Abstract
Double-stranded RNA (dsRNA) directs gene-specific, post-transcriptional silencing in many organisms, including vertebrates, and has provided a new tool for studying gene function. The biochemical mechanisms underlying this dsRNA interference (RNAi) are unknown. Here we report the development of a cell-free system from syncytial blastoderm Drosophila embryos that recapitulates many of the features of RNAi. The interference observed in this reaction is sequence specific, is promoted by dsRNA but not single-stranded RNA, functions by specific mRNA degradation, and requires a minimum length of dsRNA. Furthermore, preincubation of dsRNA potentiates its activity. These results demonstrate that RNAi can be mediated by sequence-specific processes in soluble reactions.
Keywords: RNAi, post-transcriptional gene silencing, dsRNA
Post-transcriptional gene silencing by double-stranded RNA (dsRNA), or RNA interference (RNAi), is a new tool for studying gene function in an increasing number of organisms (for reviews, see Montgomery and Fire et al. 1998; Fire 1999; Hunter 1999; Sharp 1999) including nematodes (Fire et al. 1998; Montgomery et al. 1998), fruit flies (Kennerdell and Carthew 1998; Misquitta and Paterson 1999), trypanosomes (Ngo et al. 1998), plants (Waterhouse et al. 1998), planaria (Sánchez-Alvarado and Newmark 1999), hydra (Lohmann et al. 1999), and zebrafish (Wargelius et al. 1999). The post-transcriptional silencing of endogenous genes following introduction of transgenes into plants (cosuppression; Vaucheret et al. 1998; Waterhouse et al. 1998; Baulcombe 1999), the fungus Neurospora (quelling; Cogoni et al. 1996; Cogoni and Macino 1999), flies (Pal-Bhadra et al. 1997, 1999), and mice (Bahramian and Zarbl 1999) may also be related to RNAi because antisense transcripts may be produced from transgenes, resulting in dsRNA formation.
The hallmark of RNAi is its specificity. dsRNA reduces expression of the gene from which the dsRNA sequence is derived, without detectable effect on the expression of genes unrelated in sequence (Fire et al. 1998; Montgomery et al. 1998). The function of RNAi is not known, but it may represent a cellular defense against viral infection, or perhaps a post-transcriptional mechanism for regulating gene expression in response to dsRNA formed from nuclear transcripts.
The gene silencing induced by RNAi is reversible and thus does not appear to reflect a genetic change (Fire et al. 1998). Evidence that RNAi functions post-transcriptionally is as follows: dsRNA corresponding to intron sequences does not produce RNAi (Montgomery et al. 1998), and dsRNA corresponding to exon sequences does not affect pre-mRNA levels (Ngo et al. 1998). In Caenorhabditis elegans, dsRNA targeting one gene within an operon does not effect the expression of a second gene within that operon, indicating that RNAi occurs after transcription of the nuclear polycistronic RNA (Montgomery et al. 1998). In situ hybridization experiments show that dsRNA causes a specific reduction in target mRNA levels (Fire et al. 1998; Kennerdell and Carthew 1998; Misquitta and Paterson 1999; Sánchez-Alvarado and Newmark 1999). The reduced level of the mRNA targeted by dsRNA is presumed to underlie the reduction of specific gene function produced by RNAi. However, it is possible that dsRNA exerts distinct effects on mRNA translation and stability in vivo. Quantitative analyses suggest that dsRNA can specifically decrease the concentration of an mRNA by as much as 90% (Ngo et al. 1998; Lohmann et al. 1999), although smaller effects are observed in some organisms or for particular genes (Wargelius et al. 1999). In C. elegans, RNAi has been shown to function independently of the SMG system, which was initially identified by its role in degrading translationally aberrant mRNAs (Montgomery et al. 1998).
Only a few molecules of dsRNA per cell are required to produce RNAi (Fire et al. 1998; Kennerdell and Carthew 1998). The small amount of dsRNA required for silencing and the spreading of the silencing through a broad region of the organism suggests that the dsRNA either acts catalytically or is amplified (Fire 1999). Amplification of dsRNA may occur in Neurospora, in which a gene that is similar to an RNA-dependent RNA polymerase has been shown to be required for quelling (Cogoni and Macino 1999). However, in C. elegans, replication of the dsRNA has not been detected, leading to the suggestion that the dsRNA functions catalytically (Montgomery et al. 1998). At least in C. elegans, dsRNA is efficiently transported throughout the entire organism. Remarkably, dsRNA that is fed to worms produces specific interference (Timmons and Fire 1998).
The molecular mechanisms by which dsRNA generates the RNAi effect are unknown. The recapitulation of the essential features of RNAi in vitro is a prerequisite for a biochemical analysis of the phenomenon. Here we describe gene-specific, dsRNA-mediated interference in a cell-free system derived from syncytial blastoderm Drosophila embryos. The in vitro system should complement genetic approaches to dissecting the molecular basis of RNAi.
Results and Discussion
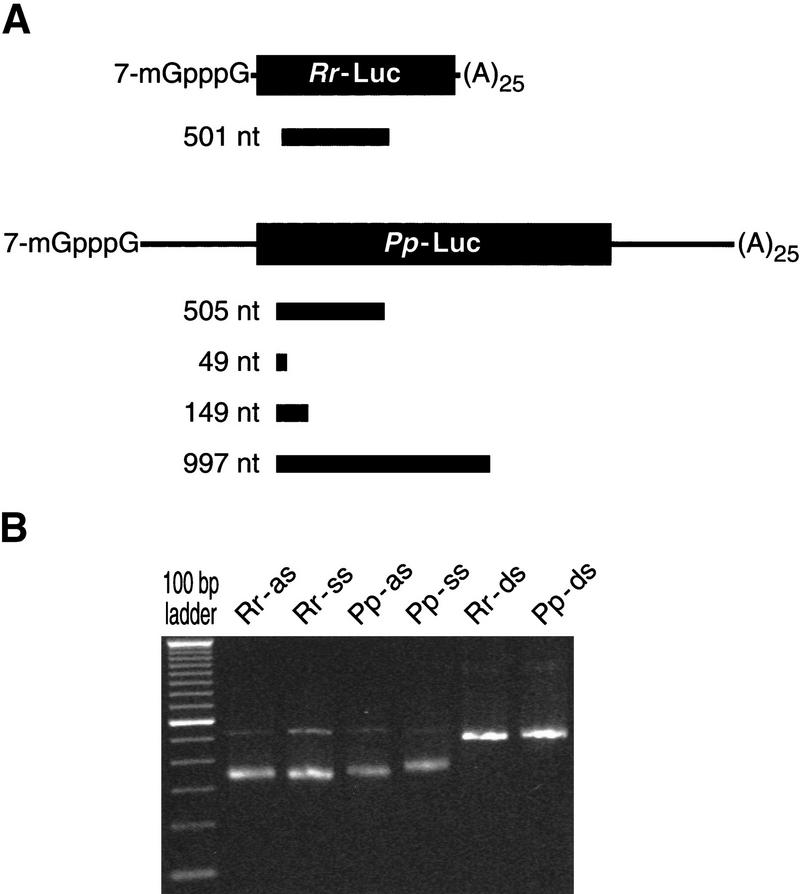
To evaluate whether dsRNA could specifically block gene expression in vitro, we used reporter mRNAs derived from two different luciferase genes, Renilla reniformis (sea pansy) luciferase (Rr-Luc) and Photinus pyralis (firefly) luciferase (Pp-Luc), that are unrelated both in sequence and in luciferin substrate specificity. dsRNA generated from one gene was used to target that luciferase mRNA, whereas the other luciferase mRNA was an internal control cotranslated in the same reaction.dsRNAs of ∼500 bp were prepared by transcription of PCR products from the Rr-Luc and Pp-Luc genes. Each dsRNA began ∼100 bp downstream of the start of translation (Fig. 1A). Sense (ss) and anti-sense (as) RNA were transcribed in vitro and annealed to each other to produce the dsRNA (Fig. 1B). The ssRNA, asRNA, and dsRNAs were each tested for their ability to block specifically expression of their cognate mRNA but not the expression of the unrelated internal control mRNA.
