Abstract
For almost 50 years, Escherichia coli has been the model for understanding how bacteria orient their movement in response to chemical cues, but recent studies of chemotaxis in other bacteria have revealed interesting variations from prevailing paradigms. Investigating the human pathogen Helicobacter pylori, Amieva and colleagues [mBio 2(4):e00098-11, 2011] discovered a new chemotaxis regulator, ChePep, which modulates swimming behavior through the canonical histidine-aspartate phosphorelay system. Functionally conserved among the epsilonproteobacteria, ChePep is essential for H. pylori to navigate deep into the stomach’s gastric glands and may be an attractive target for novel antibiotics.
Commentary
Bacterial chemotaxis is one of the best-studied signal networks in biology, having been exhaustively investigated with genetics, biochemistry, and mathematical modeling (1). Hence, it is exciting when a brand-new player in the process is discovered. In the July/August issue of mBio, Howitt and colleagues (2) described a new chemotaxis regulator, ChePep, that they discovered in the gastric pathogen Helicobacter pylori and showed to be functionally conserved among the epsilonproteobacteria.
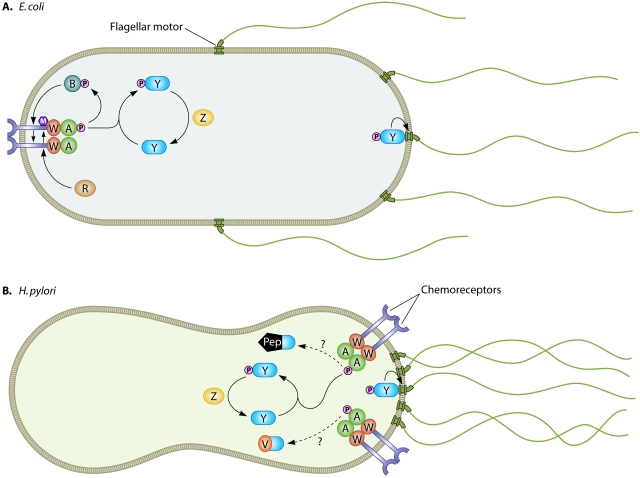
Chemotaxis is the directed movement of cells in response to chemical cues. In bacterial cells, chemoreceptors perceive chemical ligands and, through conformational changes, transduce information about their ligand-bound state to direct the activity of the histidine kinase CheA. CheA activity modulates the phosphorylation state of the response regulator CheY, which directly interacts with the flagellar motor machinery to control swimming behavior (Fig. 1).
FIG 1 .
Schematic representation of chemotaxis signaling in E. coli (A) and H. pylori (B). Chemoreceptors are shown in purple and flagellar motors in green spanning the cell membrane. Cytoplasmic chemotaxis proteins CheA, CheW, CheY, CheZ, CheR, CheB, CheV, and ChePep are labeled. Protein modifications are shown as pink circles for phosphorylation and purple hexagons for methylation. Activating interactions between signaling pathway components are indicated by arrows, and speculative interactions are indicated by dotted lines.
Bacterial cells are so small that as they move through a chemical gradient, at any moment in time a single cell experiences a fixed concentration of chemical, but over time it will experience different concentrations. Chemotaxis pathway signaling allows bacterial cells to convert temporal information of chemical exposure into spatial information about chemical concentrations and move up or down chemical gradients. Crucial to this directed motion is the process of adaptation, whereby chemoreceptors become desensitized to past ligand exposure so that they can perceive changes in ligand concentration across a wide range of concentrations. Escherichia coli, the poster child of bacterial chemotaxis research, carries out adaptation by methylating its chemoreceptors in a temporally and ligand-regulated manner to modulate their responsiveness (Fig. 1A).
As more bacterial chemoreception systems have been studied and more bacterial genomes have been sequenced, it has become increasingly clear that E. coli’s strategy for adaptation is not universally conserved (3). Many bacteria possess additional adaptation pathways that appear to control the efficiency of phosphorelay to CheY. One example is the CheV protein, absent in E. coli but widely distributed across the bacterial kingdom, which contains domains homologous to the CheA adaptor protein CheW and to the phosphorylatable response regulator domain of CheY. The H. pylori genome encodes three CheV proteins that are all involved in chemotaxis and appear to have distinct functions (4). In addition, a few bacterial genomes, including H. pylori’s, lack homologues of the chemoreceptor methylase CheR and demethylase CheB involved in adaptation in E. coli, and the H. pylori chemoreceptors lack conserved methylation sites. How H. pylori accomplishes adaptation is unknown, but ChePep may provide part of the answer (Fig. 1B).
