Abstract
Cocrystal structures of wild-type TATA box-binding protein (TBP) recognizing 10 naturally occurring TATA elements have been determined at 2.3–1.8 Å resolution, and compared with our 1.9 Å resolution structure of TBP bound to the Adenovirus major late promoter (AdMLP) TATA box (5′-TATAAAAG-3′). Minor-groove recognition by the saddle-shaped protein induces the same conformational change in each of these oligonucleotides, despite variations in promoter sequence that reduce the efficiency of transcription initiation. Three molecular mechanisms explain assembly of diverse TBP–TATA element complexes. (1) T → A and A → T transversions leave the minor-groove face unchanged, permitting formation of TBP–DNA complexes on many A/T-rich core promoter sequences. (2) Cavities in the interface between TBP and the minor-groove face of the AdMLP TATA box accommodate the exocyclic NH2 groups of G in a TACA box and in a TATAAG box. (3) Formation of a C:G Hoogsteen basepair in a TATAAAC box eliminates steric clashes that would be produced by the Watson–Crick base pair. We conclude that the structure of the TBP–TATA box complex found at the heart of the polymerase II (pol II) transcription machinery has remained constant over the course of evolution, despite variations in TBP and its DNA targets.
Keywords: TATA box, transcription, TBP–TATA complex, Pol II
In eukaryotes RNA polymerase II (Pol II) is responsible for transcribing nuclear genes encoding the mRNAs and several small nuclear RNAs. Like RNA Pol I and Pol III, Pol II cannot recognize its target promoter directly and initiate transcription without accessory proteins. Instead, this large multisubunit enzyme relies on both general transcription factors (GTFs) and transcriptional activators and coactivators (both positive and negative) to regulate transcription from class-II nuclear gene promoters (for review, see Roeder 1996). The primary DNA anchor of this complicated macromolecular machine is transcription factor IID (TFIID), a 700-kD complex composed of the TATA box-binding protein (TBP) and a set of phylogenetically conserved, Pol II-specific TBP-associated factors (for review, see Burley and Roeder 1996). DNA binding by human TFIID was first demonstrated with the adenovirus major-late promoter (AdMLP). DNase I footprinting studies of the AdMLP and selected human gene promoters revealed sequence-specific interactions with the TATA element, which are primarily mediated by TBP. Protection outside of the TATA box displays a nucleosome-like pattern of DNase I hypersensitivity, varies radically among promoters, and can be induced by some activators (for review, see Burley and Roeder 1996).
Genes encoding TBPs have been cloned from organisms ranging from archaea to human. The molecules share a phylogenetically conserved 180-residue carboxy-terminal or core segment, which contains two imperfect direct repeats and supports all of the protein's biochemically important functions in Pol II transcription (for review, see Burley and Roeder 1996). Because of the original purification and characterization of Saccharomyces cerevisiae TBP, there has been considerable progress toward understanding its mechanisms of action. Specific nanomolar-affinity (Hahn et al. 1989) binding to a TATA element entails DNA bending (Horikoshi et al. 1992) and occurs exclusively via minor groove interactions (Lee et al. 1991; Starr and Hawley 1991). Three-dimensional structures of a full-length TBP from Arabidopsis thaliana (Nikolov et al. 1992; Nikolov and Burley 1994), yeast core TBP (Chasman et al. 1993), and full-length TBP from the archaeon Pyrococcus woesei (DeDecker et al. 1996) have been determined. The monomeric protein consists of two nearly identical domains and adopts a quasisymmetric α/β structure, resembling a molecular saddle complete with stirrups (Fig. 1). The concave underside of the saddle is a highly curved, 10-stranded, antiparallel β-sheet, containing the amino acids involved in DNA binding (Fig. 2). The convex upper surface of the saddle consists of four α-helices, which interact with other transcription factors (for review, see Nikolov and Burley 1994). Cocrystal structure determinations of A. thaliana, yeast core, and human core TBPs interacting with similar TATA elements (Kim et al. 1993a,b; Kim and Burley 1994; Juo et al. 1996; Nikolov et al. 1996) revealed an unusual protein–DNA complex, characterized by extensive hydrophobic interactions with the minor groove, severely distorted DNA, and phenylalanine side chains kinking DNA by insertion between base pairs at the 5′ and 3′ ends of the TATA box. The same conformational change has been observed in triple-complex cocrystal structures of TBP plus DNA with human TFIIB (Nikolov et al. 1995), an archaeal homolog of TFIIB (Kosa et al. 1997), and yeast TFIIA (Geiger et al. 1996; Tan et al. 1996). Further relevant work on TBP includes examination of the kinetics and thermodynamics of TATA element binding (Hoopes et al. 1992; Petri et al. 1995, 1998; Parkhurst et al. 1996), and studies of the effects of prebending promoter DNA (Parvin et al. 1995) and TBP-induced DNA deformation (Sun and Hurley 1995; L. Hurley, unpubl.).
Figure 1.
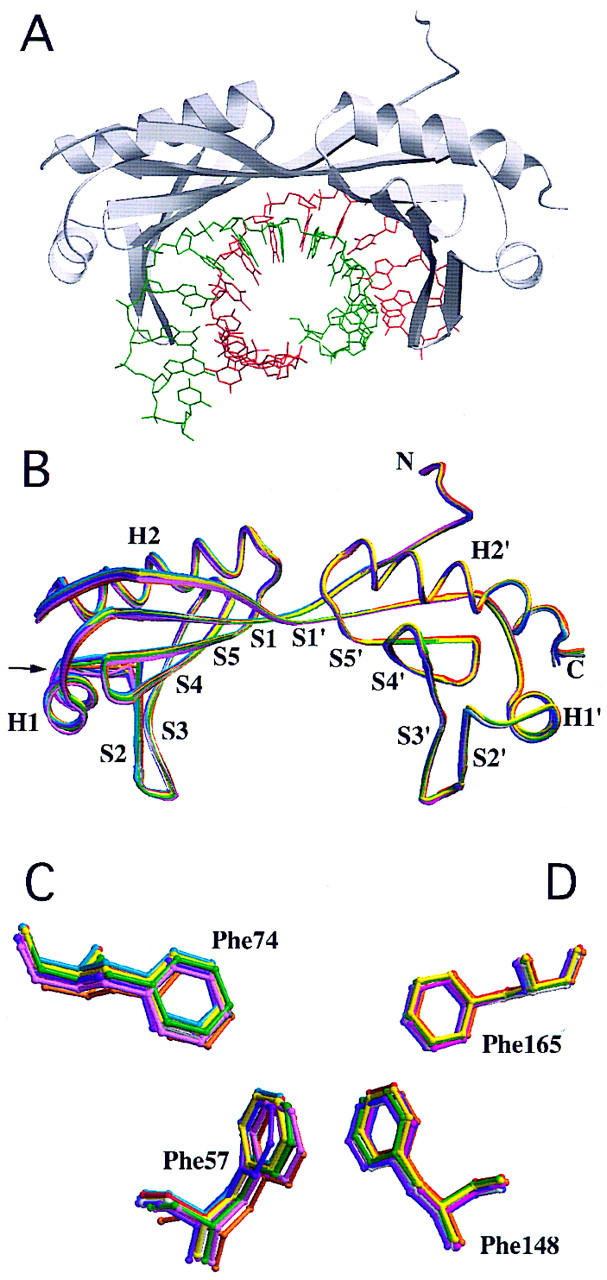
Structure of TBP bound to the TATA element. (A) MOLSCRIPT drawing of the TBP–TATA box complex (coding strand shown in green) viewed perpendicular to the twofold quasisymmetry axis. (B) Overlay of the polypeptide backbones of 11 TBP–DNA complexes, by use of a spaghetti representation with a different color for each cocrystal structure and the view shown in A. Atomic stick figure representations of the amino-terminal (Phe-57 and Phe-74) (C) and carboxy-terminal (Phe-148 and Phe-165) (D) phenylalanine pairs from the 11 TBP–DNA complexes, by use of the color coding in B. Overlay was performed by least-squares superposition of the α-carbons only.
Figure 2.
