Abstract
Hepatitis C virus (HCV) infection is the most common blood borne infection in the U.S. with estimates of 4 million HCV-infected individuals in the U.S. and 170 million worldwide1. The majority (70%–80%) of HCV infections persist and about 30% of individuals with persistent infection develop chronic liver disease, including cirrhosis and hepatocellular carcinoma2. Epidemiological, viral, and host factors have been associated with the differences in HCV clearance or persistence and studies have demonstrated that a strong host immune response against HCV favors viral clearance3,4. Thus, variation in genes involved in the immune response may contribute to the ability to clear the virus. In a recent genome-wide association study, a single nucleotide polymorphism (rs12979860) 3kb upstream of the IL28B gene, which encodes the type III interferon IFN-λ3, was shown to associate strongly with more than a 2-fold difference in response to HCV drug treatment5. To determine the potential effect of rs12979860 variation on outcome to HCV infection in a natural history setting, we genotyped this variant in HCV cohorts comprised of individuals who spontaneously cleared the virus (N = 388) or had persistent infection (N = 620). We show that the C/C genotype strongly enhances resolution of HCV infection amongst individuals of both European and African ancestry (European: OR = 0.38, p = 10−7; African: OR = 0.32, p = 10−4; combined: OR = 0.33, p <10−12). To date, this is the strongest and most significant genetic effect associated with natural clearance of HCV, and these results implicate a primary role for IL28B in resolution of HCV infection.
Approximately 30 percent of individuals spontaneously clear acute hepatitis C infection. Host genetic variation is assumed to explain the heterogeneity in HCV clearance across individuals because such differences occur even after exposure to the same HCV inoculum and because there are ethnic differences in clearance frequency6,7. Variation in genes involved in the immune response has already been linked to outcome of acute HCV infection8,9, presumably due to alteration in the strength and quality of the immune response. However, most variability in spontaneous HCV clearance remains unexplained.
A recent genome wide association study (GWAS) of >1600 individuals chronically infected with hepatitis C participating in a clinical treatment trial with pegylated interferon alpha and ribavirin identified a single nucleotide polymorphism (SNP) on chromosome 19q13, rs12979860, that was strongly associated with sustained virological response (SVR)5,10. This SNP maps 3 Kb upstream of the IL28B gene which encodes the type III interferon IFN-λ3. The C/C genotype was associated with 2.5 or greater rate (depending on ethnicity) of SVR compared to the T/T genotype and the C allele was over-represented in a random multi-ethnic population as compared to the chronically-infected study cohort, raising the possibility that the C allele may favor spontaneous clearance of HCV.
In order to address directly the role of the rs12979860 SNP in HCV clearance, we genotyped 1008 individuals from 6 independent HCV cohorts composed of individuals who cleared virus (N = 388) and individuals with persistent infection (N = 620). Genotypes were in Hardy-Weinberg equilibrium in both blacks and whites (p = 0.47 and 0.77, respectively). The frequency of the C allele was significantly greater among whites than blacks in both the clearance (p = 3×10−10) and persistence groups (p = 1×10−21) (Table 1). In both blacks and whites, however, there were significant differences in allele frequencies (C vs. T) between the clearance and persistence groups, where the C allele showed greater frequencies in the clearance group than in the persistence group (80.3% vs. 66.7% respectively in whites, p = 7×10−8; 56.2% vs. 37% respectively in blacks, p = 1×10−5).
Table 1.
Characteristics of study subjectsa
| Characteristic | Clearance (N = 388) | Persistence (N = 620) |
|---|---|---|
| Mean age, yearsb | 33.9 | 32.0 |
| Male, % (N) | 78.6 (305) | 80.2 (497) |
| White, % (N) | 67.3 (261) | 61.5 (381) |
| Black, % (N) | 25 (97) | 31.1 (193) |
| Other, % (N) | 7.7 (30) | 7.4 (46) |
| HBsAg status, % positive (N)c | 9.7 (30) | 3.0 (18) |
| HIV status, % positive (N) | 19.3 (75) | 24.4 (151) |
| rs12979860, allele frequency, % | ||
| C | whites = 80.3 blacks = 56.2 |
whites = 66.7 blacks = 37 |
| T | whites = 19.7 blacks = 43.8 |
whites = 33.3 blacks = 63 |
68.7% of study subjects were derived from cohorts that were matched on HIV status, gender and ethnicity.
