyzed as in panel A. (C) Representation of the regions of blpA analyzed in panels A and B drawn to scale. The small arrow designates the location of the repeat sequence.
DISCUSSION
Bacteriocin production provides organisms with a means to outcompete sensitive members of the surrounding flora. We were interested in the observation that many tested clinical isolates lack any appreciable inhibitory activity despite containing an apparently intact blp locus (1, 2). Our current study of an isolate collection from an area with a high rate of colonization and disease, where the bacterial isolates would be expected to have the most competitive phenotypes, allowed us to dissect the various strategies that pneumococci have adapted to take advantage of bacteriocin production and immunity. In studying this collection, we have identified a common cheater phenotype, where some strains bypass costly endogenous pheromone and bacteriocin secretion while maintaining the ability to express immunity in the presence of signal pheromone produced by surrounding strains. We have identified that the mechanism for this cheater phenotype occurs via a 4-bp insertion in the gene encoding the transporter BlpA, which is responsible for pheromone and bacteriocin secretion.
It is clear that the 4-bp insertion is present in a significant number of pneumococcal strains. A compilation of the 28 unique fully sequenced blp loci from the database demonstrates that 11/28 (39%) would be predicted to encode a functional full-length BlpA protein, 12/28 (43%) have the 4-bp frameshift repeat in blpA, and the remaining 5/28 (18%) have other frameshift mutations or large deletions in blpA. BlpA sequencing and phenotypic analysis have demonstrated that our South African isolate collection closely mimics these numbers: 26/51 (51%) isolates have the 4-bp repeat or a deletion in blpA, while 25/51 (49%) lack the 4-bp repeat (although the presence of mutations 3′ to the repeat region was not assessed in all cases).
Phylogenetic analysis of blpA sequences demonstrates tight clustering of repeat-containing strains only when sequences flanking the repeat region are analyzed. This suggests that repeats are gained and lost via horizontal transfer by homologous recombination with foreign DNA rather than via an endogenous source such as slipped-strand mispairing or intragenetic recombination. Repeat-containing blpA genes are found in strains in 4/6 major pneumococcal lineages defined by Donati et al. (15), and intact blpA genes are found in 5/6 lineages, further supporting the hypothesis that repeat acquisition and loss are a dynamic process rather than the result of expansion of a single ancestral strain. This is likely facilitated by the natural competence of this organism, which may allow individual isolates to sample different strategies for survival. Although other frameshift mutations can be found in the blpA gene, the 4-bp repeat is the predominant strategy for inactivation. Why this mutation has been selected for widespread dissemination over others is not known. Although theoretical, it is possible that the truncated N-terminal portion of BlpA that results from the frameshift plays a role in bacterial fitness.
It should be noted that, in the article by Lux et al., inhibitory activity attributable to the blp locus was noted in strains with frameshifted BlpA proteins (2). We have analyzed our isolate collection using identical assay conditions and have failed to note inhibitory activity or pheromone secretion in any isolate with an interrupted BlpA protein (data not shown).
It is unclear what selects for production over immunity only (cheaters) in pneumococcal populations. Populations of inhibitory, immune, and sensitive strains probably exist in constant flux, as has been described for colicin-producing populations due to a balance of energetic cost to competitive advantage via direct growth inhibition (16). The energetic cost of bacteriocin and pheromone secretion and the potential for activation of the locus even in the absence of competitors likely drive the selection of cheater strains that benefit from their neighbor’s bacteriocin production. Cheater strains run the risk of elimination if they encounter a producing strain that secretes a pheromone that they cannot sense. We have been unable to test this hypothesis experimentally by looking at growth kinetics or competition because the locus is only spontaneously activated when bacteria are grown on solid media. The fact that the entire locus, including the two-component regulatory genes, is maintained in every available sequenced strain, including those that lack the capacity for self-regulation, argues that the ability to respond to exogenous pheromone is an important survival strategy.