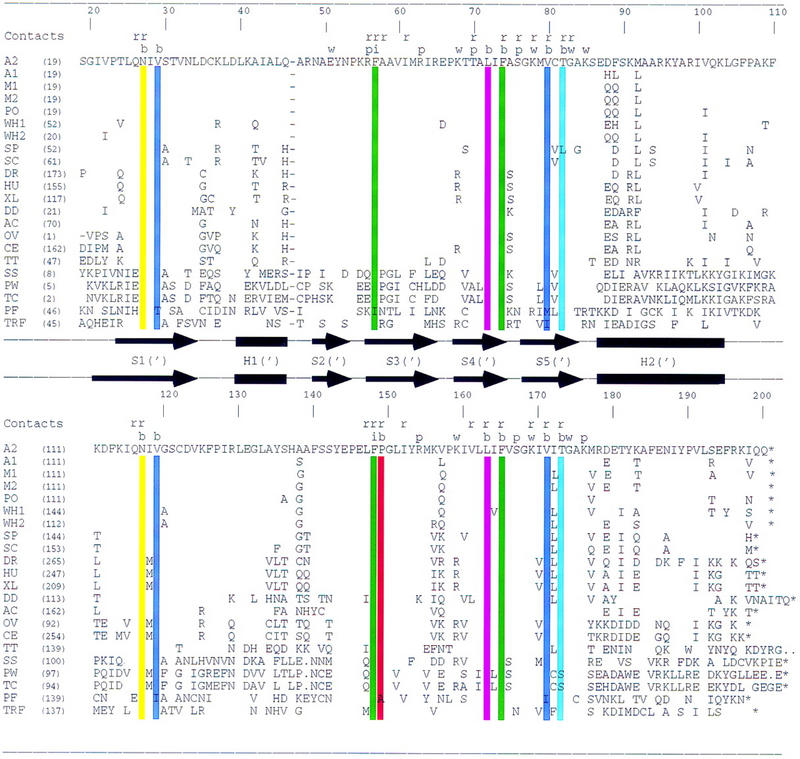
TBP sequence alignments. Sequences of 21 TBPs and D. melanogaster TBP-related factor aligned by use of the three-dimensional structure of A. thaliana TBP2. (A1) A. thaliana TBP isoform 1; (A2) A. thaliana TBP isoform 2; (M1) maize TBP isoform 1; (M2) maize TBP isoform 2; (PO) potato TBP; (W1) wheat TBP isoform 1; (W2) wheat TBP isoform 2; (SP) S. pombe TBP; (SC) S. cerevisiae TBP; (DM) D. melanogaster TBP; (HS) human TBP; (XL) Xenopus laevis TBP; (DD) Dictyostelium discoideum TBP; (AC) Acanthamoeba castellanii TBP; (OV) Onchocerca volvulus TBP; (CE) Caenorhabditis elegans TBP; (TT) Tetrahymena thermophilus TBP; (SS) Sulfolobus shibatae TBP; (PW) P. woesei TBP; (TC) T. celer TBP; (PF) P. falciparum TBP; (TRF) D. melanogaster TRF. The secondary structure is denoted with arrows (β-strands) and solid bars (α-helices). Scheme for identifying protein–DNA contacts is as follows: (r) ribose; (p) phosphate; (w) water-mediated backbone; (b) base; (i) intercalating phenylalanine. Color coding of residues interacting with bases is identical to that used in Fig. 4.
Computational studies of the eukaryotic promoter database (EPD) have also yielded important insights into the function of TBP (see Table 1 for a summary of results available on the internet via http://www.epd.isb-sib.ch/promoter_elements). An exhaustive statistical survey documented that the TATA box is an A/T-rich 8-bp segment often flanked by G/C-rich sequences (Bucher 1990). Despite the marked preference for A:T and T:A base pairs, C:G and/or G:C base pairs occur frequently at five of the eight positions. Thus, TBP can bind productively to a large number of diverse TATA elements, some of which bear little resemblance to the optimal TATA sequence (5′-TATATAAG-3′), identified by an in vitro binding-site selection experiment with Acanthamoeba TBP (Wong and Bateman 1994).
Table 1.
TATA element variants
| Name
|
Oligonucleotide
|
cUAS (%)
|
MEL1 (%)
|
Observed frequency
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AdMLP | 5′ | −31 | −24 | 100 | 100 | A/C/G/T | |||||||||||||
| G | C | T | A | T | A | A | A | A | G | G | G | C | A | ||||||
| C | G | A | T | A | T | T | T | T | C | C | C | G | T | ||||||
| −31‘ | −24‘ | 5′ | |||||||||||||||||
| A(−31) | A | A | T | A | A | A | A | G | 16 | 11 | 5.0/11.0/4.5/79.5 | ||||||||
| T | T | A | T | T | T | T | C | ||||||||||||
| T(−30) | T | T | T | A | A | A | A | G | 29 | 20 | 83.5/1.3/1.3/13.9 | ||||||||
| A | A | A | T | T | T | T | C | ||||||||||||
| C(−29) | T | A | C | A | A | A | A | G | 24 | 15 | 4.4/3.3/0.9/91.4 | ||||||||
| A | T | G | T | T | T | T | C | ||||||||||||
| T(−28) | T | A | T | T | A | A | A | G | N.D. | 14 | 89.2/0.8/1.7/8.4 | ||||||||
| A | T | A | A | T | T | T | C | ||||||||||||
| T(−27) | T | A | T | A | T | A | A | G | 43 | 25 | 71.0/0.8/0.5/27.7 | ||||||||
| A | T | A | T | A | T | T | C | ||||||||||||
| T(−26) | T | A | T | A | A | T | A | G | 6 | 6 | 84.8/2.9/9.5/2.8 | ||||||||
| A | T | A | T | T | A | T | C | ||||||||||||
| G(−26) | T | A | T | A | A | G | A | G | N.D. | 18 | 84.8/2.9/9.5/2.8 | ||||||||
| A | T | A | T | T | C | T | C | ||||||||||||
| T(−25) | T | A | T | A | A | A | T | G | 110 | 90 | 45.0/3.4/16.4/35.2 | ||||||||
| A | T | A | T | T | T | A | C | ||||||||||||
| C(−25) | T | A | T | A | A | A | C | G | 5 | 6 | 45.0/3.4/16.4/35.2 | ||||||||
| A | T | A | T | T | T | G | C | ||||||||||||
| T(−24) | T | A | T | A | A | A | A | T | N.D. | N.D. | 35.8/14.0/38.4/11.8 | ||||||||
| A | T | A | T | T | T | T | A | ||||||||||||
Crystallization oligonucleotide and two assays of transcriptional efficiency [(N.D.) not done] are given for each cocrystal structure of wild-type TBP bound to a variant of the AdMLP TATA element. Eukaryotic Promoter Database estimated frequency statistics are given for each of the eight positions in the TATA element (http://www.epd.isb-sib.ch/promoter_elements). The estimated frequencies corresponding to the AdMLP and the TATA variant base pairs are denoted with boldface type and underlining, respectively. Values of ∼1% or less are incompatible with the structural data and are thought to reflect noise in the computational process used to identify and align TATA elements in the EPD (P. Bucher, pers. comm.).
In an effort to understand how TBP can support Pol II transcription initiation from many different TATA boxes in class-II nuclear gene promoters, we have made a systematic X-ray crystallographic study of a canonical wild-type TBP (A. thaliana TBP isoform 2) recognizing 10 naturally occurring variants of the AdMLP TATA element. The structure of the TBP–DNA complex is essentially independent of TATA element sequence. Three distinct molecular mechanisms allow TBP to induce the same DNA deformation, one of which uses Hoogsteen base pairs adjacent to the 3′ kink site. Detailed analyses of these high-resolution cocrystal structures document that TATA element recognition has remained constant over the course of evolution.
Results and Discussion
Study design and functional characterization of TATA element variants
Table 1 illustrates the oligonucleotides used for crystallization with wild-type TBP. The 10 sequences represent naturally occurring single-base variants of the AdMLP TATA element (5′-TATAAAAG-3′), including at least one substitution at each position. Diffraction-quality cocrystals were obtained with C:G or G:C base pairs at four of the five TATA box positions, where they occur with non-zero estimated frequency in the EPD (ranging from 2.9% to 38.4%). Defining the intrinsic strength of the AdMLP TATA box to be 100% (see Materials and Methods), the variants show transcription activity levels ranging between 5% and 110%. (Similar cocrystallization trials were unsuccessful with oligonucleotides that do not support transcription initiation or TBP binding, including 5′-TGTAAAAG-3′, 5′-TCTAAAAG-3′, 5′-TAAAAAAG-3′, 5′-TATGAAAG-3′, and 5′-TATACAAG-3′.) Table 2 provides a summary of the crystallographic statistics, showing that all 10 newly determined cocrystal structures are of the highest quality. The presence of two protein–DNA complexes/asymmetric units in each case permits an estimate of the precision of each set of atomic coordinates. Root-mean-square deviations (rmsds) between α-carbons range between 0.2 and 0.4 Å, which is comparable to the precision of the X-ray method at resolution limits between 1.8 and 2.3 Å (Brünger and Rice 1997).
Table 2.
