Abstract
Endometriosis is a common, chronic gynaecological disease affecting up to 10% of women in their reproductive years. Its aetiology still remains unclear, but evidence indicates genetic factors play a role. We previously identified a region of significant linkage on chromosome 7 in 52 families comprising at least three affected women, stretching ∼6.4 Mb. We screened coding regions and parts of the regulatory regions of three candidate genes with a known role in endometrial development and function—INHBA, SFRP4 and HOXA10—located under or very near the linkage peak, for potential causal mutations using Sanger sequencing. Sequencing was conducted in 47 cases from the 15 families contributing most to the linkage signal (Zmean ≥ 1). Minor allele frequencies (MAFs) of observed variants were compared with MAFs from two publicly available reference populations of European ancestry: 60 individuals in HapMap and 150 individuals in the 1000 Genomes Project. A total of 11 variants were found, 5 (45%) of which were common (MAF > 0.05) among the 15 case families and the reference populations (P-values for MAF difference: 0.88–1.00). The remaining six were rare and unlikely to be individually or cumulatively responsible for the linkage signal. The results indicate that the coding regions of these three genes do not harbour mutations responsible for linkage to endometriosis in these families.
Keywords: endometriosis, aetiology, PCR sequencing, candidate gene, coding region
Introduction
Endometriosis is a benign, estrogen-dependent gynaecological disease that affects up to 10% of women of reproductive age and 35–50% of women seeking medical advice for infertility and/or pelvic pain (Giudice and Kao, 2004). The aetiology and pathophysiology of endometriosis remain unclear, but the prevailing hypothesis for peritoneal disease is that it originates from exposure to retrograde menstruation (Sampson, 1927). However, considering that retrograde menstruation occurs in 76–90% of women (Seli and Arici, 2003) and only 5–15% of women in the general population are affected, additional factors must be present that determine the susceptibility to, and the spontaneous evolution of, endometriosis.
There is now considerable evidence that genetic factors play a role in the aetiology of endometriosis. This includes studies showing familial clustering in humans (Kennedy et al., 1995) and non-human primates (Zondervan et al., 2004); a higher prevalence among first-degree relatives of patients with endometriosis (Simpson et al., 1980; Moen and Magnus, 1993; Matalliotakis et al., 2008); a higher concordance rate in monozygotic compared with dizygotic twins (Treloar et al., 1999), and a higher kinship coefficient between cases compared with age- and sex-matched controls in a large genealogical database in Iceland (Stefansson et al., 2002).
In an effort to identify chromosomal regions harbouring genes implicated in the aetiology of endometriosis, the International Endogene Study (IES) previously conducted a whole genome linkage analysis involving 1176 families with affected sister-pairs, which showed significant linkage to chromosome 10q26 (Treloar et al., 2005). In a subsequent study involving a subset of 248 families with three or more affected members, a second region of significant linkage was found on chromosome 7 (7p13–15) following a near-Mendelian pattern of inheritance (Zondervan et al., 2007). Parametric linkage analyses showed that the causal allele was most likely to be moderately rare (minor allele frequency = 0.02–0.04) with the two contributing datasets suggesting either a reduced penetrance dominant model, or a near-complete penetrance recessive model.
The present study aimed to investigate the putative role in endometriosis of three candidate genes in the region of linkage: Inhibin Beta A subunit (INHBA, OMIM 147290), Secreted Frizzle-Related Protein 4 (SFRP4, OMIM606570) and Homeobox gene A10 (HOXA10, OMIM142597). These three genes were chosen for mutation screening because of their putative role in endometrial function. INHBA is a multifunctional molecule, of which the homo-dimer forms activin A, and the hetero-dimer combines with the inhibin alpha subunit to form inhibin A. Both activins and inhibins are members of the transforming growth factor beta superfamily. Activins regulate a wide variety of cellular events, such as cell proliferation, differentiation and apoptosis (Chen et al., 2002). Altered expression of inhibins/activins has been reported in endometriotic cysts and the eutopic endometrium of women with endometriosis (Reis et al., 2001; Rombauts et al., 2006). The abnormal expression of activin A in conditions such as endometrial hyperplasia/adenocarcinoma and endometriosis further supports a possible role for activin A in endometrial pathophysiology (Florio et al., 2003). SFRP4, a member of the frizzled-related protein family, is expressed in human endometrium (Abu-Jawdeh et al., 1999). Frizzled-related proteins are antagonistic molecules of the WNT signalling pathway, involved in the mediation of cell–cell and cell–extracellular matrix interactions and the regulation of cytoskeletal rearrangements, apoptosis and proliferation (Abu-Jawdeh et al., 1999). Two recently published genome-wide association studies (GWAS) have reported a possible association between endometriosis and rs7521902, a SNP in an LD block including the WNT4 gene (Uno et al., 2010; Painter et al., 2011), which also suggested the WNT signalling pathway might play a role in the development of endometriosis. Although SFRP4 has not been investigated in endometriosis, its function in human endometrium has been studied. Expression of SFRP4 is up-regulated (Tulac et al., 2003) and co-localized with estrogen receptor-α (Fujita et al., 2002) in the proliferative phase of the menstrual cycle, and up-regulated in human endometrial and breast carcinomas compared with normal endometrium (Abu-Jawdeh et al., 1999), implicating SFRP4 in endometrial physiology and tumour formation. HOX genes, the principal regulators of tissue differentiation in the embryo, play an important role in endometrial development and receptivity. Aberrant HOX gene expression in endometrium from endometriosis cases versus controls shows that altered development of endometrium at the molecular level may contribute to the aetiology of infertility in patients with endometriosis (Taylor et al., 1999). Stromal levels of expression of HOXA10 are less in endometriotic tissue than in eutopic endometrium (Browne and Taylor, 2006) and less in the eutopic endometrium of endometriosis cases versus controls (Gui et al., 1999). Whether or not the decrease in HOXA10 levels seen in women with endometriosis is causal or reactive (part of the altered uterine environment) remains unclear (Lee et al., 2007).