In this study, we also demonstrate that pherotype, blpA sequence, and BIR can vary independently of each other despite a separation of less than 4 kb on the genome. This suggests that separate selective pressures drive the diversity of BlpC type, BIR content, and activation status via blpA reading frame alterations. The observation that many strains exist in a transporter-deficient state, which only allows for immunity when bacteria are activated by a neighbor secreting their cognate BlpC type, may explain the selective pressure on pherotype. Strains that have an active blp locus likely benefit from variability in their BIR content by altering their arsenal of bacteriocin peptides, while the requirement for bacteriocin-specific immunity may drive variability of the BIR even in inactive strains. The striking variability in this locus provides us with a snapshot view of the effects of the pressure exerted on this organism by other members of the nasopharyngeal flora.
MATERIALS AND METHODS
The collection of colonizing and invasive disease isolates was approved by the Human Research Ethics Committee, University of the Witwatersrand, South Africa. Written informed consent was obtained from colonization study participants or their legal guardians. Invasive isolates were acquired as part of routine clinical care, and no patient information was recorded. All animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (17). The protocol was approved by the Committee on the Use and Care of Animals at the University of Michigan.
Bacterial strains, serotyping, and growth conditions.
Pneumococcal isolates were derived from ongoing colonization studies or invasive disease surveillance in South Africa and were derived from individuals who had colonization or disease with one or more serotypes (see Table S1 in the supplemental material). Pneumococci were serotyped by the Quellung method using specific antisera, including the new 6C antiserum (Statens Serum Institut, Copenhagen, Denmark). Pneumococci were grown at 37°C in Todd-Hewitt medium with 0.5% yeast extract (THY) or on tryptic soy agar (TSA) plates supplemented with 4,741 U of catalase (Worthington, Lakewood, NJ) or 5% sheep blood (SBA). Plates containing pneumococci were grown in 5% CO2. Escherichia coli was grown in Luria broth (LB) or LB agar with appropriate antibiotics. The following antibiotic concentrations were used: for S. pneumoniae, 500 µg/ml kanamycin, 200 µg/ml streptomycin, 2 µg/ml chloramphenicol, 1 µg/ml erythromycin, and 100 µg/ml spectinomycin; and for E. coli, 50 µg/ml kanamycin, 20 µg/ml chloramphenicol, 100 µg/ml erythromycin, and 100 µg/ml spectinomycin.
Overlay assays.
Strains to be tested for inhibitory activity were grown up initially on SBA plates overnight. Colonies were scraped from SBA plates and inserted into TSA plates with catalase using a 48-pin replicator or pipette tip. Growth was allowed to continue for 6 h at 37°C before the overlay was applied. Overlay strains were grown to an optical density at 620 nm (OD620) of 0.3 to 0.5 in THY medium at 37°C. Two hundred microliters of broth culture was added to 7 ml of molten TSA containing 0.5% agar and catalase and quickly applied to the top of the plate containing the pregrown isolates. Plates were incubated overnight at 37°C before being examined for zones of clearing. Activity overlay assays were performed in an identical manner, except 50 µl of X-Gal at 40 mg/ml was added to the soft agar before pouring.
Construction of gain- and loss-of-function strains.