There was 1 individual in the clearance group for which age was not available.
Information for HBsAg status was unavailable for 80 individuals in the clearance group and 26 in the persistence group.
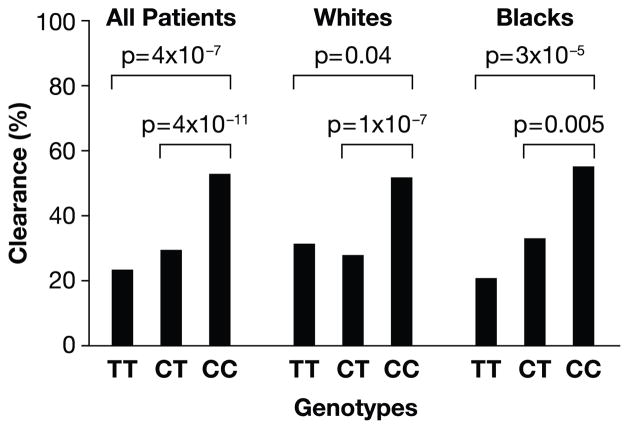
More striking differences were observed in an analysis of genotype frequencies where patients with the C/C genotype were three times more likely to clear HCV relative to patients with the C/T and T/T genotypes combined (OR = 0.33, p < 10−12 for combined ethnic groups; Table 2 and Figure 1). Stratification of this analysis by ethnicity indicated that the strength of the protective C/C effect was similar in blacks and whites (OR = 0.32, p = 1 × 10−4 and OR = 0.38, p = 1 × 10−7, respectively). However, a comparison of C/C to the T/T group alone suggested stronger protection conferred by C/C in blacks (OR = 0.21, p = 3 × 10−5) relative to that in whites (OR = 0.50, p = 0.04), though our power to detect a true difference is limited due to small sample sizes in some groups. Overall, the protective effect of C appears to be primarily recessive, as no significant difference was observed between the C/T and T/T genotypes in blacks, whites, or combined ethnic groups for clearance of HCV (data not shown), and C/C was consistently protective relative to C/T and/or T/T (Table 2 and Figure 1). The proportion of individuals with C/T or T/T genotypes who cleared the virus (28% for combined genotypes) was similar to a ‘general population’ expectation, since HCV clearance occurs in approximately 30% of those infected. In contrast, HCV clearance was observed much more frequently than expected (53%) in the C/C group. These results nicely mirror the protective effect of the C/C genotype on SVR after HCV treatment observed previously5, where the protection conferred by the C allele also appeared to be recessive in both their Caucasian and African American patients.
Table 2.
Effect of IL28B rs12979860 genotype on clearance of hepatitis C virus
| Adjusted by cohort and ethnicity | |||||
|---|---|---|---|---|---|
| All subjects, genotype | Frequency of clearance, %(N) | Frequency of persistence, %(N) | Comparison | OR (95% CI) | P value |
| TT | 23.4 (37) | 76.6 (121) | CCvsTT | 0.29 (0.18–0.47) | 4×10−7 |
| CT | 29.5 (124) | 70.5 (297) | CCvsCT | 0.35 (0.25–0.48) | 4×10−11 |
| CT+TT | 28 (161) | 72 (418) | CCvsCT+TT | 0.33 (0.25–0.45) | 3×10−13 |
| CC | 53 (227) | 47 (202) | |||
| White subjects, genotype | Adjusted by cohort | ||||
| TT | 31.4 (16) | 68.6 (35) | CCvsTT | 0.50 (0.25–0.98) | 0.04 |
| CT | 27.8 (71) | 72.2 (184) | CCvsCT | 0.36 (0.24–0.52) | 1×10−7 |
| CT+TT | 28.4 (87) | 71.6 (219) | CCvsCT+TT | 0.38 (0.26–0.54) | 1×10−7 |
| CC | 51.8 (174) | 48.2 (162) | |||
| Black subjects, genotype | Adjusted by cohort | ||||
| TT | 20.8 (20) | 79.2 (76) | CCvsTT | 0.21 (0.10–0.44) | 3×10−5 |
| CT | 33 (45) | 67 (91) | CCvsCT | 0.40 (0.21–0.75) | 0.005 |
| CT+TT | 28 (65) | 72 (167) | CCvsCT+TT | 0.32 (0.17–0.57) | 1×10−4 |
| CC | 55.2 (32) | 44.8 (26) | |||
OR = odds ratio; CI = confidence interval
Figure 1.