Figure 1.
Reporter mRNAs and dsRNAs. (A) RNAs used in this study. Lengths and positions of the ssRNA, asRNA, and dsRNAs are shown as black bars relative to the Rr-Luc and Pp-Luc reporter mRNA sequences. Black rectangles indicate the two unrelated luciferase coding sequences, lines correspond to the 5′ and 3′ UTRs of the mRNAs. (B) Native gel electrophoresis of the individual Rr 501 nt and Pp 505 nt asRNAs and ssRNAs used to form the Rr and Pp dsRNAs.
The ssRNA, asRNA, or dsRNA was incubated for 10 min in a reaction containing Drosophila embryo lysate, then both Pp-Luc and Rr-Luc mRNAs were added and the incubation continued for an additional 60 min. The Drosophila embryo lysate efficiently translates exogenously transcribed mRNA under the conditions used. The amounts of Pp-Luc and Rr-Luc enzyme activities were measured and were used to calculate ratios of either Pp-Luc/Rr-Luc (Fig. 2A) or Rr-Luc/Pp-Luc (Fig. 2B). To facilitate comparison of different experiments, the ratios from each experiment were normalized to the ratio observed for a control in which buffer was added to the reaction in place of ssRNA, asRNA, or dsRNA.
Figure 2.
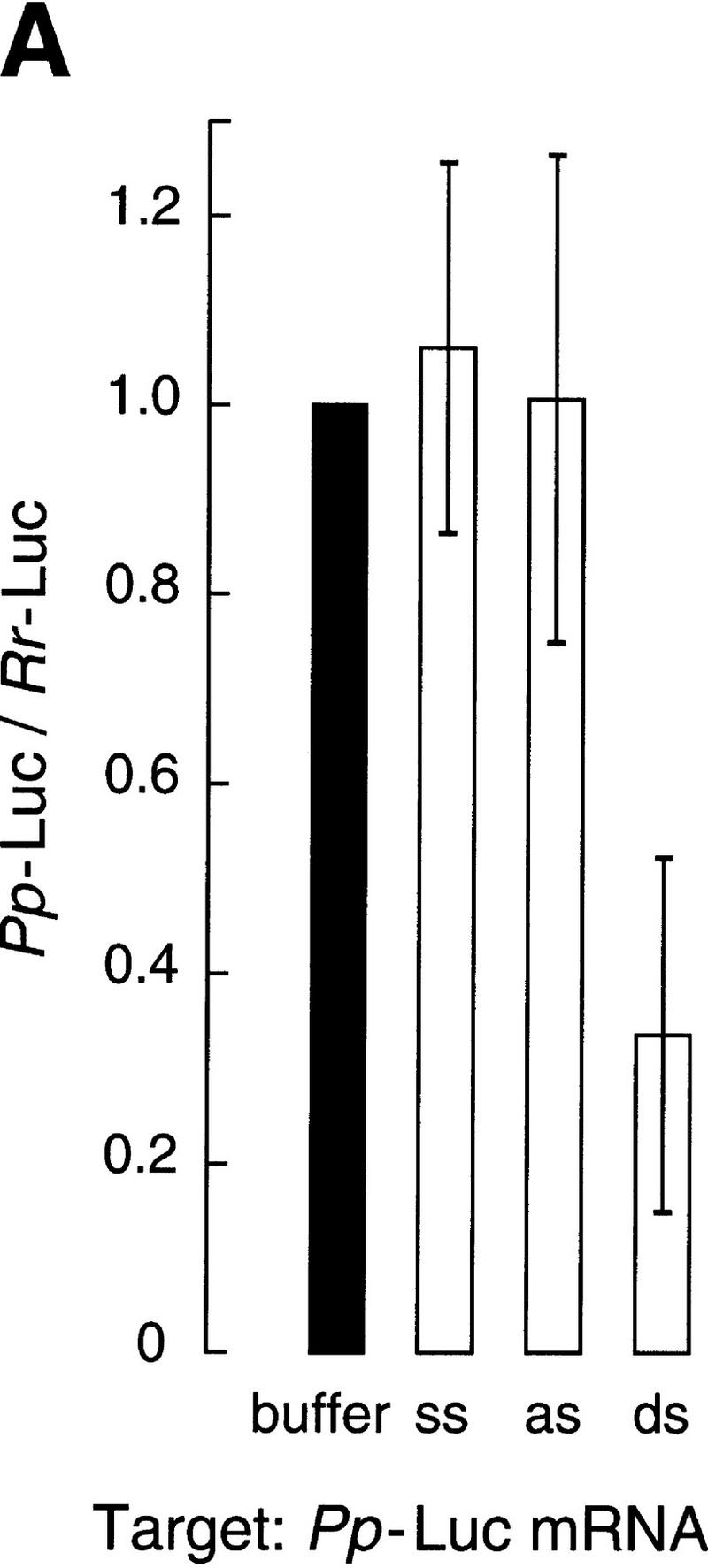
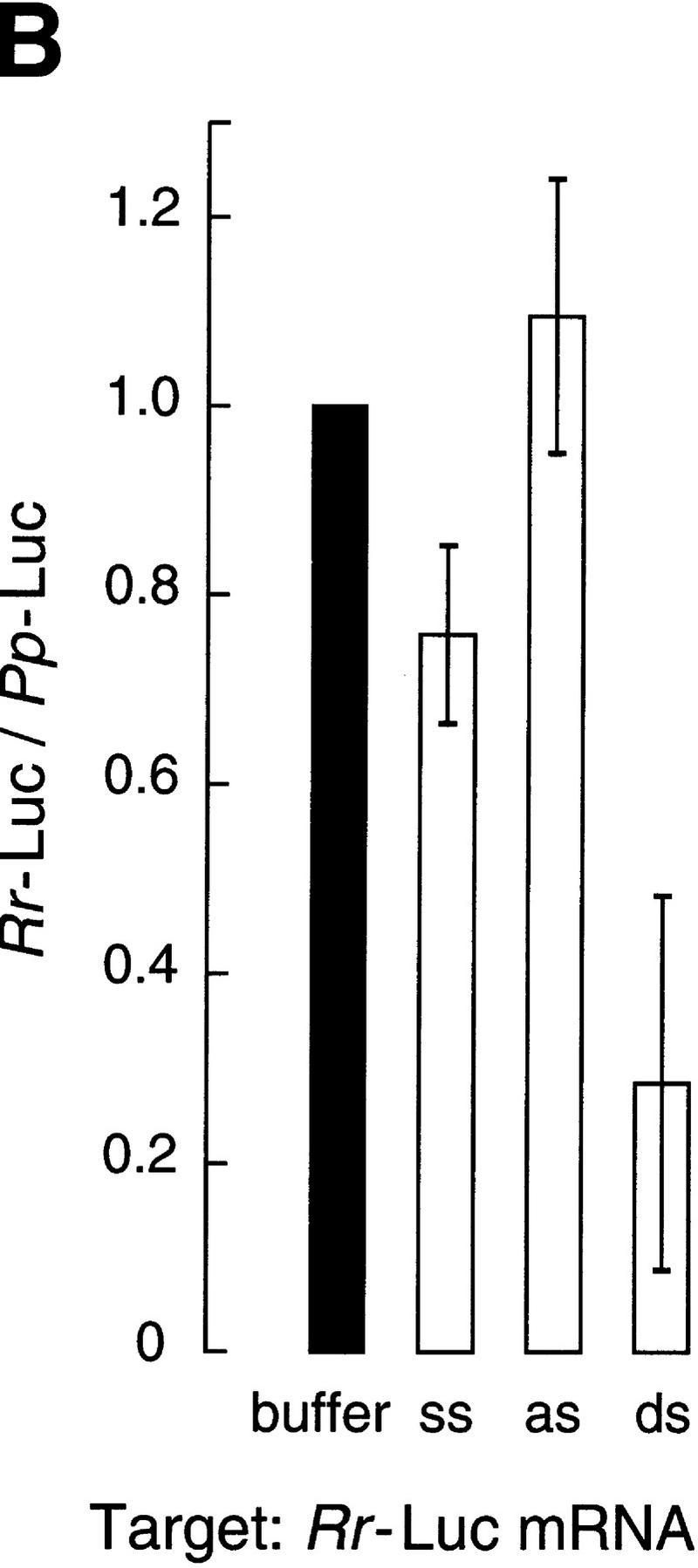
Gene-specific interference by dsRNA in vitro. (A) Ratio of luciferase activities after targeting 50 pm Pp-Luc mRNA with 10 nm ssRNA, asRNA, or dsRNA from the 505-bp segment of the Pp-Luc gene. The data are the average values of seven trials ± standard deviation. Four independently prepared lysates were used. Luciferase activity was normalized to the buffer control and so a ratio equal to one indicates no gene-specific interference. (B) Ratio of luciferase activities after targeting 50 pm Rr-Luc mRNA with 10 nm ssRNA, asRNA, or dsRNA from the 501-bp segment of the Rr-Luc gene. The data are the average values of six trials ± standard deviation. A Rr-Luc/Pp-Luc ratio equal to one indicates no gene-specific interference.