Pneumococcal transformations were performed as previously described (1, 18). Serotype 19A replacements were performed by using the strain 19AΔBIR, which is a streptomycin-resistant strain that contains an exchangeable Kanr rpsL+ cassette encoding kanamycin resistance and streptomycin sensitivity (Janus cassette) (19) in place of the BIR. This strain was transformed with genomic DNA from the inhibitory isolates P133, P140, and P164, and transformants were isolated on TSA containing streptomycin. Each isolate was then back-transformed once, and the inhibitory phenotype was confirmed by overlay assay. A similar strategy was used to create the transformant containing the P133 BIR but the original type 3-producing upstream region. In this case, a type 6A strain carrying a Janus cassette in its BIR was transformed with P133 genomic DNA. Streptomycin-resistant colonies were screened for secretion of a type 3 blpC using reporter overlays with the type 3 reporter strain, P1802. Insertion of the P133 BIR was confirmed by RFLP analysis as described below. The entire blp locus from this strain was then moved into the 19Ablp deletion strain, P690. blpRH deletions were introduced into isolates P133, P140, and P164 by transformation with genomic DNA from strain 6AΔblpR by selection on erythromycin. Mutations were confirmed by PCR of the blpRH region. This strategy could not be used for P174 because it was resistant to erythromycin at baseline. To make a deletion in this isolate, whole-blp-locus deletions were constructed by joining the genomic region upstream of blpT in the TIGR4 strain to the region just upstream of blpX in the pCR2.1 vector by sequential cloning steps using primers 8 and 9 for the upstream portion and 10 and 11 for the downstream portion (see Table S5 in the supplemental material). The two regions were separated by a unique ClaI site. The upstream region and downstream region were separated by digestion with ClaI, and the Janus cassette with engineered ClaI sites generated with primers 6 and 7 was ligated to this plasmid. The resulting ligation was used to transform strain P376, and the resulting transformants were selected on TSA with kanamycin. The resulting strain, P537, was exquisitely sensitive to blp bacteriocin-mediated inhibition and was used for overlay assays to demonstrate inhibitory activity. This mutation was moved into P174 using genomic DNA. All constructs to be tested in mice were moved into the type 19A background, which colonizes mice at high levels without evidence of invasive disease. The streptomycin-resistant version of the type 19A strain, 19AstR, was transformed with genomic DNA from the blp deletion strain P537 and selected on kanamycin. After a single back-transformation to remove unlinked DNA, this strain was then transformed with P174 genomic DNA to create 19Ablp174 for colonization experiments. Transformants were selected on TSA containing streptomycin. RFLP analysis and overlay assays to assess inhibition and immunity were used to confirm transfer of the blp phenotype from the donor strain.
Mouse competitive colonization.
Six- to 7-week-old female BALB/c mice were inoculated intranasally with 0.5 × 107 to 1.0 × 107 CFU/10 µl of bacteria resuspended in phosphate-buffered saline (PBS). Each strain was given alone and in combination to five mice per group. Mice were sacrificed by CO2 asphyxiation, and nasal washes were performed and plated as previously described (1). Statistical analysis was performed by Mann-Whitney analysis. Colonization by 19Ablp133CR6 in the 19AΔBIR-19Ablp133CR6 dually colonized mice was determined by subtracting the kanamycin-resistant colonies from the colony count on nonselective media. Because there was no selective marker to distinguish 19Ablp133CR6 from 19Ablp174, colonies were picked from nonselective plates and allowed to grow in a 96-well plate containing THY medium for 6 h. Ninety-six colonies were chosen from each cocolonized mouse. Sixteen colonies were tested from each singly colonized mouse. The resulting cultures were replica plated on SBA plates, and overnight growth from these plates was stabbed into TSA plates. Overlay assays with the sensitive strain P537 was then performed. Colonies with inhibition were scored as by 19Ablp133CR6; those without inhibition were scored as 19Ablp174. Interpretability of the results was verified by confirming that 100% of organisms derived from singly colonized animals had the expected phenotype. Colony counts were obtained by multiplying the percentage of each strain determined by this method by the total colony counts on nonselective media. A competitive index was determined using the method of Monk et al. (20). P values for the competitive index were determined by Mann-Whitney analysis.
Construction of P174on and P174off strains.
The plasmid pE56 was constructed by amplifying a fragment containing the 5′ region of blpA and the divergent promoters of blpA and blpI plus 54 nucleotides (nt) of blpI coding sequence flanked by NsiI and XbaI sites off of the inhibitory strain P133, using primers 3 and 4 (see Table S5). This sequence was cloned into NsiI-XbaI-cut reporter plasmid pEVP3 (21) creating a transcriptional fusion of the blpI promoter to the lacZ gene. This plasmid was made kanamycin resistant by inserting the kanamycin gene amplified with primers 14 and 15 into a unique EcoRI site in the plasmid. The resulting plasmid, pE65, was transformed into P174, and transformants were selected on TSA containing kanamycin. Because the insertion event would result in a duplication of the inserted region, including the frameshift 4-bp repeat sequence found in P174 but not P133, the intact version of blpA in each transformant was determined by amplifying the blpA sequence in closest proximity to the lacZ gene using primers 3 and 17 and then sequencing this product using primer 16. Overlay assays evaluating for bacteriocin production with these strains were performed with strains carrying a deletion in a novel non-blp bacteriocin locus (lanC) to remove the possible impact of this locus on the results (data not shown).