Percentage of HCV clearance by rs12979860 genotype. Data are shown for all patients, as well as whites and blacks separately.
Some individuals used in this study of HCV were co-infected with hepatitis B virus (HBV) and/or HIV. In order to eliminate the possibility that co-infection with these viruses may confound the effect of rs12979860 on HCV outcome, analyses were performed using a multivariate model that included hepatitis B surface antigen status as a co-variate or stratifying by HIV status. Neither of these two chronic viral infections altered the effect of this locus on outcome of an acute HCV infection (Suppl. Table 1 and 2). We also tested whether there were any differences in the effect of the protective rs12979860 C allele as a function of the route of HCV acquisition (plasma products vs. injection drug use), but found no significant differences between the two groups (data not shown). Finally, adjusting by other host genetic factors that associate with clearance of HCV did not alter the protection conferred by the CC genotype (Supplementary information I).
Patients with lower baseline HCV viral load (VL) respond more favorably to interferon alpha treatment11,12. However, little is known regarding the impact of VL during acute infection on spontaneous HCV clearance because very few HCV-infected individuals are identified and studied at this early phase13. We reasoned that the mechanism of protection of the CC genotype might also extend to greater control of VL in the chronic phase, but there was no correlation between rs12979860 genotype and VL (Supplementary Information II and Supplementary Figure 1). However, differences in viral load assays used in the various cohorts may mask a small correlation, even after conversion to international units.
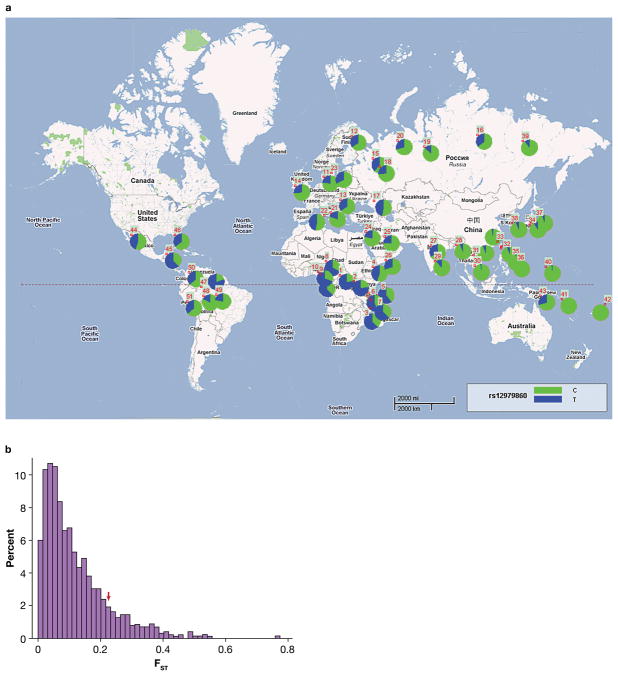
The frequency of HCV clearance varies markedly across ethnic groups9 and differences in allele frequencies for the rs12979860 SNP were observed in the present study (Table 1) and in Ge et al5. Indeed, the observation that the C allele is less frequent among individuals of African descent relative to those of European descent might explain in part the observed discrepancy in the frequency of viral clearance in these two ethnic groups, where clearance occurs in 36.4% of HCV infections in nonblacks, but only 9.3% in blacks7. In order to gain greater insight into the geographic frequency distribution of the protective C allele, we genotyped 2371 individuals from 51 populations worldwide (Table 3 and Figure 2A).