Figure 2A shows that a 10-nm concentration of the 505-bp dsRNA identical to a portion of the sequence of the Pp-Luc gene specifically inhibited expression of the Pp-Luc mRNA but did not affect expression of the Rr-Luc internal control. Neither ssRNA nor asRNA affected expression of Pp-Luc or the Rr-Luc internal control. Thus, Pp-Luc expression was specifically inhibited by its cognate dsRNA. Conversely, a 10 nm concentration of the 501-bp dsRNA directed against the Rr-Luc mRNA specifically inhibited Rr-Luc expression but not that of the Pp-Luc internal control (Fig. 2B). Again, comparable levels of ssRNA or asRNA had little or no effect on expression of either reporter mRNA. On average, dsRNA reduced specific luciferase expression by 70% in these experiments, in which luciferase activity was measured after a 1 hr incubation. In other experiments in which the translational capacity of the reaction was replenished by the addition of fresh lysate and reaction components, we observed a further reduction in targeted luciferase activity relative to the internal control (data not shown).
The ability of dsRNA but not asRNA to inhibit gene expression in these lysates is not merely a consequence of the greater stability of the dsRNA (half-life ≥2 hr) relative to the single-stranded RNAs (half-life ∼10 min). ssRNA and asRNA transcribed with a 7-methyl guanosine cap were as stable in the lysate as uncapped dsRNA, but do not inhibit gene expression (data not shown). In contrast, dsRNA formed from the capped ssRNA and asRNA specifically blocks expression of the targeted mRNA (data not shown).