Summary of crystallographic statistics
|
|
AdMLP
|
A(−31)
|
T(−30)
|
C(−29)
|
T(−28)
|
T(−27)
|
T(−26)
|
G(−26)
|
T(−25)
|
C(−25)
|
T(−24)
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| Data collection | |||||||||||
| Source/detector | X25/IP | A1/CCD | F1/IP | A1/CCD | A1/CCD | F1/IP | A1/CCD | X25/IP | X25/IP | F1/IP | F1/IP |
| Resolution (Å) | 20–1.88 | 15–2.29 | 15–1.79 | 15–1.90 | 15–2.09 | 15–2.3 | 15–2.09 | 15–1.93 | 15–2.23 | 15–1.95 | 15–1.86 |
| Rmerge (I) | 0.052 | 0.053 | 0.051 | 0.037 | 0.045 | 0.058 | 0.060 | 0.050 | 0.072 | 0.054 | 0.042 |
| Completeness (%) | 93.2 | 90.9 | 98.1 | 86.8 | 97.0 | 96.2 | 89.5 | 98.7 | 98.5 | 99.5 | 96.5 |
| Refinement statistics | |||||||||||
| Resolution (Å) | 6–1.9 | 6–2.3 | 6–1.8 | 6–1.9 | 6–2.1 | 6–2.3 | 6–2.1 | 6–1.93 | 6–2.23 | 6–1.95 | 6–1.86 |
| Rcryst (F) | 0.197 | 0.182 | 0.193 | 0.209 | 0.194 | 0.194 | 0.193 | 0.196 | 0.191 | 0.199 | 0.210 |
| Rfree (F) | 0.265 | 0.257 | 0.239 | 0.275 | 0.266 | 0.263 | 0.251 | 0.252 | 0.267 | 0.255 | 0.264 |
| ΔBonds (Å) | 0.013 | 0.009 | 0.008 | 0.011 | 0.009 | 0.009 | 0.009 | 0.008 | 0.009 | 0.009 | 0.010 |
| ΔAngles (°) | 1.7 | 1.5 | 1.4 | 1.6 | 1.5 | 1.4 | 1.4 | 1.4 | 1.5 | 1.5 | 1.5 |
| ΔB-factors (Å2) | 1.8 | 2.0 | 1.9 | 2.0 | 1.9 | 2.0 | 1.9 | 2.0 | 1.9 | 1.9 | 1.9 |
| No. of water molecules | 520 | 269 | 478 | 350 | 350 | 263 | 378 | 502 | 329 | 472 | 439 |
| 〈B〉 protein (Å2) | 26 | 26 | 25 | 31 | 28 | 29 | 26 | 26 | 30 | 25 | 26 |
| 〈B〉 DNA (Å2) | 31 | 31 | 28 | 36 | 31 | 36 | 32 | 31 | 36 | 30 | 32 |
| 〈B〉 water (Å2) | 40 | 39 | 39 | 44 | 40 | 41 | 39 | 41 | 42 | 37 | 40 |
| PDB code | 1QNE | 1QNC | 1QNA | 1QN9 | 1QN8 | 1QN7 | 1QN6 | 1QN5 | 1QNB | 1QN3 | 1QN4 |
Rmerge (I) = Σ|I − 〈I〉 |/ΣI, where I = observed intensity and 〈I〉 = average intensity obtained from multiple observations of symmetry related reflections. ΔBonds and ΔAngles are the respective rmsds from ideal values. ΔB-factors is the rmsd between the B values of covalently bonded atomic pairs. Free R-factor was calculated with 10% of the data omitted from the structure refinement. Source/detector denotes beamline (X25 at National Synchrotron Light Source; A1 and F1 at Cornell High Energy Synchrotron Source) and detector [(IP) image plate; (CCD) charge-coupled device].
TBP structure is not affected by TATA element sequence variation
Figure 1B illustrates the structures of TBP extracted from each cocrystal structure and overlaid by least-squares superposition on our original AdMLP cocrystal structure (Kim et al. 1993a; Kim and Burley 1994). With the exception of the short segment connecting α-helix H1 to β-strand S2, TBP structural variation as a function of TATA element sequence is comparable to the precision of the atomic coordinates (α-carbon rmsds = 0.2–0.5 Å). All subsequent structural comparisons are based on the superpositions in Figure 1B. The phenylalanine pairs (Phe-148 and Phe-165, Phe-57, and Phe-74) responsible for kinking the TATA element at its 5′ and 3′ ends also show no significant structural variation (Fig. 1C,D).
Trajectory of the DNA double-helix axis is not affected by TATA element sequence variation
Figure 3 illustrates only minimal variation in the trajectory of the DNA double-helix axis as a function of TATA element sequence. This similarity documents that TBP binding induces the same deformation in the core promoter, independent of the precise sequence and transcription activity of the TATA element. Detailed analyses of the DNA structural parameters (Lavery and Sklenar 1989; Stofer and Lavery 1993) for each TATA element (data not shown) reveal only minor differences when compared with the AdMLP TATA box bound to the same protein (Kim and Burley 1994).
Figure 3.
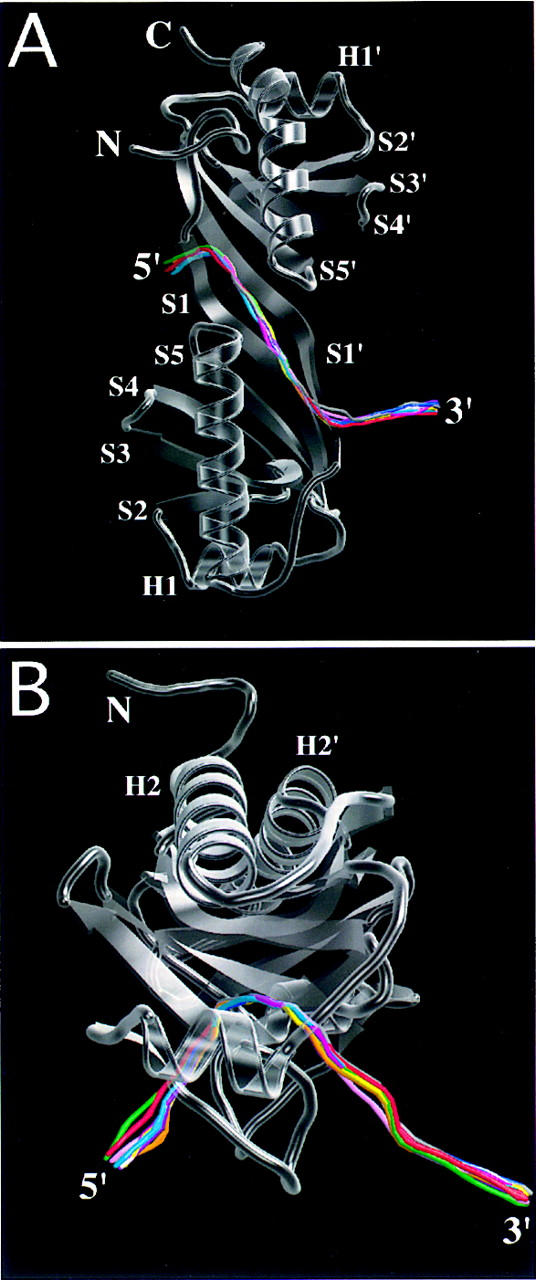
DNA deformation in the TBP–TATA element complex. Composite drawings showing a semitransparent ribbon representation of TBP with a line representation of the trajectory of the DNA double-helix axis for each cocrystal structure, calculated by use of CURVES (Stofer and Lavery 1993). The color coding of each double-helix axis corresponds to those used in Fig. 1. (A) Viewed along the twofold quasisymmetry axis; (B) viewed through the stirrups of the molecular saddle. Overlay was performed by least-squares superposition of the protein structures only.
Structure of the TBP–DNA complex is not affected by TATA element sequence variation
Pairwise comparisons of our TBP–DNA cocrystal structures document that even the detailed structure of the protein–DNA complex is not significantly affected by variations in the TATA element (rmsds for core α-carbon and TATA element C1′ atoms = 0.2–0.5 Å). A comparison of plant and archaeal TBP–DNA complexes reveals a very similar protein–DNA interface (rmsds for TATA element C1′ atoms = 1.1 Å), despite the fact that the upper surfaces of the two molecular saddles are somewhat different (data not shown). We conclude that the structure of the TBP–TATA element complex is independent of the sequence of the TATA element.
Figure 4 provides a schematic view of the protein surface responsible for TATA element recognition. In all available crystal structures containing TBP and DNA, only 15 highly conserved residues contribute to interactions with the minor-groove edges of the bases. Excluding Plasmodium falciparum TBP and Drosophila melanogaster TBP-related factor (TRF) from the sequence comparison, these 15 residues are almost absolutely invariant (Fig. 2). The only exceptions are a Thr-82 → Leu substitution in Schizosaccharomyces pombe TBP, and Thr-173 → Ser substitutions in P. woesei and Thermococcus celer TBPs. The sequence of P. falciparum TBP displays six conservative substitutions, including Val-29 → Thr, Phe-57 → Ile, Val-80 → Met (which is an Ile in Drosophila TRF), Val-119 → Ile, Pro-149 → Ala, and Val-171 → Ile. The residues responsible for direct and water-mediated interactions with the DNA backbone are also highly conserved, showing minimal variation particularly in the carboxy-terminal half of the protein (Fig. 2). Thus, it is not surprising that the three-dimensional structure of TBP and its interactions with DNA have remained unchanged throughout evolution.
Figure 4.
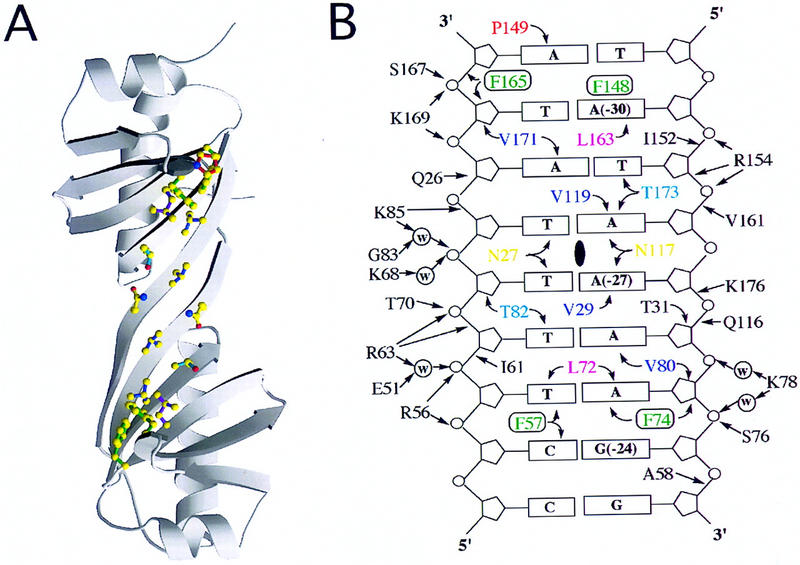
Complementary protein and DNA-binding surfaces. (A) MOLSCRIPT drawing of the DNA-binding surface of TBP viewed along the twofold quasisymmetry axis. Amino acid side chains making base contacts are drawn as stick figures. Atom color codes: yellow, carbon; blue, nitrogen; red, oxygen. side chain color codes: red, Pro149; green, intercalating phenylalanines Phe-148, Phe-165, Phe-57, Phe-74; magenta, Leu-163, Leu-72; dark blue, Val-171, Val-119, Val-80, Val-29; light blue, Thr-173, Thr-82; yellow, Asn-27, Asn-117. Color coding of residues interacting with bases is identical to that used in Fig. 2. (B) Schematic drawing of the minor-groove surface of the crystallization oligonucleotide showing all direct and water-mediated contacts. The location of the twofold quasisymmetry axis is denoted by a solid, vertical oval. A and B have been prepared as if the image of the underside of TBP was on one page of an open book facing an image of the minor groove of the AdMLP TATA element.