Table 3.
rs12979860 C allele frequency in worldwide populations
| No. | Population | Region | N | C allele frequency, % |
|---|---|---|---|---|
| 1 | Biaka Pygmies | Africa | 66 | 23.5 |
| 2 | Mbuti Pygmies | Africa* | 39 | 23.1 |
| 3 | Chagga | Africa* | 44 | 37.5 |
| 4 | Ethiopian Jews | Africa* | 21 | 54.8 |
| 5 | Masai | Africa* | 20 | 40.0 |
| 6 | Sandawe | Africa* | 37 | 44.6 |
| 7 | Zaramo | Africa | 39 | 37.2 |
| 8 | Hausa | Africa* | 38 | 31.6 |
| 9 | Ibo | Africa* | 47 | 38.3 |
| 10 | Yoruba | Africa* | 77 | 31.2 |
| 11 | Danish | Europe* | 51 | 76.5 |
| 12 | Finns | Europe* | 33 | 65.2 |
| 13 | Hungarians | Europe* | 142 | 65.1 |
| 14 | Irish | Europe* | 113 | 73.9 |
| 15 | Russians, Vologda | Europe* | 48 | 61.4 |
| 16 | Russians | Europe* | 32 | 64.1 |
| 17 | Adygei | Europe* | 53 | 52.8 |
| 18 | Chuvash | Europe* | 40 | 73.7 |
| 19 | Khanty | Europe | 49 | 85.7 |
| 20 | Komi | Europe* | 47 | 70.2 |
| 21 | Roman Jews | Europe* | 27 | 79.6 |
| 22 | Sardinians | Europe | 34 | 52.9 |
| 23 | European American | Europe | 92 | 67.4 |
| 24 | Druze | Southwest Asia | 96 | 77.6 |
| 25 | Kuwaitis | Southwest Asia | 16 | 75.0 |
| 26 | Yemenite Jews | Southwest Asia | 41 | 69.5 |
| 27 | Indians | South Asia* | 29 | 65.5 |
| 28 | Kachari | South Asia* | 17 | 94.1 |
| 29 | Thoti | South Asia* | 14 | 89.3 |
| 30 | Cambodians | Southeast Asia* | 24 | 97.9 |
| 31 | Laotians | Southeast Asia* | 118 | 93.6 |
| 32 | Chinese, Taiwan | East Asia* | 47 | 93.6 |
| 33 | Chinese, San Francisco | East Asia* | 59 | 97.5 |
| 34 | Hakka | East Asia* | 40 | 95.0 |
| 35 | Ami | East Asia | 40 | 98.8 |
| 36 | Atayal | East Asia | 40 | 100.0 |
| 37 | Japanese | East Asia* | 50 | 91.0 |
| 38 | Koreans | East Asia* | 54 | 93.5 |
| 39 | Yakut | East Asia* | 50 | 90.0 |
| 40 | Micronesians | Oceania* | 36 | 98.6 |
| 41 | Nasioi | Oceania* | 23 | 100.0 |
| 42 | Samoans | Oceania* | 8 | 100.0 |
| 43 | Papua New Guineans | Oceania | 22 | 70.4 |
| 44 | Pima, Mexico | North America | 99 | 55.5 |
| 45 | Mayans | North America | 52 | 37.5 |
| 46 | Muscogees | North America | 10 | 65.0 |
| 47 | Ticuna | South America | 62 | 20.2 |
| 48 | Karitiana | South America | 54 | 82.4 |
| 49 | Surui | South America | 47 | 77.7 |
| 50 | Guihiba speakers | South America | 12 | 62.5 |
| 51 | Quechua | South America | 22 | 63.6 |
indicates samples used in FST estimation.
Figure 2.