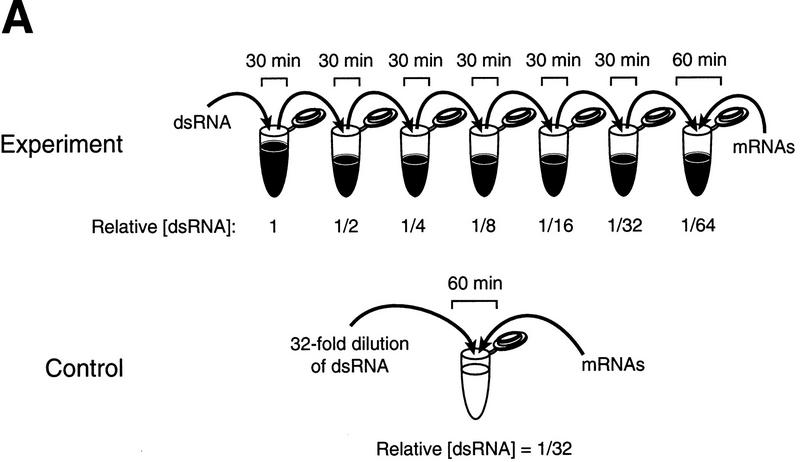
Effective RNAi in Drosophila requires the injection of ∼0.2 fmole of dsRNA into a syncytial blastoderm embryo (Kennerdell and Carthew 1998; Carthew 1999). Because the average volume of a Drosophila embryo is ∼7.3 nl, this corresponds to an intracellular concentration of ∼25 nm (Mazur et al. 1988). Gene expression in the Drosophila lysate was inhibited by a comparable concentration of dsRNA (10 nm), but lowering the dsRNA concentration 10-fold decreased the amount of specific interference (data not shown). Ten nanomolar dsRNA corresponds to a 200-fold excess of dsRNA over target mRNA added to the lysate. To test whether this excess of dsRNA might reflect a time- and/or concentration-dependent step in which the input dsRNA was converted to a form active for gene-specific interference, the effect of preincubation of the dsRNA on its ability to inhibit expression of its cognate mRNA was examined. Because the translational capacity of the lysates is significantly reduced after 30 min of incubation at 25°C (our unpublished observations), we wished to ensure that all factors necessary for RNAi remained active throughout the preincubation period. Therefore, every 30 min, a reaction containing dsRNA and lysate was mixed with a fresh reaction containing unincubated lysate (Fig. 3A). After six successive serial transfers spanning 3 hr of preincubation, the dsRNA, now diluted 64-fold relative to its original concentration, was incubated with lysate and 50 pm of target mRNA for 60 min. Finally, the Pp-Luc and Rr-Luc enzyme levels were measured. For comparison, the input amount of dsRNA (10 nm) was diluted 32-fold in buffer, and its capacity to generate gene-specific dsRNA interference in the absence of any preincubation step was assessed.
Figure 3.
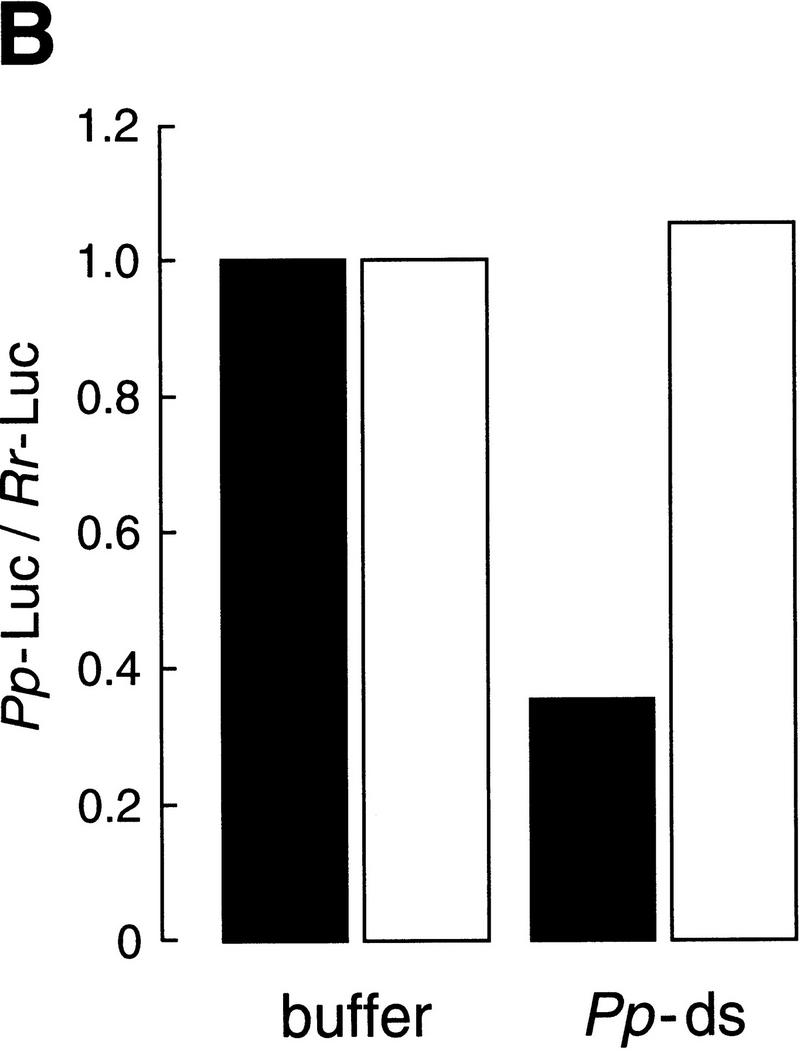
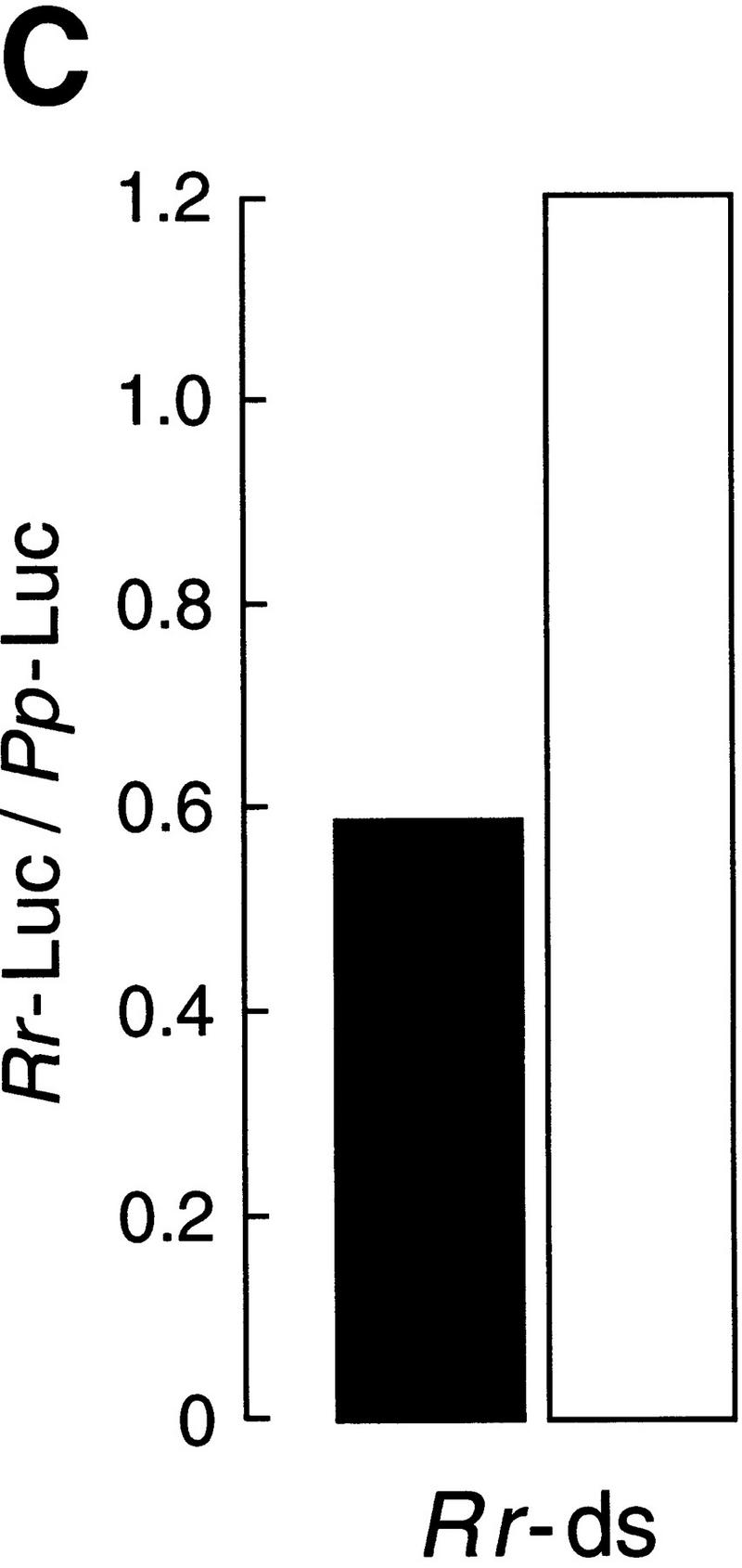
Incubation in the Drosophila embryo lysate potentiates dsRNA for gene-specific interference. (A) Experimental strategy. The same dsRNAs used in Fig. 2 (or buffer) was serially preincubated with twofold dilutions in six successive reactions with Drosophila embryo lysate, then tested for its capacity to block mRNA expression. As a control, the same amount of dsRNA (10 nm) or buffer was diluted directly in buffer and incubated with Pp-Luc and Rr-Luc mRNAs and lysate. (B) Potentiation when targeting Pp-Luc mRNA. Black columns indicate the dsRNA or the buffer was serially preincubated; white columns correspond to a direct 32-fold dilution of the dsRNA. Values were normalized to those of the buffer controls. (C) Potentiation when targeting Rr-Luc mRNA. The corresponding buffer control is shown in B.