Three mechanisms explain how TBP exploits the same induced fit strategy to recognize all 10 variants of the AdMLP TATA element
The major-groove face of B-DNA is varied chemically as a function of sequence, which explains why most sequence-specific DNA-binding proteins interact with the major-groove edges of base pairs in their recognition elements (for review, see Patikoglou and Burley 1997). In contrast, the minor-groove face is chemically monotonous as a function of sequence (Seeman et al. 1976). T → A and A → T transversions leave the structure of the minor-groove face essentially unchanged, because both A:T and T:A display similarly positioned pairs of hydrogen bond acceptors on their minor-groove edges. G:C and C:G, on the other hand, provide some relative chemical variation. Minor-groove differences between G:C and A:T or C:G and T:A arise from an exocyclic NH2 protruding from G. It is not surprising that DNA-binding proteins exhibiting little or no sequence specificity interact primarily with the minor groove (for review, see Patikoglou and Burley 1997). TBP exploits an induced-fit mechanism that relies, in part, on the chemical monotony of the minor groove to recognize many different core promoters during Pol II transcription initiation.
Minor-groove structural degeneracy of A:T and T:A permits TBP–DNA complex formation on many A/T-rich promoter sequences, albeit with dramatically reduced transcriptional efficiency in some cases (Table 1). Figure 5 depicts the consequences of changing the first base pair of the TATA element from T:A to A:T in our A(−31) cocrystal structure of an AATA box (5′-AATAAAAG-3′, frequency = 5.0%, activity = 11%–16%), and the second base pair from A:T to T:A in our T(−30) structure of a TTTA box (5′-TTTAAAAG-3′, frequency = 13.9%, activity = 20%–29%). In both cases, the two cocrystal structures are essentially identical because the protein cannot readily distinguish T:A from A:T. We obtained similar findings for cocrystal structures of T(−28) [TATT box (5′-TATTAAAG-3′, frequency = 8.4%, activity = 14%)] and T(−27) [TATAT box (5′-TATATAAG-3′, frequency = 27.7%, activity = 25%–43%)], and T(−25) [TATAAAT box (5′-TATAAATG-3′, frequency = 35.2%, activity = 90%–110%)] and T(−24) [TATAAAAT box (5′-TATAAAAT-3′, frequency = 11.8%, activity not measured)] (Figs. 6 and 7). Although TBP cannot readily distinguish T:A from A:T, work by the laboratories of Dervan and Rees has demonstrated that a class of small molecules can do so (Kielkopf et al. 1998).
Figure 5.
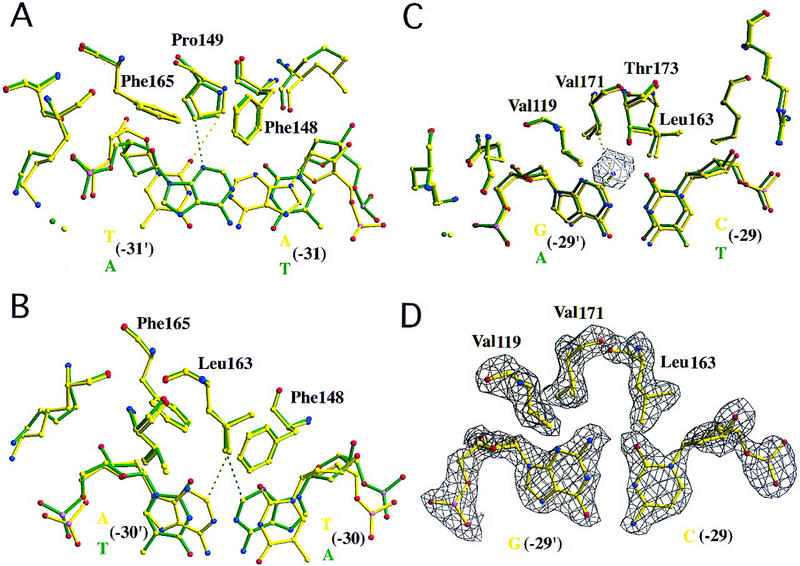
A(−31), T(−30), and C(−29) TATA variants bound to wild-type TBP. Overlays of the structure of TBP bound to the AdMLP (green) and variant TATA elements (yellow), calculated with least-squares superposition of the protein structures. Each composite view shows only the base pair differing between the two crystallization oligonucleotides, surrounding protein residues and bridging water molecules. Protein and DNA are illustrated as atomic stick figures, with color coding for atom type. (C) Yellow; (O) red; (N) blue; (P) pink. Prime denotes bases of the noncoding strand (color coded red in Fig. 1A). Electron density difference (|Fobserved| − |Fcalculated|) maps were calculated with simulated annealing (maximum temperature = 1000 K) after omitting the illustrated base pair from the phase calculation and fixing surrounding atoms within a 3 Å shell. This procedure eliminates model bias, yielding experimental electron density for the structural differences observed between AdMLP and the variant TATA element. The same image format and color-coding scheme is used for Figs. 6–8. (A) A(−31) variant (5′-AATAAAAG-3′, frequency = 5.0%, activity = 11%–16%). Substitution of A:T for T:A yields similar contacts between Pro-149 and the minor-groove edge of the first base pair. Broken lines denote van der Waals contacts between Pro-149 and O2 of T(−31′) (yellow) and N3 of A(−31′) (green) with distances of 3.1 and 3.3 Å, respectively. (B) T(−30) variant (5′-TTTAAAAG-3′, frequency = 13.9%, activity = 20%–29%). The C2 atoms of A(−30′) (yellow) and A(−30) (green) are similarly close to Leu-163 (3.7 and 3.6 Å, respectively). (C) C(−29) variant (5′-TACAAAAG-3′, frequency = 3.3%, activity = 15%–24%). Exocyclic NH2 of G(−29′) is accommodated in a pocket, formed by the side chains of Val-119, Val-171, Thr-173, and Leu-163. The closest G(−29′) NH2 contact occurs with Val-171 [3.6 Å]. Inspection of our original AdMLP cocrystal structure reveals a cavity (denoted by a meshwork polygon) between A(29′) and the residues forming the exocyclic NH2 pocket. (D) Simulated annealing electron density map corresponding to the view illustrated in C, contoured at 2.5ς. The map shows clear density for the exocyclic NH2 of G(−29′).
Figure 6.
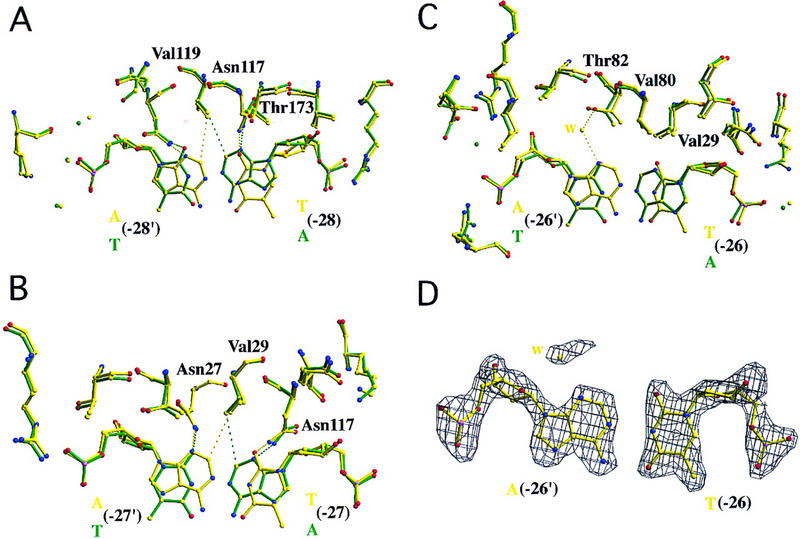
T(−28), T(−27), and T(−26) TATA variants bound to wild-type TBP. (A) T(−28) variant (5′-TATTAAAG-3′, frequency = 8.4%, activity = 14%). This single base pair change places hydrogen bond acceptors in a similar position as in the AdMLP cocrystal structure, satisfying the hydrogen bond donors of Asn-27 and Asn-117 [A(−28′) N3–Asn-27 ND2 = 3.2 Å; T(−28′) O2–Asn-27 ND2 = 3.2 Å; T(−28) O2–Asn-117 ND2 = 2.9 Å; A(−28) N3–Asn-117 ND2 = 3.4 Å]. No unfavorable steric contacts are introduced [A(−28′) C2–Val-119 CG2 = 3.7 Å vs. A(−28) C2–Val-119 CG2 = 3.7 Å in the AdMLP cocrystal structure]. The label for Asn-27 has been omitted for clarity. (B) T(−27) variant (5′-TATATAAG-3′, frequency = 27.7%, activity = 25%–43%). This substitution has the same effect as the T(−28) variant [A(−27′) N3–Asn-27 ND2 = 3.1 Å; T(−27′) O2–Asn-27 ND2 = 2.8 Å; T(−27) O2–Asn-117 ND2 = 3.1 Å; A(−27) N3–Asn-117 ND2 = 3.2 Å]. Again, a valine side chain (Val-29) is in van der Waals contact with the minor-groove face of the A [A(−27′) C2–Val-29 CG2 = 3.6 Å vs. A(−27) C2–Val-29 CG2 = 3.7 Å in the AdMLP cocrystal structure]. (C) T(−26) variant (5′-TATAATAG-3′, frequency = 2.8%, activity = 6%). This A:T to T:A substitution yields subtle changes in the structure of the protein–DNA interface, permitting entry of a water molecule (yellow sphere) bridging between the minor groove edge of the A and a threonine side chain [A(−26′) N3–H2O = 2.9 Å; H2O–Thr-82 OG1 = 2.6 Å]. (D) Simulated annealing electron density map corresponding to the view illustrated in C, contoured at 2.5ς. The map shows clear electron density for the variant T:A base pair and interfacial water molecule.