Sampling locations, allele frequencies and degree of regional differentiation of the rs12979860 C allele. (A) The numbers identifying populations are given in Table 3. The pie charts show the frequency of the C (green) and T (blue) alleles in each population sampled. (B) Frequency distribution of FST values for 1062 SNPs from 32 of the samples grouped into six regions (Africa, Europe, South Asia, Southeast Asia, East Asia, Oceania). The red arrow indicates the position of the estimated FST for rs12979860.
The global pattern of allele frequencies shows a striking pattern in which the allele leading to greater natural HCV clearance is nearly fixed throughout East Asia, has an intermediate frequency in Europe, and is the minor allele in Africa (Figure 2A). A comparison of the rs12979860 allele frequency diversity across 32 world populations (as measured by FST) with that for 1062 SNPs typed in these same samples shows that the rs12979860 polymorphism has a greater differential frequency (FST = 0.23) than most of the other polymorphisms (mean FST = 0.12, standard deviation = 0.1), falling within the upper 12.5 percentile of the distribution of FST values (Figure 2B). Notably, the high frequencies of the C allele found in North and Eastern Asian populations are not reflected in correspondingly high frequencies in their American relatives. Thus, if this locus has been under selection pressure, changes in the selective force that may be dependent on geographical locale, likely occurred after the colonization of the New World. That a common variant has such a strong impact on hepatitis C may indicate that it has actually been under selection, adding to an emerging interpretation of genome-wide association studies whereby common variants rarely have large effects unless they were selected to do so14.
The rs12979860 SNP is only 3 Kb upstream of the IL28B gene, which encodes the type III interferon IFN-λ3, and this SNP is in strong linkage disequilibrium (r2>0.85) with a nonsynonymous coding variant in the IL28B gene (213A>G, K70R; rs8103142)5. Thus, it is possible that this 213A>G change alters the function of IFN-λ3 and explains the genetic data described herein, but functional data will be essential to defining the precise biological mechanism. Type III interferons include three members, IFN-λ1, IFN-λ2 and IFN-λ3, and the genes encoding these molecules are clustered on human chromosome 19q1315,16. They are structurally related to the IL-10 superfamily of cytokines, but share functional characteristics with the type I interferons (IFN-α and IFN-β) in that they are induced by viral infections, signal through the JAK-STAT pathway, and exhibit anti-viral activity in vitro15,16. IFN-λ1 has been shown to exhibit dose- and time-dependent HCV inhibition, induce increases in levels of interferon stimulated genes, and enhance the antiviral efficacy of IFN-α17. It is possible that IFN-λ3 works through a similar mechanism. In vitro, IFN-λ3 is at least as potent as IFN-λ1 in terms of protecting HepG2 cells from lysis after infection with encephalomyocarditis virus18. Severe side effects in HCV treatment have been observed with IFN-α therapy19, whereas IFN-λ treatment may exhibit less “interferon-like” adverse effects because their receptor is only expressed on a limited number of cell types20. Whether IFN-λ may serve as an alternative treatment modality for HCV infection is under investigation.
In conclusion, we have shown that the rs12979860 polymorphism upstream of IL28B which was previously associated with HCV treatment response also has a dramatic impact on natural clearance of HCV and may have been under selection in human history. It is now a priority to determine the mechanisms through which IL28B promotes viral defense, and the full range of viruses affected by these mechanisms.
METHODS SUMMARY
Subjects in this study were participants in one of 6 studies: (i) AIDS Link to Intravenous experience (ALIVE)21 (N = 281); (ii) Multicenter Hemophilia Cohort Study (MHCS)22 (N = 305); (iii) Hemophilia Growth and Development Study (HGDS)23 (N = 106); (iv) Correlates of Resolved Versus Low Level Viremic Hepatitis C Infection in Blood Donors study (REVELL) (N = 85); (v) an HCV clinic cohort in Portland, Oregon (N = 51); (vi) a cohort of injection drug users from the UK (N = 180) (see Methods for details). Fifty one worldwide populations (N = 2371) from the ALlele FREquency Database (ALFRED)24, were also genotyped in this study. Details of sampling and ethnographic information for these populations can be found at http://alfred.med.yale.edu/. All populations were in Hardy-Weinberg equilibrium with the exception of one, the Finnish sample (n = 33, p = 0.05). Genotyping was performed using the ABI TaqMan allelic discrimination kit and the ABI7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). SAS 9.1 (SAS Institute) was used for statistical analyses.