The preincubation of the dsRNA in lysate significantly potentiated its capacity to inhibit specific gene expression. Whereas the dsRNA diluted 32-fold showed no effect, the preincubated dsRNA was, within experimental error, as potent as undiluted dsRNA, despite having undergone a 64-fold dilution. Potentiation of the dsRNA by preincubation was observed for dsRNAs targeting both the Pp-Luc mRNA (Fig. 3B) and the Rr-Luc mRNA (Fig. 3C). Taking into account the 64-fold dilution, the activation conferred by preincubation allowed a 156-pm concentration of dsRNA to inhibit 50 pm target mRNA. Higher dilutions of the activated dsRNA may be effective, but have not been tested. Although both dsRNAs tested were activated by the preincubation procedure, each fully retained its specificity to interfere with expression only of the mRNA to which it is homologous. Further study of the reactions may provide a route to identifying the mechanism of dsRNA potentiation.
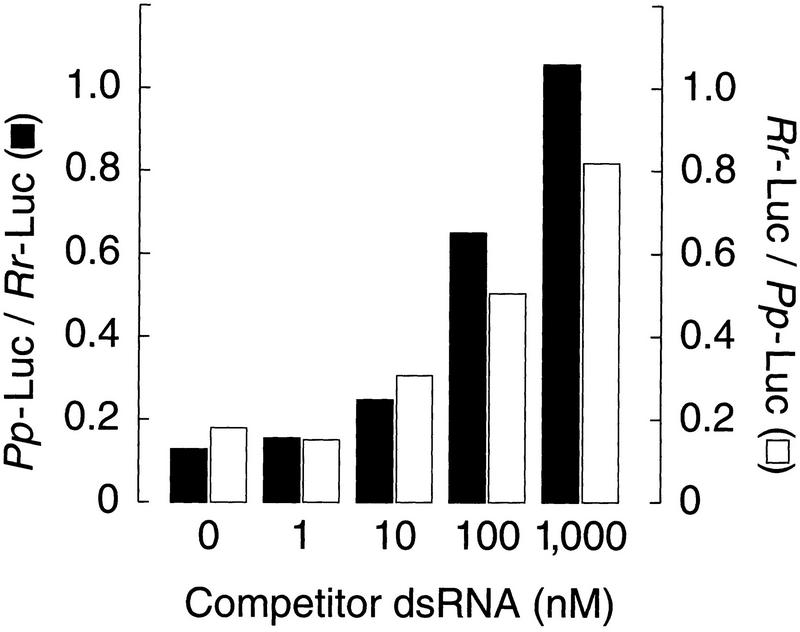
One possible explanation for the observation that preincubation of the dsRNA enhances its capacity to inhibit gene expression in these lysates is that specific factors either modify and/or associate with the dsRNA. Accordingly, the addition of increasing amounts of dsRNA to the reaction might titrate such factors and decrease the amount of gene-specific interference caused by a second dsRNA of unrelated sequence. For both Pp-Luc mRNA and Rr-Luc mRNA, addition of increasing concentrations of the unrelated Drosophila nanos dsRNA to the reaction decreased the amount of gene-specific interference caused by dsRNA targeting the reporter mRNA (Fig. 4). None of the tested concentrations of nanos dsRNA affected the levels of translation of the untargeted mRNA, demonstrating that the nanos dsRNA specifically titrated factors involved in gene-specific interference and not components of the translational machinery. The limiting factor(s) was titrated by addition of ∼1000 nm dsRNA, a 200-fold excess over the 5 nm of dsRNA used to produce specific interference.
Figure 4.
Effect of competitor dsRNA on gene-specific interference. Increasing concentrations of nanos dsRNA (508 bp) were added to reactions containing 5 nm dsRNA (the same dsRNAs used in Fig. 2) targeting Pp-Luc mRNA (black columns, left axis) or Rr-Luc mRNA (white columns, right axis). Each reaction contained both a target mRNA (Pp-Luc for the black columns, Rr-Luc for the white) and an unrelated control mRNA (Rr-Luc for the black columns, Pp-Luc for the white). Values were normalized to the buffer control (not shown). The reactions were incubated under standard conditions (see Materials and Methods).