Figure 7.

G(−26), T(−25), and T(−24) TATA variants bound to wild-type TBP. (A) G(−26) variant (5′-TATAAGAG-3′, frequency = 9.5%, activity = 18%). Exocyclic NH2 of G(−26) is accommodated in a pocket, formed by the side chains of Val-29, Val-80, Thr-82, and Leu-72 (omitted for clarity). The closest G(−26) NH2 contact occurs with Val-80 (3.7 Å). Inspection of our original AdMLP cocrystal structure reveals a cavity (denoted by a meshwork polygon) between A(26) and the residues forming the exocyclic NH2 pocket. (B) Simulated annealing electron density map corresponding to the view illustrated in A, contoured at 2.5ς. The map shows clear density for the exocyclic NH2 of G(−26). (C) T(−25) variant (5′-TATAAATG-3′, frequency = 35.2%, activity = 90%–110%). The van der Waals contact between A C2 and Leu-72 is preserved [A(−25′) C2–Leu-72 CD1 = 3.6 Å vs. A(−25) C2–Leu-72 CD1 = 3.7 Å in the AdMLP cocrystal structure]. This panel is very similar to the view illustrated in Fig. 5B, in which A:T and T:A base pairs make van der Waals contacts with another leucine side chain related by twofold quasisymmetry (Leu-163). (D) T(−24) variant (5′-TATAAAAT-3′, frequency = 11.8%, activity = ND). Like position 1 (Fig. 5A), all four base pairs are tolerated at this position. In the AdMLP cocrystal structure, the closest contact between protein and DNA involves CE1 of Phe-57 and the exocyclic NH2 of G(−24) (3.6 Å). Substitution of G:C with T:A yields a similar van der Waals contact [A(−24′) C2–Phe-57 CE1 = 3.4 Å].
Shape complementarity is not sufficient, however, to ensure that a given sequence will bind productively to TBP. For example, a TAAAAAA box is inactive in Pol II transcription (Wobbe and Struhl 1990) and does not form a stable complex with TBP (Starr et al. 1995). Model building suggests that the TAAAAAA box should function (data not shown), but the A tract is probably too rigid (DiGabriele and Steitz 1993) to undergo the deformation characteristic of all known TBP–DNA complexes. Crystallization attempts with oligonucleotides bearing TAAAAAA were entirely unsuccessful.
The second mechanism underlying relaxed DNA-binding specificity allows TBP to accommodate the exocyclic NH2 of G in the C:G base pair of a TACA box [C(−29) (5′-TACAAAAG-3′, frequency = 3.3%, activity = 15%–24%)] and in the G:C base pair of a TATAAG box [G(−26) (5′-TATAAGAG-3′, frequency = 9.5%, activity = 18%)]. Inspection of our AdMLP cocrystal structure (5′-TATAAAAG-3′) reveals two cavities between TBP and DNA. The first cavity receives the exocyclic NH2 of G in a TACA box, explaining why a C:G base pair at the third position can be accommodated without any structural changes in protein or the DNA (Fig. 5C). The exocyclic NH2 of G at position six in a TATAAG box is found in the second cavity (Fig. 7A), again allowing for productive PIC formation and Pol II transcription initiation (Table 1). The quasispherical cavity at position 3 does not appear to be able to accommodate the exocyclic NH2 of G in a hypothetical TAGA box, which almost never appears in the EPD (frequency <1%). The same constraints may not apply to a hypothetical TATAAC box (frequency = 2.9%), because the cavity at position 6 is somewhat elongated and may receive the protruding amino group from the bottom strand of the promoter. Further analyses of all available TBP–TATA element interfaces do not reveal the existence of any other sizable cavities (data not shown), and we believe that the cavity mechanism of specificity broadening is restricted to positions 3 and 6.
The final contributor to relaxed DNA-binding specificity involves TBP-induced formation of Hoogsteen base pairs (Hoogsteen 1963), which have not been described previously in protein–DNA complexes. Inspection of our AdMLP cocrystal structure suggests that TBP should not tolerate C:G or G:C base pairs at position 7, because the exocyclic NH2 would clash with the side chain of Leu-72 (see the structure of TBP bound to 5′-TATAAATG-3′, illustrated in Fig. 7C). Bucher's studies of the EPD, however, yielded non-zero estimates for the frequencies of C:G and G:C base pairs at position 7 (3.4% and 16.4%, respectively). Cocrystallization of TBP with the oligonucleotide corresponding to a TATAAAC box [C(−25) (5′-TATAAACG-3′, frequency = 3.4%, activity = 5%–6%)] allowed us to uncover a remarkable explanation for broadened DNA-binding specificity at position 7. A 180° torsion angle change about the C1′–N9 bond (Fig. 8), giving a syn instead of the normal anti conformation, creates a C:G Hoogsteen base pair that is stabilized by interstrand hydrogen bonding (C N4–G O6 = 2.9 Å and possibly C N3–G N7 = 3.0 Å) plus an intrastrand hydrogen bond with the backbone (G N2–phosphate O = 2.9 Å). This additional DNA deformation prevents a steric clash between the exocyclic NH2 of G and Leu-72 that would be produced by the corresponding Watson–Crick base pair (data not shown). At the same time, the Hoogsteen base pair preserves many of the van der Waals interactions with Phe-57 and Phe-74, which are largely responsible for the DNA kink at the 3′ end of the TATA box. Similar steric arguments apply to the problem of accommodating a G:C base pair at this position, and model building suggests that TATA elements bearing a G at position 7 exploit an analogous G:C Hoogsteen base pair (data not shown). The corresponding TATAAAG box occurs often in eukaryotic promoters (frequency = 16.4%) and is capable of supporting Pol II transcription initiation both in vitro and in vivo (Wobbe and Struhl 1990).
Figure 8.
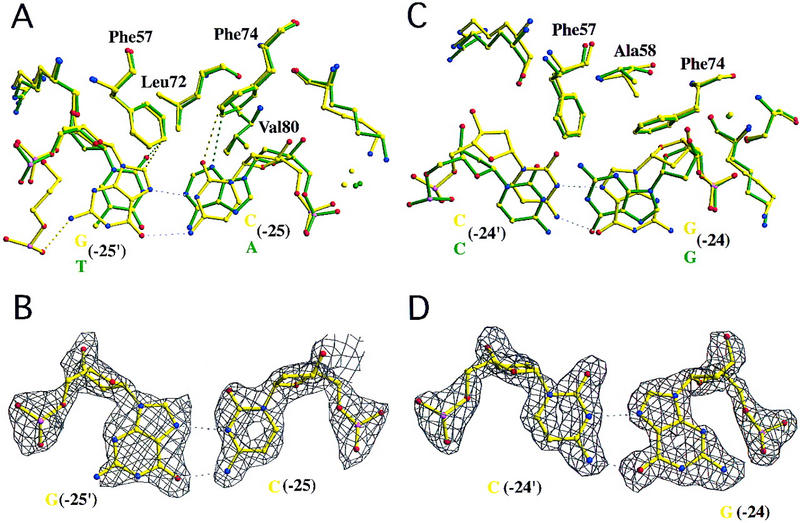
Hoogsteen base pairs in two different TBP–DNA complexes. (A) C(−25) variant (5′-TATAAACG-3′, frequency = 3.4%, activity = 5%–6%). A C:G Watson–Crick base pair at this position would create a steric clash between the exocyclic NH2 of G(−25′) and the side chain of Leu-72. The C(−25) variant of the AdMLP TATA element avoids this steric clash by undergoing a 180° rotation about the C1′-N9 glycosidic bond, going from the normal anti to the syn conformation. The resulting Hoogsteen base pair (hydrogen bonds denoted with white broken lines) is stabilized by a G(−25′) NH2–C(−24′) O1P hydrogen bond (2.9 Å) between the base and the DNA backbone. (B) Simulated annealing electron density map corresponding to the view illustrated in A, contoured at 2.5ς. The map shows excellent electron density for C in the Hoogsteen base pair. The electron density for G is not as well defined. (C) G:C Hoogsteen base pair at position 8 in the cocrystal structure of the T(−30) variant, superimposed on the corresponding base pair of the AdMLP cocrystal structure. Unlike the Hoogsteen base pair depicted in A and B, this unusual DNA structure is the result of a stabilizing interaction with a neighboring protein–DNA complex in the crystal lattice. (D) Simulated annealing electron density map corresponding to the view illustrated in C, contoured at 2.5ς. The map shows excellent electron density for the entire Hoogsteen base pair.