We screened the coding regions of INHBA, SFRP4 and HOXA10 for rare variants through Sanger (PCR-based) sequencing of DNA from 47 cases in the 15 families contributing most to the chromosome 7 linkage signal, to investigate if an accumulation of variants in these genes could account for the signal.
Materials and Methods
Samples
In our previous study, we reported a linkage region on chromosome 7 in 248 Caucasian families. Most of the linkage evidence came from 52 extended families collected by the Oxford group. Analysis of these 52 families alone showed a significant linkage signal in the same region under a dominant model with reduced penetrance (Zondervan et al., 2007). Our sequencing study therefore focused solely on these 52 families. All affected members (a total of 47 women) from the 15 out of 52 families that contributed most to the non-parametric linkage peak on chromosome 7 (with a non-parametric linkage Zmean ≥ 1) were selected. Genomic DNA was extracted from peripheral blood using the QIAamp® DNA Blood Maxi Kit (Qiagen Ltd, West Sussex, UK), according to the manufacturer's protocol.
PCR sequencing and mutation detection
The 5′ untranslated regions (UTRs), exons and ∼500 bp of the upstream sequence of INHBA, SFRP4 and HOXA10 were screened for any deletion and/or loss of function mutations. The primers of all amplicons used for PCR and their annealing temperatures are listed in Supplementary data, Table S1. A 15 µl reaction system contains 7.5 µl Taq Master Mix from Qiagen, 5 µM of each primer and about 10 ng genomic DNA. The protocol for amplification of the first fragment of exon 1 of HOXA10 was modified by adding 5% of dimethyl sulphoxide in the reaction system to increase the amplification specificity. PCR reactions were performed by denaturing at 94°C for 10 min followed by 35 cycles of (i) denaturing at 94°C for 45 s; (ii) annealing for 45 s at the corresponding temperature listed in Supplementary data, Table S1; and (iii) elongating at 72°C for 45 s. A final elongation step at 72°C for 10 min followed cycling.
All PCR products were sequenced using a Sanger sequencing protocol. In brief, PCR products were treated with a PCR Product Pre-Sequencing Kit (USB® Corporation, Cleveland, USA) to clean up the redundant primers and dNTP. Next, cycle sequencing reactions were conducted on the treated PCR products using the same primers as for amplification. For amplicons with sizes around or larger than 1 kb, two inner sequencing primers, one for forward and the other for reverse, were used so that the whole amplicon could be covered from both directions. The sequences of these primers are listed in Supplementary data, Table S2. The extension products were purified by either ethanol/EDTA precipitation or PERFORMA® DTRV3 96-well short plates (Edge BioSystems, Gaithersburg, USA). The purified products were run on an ABI 3700; genotypes were called using proprietary sequence analysis software (Applied Biosystems, Foster, USA). Mutation variants in DNA sequences were identified using the Staden package version 1.6.0 (Staden, 1996).
Statistical methods
Using sequencing data from cases in each pedigree, allele frequencies of all variants identified were estimated from pedigree data using a maximum likelihood method implemented in the programme MENDEL v10.0 (Model 1 was used, which adjusts estimates for familial relationships; Lange et al., 2001). Where available, the allele frequencies of known SNPs were compared with their corresponding population-based frequencies in 60 individuals from the CEU—CEPH (Centre d’Étude du Polymorphisme Humain) Utah residents with Northern and Western European ancestry—HapMap sample (http://www.hapmap.org) and 150 individuals sequenced as part of the 1000 Genomes (1000G) Project (http://www.1000genomes.org/, the final release of the pilot data in July 2010). Statistical differences in allele frequencies between individuals from case pedigrees and population-based individuals were calculated using Fisher's exact tests in SPSSv18.