Howitt et al. discovered ChePep serendipitously as a protein localized to H. pylori’s polar tuft of flagella (2). When they deleted chePep, they found that the mutant cells exhibited a hyperreversal phenotype similar to the hypertumbling phenotype observed in E. coli ΔcheB adaptation mutants (5). Howitt et al. visualized changes in bacterial swimming in response to microinjection of the chemorepellant hydrochloric acid to show that ΔchePep mutants are less efficient than wild-type cells at avoiding a noxious chemical (2). Similarly, E. coli cells that lack the ability to adapt (ΔcheR ΔcheB double mutants) exhibit only transient responsiveness to chemical stimuli (5). To tease apart where ChePep functions in the chemotransduction pathway, Howitt et al. performed epistasis experiments with other chemotaxis mutants (2). They found that a ΔchePep ΔcheY double mutant resembles the smooth-swimming ΔcheY single mutant; in other words, without the core signal transduction machinery, the absence of ChePep is immaterial. This suggests that ChePep’s activity is required for signaling events that modulate CheY function, as opposed to influencing flagellar motor activity directly, and is consistent with a role for ChePep in adaptation. The localization of ChePep to the flagellar pole may be consistent with a role in regulating chemoreceptors, since in H. pylori’s close relative Helicobacter hepaticus, the chemoreceptors are located adjacent to the polar flagella, in contrast to E. coli’s distribution of chemoreceptor in a “nose” distal from its lateral and polar flagella (6) (Fig. 1).
Che Pep is a novel, highly negatively charged protein, but its N terminus contains a putative response regulator domain homologous to that of CheY, including the invariant aspartate that is phosphorylated by histidine kinases such as CheA. Howitt et al. found ChePep sequences exclusively among the epsilonproteobacteria and showed that ChePep homologues from both Campylobacter jejuni (another human pathogen) and Caminibacter mediatlanticus (a hydrothermal vent resident), which had limited overall sequence similarity to H. pylori ChePep but conserved N-terminal response regulator domains, could rescue the H. pylori ΔchePep mutant (2). Based on the ΔchePep mutant phenotype and the presence of a CheY-type response regulator domain, it is tempting to speculate that ChePep functions in the process of chemoreceptor adaptation, possibly serving as a “phospho sink” that diverts phosphotransfer from the kinase CheA away from the response regulator CheY (Fig. 1B). Consistent with this model, the ChePep mutant behaves as if it has a surplus of phosphorylated CheY by reversing directions excessively. It will be interesting to learn which proteins ChePep interacts with in H. pylori cells and whether it can be phosphorylated by CheA.
The paper of Howitt et al. also sheds new light on H. pylori’s gastric habitat and the importance of chemotaxis in the bacterium’s ability to set up residence in the stomach (2). Ottemann and colleagues had previously shown that chemotaxis is required for H. pylori to colonize the mouse stomach to maximal levels (7) and that chemotaxis mutants interact less intimately with gastric mucosal cells than wild-type H. pylori does (8). Furthermore, they found that chemotaxis-deficient mutants (such as ΔcheY mutants) are preferentially eliminated from the stomach when paired with wild-type cells (7), suggesting that competition for stable residence in this organ is fierce. Howitt et al. found similar dynamics with their ΔchePep mutant; on its own, it colonizes the mouse stomach about an order of magnitude less than the wild-type strain but it is readily displaced by wild-type cells in coinfections (2). Using confocal microscopy, Howitt and colleagues expanded our view of H. pylori’s existence in the mouse stomach and niche competition. Their images reveal that while both wild-type and ΔchePep mutant bacteria can reside in the superficial mucus lining of the stomach epithelium, only the wild-type strain is found in dense colonies deep in the gastric glands. The exclusion of the ΔchePep mutants from the gastric glands indicates that H. pylori cells must navigate chemical gradients to infect these structures and, in conjunction with the coinfection data, suggests that residence within these glands provides a strong selective advantage for maintaining residence in the stomach. Possibly, these gland-inhabiting H. pylori cells serve as a reservoir for repopulating the superficial mucosa, which is subject to continual flow forces and epithelial cell sloughing.
The images of H. pylori in gastric glands taken by Howlitt et al. remind us that a bacterium’s pathogenicity depends on its context. H. pylori can be resident in asymptomatic hosts for decades, possibly conferring protection against esophageal adenocarcinoma and even obesity, but it can also drive pathological events in the stomach that result in cancer (9). The importance of ChePep in H. pylori’s ability to insinuate itself deep into the gastric crypts, along with its specificity to the epsilonproteobacteria, makes it an intriguing new target for antibiotics that could be used to change the context of the bacterium-host interaction by allowing H. pylori to colonize but keeping it from getting too cozy.
Footnotes
Citation Sweeney EG, Guillemin K. 2011. A gastric pathogen moves chemotaxis in a new direction. mBio 2(5):e00201-11. doi:10.1128/mBio.00201-11.
REFERENCES
- 1. Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat. Rev. Microbiol. 9:153–165 [DOI] [PubMed] [Google Scholar]
- 2. Howitt MR, et al. 2011. ChePep controls Helicobacter pylori infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2(4):e00098-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. 2010. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 18:494–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lowenthal AC, et al. 2009. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 155:1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parkinson JS, Houts SE. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Briegel A, et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. U. S. A. 106:17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams SM, et al. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119:2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]