We believe that Hoogsteen base pairs can form at position 7 because the energy barrier for the anti to syn glycosidic torsion angle change is very low for unstacked DNA bases (Ornstein et al. 1978), which is precisely the case in the vicinity of the 3′ kink. Steric clashes between TBP and Watson–Crick C:G or G:C base pairs at position 7 preclude assembly of stable protein–DNA complexes in the absence of further conformational changes in the nucleic acid. TBP has solved this problem by exploiting the phenylalanine-induced kink between positions 7 and 8 to allow rotation of the position 7 G base about the C1′–N9 bond. Noncrystallographic symmetry allows us to rule out some lattice packing artifact in C(−25), because we see the same behavior in both crystallographically independent TBP–TATAAAC box cocrystal structures comprising the asymmetric unit.
Although Hoogsteen base pairs have not been described in protein–DNA complexes, there is extensive literature on Hoogsteen base pairing in drug–DNA complexes (for review, see Chen and Patel 1995). C:G Hoogsteen base pairs were detected in a cocrystal structure of Triostin A bound to a self-complementary duplex oligonucleotide with sequence 5′-GCGTACGC-3′ (Wang et al. 1986). Like our observation at position 7 of the TATAAAC box, these Hoogsteen base pairs flank drug intercalation sites where unstacking occurs. Formation of a C:G (or G:C) Hoogsteen base pair stabilized by two hydrogen bonds (Fig. 8) requires protonation of C at position N3 (pKa = 4.6 in the absence of environmental effects). X-ray crystallography at 1.95 Å resolution cannot reveal the protonation state of a particular titratable group, but we presume that the pKa of N3 is shifted toward neutral in our Hoogsteen base pairs, as observed by NMR in drug–DNA complexes (for review, see Escude et al. 1996).
We also detected a G:C Hoogsteen base pair on the other side of the 3′ kink, created by insertion of Phe-57 and Phe-74. Figure 8, C and D, illustrates a portion of the TTTA box cocrystal structure [T(−30) 5′-TTTAAAAG-3′, frequency = 13.9%, activity = 20%–29%]. Bucher's (1990) studies of the EPD reveal no substantial base pair preference (Table 1), and our model building results suggest that all four Watson–Crick base pairs can be accommodated at position 8 (data not shown). In this case, we are confident that the observed Hoogsteen base pair actually is an artifact of lattice packing within this particular protein–DNA cocrystal (each of the 10 newly determined cocrystal structures displays a unique lattice packing arrangement). Arg-65, protruding from a nearby TBP–DNA complex in the crystal lattice, makes two coplanar hydrogen bonds with G(−23) O6 and N7 (both 2.9 Å) and stacks on the π electron cloud of G(−24) (3.4 Å interplanar distance), thereby stabilizing the observed Hoogsteen base pair (Fig. 8C,D).
Detection of Hoogsteen base pairs adjacent to the 3′ kink site in the TATA element suggests that a similar situation might be obtained at the 5′ end of the recognition element, where the DNA is equally kinked. Many TATA boxes are quasisymmetric and the underside of the TBP saddle displays approximate twofold symmetry about an axis passing between β-strands S1 and S1′ and running perpendicular to the β-sheet (Fig. 4). Leu-72 even has a counterpart, Leu-163, which is related by twofold quasisymmetry. On this side of the saddle, however, Leu-163 really does preclude G:C or C:G base pairs at position 2 (frequencies = 1%), and wild-type TBP does not make productive complexes with either TCTA or TGTA boxes (Wobbe and Struhl 1990). Site-directed mutagenesis of Leu-163 → Val (with an accompanying Ile → Phe substitution) yielded a mutant TBP capable of directing Pol II transcription initiation in vivo from both TATA and TGTA boxes (Strubin and Struhl 1992). Thus, the TGTA box must be able to undergo the obligate TBP-induced DNA deformation when it is bound to the mutant protein. Our attempts to cocrystallize wild-type TBP with oligonucleotides containing TCTA or TGTA boxes were unsuccessful (data not shown), and we believe that Hoogsteen base pair formation does not occur in the vicinity of the 5′ kink site.
This functional asymmetry between the 5′ and 3′ ends of the TATA element has been detected independently by Hurley and coworkers (Sun and Hurley 1995; L. Hurley, unpubl.). Their chemical modification studies with pluramycin, a probe for deformed DNA, demonstrated that TBP-dependent reactivity extends beyond the confines of the 3′ end of 5′-TATAAAAA-3′ but never 5′ of the T:A base pair at position 1. In contrast, a perfectly symmetric TATA element (5′-TATATATA-3′) shows equal TBP-dependent pluramycin reactivity at both ends of the oligonucleotide. Thus, subtle asymmetry in the deformability of various TATA element sequences may contribute to asymmetric behavior during complex formation. If true, this phenomenon could help dictate the polarity of TBP binding, which continues to elude definitive explanation. This intrinsic effect of the TATA element would be independent of any polarity-determining role that TFIIB may play, via interactions with the promoter upstream of the TATA box (Cox et al. 1997; Lagrange et al. 1998; Qureshi and Jackson 1998). It is remarkable that the TBP-dependent effects of pluramycin reactivity are not influenced by addition of TFIIB (L. Hurley, unpubl.).
Not surprisingly, all 10 newly determined cocrystal structures display subtle changes at the level of both TBP and DNA, when compared with our AdMLP cocrystal structure. The most remarkable difference was observed in our TATAAT box cocrystal structure [T(−26) (5′-TATAATAG-3′, frequency = 2.8%, activity = 6%)]. Modest structural rearrangements created a void in the protein–DNA interface that is occupied by a water molecule (Fig. 6C,D) in both halves of the asymmetric unit. This finding was somewhat unexpected, because there is no precedent for an ordered water molecule being found between TBP and DNA. It is made even more interesting by the fact that this particular TATA box is almost 20-fold weaker than its AdMLP counterpart, which differs by only 1 bp (A:T → T:A).
Functional definition of the TATA element
The wealth of information available on TBP–DNA complexes makes a functional definition of the TATA element possible. Inspection of extant cocrystal structures plus the results of model building exercises allow us to predict which of the four possible base pairs are tolerated at each of the eight positions comprising a TATA box. Specifically, we can predict which octameric sequences can present a minor groove surface that is complementary to the underside of the molecular saddle. Regrettably, we cannot make quantitative judgments regarding whether or not a particular combination of allowed base pairs will yield a sequence capable of undergoing the obligate conformational change on binding to TBP. We have already discussed the example of an A tract, but there are bound to be other sequence-specific effects that preclude certain combinations of allowed base pairs. For reviews on sequence-dependent effects in protein–DNA complexes see Dickerson (1998) and Olson et al. (1998).
Assuming that all functional TATA elements must present a complementary minor-groove surface to TBP after they undergo the same DNA deformation and taking each of the eight positions in turn, we derived a structure-based definition for a TATA element that is also consistent with the results of Bucher's (1990) studies of the EPD.
TATA definition
T >> c > a ≅ g/A >> t/T >> a ≅ c/A >> t/T >> a/A >> g > c ≅ t/A ≅ T > g > c/G ≅ A > c ≅ t.
Position 1 (T >> c > a ≅ g)
All four base pairs are observed in nature and are compatible with our structural insights. The EPD survey identifies a marked preference for T (frequency = 79.5%), which may be related to the fact that the energy of T:A stacking on the adjacent base pair is relatively small (except for T:A stacking on C:G) (Ornstein et al. 1978) and easy to overcome by intercalation of Phe-148 and Phe-165.
Position 2 (A >> t)
A:T and T:A appear to be equally acceptable from the structural standpoint. Both G:C and C:G are forbidden by steric clashes with Leu-163. The EPD analysis confirms that G:C and C:G are vanishingly rare (frequencies = 1%). There is a marked preference for A:T over T:A (frequencies = 83.5% and 13.9%, respectively), which is probably correlated with the frequency of T:A in position 1. T:A on A:T stacking energy is the lowest of all possible combinations (Ornstein et al. 1978), again favoring unstacking of the first two base pairs by Phe-148 and Phe-165.
Position 3 (T >> a ≅ c)
T:A, A:T and C:G are all structurally compatible at this position, whereas the exocyclic NH2 of G:C would clash with Val-119. T:A dominates (frequency = 91.4%), which may reflect the preference for T:A in position 1 followed by A:T in position 2. An A:T in position 3 (frequency = 4.4%) would create an A tract.
Position 4 (A >> t)
Our structures suggest that only A:T and T:A are permitted, which is consistent with database findings (frequencies = 89.2% and 8.4%, respectively). Val-119 precludes G:C and C:G (both frequencies = 1%).
Position 5 (T >> a)
Like position 4, A:T and T:A are structurally permitted and observed in nature (frequencies = 71.0% and 27.7%, respectively), whereas G:C and C:G are precluded by Val-29 (both frequencies <1%). We infer that mutation of either of these critical valine residues (29 or 119) to alanine would greatly broaden the specificity of TBP binding.