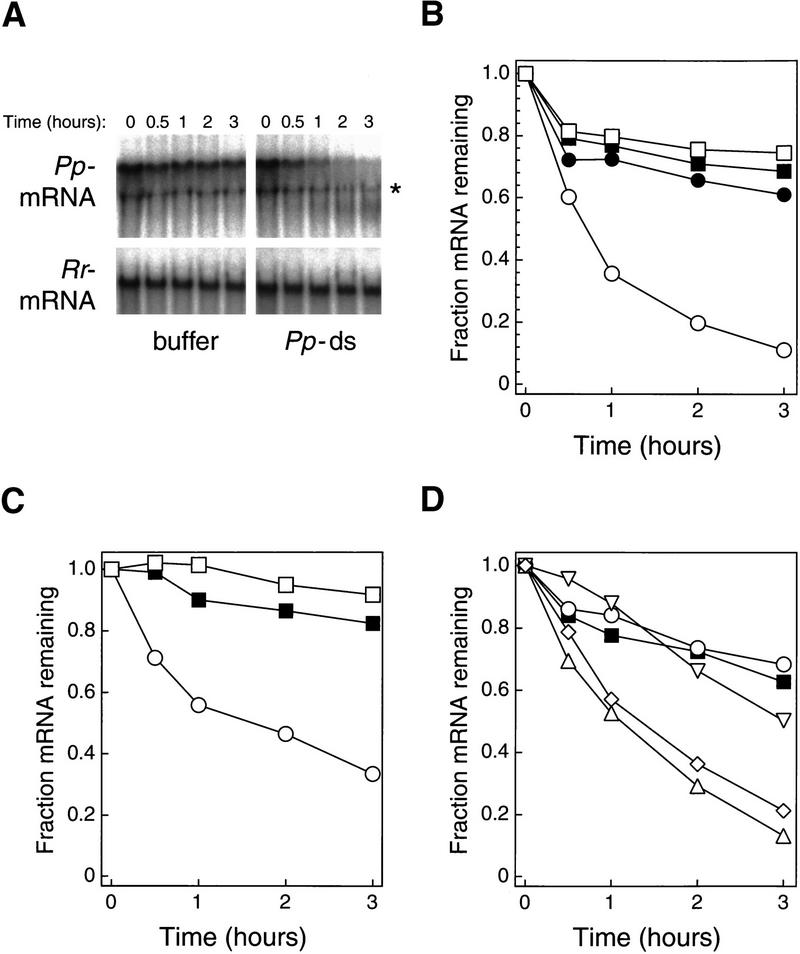
Interference in vitro might reflect either a specific inhibition of mRNA translation or the targeted destruction of the specific mRNA. To distinguish these two possibilities, the fates of the Pp-Luc and Rr-Luc mRNAs were examined directly with 32P-radiolabeled substrates. In the absence of dsRNA, both the Pp-Luc and Rr-Luc mRNAs were stable in the lysates, with ∼75% of the input mRNA remaining after 3 hr of incubation. (About 25% of the input mRNA is rapidly degraded in the reaction and likely represents uncapped mRNA generated by the in vitro transcription process.) In the presence of dsRNA (10 nm, 505 bp) targeting the Pp-Luc mRNA, <15% of the Pp-Luc mRNA remained after 3 hr (Fig. 5A,B). As expected, the Rr-Luc mRNA remained stable in the presence of the dsRNA targeting Pp-Luc mRNA. Conversely, dsRNA (10 nm, 501 bp) targeting the Rr-Luc mRNA caused the destruction of the Rr-Luc mRNA but had no effect on the stability of Pp-Luc mRNA (Fig. 5C). For both mRNAs, capped asRNA has a very small effect on the stability of the target (data not shown). This effect may be caused by a small amount of dsRNA contaminating the asRNA. Low levels of dsRNA that form during in vitro transcription of asRNA cause RNAi in vivo (Fire et al. 1998). Alternatively, a small fraction of the capped asRNA could have annealed to mRNA in the reaction, creating dsRNA.
Figure 5.
Effect of dsRNA on mRNA stability. (A) Stability of 10 nm Pp-Luc mRNA or Rr-Luc mRNA incubated in lysate with either buffer or 505-bp Pp-dsRNA (10 nm). Samples were deproteinized after the indicated times and the 32P-radiolabeled mRNAs were then resolved by denaturing gel electrophoresis. The band marked with an asterisk likely results from radioactivity being swept ahead of the abundant ribosomal RNA in the lysate. (B) Quantitation of the data in A. (Circles) Pp-Luc mRNA; (boxes) Rr-Luc mRNA; (filled symbols) buffer incubation; (open symbols) incubation with Pp-dsRNA. (C) Stability of Rr-Luc mRNA incubated with Rr-dsRNA or Pp-dsRNA. (█) buffer; (□) Pp-dsRNA (10 nm); (○) Rr-dsRNA (10 nm). (D) Dependence on dsRNA length. The stability of the Pp-Luc mRNA was assessed after incubation in lysate in the presence of buffer or dsRNAs of different lengths. (█) Buffer; (○) 49-bp dsRNA (10 nm); (▿) 149-bp dsRNA (10 nm); (▵) 505-bp dsRNA (10 nm); (⋄) 997-bp dsRNA (10 nm). Reactions were incubated under standard conditions (see Materials and Methods).
In the in vitro reaction, dsRNA specifically caused accelerated decay of the mRNA to which it is homologous, with no effect on the stability of the unrelated control mRNA. The in vitro results suggest that in vivo, at least in Drosophila, the effect of dsRNA is to destabilize the target mRNA directly, not to change the subcellular localization of the mRNA, for example, by causing it to be specifically retained in the nucleus, resulting in subsequent, nonspecific degradation.
These results are consistent with the observation that RNAi leads to reduced cytoplasmic mRNA levels in vivo, as measured by in situ hybridization (Montgomery et al. 1998) and Northern blotting (Ngo et al. 1998). Northern blot analyses in trypanosomes and hydra suggest that dsRNA typically decreases mRNA levels by <90% (Ngo et al. 1998; Lohmann et al. 1999). The data presented here show that in vitro mRNA levels are reduced 65%–85% after 3 hr incubation, an effect comparable with observations in vivo. They also agree with the finding that RNAi in C. elegans is post-transcriptional (Montgomery et al. 1998). The simplest explanation for the specific effects on protein synthesis is that it reflects the accelerated rate of RNA decay. However, the results do not exclude independent but specific effects on translation as well as stability.