Methods
Study subjects
The AIDS Link to Intravenous experience (ALIVE) is an ongoing study of injection drug users21. The Multicenter Hemophilia Cohort Study (MHCS) is a prospectively followed cohort of patients with coagulation disorders from 16 treatment centers22. The Hemophilia Growth and Development Study (HGDS) is a continuing study of children and adolescents with hemophilia23. The REVELL Study draws both resolved and chronic HCV infections from a large blood bank network consisting of 17 blood centers in the western and southern United States. Individuals with viral clearance in the ALIVE, MHCS and HGDS cohorts were matched to two individuals with viral persistence within the same cohort based on HIV status, gender and ethnicity (African American, European American, Other). All of the individuals in the HCV clinic cohort cleared the virus, so they were not matched. Participants in the REVELL cohort were all HIV negative and HBV negative. Participants in the UK cohort were individuals referred to hepatology clinics. They were all Caucasian, HBV negative, and HIV negative. There was no significant difference in the results when matched and unmatched cohorts were analyzed separately (Table S3)
HCV testing
HCV infection was established by a second or third generation enzyme immunoassay (EIA) (Ortho Diagnostics Systems, Raritan, NJ). Individuals with subsequent negative HCV RNA tests were confirmed anti-HCV positive by a third generation EIA test or a recombinant immunoblot assay (RIBA) that was separated from the first by a minimum of 6 months. HCV RNA was assessed by a branched DNA (bDNA) assay (Quantiplex HCV RNA 2.0 assay; Chiron Corporation, Emeryville, CA), a qualitative HCV COBAS AMPLICOR system (COBAS AMPLICOR HCV; Roche Diagnostics, Branchburg, N.J.), or by transcription-mediated amplification (TMA) (Novartis, Emeryville and Gen Probe Inc., San Diego, CA. Those subjects with a sample below the limit of detection by the bDNA assay (potential subjects with HCV recovery) had a sample separated by 6 months from the first one tested with the qualitative COBAS. HCV infection in blood donors (REVELL study) was established by third generation EIA and confirmed using RIBA. HCV RNA status was established using nucleic acid amplification testing (NAT) of minipools representing sixteen donation samples (Procleix HIV-1/HCV Assay, Gen-Probe, San Diego/Novartis, Emeryville, CA). A reactive minipool result triggered NAT of the individual donations comprising the pool in order to identify the NAT-reactive donation. Residual volume after NAT screening from all antibody positive/RNA negative donors was retested by duplicate undiluted HCV RNA testing using TMA. Individuals with HCV clearance had undetectable HCV RNA in serum or plasma at two time points separated by a minimum of six months. Persistently infected individuals had detectable HCV RNA in serum or plasma at two time points separated by a minimum of six months.
Statistical analysis
SAS 9.1 (SAS Institute) PROC FREQ was used to compute frequencies and Fisher’s exact test P valueson categorical variables. PROC LOGISTIC was used to obtain odds ratios and 95% confidence intervals. PROC MEANS was used to calculate mean age. Analyses were performed with all ethnic groups combined and whites and blacks separately. All analyses were adjusted for study groups, and for ethnicity in three categories (whites, blacks and others) when all ethnic groups were combined. Statistical significance refers to two-sided P values of <0.05.