Position 6 (A >> g > c ≅ t)
Our collection of cocrystal structures includes A:T, T:A and G:C, all of which function in transcription (Wobbe and Struhl 1990). The remaining possibility, C:G, is present at a low but statistically significant level (frequency = 2.9%) and is active in transcription (Wobbe and Struhl 1990).
Position 7 (A ≅ T > g > c)
The EPD contains examples of all four base pairs at this position. Our structural study focused on C:G (frequency = 3.4%) revealed the most significant structural ing with G in the syn conformation, thereby avoiding a steric clash with Leu-72.
Position 8 (G ≅ A > c ≅ t)
All four base pairs are found in the EPD and are compatible with the structural data.
Implications for promoter sequence conservation and regulation of Pol II transcription initiation
Our functional definition of the TATA element implies that 6144 of the 65,536 possible octameric sequences could present complementary minor-groove surfaces to the underside of the molecular saddle. The actual number must be lower, because some combinations of individually allowed base pairs cannot undergo the required conformational change during TBP binding. In advance of a systematic study, it is impossible to know how many putative TATA elements bind TBP productively, but it probably numbers in the thousands. Typical sequence-specific DNA-binding proteins could not bind such large ensembles of sequences with high affinity, because they interact with the major groove. Minor-groove recognition of TATA elements by TBP, on the other hand, represents an architecturally elegant solution to two potential problems arising from errors occurring during DNA replication. First, random point mutants in TATA boxes controlling expression of essential genes are not invariably lethal. Second, variations in TATA box sequence do not significantly perturb later steps in PIC assembly. TATA element deformation by TBP creates a structurally invariant nucleoprotein complex that serves as the receptor for TFIIB and TFIIA. Subsequent entrants to the PIC (Pol II/TFIIF, TFIIE, TFIIH) would also see the same multiprotein–DNA complex no matter what TATA box was actually present in the promoter. It is, of course, possible that one or more factors could gain access to the major-groove face of the TATA box while it is bound to TBP and affect transcription initiation from specific promoters, as suggested by the results of Lee and Roeder (1997).
Transcription activation in vivo is usually thought of as being composed of two distinct subprocesses, antirepression and true activation. We have already discussed the connection between TBP binding to the minor-groove face of the TATA box and the repressive effects of chromatin (Kim et al. 1993a), and now go on to consider true activation. Our crystallographic analysis of DNA recognition by TBP does not specifically address the issue of how transcriptional activators work, but the structural and functional data do provide some mechanistic insights. The fact that the cocrystal structures are essentially identical, despite 20-fold differences in transcription activity (all other things being equal in two different in vivo assays), immediately tells us that thermodynamic, kinetic, or dynamic differences must be responsible for the observed variations in transcriptional efficiency.
Preliminary biophysical observations demonstrate that our transcriptionally weaker TATA element variants bind to TBP with lower affinities when compared with the AdMLP (A.K. Mollah, B. Gilden, E. Jamison, M.D. Librizzi, S.K. Burley, I.M. Willis, and M. Brenowitz, in prep.). Variations in association (kon) and/or dissociation (koff) rates could reduce both binding affinity (KD = koff/kon) and the efficiency of transcription initiation. If the obligate, TBP-induced conformational change takes the same amount of time (i.e., kon remains unchanged) for some TATA elements, the half-lives of the corresponding TBP–DNA complexes must be decreased in the weaker TATA boxes (i.e., koff increases). Alternatively, changes in TATA-box sequence could also slow the rate of TBP–DNA complex formation. During Hoogsteen base pair formation at position 7, we know that kon is reduced (Mollah, S.K. Burley, and M. Brenowitz, in prep.), presumably because it takes longer to form a biochemically productive TBP–DNA complex dependent on rotation about the glycosidic bond. We suggest that some transcriptional activators will exert their positive effects on mRNA production by increasing the half-life of the foundation on which the PIC is assembled on a specific promoter. Presumably, this strategy allows regulatory proteins bound upstream to overcome the effects of an intrinsically weak TATA element, as seen for the Zta trans-activator protein (Lieberman and Berk 1991). A transcriptional activator could also up-regulate gene expression by increasing TBP (or TFIID) recruitment and increasing kon, which may well be the case with the artificial lex–TBP fusion transcription system (Chatterjee and Struhl 1995), the Zta trans-activator (Lieberman and Berk 1994), and other experimental systems cited in Chi et al. (1995).
Regrettably, our study does not provide any direct insight into the precise molecular mechanisms responsible for transcription initiation from the so-called TATA-less promoters (for review, see Smale et al. 1998). However, we do believe that the functional definition of the TATA element discussed earlier will serve as a useful tool for identifying bona fide TATA-less promoters (i.e., sequences that are not capable of forming the protein–DNA complex illustrated in Fig. 1A). This definition does not preclude TBP binding to G/C-rich sequences upstream of transcription start sites with a different molecular recognition strategy. Although such a scenario represents a formal possibility, it seems more likely that other components of TFIID interact with the core promoters of class-II nuclear genes in the absence of a functional TATA element. Alternatively, initiator element-binding proteins could recruit components of the Pol II transcription machinery to a TATA-less promoter.
Conclusion
This paper presents an atomic resolution analysis of the molecular mechanisms responsible for TATA element recognition by TBP during Pol II transcription initiation. Our work provides a detailed picture of how TBP exploits minor-groove interactions and formation of Hoogsteen base pairs to recognize thousands of octameric sequences while inducing a dramatic distortion in the DNA double helix. The structure of TBP bound to the deformed core promoter is independent of the origin of TBP and of the sequence of the TATA box, demonstrating that the Pol II PIC is assembled on a nucleoprotein foundation that has remained unchanged throughout evolution.
Materials and methods
Reagent preparation and crystallization
Wild-type TBP isoform 2 from A. thaliana was overexpressed in Escherichia coli and purified to homogeneity (Nikolov et al. 1992). The 14-bp oligonucleotides containing variants of the AdMLP TATA box (Table 1) were prepared as described previously (Kim et al. 1993a). Cocrystals were obtained by mixing an equimolar ratio of DNA and TBP to form the complex at a final concentration of ∼0.5 mm in 40 mm 2-(N-morpholino)ethane sulfonic acid (MES) at pH 5.9, 60 mm or 100 mm KCl (depending on the oligonucleotide), 4 mm MgCl2, 14% (vol/vol) glycerol, 300 mm ammonium acetate, and 10 mm DTT and equilibrating against a reservoir containing 12% (vol/vol) glycerol, 25 mm MES (pH 5.9), and 10 mm DTT with sitting drop vapor diffusion at 4°C. Following seeding, plate-like cocrystals (0.8 × 0.8 × 0.1 mm) grew in weeks.
X-ray data collection, structure determination, and refinement
Diffraction data were obtained from flash-frozen cocrystals via the oscillation method and integrated, scaled, and merged with DENZO/SCALEPACK (Otwinowski and Minor 1997). Most of the cocrystals (8/10) were isomorphous with our AdMLP cocrystals (Kim et al. 1993a) (P21: a = 41.8, b = 146.7, c = 57.4, β = 90.5°, two complexes/asymmetric unit). The remaining two, A(−31) and T(−24), adopted a similar but different lattice packing arrangement (P21: a = 42, b = 57, c = 147, β = 96°, two complexes per asymmetric unit). Initial phases for the A(−31) and T(−24) structures were determined by molecular replacement with the AdMLP cocrystal structure (Kim et al. 1993a) as the search model, after removal of the altered base pair and nearby protein residues. The remaining structures were phased directly with the search model.
XPLOR refinement (Brünger 1992b) for each structure converged, giving crystallographic R-factors of 18.2%–21.0% and free R-factors of 23.9%–27.5%, with excellent stereochemistry (Table 2). At the final resolution limits, no restraints were placed on sugar pucker, DNA backbone torsion angles, or hydrogen bonding of the altered base pair. The electron density for the polypeptide backbones is continuous everywhere at 1.3ς in (2|Fobserved| − |Fcalculated|) difference Fourier syntheses (data not shown). PROCHECK (Laskowski et al. 1993) revealed no more than one or two unfavorable (φ,ϑ) combinations in any given structure, and main-chain and side-chain structural parameters consistently better than those expected at these resolution limits (overall G-factor = 0.2–0.3). Atomic coordinates have been submitted to the Protein Data Bank (PDB).
Construction of reporter gene plasmids with variant TATA elements
A synthetic promoter was constructed by introducing the AdMLP TATA element into the yeast Gal1 promoter with a Gal4-binding site [consensus UAS (5′-CGGAGGACTGTCCTCCG-3′) or the MEL1 UAS (5′-CGGCCATATGTCTTCCG-3′)] located 200 bp upstream. The synthetic promoters were placed immediately upstream of a lacZ reporter gene, yielding two reporter plasmids (pLS1 and pLS2), which differ only in the sequence of the Gal4-binding site. By use of pLS1 and pLS2 as parent plasmids, other constructs containing nine variant TATA elements were made by site-directed mutagenesis.
Reporter gene experiments
The reporter plasmids described above were transformed into yeast strain Sc18 (GAL4, gal80, ura3-52, leu2-3,112, his3, trp, MEL1). Transformed cells were grown on minimal media lacking uracil to mid-log phase and then harvested as described previously (Vashee and Kodadek 1995). The carbon source was 2% galactose, 3% glycerol, and 2% lactic acid. Reported β-galactosidase values are accurate to ±15% and are the result of at least three independent, replicate measurements. For AdMLP, the absolute levels of β-galactosidase activity supported by the consensus and MEL1 UAS Gal4-binding sites differed by an order of magnitude. In Table 1, each is listed as 100% and the activity levels obtained with the other TATA elements are appropriately normalized.