RFLP analysis of the BIR, blpC, and blpA sequencing.
The BIRs of all isolates were analyzed by PCR amplification using conserved primers 1 and 2. PCR products were purified and digested with either AseI or MfeI. The resulting digestion patterns were compared after ethidium bromide (EtBr) staining with 1% agarose gels. Isolates with identical patterns in both digests were assigned to the same group. The blpC sequence was determined by sequencing the PCR fragment produced by amplification with primers 12 and 13. The blpA gene of selected isolates was sequenced in its entirety after amplification with primers 18 and 19. The repeat-containing region of the remaining isolates was amplified and sequenced using primers 3 and 16.
The BlpC type P164 and R6 pheromone reporter strains were constructed as follows. A fragment containing 700 bp upstream of the first bacteriocin gene, blpQ, in addition to the first 43 nt of coding sequence, was amplified from the type 6B strain P4 determined by sequencing to carry a type P164 blpC gene, using primers 3 and 5. The fragment was cloned into the reporter plasmid pEVP3 as described for the construction of pE56, creating plasmid pE57. This plasmid was transformed into the type 6B strain and selected on TSA plates with chloramphenicol. Insertion of the plasmid into the expected site in the blp locus was confirmed by PCR using primers 20 and 17. In order to facilitate further manipulation, genomic DNA from this strain was moved into the laboratory strain, R6x, which carries an R6 blpC gene. Transformants were selected on on TSA with chloramphicol, and the colonies were screened for responsiveness to type P164 or type R6 synthetic peptides by plating on TSA plates supplemented with X-Gal and 100 ng/ml of synthetic peptide (Genscript, Piscataway, NJ). One colony of each was chosen, and the blpC sequence verified by sequencing using primers 12 and 13. To remove the impact of endogenous BlpC secretion on these strains, the blpA gene was disrupted by transforming strains with the plasmid pE49, which contains the interrupted blpA gene from strain P1802, allelic exchange was verified using primers 21 and 23. The type 6A reporter strain, P1802, was previously constructed (7).
Construction of a phylogenetic tree of blpA nucleotide sequences surrounding and distant from the repeat region.
blpA sequences were obtained from the publicly accessible databases and from the strains in our collection in which the blpA gene was fully sequenced. Sequences in the 500 bp flanking the repeat region and a 500-bp fragment 933 bp away were aligned and evaluated using MEGA 5 (22). The evolutionary history was inferred using the neighbor-joining method (23). Bootstrap analysis was performed on 5,000 replicates. The evolutionary distances were computed using the maximum composite likelihood method (24). All positions containing gaps and missing data were eliminated, including the repeat sequence and a 27-nt duplicated sequence. Two genome sequence strains with large deletions in the blpA gene were not included in the analysis (Taiwan 19A and TCH8431/19A).
Nucleotide sequence accession number.
Newly derived sequences were submitted to GenBank under the following accession numbers: P133 BIR, HQ702850; P132 BIR, HQ676608; P140 BIR, HQ668083; P155 BIR, HQ668085, and P164 BIR, HQ668084. The accession numbers of the BlpA full sequence are as follows: P124, HQ690079; P131, HQ690080; P132, HQ690081; P140, HQ690082; P141, HQ690083; P144, HQ690084; P147, HQ690085; P155, HQ693884; P158, HQ693885; P162, HQ693886; P163, HQ693887; P164, HQ693888; P173, HQ693889; P174, HQ693890; and P155 BlpC, HQ668086.