In vivo, RNAi requires a minimum length of dsRNA (Ngo et al. 1998). The ability of RNA duplexes of lengths 49, 149, 505, and 997 bp (diagrammed in Fig. 1A) to target the degradation of the Pp-Luc mRNA in vitro was assessed. In good agreement with in vivo observations, the 49-bp dsRNA was ineffective in vitro, whereas the 149-bp dsRNA enhanced mRNA decay only slightly, and both the 505- and 997-bp dsRNAs caused robust mRNA degradation (Fig. 5D).
We asked whether the gene-specific interference observed in Drosophila lysates was a general property of cell-free translation systems. The effects of dsRNAs on expression of Pp-Luc and Rr-Luc mRNA were examined in commercially available wheat germ extracts and rabbit reticulocyte lysates. There was no effect of addition of 10 nm of either ssRNA, asRNA, or dsRNA on the expression of either mRNA reporter in wheat germ extracts (data not shown). In contrast, the addition of 10 nm of dsRNA to the rabbit reticulocyte lysate caused a profound and rapid, nonspecific decrease in mRNA stability (data not shown). For example, addition of Rr-Luc dsRNA caused degradation of both Rr-Luc and Pp-Luc mRNAs within 15 min. The same nonspecific effect was observed on addition of Pp-Luc dsRNA. The nonspecific destruction of mRNA induced by the addition of dsRNA to the rabbit reticulocyte lysate presumably reflects the previously observed activation of RNase L by dsRNA (Clemens and Williams 1978; Williams et al. 1979; Zhou et al. 1993; Matthews 1996). Mouse cell lines lacking dsRNA-induced anti-viral pathways have been described recently (Zhou et al. 1999) and may be useful in the search for mammalian RNAi. If RNAi exists in mammals, as might be predicted from the presence of RNAi-like phenomena in invertebrates, plants, and fungi, as well as the recent report of RNAi in the vertebrate Danio rerio (zebrafish; Wargelius et al. 1999), it is likely obscured by the rapid induction by dsRNA of nonspecific antiviral responses.
dsRNA-targeted destruction of specific mRNA is characteristic of RNAi, which has been observed in vivo in many organisms, including Drosophila. The system described above recapitulates in a reaction in vitro many aspects of RNAi. The targeted mRNA is specifically degraded, whereas unrelated control mRNAs present in the same solution are not affected. The process is most efficient with dsRNAs >150 bp in length. The dsRNA-specific degradation reaction in vitro is probably general to many, if not all, mRNAs, as it was observed by use of two unrelated genes.
The magnitude of the effects we observe on mRNA stability in vitro are comparable with those reported in vivo (Ngo et al. 1998; Lohmann et al. 1999). However, the reaction in vitro requires an excess of dsRNA relative to mRNA. In contrast, a few molecules of dsRNA per cell can inhibit gene expression in vivo (Fire et al. 1998; Kennerdell and Carthew 1998). The difference between the stoichiometry of dsRNA to target mRNA in vivo and in vitro should not be surprising in that most in vitro reactions are less efficient than their corresponding in vivo processes. Interestingly, incubation of the dsRNA in the lysate greatly potentiated its activity for RNAi, indicating that it is either modified or becomes associated with other factors or both. Perhaps a small number of molecules is effective in inhibiting the targeted mRNA in vivo because the injected dsRNA has been activated by a process similar to that reported here for RNAi in Drosophila lysates. The nature of this activation process, the mechanism of destruction of the targeted mRNAs, and the identification of cellular factors essential for RNAi await further experiments.
Materials and methods
RNAs
Rr-Luc mRNA consisted of the 926-nucleotide Rr luciferase coding sequence flanked by 25 nucleotides of 5′ untranslated sequence from the pSP64 plasmid polylinker and 25 nucleotides of 3′ untranslated sequence consisting of 19 nucleotides of pSP64 plasmid polylinker sequence followed by a 6-nt SacI site. PP-Luc mRNA contained the 1653-nt Pp luciferase coding sequence with a KpnI site introduced immediately before the Pp luciferase stop codon. The Pp coding sequence was flanked by 5′ untranslated sequences consisting of 21 nt of pSP64 plasmid polylinker followed by 512 nt of the 5′ untranslated region (UTR) from the Drosophila hunchback mRNA and 3′ untranslated sequences consisting of the 562-nt hunchback 3′ UTR followed by a 6-nt SacI site. The hunchback 3′ UTR sequences used contained six G-to-U mutations that disrupt function of the Nanos Response Elements in vivo and in vitro (D. Chagnovich, P.D. Zamore, R. Lehman, and D.P. Bartel, unpubl.). Both reporter mRNAs terminated in a 25-nt poly(A) tail encoded in the transcribed plasmid. For both Rr-Luc and Pp-Luc mRNAs, the transcripts were generated by run-off transcription from plasmid templates cleaved at an NsiI site that immediately followed the 25-nt-encoded poly(A) tail. To ensure that the transcripts ended with a poly(A) tail, the NsiI-cleaved transcription templates were resected with T4 DNA Polymerase in the presence of dNTPs. The SP6 mMessage mMachine kit (Ambion) was used for in vitro transcription. With this kit, ∼80% of the resulting transcripts are 7-methyl guanosine capped. 32P-radiolabeling was accomplished by including [α-32P]UTP in the transcription reaction.
For Pp-Luc, ssRNA, asRNA, and dsRNA corresponded to positions 93–597 relative to the start of translation, yielding a 505-bp dsRNA. For Rr-Luc, asRNA, ssRNA, and dsRNA corresponded to positions 118–618 relative to the start of translation, yielding a 501-bp dsRNA. The Drosophila nanos competitor dsRNA corresponded to positions 122–629 relative to the start of translation, yielding a 508-bp dsRNA. ssRNA, asRNA, and dsRNA (diagrammed in Fig. 1A) were transcribed in vitro with T7 RNA polymerase from templates generated by the PCR. After gel purification of the T7 RNA transcripts, residual DNA template was removed by treatment with RQ1 DNase (Promega). The RNA was extracted with phenol and chloroform, and then precipitated and dissolved in water.
RNA annealing and native gel electrophoresis
ssRNA and asRNA (0.5 μm) in 10 mm Tris-HCl (pH 7.5) with 20 mm NaCl were heated to 95°C for 1 min, then cooled and annealed at room temperature for 12–16 hr. The RNAs were precipitated and resuspended in lysis buffer (below). To monitor annealing, RNAs were electrophoresed in a 2% agarose gel in TBE buffer and stained with ethidium bromide (Sambrook et al. 1989).