Acknowledgments
We thank Drs. L. Berman, M. Capel, and R.M. Sweet for help at beamline X25 at the National Synchrotron Light Source; Drs. S. Ealick and D. Thiel and the MacCHESS staff for help at beamlines A1 and F1 at the Cornell High Energy Synchrotron Source; E. Halay for help with DNA production; Drs. J. Bonanno, D. Jeruzalmi, J. Marcotrigiano, D.B. Nikolov, S.K. Nair, and X. Xie for useful discussions and much help with computing and figure preparation. For their many useful suggestions, we are grateful to Drs. K. Arndt, M. Brenowitz, P. Bucher, R.E. Dickerson, J. Kahn, L. Hurley, J. Kuriyan, W.K. Olson, G.A. Petsko, R.G. Roeder, P.B. Sigler, and M. Vasseur. This work was supported by the Howard Hughes Medical Institute (S.K.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
Dedicated to the memory of Nikolaos Patikoglou.
E-MAIL burley@rockvax.rockefeller.edu; FAX (212) 327-8337.
References
- Brünger AT. X-PLOR v. 3.1 manual. New Haven, CT: Yale University; 1992. [Google Scholar]
- Brünger AT, Rice LM. Crystallographic refinement by simulated annealing: Methods and applications. Methods Enzymol. 1997;277:243–269. doi: 10.1016/s0076-6879(97)77015-0. [DOI] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–568. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID. Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Chasman D, Flaherty K, Sharp P, Kornberg R. Crystal structure of yeast TATA-binding protein and a model for interaction with DNA. Proc Natl Acad Sci. 1993;90:8174–8178. doi: 10.1073/pnas.90.17.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- Chen H, Patel DJ. Solution structure of a quinomycin bisintercalator-DNA complex. J Mol Biol. 1995;246:164–179. doi: 10.1006/jmbi.1994.0074. [DOI] [PubMed] [Google Scholar]
- Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- Cox JM, Hayward MM, Sanchez JF, Gegnas LD, van der Zee S, Dennis JH, Sigler PB, Shepartz A. Bidirectional binding of the TATA box binding protein to the TATA box. Proc Natl Acad Sci. 1997;84:13475–13480. doi: 10.1073/pnas.94.25.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDecker BS, O'Brien R, Fleming PJ, Geiger JH, Jackson SP, Sigler PB. The crystal structure of a hyperthermophilic archael Tata-box binding protein. J Mol Biol. 1996;264:1072–1084. doi: 10.1006/jmbi.1996.0697. [DOI] [PubMed] [Google Scholar]
- Dickerson RE. Sequence-dependent B-DNA conformation in crystals and in protein complexes. In: Sarma RH, Sarma MH, editors. Structure, motion, interaction, and expression of biological macromolecules, Proceedings of the Tenth Conference in Albany, NY. Albany, NY: Adenine Press; 1998. pp. 17–30. [Google Scholar]
- DiGabriele AD, Steitz TA. A DNA dodecamer containing an adenine tract crystallizes in a unique lattice and exhibits a new bend. J Mol Biol. 1993;231:1024–1039. doi: 10.1006/jmbi.1993.1349. [DOI] [PubMed] [Google Scholar]
- Escude C, Mohammadi S, Sun J-S, Nguyen C-H, Bisagni E, Liquier J, Taillandier E, Garestier T, Helene C. Ligand-induced formation of Hoogsteen-paired parallel DNA. Chem Biol. 1996;3:57–65. doi: 10.1016/s1074-5521(96)90085-x. [DOI] [PubMed] [Google Scholar]
- Geiger JH, Hahn S, Lee S, Sigler PB. The crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- Hahn S, Buratowski S, Sharp PA, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogsteen K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1963;16:907–916. [Google Scholar]
- Hoopes B, LeBlanc J, Hawley D. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J Biol Chem. 1992;267:11539–11546. [PubMed] [Google Scholar]
- Horikoshi M, Bertuccioli C, Takada R, Wang J, Yamamoto T, Roeder RG. Transcription Factor TFIID induces DNA bending upon binding to the TATA element. Proc Natl Acad Sci. 1992;89:1060–1064. doi: 10.1073/pnas.89.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo ZS, Chiu TK, Lieberman PM, Baikalov I, Berk AJ, Dickerson RE. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- Kielkopf CL, White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB, Rees DC. A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science. 1998;282:111–115. doi: 10.1126/science.282.5386.111. [DOI] [PubMed] [Google Scholar]
- Kim JL, Burley SK. 1.9 Å resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nat Struct Biol. 1994;1:638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993a;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993b;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Kosa PF, Ghosh G, DeDecker BS, Sigler PB. The 2.1 Å crystal structure of an arachael preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes & Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RJ, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–290. [Google Scholar]
- Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: Conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Lee DK, Roeder RG. Functional significance of the TATA element major groove in transcription initiation by RNA polymerase II. Nucleic Acids Res. 1997;25:4338–4345. doi: 10.1093/nar/25.21.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Horikoshi M, Roeder RG. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- Lieberman PM, Berk AJ. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interactions. Genes & Dev. 1991;5:2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- ————— A mechanism for TAFs in transcriptional activation: Activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes & Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Burley SK. 2.1 Å Resolution refined structure of a TATA box-binding protein (TBP) Nat Struct Biol. 1994;1:621–637. doi: 10.1038/nsb0994-621. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Hu S-H, Lin J, Gasch A, Hoffmann A, Horikoshi M, Chua N-H, Roeder RG, Burley SK. Crystal structure of TFIID TATA-box binding protein. Nature. 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay E, Usheva A, Hisatake K, Lee D, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Hoffmann A, Roeder RG, Burley SK. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci. 1996;93:4956–4961. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson WK, Gorin AA, Lu X-J, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced form protein-DNA crystal complexes. Proc Natl Acad Sci. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornstein RL, Rein R, Breen DL, MacElroy RD. An optimized potential function for the calculation of nucleic acid interaction energies. I. Base stacking. Biopolymers. 1978;17:2341–2361. doi: 10.1002/bip.1978.360171005. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parkhurst K, Brenowitz M, Parkhurst L. Simultaneous binding and bending of promoter DNA by TBP: Real time kinetic measurements. Biochemistry. 1996;35:7459–7465. doi: 10.1021/bi9530301. [DOI] [PubMed] [Google Scholar]
- Parvin JD, McCormick RJ, Sharp PA, Fisher DE. Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature. 1995;273:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- Patikoglou G, Burley SK. Eukaryotic transcription factor-DNA complexes. Annu Rev Biophys Biomol Struct. 1997;26:289–325. doi: 10.1146/annurev.biophys.26.1.289. [DOI] [PubMed] [Google Scholar]
- Petri V, Hsieh M, Brenowitz M. Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry. 1995;34:9977–9984. doi: 10.1021/bi00031a020. [DOI] [PubMed] [Google Scholar]
- Petri V, Hsieh M, Jamison E, Brenowitz M. DNA sequence specific recognition by the TATA binding protein: Promoter dependent differences in the thermodynamics and kinetics. Biochemistry. 1998;37:15842–15849. doi: 10.1021/bi981072u. [DOI] [PubMed] [Google Scholar]
- Qureshi SA, Jackson SP. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST, Jain A, Kaufmann J, Emami KH, Lo K, Garraway IP. The initiator element: A paradigm for core promoter heterogeneity within metazoan protein-coding genes. Symp Quant Biol. 1998;63:21–31. doi: 10.1101/sqb.1998.63.21. [DOI] [PubMed] [Google Scholar]
- Starr DB, Hawley DK. TFIID binds in the minor groove of the TATA box. Cell. 1991;67:1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- Starr D, Hoopes B, Hawley D. DNA bending is an important component of site-specific recognition by the TATA binding protein. J Mol Biol. 1995;250:434–446. doi: 10.1006/jmbi.1995.0388. [DOI] [PubMed] [Google Scholar]
- Stofer E, Lavery R. Measuring the geometry of DNA grooves. Biopolymers. 1993;34:337–346. doi: 10.1002/bip.360340305. [DOI] [PubMed] [Google Scholar]
- Strubin M, Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- Sun D, Hurley L. TBP unwinding of the TATA box induces a specific downstream unwinding site that is targeted by pluramycin. Chem Biol. 1995;2:457–469. doi: 10.1016/1074-5521(95)90263-5. [DOI] [PubMed] [Google Scholar]
- Tan S, Hunziker Y, Sargent DF, Richmond TJ. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- Vashee S, Kodadek T. The activation domain of GAL4 protein mediates cooperative promoter binding with general transcription factors in vivo. Proc Natl Acad Sci. 1995;92:10683–10687. doi: 10.1073/pnas.92.23.10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AH-J, Ughetto G, Quigley GJ, Rich A. Interactions of antibiotic and DNA: The molecular structure of triostin A-d(GCGTACGC) complex. J Biomol Struct Dyn. 1986;4:319–342. doi: 10.1080/07391102.1986.10506353. [DOI] [PubMed] [Google Scholar]
- Wobbe C, Struhl K. Yeast and human TATA-binding proteins have nearly identical sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Bateman E. TBP-DNA interactions in the minor groove discriminate between A:T and T:A base pairs. Nucleic Acids Res. 1994;22:1890–1896. doi: 10.1093/nar/22.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]