SUPPLEMENTAL MATERIAL
RFLP digests of the BIR from the TIGR4 group and a seletion of the South African strain collection. The BIR was amplified with primers in the conserved flanking regions of blpA and blpY. PCR products were purified and digested with AseI or MfeI. Digests were separated on a 1% agarose gel and stained with EtBr. (A) Digestion of the BIR fragment from isolates with a TIGR4-like BIR. (B) AseI digest of the BIR from the 10 most common patterns. Numbers refer to group numbers designated in Table 1. Download Figure S1, PDF file, 1.2 MB.
Bacterial strains and plasmids used in this study.
Detailed summary of findings for South African isolates.
Compilation of blp locus information from available database entries.
BlpC type and amino acid sequence.
Primers used in this study.
ACKNOWLEDGMENTS
We acknowledge the Group for Enteric, Respiratory and Meningeal Disease Surveillance in South Africa (GERMS-SA) for their efforts in collecting the invasive isolates used in this study and Ankur Dalia and Jason Weinberg for careful reading of the manuscript.
This work was supported by grants from the NIH R01AI078538 (J.N.W. and S.D.) and K08AI071090 (S.D.) and by a generous donation from the AvFuel Corporation (Ann Arbor, MI).
Footnotes
Citation Son MR, et al. 2011. Conserved mutations in the pneumococcal bacteriocin transporter gene, blpA, result in a complex population consisting of producers and cheaters. mBio 2(5):e00179-11. doi:10.1128/mBio.00179-11.
REFERENCES
- 1. Dawid S, Roche AM, Weiser JN. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75:443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lux T, Nuhn M, Hakenbeck R, Reichmann P. 2007. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J. Bacteriol. 189:7741–7751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Saizieu A, et al. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Håvarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229–240 [DOI] [PubMed] [Google Scholar]
- 5. Pestova EV, Håvarstein LS, Morrison DA. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853–862 [DOI] [PubMed] [Google Scholar]
- 6. Reichmann P, Hakenbeck R. 2000. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol. Lett. 190:231–236 [DOI] [PubMed] [Google Scholar]
- 7. Dawid S, Sebert ME, Weiser JN. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191:1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Croucher NJ, et al. 2009. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniae Spain23F ST81. J. Bacteriol. 191:1480–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding F, et al. 2009. Genome evolution driven by host adaptations results in a more virulent and antimicrobial-resistant Streptococcus pneumoniae serotype 14. BMC Genomics 10:158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dopazo J, et al. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 7:99–125 [DOI] [PubMed] [Google Scholar]
- 11. Hiller NL, et al. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog. 6:e1001108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiller NL, et al. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J. Bacteriol. 189:8186–8195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tettelin H, et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506 [DOI] [PubMed] [Google Scholar]
- 14. Håvarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donati C, et al. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 11:R107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirkup BC, Riley MA. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412–414 [DOI] [PubMed] [Google Scholar]
- 17. National Research Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 18. Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39:508–518 [DOI] [PubMed] [Google Scholar]
- 19. Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67:5190–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monk IR, Casey PG, Cronin M, Gahan CG, Hill C. 2008. Development of multiple strain competitive index assays for Listeria monocytogenes using pIMC; a new site-specific integrative vector. BMC Microbiol. 8:96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123–128 [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 23. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 24. Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RFLP digests of the BIR from the TIGR4 group and a seletion of the South African strain collection. The BIR was amplified with primers in the conserved flanking regions of blpA and blpY. PCR products were purified and digested with AseI or MfeI. Digests were separated on a 1% agarose gel and stained with EtBr. (A) Digestion of the BIR fragment from isolates with a TIGR4-like BIR. (B) AseI digest of the BIR from the 10 most common patterns. Numbers refer to group numbers designated in Table 1. Download Figure S1, PDF file, 1.2 MB.
Bacterial strains and plasmids used in this study.
Detailed summary of findings for South African isolates.
Compilation of blp locus information from available database entries.
BlpC type and amino acid sequence.
Primers used in this study.