Lysate preparation
Zero- to 2-hr-old embryos from Oregon R flies were collected on yeasted molasses agar at 25°C. Embryos were dechorionated for 4–5 min in 50% (vol/vol) bleach, washed with water, blotted dry, and transferred to a chilled Potter-Elvehjem tissue grinder (Kontes). Embryos were lysed at 4°C in 1 ml of lysis buffer (100 mm potassium acetate, 30 mm HEPES-KOH at pH 7.4, 2 mm magnesium acetate) containing 5 mm dithiothreitol (DTT) and 1 mg/ml Pefabloc SC (Boehringer Mannheim) per gram of damp embryos. The lysate was centrifuged for 25 min at 14,500g at 4°C, and the supernatant flash frozen in aliquots in liquid nitrogen and stored at −80°C.
Reaction conditions
Lysate preparation and reaction conditions were derived from those described by Hussain and Leibowitz (1986). Reactions contained 50% (vol/vol) lysate, mRNAs (10–50 pm final concentration), and 10% (vol/vol) lysis buffer containing the ssRNA, asRNA, or dsRNA (10 nm final concentration). Each reaction also contained 10 mm creatine phosphate, 10 μg/ml creatine phosphokinase, 100 μm GTP, 100 μm UTP, 100 μm CTP, 500 μm ATP, 5 mm DTT, 0.1 U/μL RNasin (Promega), and 100 μm of each amino acid. The final concentration of potassium acetate was adjusted to 100 mm. For standard conditions, the reactions were assembled on ice and then preincubated at 25°C for 10 min before adding mRNA. After adding mRNAs, the incubation was continued for an additional 60 min. The 10-min preincubation step was omitted for the experiments in Figures 3 and 5. Reactions were quenched with 4 volumes of 1.25× Passive Lysis Buffer (Promega). Pp and Rr luciferase activity was detected in a Monolight 2010 Luminometer (Analytical Luminescence Laboratory) with the Dual-Luciferase Reporter Assay System (Promega).
RNA stability
Reactions with 32P-radiolabeled mRNA were quenched by the addition of 40 volumes of 2× PK buffer (200 mm Tris-HCl at pH 7.5, 25 mm EDTA, 300 mm NaCl, 2% wt/vol sodium dodecyl sulfate). Proteinase K (E.M. Merck; dissolved in water) was added to a final concentration of 465 μg/ml. The reactions were then incubated for 15 min at 65°C, extracted with phenol/chloroform/isoamyl alcohol (25:24:1), and precipitated with an equal volume of isopropanol. Reactions were analyzed by electrophoresis in a formaldehyde/agarose (0.8% wt/vol) gel (Sambrook et al. 1989). Radioactivity was detected by exposing the agarose gel [dried under vacuum onto Nytran Plus membrane (Amersham)] to an image plate (Fujix) and quantified with a Fujix Bas 2000 and Image Gauge 3.0 (Fujix) software.
Commercial lysates
Untreated rabbit reticulocyte lysate (Ambion) and wheat germ extract (Ambion) reactions were assembled according to the manufacturer's directions. dsRNA was incubated in the lysate at 27°C (wheat germ) or 30°C (reticulocyte lysate) for 10 min prior to the addition of mRNAs.
Acknowledgments
We thank Danny Chagnovich for the generous gift of the luciferase reporter plasmids. We also thank Terri Orr-Weaver for providing fly resources, Marlene Castle for maintaining population cages, and Jen Cook-Chrysos for help in preparing the figures. P.D.Z. was supported by a Charles H. Hood fellowship. Support for this work was provided to D.P.B. by the Searle Scholars Program/The Chicago Community Trust and to P.A.S. by the National Institutes of Health through a United States Public Health Service MERIT award, by the National Science Foundation, and partially by the National Cancer Institute through a Cancer Center Support core grant.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Phillip.Zamore@umassmed.edu (PDZ) or ttuschl@mpibpc.gwdg.de (TT); FAX (617) 258-6768.
References
- Bahramian MB, Zarbl H. Transcriptional and posttranscriptional silencing of rodent alpha1(I) collagen by a homologous transcriptionally self-silenced transgene. Mol Cell Biol. 1999;19:274–283. doi: 10.1128/mcb.19.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC. Fast forward genetics based on virus-induced gene silencing. Curr Opin Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- Carthew, R. 1999. www1.pitt.edu/∼carthew/manual/RNAi_Protocol.html
- Clemens M, Williams B. Inhibition of cell-free protein synthesis by pppA2′p5′A2′5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Fire A. RNA-triggered gene silencing. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hunter CP. A touch of elegance with RNAi. Curr Biol. 1999;9:R440–442. doi: 10.1016/s0960-9822(99)80276-0. [DOI] [PubMed] [Google Scholar]
- Hussain I, Leibowitz MJ. Translation of homologous and heterologous messenger RNAs in a yeast cell-free system. Gene. 1986;46:13–23. doi: 10.1016/0378-1119(86)90162-9. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Endl I, Bosch TC. Silencing of developmental genes in hydra. Dev Biol. 1999;214:211–214. doi: 10.1006/dbio.1999.9407. [DOI] [PubMed] [Google Scholar]
- Matthews M. Interactions between viruses and the cellular machinery for protein synthesis. In: Hershey J, Mathews M, Sonenberg N, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 505–548. [Google Scholar]
- Mazur P, Schneider U, Jacobsen K, Mahowald A. Survival of intact Drosophila eggs at various stages of embryonic development as a function of the extent of dehydration and of intraembryonic freezing. Cryobiology. 1988;25:543–544. [Google Scholar]
- Misquitta L, Paterson BM. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): A role for nautilus in embryonic somatic muscle formation. Proc Natl Acad Sci. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, Fire A. Double stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression in Drosophila: Gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/s0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- ————— Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell. 1999;99:35–46. doi: 10.1016/s0092-8674(00)80060-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez-Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. RNAi and double-strand RNA. Genes & Dev. 1999;13:139–141. [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Mourrain P, Palauqui JC, Vernhettes S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- Wargelius A, Ellingsen S, Fjose A. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem Biophys Res Commun. 1999;263:156–161. doi: 10.1006/bbrc.1999.1343. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Gilbert CS, Kerr IM. The respective roles of the protein kinase and pppA2′p5′A2′p5′A-activated endonuclease in the inhibition of protein synthesis by double stranded RNA in rabbit reticulocyte lysates. Nucleic Acids Res. 1979;6:1335–1350. doi: 10.1093/nar/6.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Hassel B, Silverman R. Expression cloning of 2-5A-dependent RNAase: A uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- Zhou A, Paranjape JM, Der SD, Williams BR, Silverman RH. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology. 1999;258:435–440. doi: 10.1006/viro.1999.9738. [DOI] [PubMed] [Google Scholar]