FST analysis
Population samples were checked in ALFRED to identify all SNPs for which individuals had been typed. For 32 samples an additional 1062 SNPs were available. Data were pooled by regional affiliation (see Table 3) into 6 groupings allowing improved frequency estimation: Africa, Europe, South Asia, Southeast Asia, East Asia, and Oceania. The extent of regional differentiation, FST, was determined for each of the individual SNPs and its distribution plotted. The FST of similarly pooled rs12979860 allele frequencies was compared to that found at other loci.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This research was supported by NIH grants R01DA013324 (DT), R01DA004334 (GK), R01DA012568 (SM), R01HL076902 (MB), R01DK60590 (HR), and R01HD41224 (SD). SK is a Wellcome Trust Senior Clinical Fellow.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions D.L.T., C.L.T., J.G.M., D.B.G. and M.C. designed the study. M.C. directed the study. M.P.M. performed the genotyping. M.P.M and M.C. wrote the manuscript, with major edits by D.L.T., C.L.T. and D.B.G. The analyses were performed by Y.Q, D.G. and C.O. The samples used in the study were provided by C.L.T. J.K., K.K., S.I.K., G.A., J.J.G., G.D.K., S.M.D., H.R.R., L.H.T., M.P.B., and D.L.T. All authors contributed to preparing the final manuscript.
Author information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Centers for Disease Control and Prevention. MMWR Recomm Rep. 1998;47(RR-19):1–39. [PubMed] [Google Scholar]
- 2.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36(5 Suppl 1):S35–46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 3.Cooper S, et al. Analysis of a successful immune response against hepatitis C virus. Immunity. 1999;10(4):439–449. doi: 10.1016/s1074-7613(00)80044-8. [DOI] [PubMed] [Google Scholar]
- 4.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5 (3):215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 5.Ge D, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009 doi: 10.1038/nature08309. In Press. [DOI] [PubMed] [Google Scholar]
- 6.Kenny-Walsh E. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. Irish Hepatology Research Group. N Engl J Med. 1999;340(16):1228–1233. doi: 10.1056/NEJM199904223401602. [DOI] [PubMed] [Google Scholar]
- 7.Thomas DL, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284(4):450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- 8.Thio CL. Host genetic factors and antiviral immune responses to hepatitis C virus. Clin Liver Dis. 2008;12(3):713–726. xi. doi: 10.1016/j.cld.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thio CL, Thomas DL, Carrington M. Chronic viral hepatitis and the human genome. Hepatology. 2000;31(4):819–827. doi: 10.1053/he.2000.4316. [DOI] [PubMed] [Google Scholar]
- 10.McHutchison JG, Lawitz EJ, Shiffman ML, et al. Peginterferon alpha-2b or alpha-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361(6):24–37. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 11.Lindsay KL, et al. A randomized, double-blind trial comparing pegylated interferon alfa-2b to interferon alfa-2b as initial treatment for chronic hepatitis C. Hepatology. 2001;34(2):395–403. doi: 10.1053/jhep.2001.26371. [DOI] [PubMed] [Google Scholar]
- 12.Zeuzem S, et al. Peginterferon alfa-2a in patients with chronic hepatitis C. N Engl J Med. 2000;343(23):1666–1672. doi: 10.1056/NEJM200012073432301. [DOI] [PubMed] [Google Scholar]
- 13.Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29(3):908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 15.Kotenko SV, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 16.Sheppard P, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 17.Marcello T, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 18.Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun. 2009;10(2):125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 19.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345 (1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 20.Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leukoc Biol. 2004;76(2):314–321. doi: 10.1189/jlb.0204117. [DOI] [PubMed] [Google Scholar]
- 21.Vlahov D, et al. The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr. 1991;109:75–100. [PubMed] [Google Scholar]
- 22.Goedert JJ, et al. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med. 1989;321(17):1141–1148. doi: 10.1056/NEJM198910263211701. [DOI] [PubMed] [Google Scholar]
- 23.Hilgartner MW, et al. Hemophilia growth and development study. Design, methods, and entry data. Am J Pediatr Hematol Oncol. 1993;15(2):208–218. doi: 10.1097/00043426-199305000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Osier MV, et al. ALFRED: An allele frequency database for anthropology. Am J Phys Anthropol. 2002;119(1):77–83. doi: 10.1002/ajpa.10094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.