Results
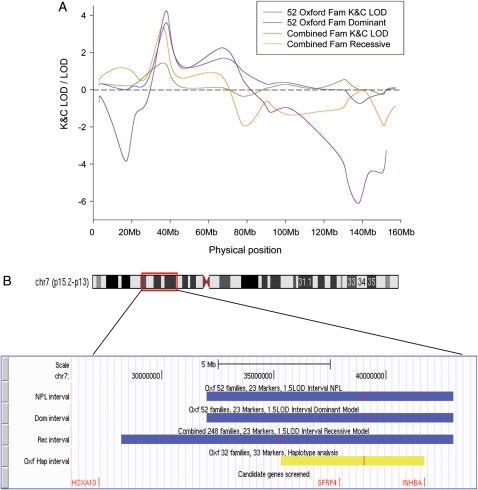
Figure 1a shows the original linkage results for the 52 Oxford families, and all 248 Oxford and Australian families combined (non-parametric K&C LOD scores and best-fitting parametric dominant and recessive models, respectively). Figure 1b shows the 1.5 LOD [logarithm (base 10) odds] support intervals of the original signal (blue) and the refined region of interest identified through fine-mapping and haplotype analysis using the original 23 and an additional 10 markers in the Oxford dataset (yellow). The most likely region of linkage for the Oxford dataset was reduced to 6.4 Mb and contained 48 genes in total (Zondervan et al., 2007). Two of the selected candidate genes, SFRP4 and INHBA, were located in this region (Fig. 1b). HOXA10 was located just outside the 1.5 LOD support and haplotype boundaries of the combined Oxford + Australian dataset; however, it was also selected because of previous evidence suggesting it has a role in endometriosis (Gui et al., 1999; Browne and Taylor, 2006).
Figure 1.
Linkage and haplotype analysis among families with at least three members affected with endometriosis. (A) Original results of non-parametric (K&C LOD) and parametric (dominant and recessive model) linkage analyses among a combined Oxford/Australia dataset of 248 families and the 52 Oxford families [Reproduced with permission from Human Reproduction (Zondervan et al., 2007)]; (B) Chromosomal location of the original linkage signal and 1.5 LOD support interval of non-parametric (NPL) and parametric (Dominant model and Recessive model) (blue); boundaries identified by haplotype analysis among Oxford families (yellow). The peak location of each interval is indicated as a red vertical bar in an interval. The positions of the three candidate genes (HOXA10, SFRP4 and INHBA) sequenced in the present study are indicated in red in the bottom.
The results of PCR sequencing of the 5′ UTRs and coding regions of INHBA, SFRP4 and HOXA10 in the 47 endometriosis cases from the 15 families are described in Table I. No variants were identified in the coding regions of INHBA. However, in the 5′ UTR, one variant, g.−199A>G(rs73100934), was identified in two samples belonging to one family with an estimated frequency of the G allele of the families of 3%. The estimated allele frequency of this SNP in the 1000G Project was 9% (no allele frequency data available in the HapMap database).
Table I.
Variants identified in three genes by PCR sequencing among 47 cases derived from 15 families contributing most to the chromosome 7 linkage signal.
| Variants | Position | SNP ID | Est. Freq.a | 1000G Freq. (P)b (n = 150) | HM Freq. (P)c (n = 60) |
|---|---|---|---|---|---|
| INHBA | |||||
| g.−199A>G | 5′UTR | rs73100934 | 0.03 | 0.09 (0.86) | N/A |
| SFRP4 | |||||
| g.−565G>T | 5′US | rs62443106 | 0.03 | 0.03 (1.00) | N/A |
| g.−415G>A | 5′US | Novel | 0.02 | N/A | N/A |
| g.−256C>T | 5′US | rs71546610 | 0.20 | 0.32 (0.85) | N/A |
| g.−163G>A | 5′UTR | Novel | 0.02 | N/A | N/A |
| g.567G>A | Exon 3 | rs1132552 | 0.38 | 0.38 (1.00) | 0.42 (0.95) |
| g.786C>T | Exon 4 | rs1132553 | 0.60 | 0.63 (0.97) | 0.58 (0.98) |
| g.958C>A | Exon 6 | rs1802073 | 0.33 | 0.33 (1.00) | 0.33 (1.00) |
| g.1019G>A | Exon 6 | rs1802074 | 0.14 | 0.22 (0.88) | 0.24 (0.86) |
| HOXA10 | |||||
| g.−351C>T | 5′UTR | Novel | 0.02 | N/A | N/A |
| g.1157G>A | Exon 2 | rs34957925 | 0.02 | 0.04 (0.93) | 0.05 (0.91) |
5′US, 5′ upstream; N/A, not available.
aEstimated frequencies of the mutated alleles in the 15 linkage pedigrees.
bPopulation-based frequency in CEU population from 1000 Genomes Project (release in July 2010) and P-value of allele frequency difference between cases and 1000G individuals.
cPopulation-based frequency in CEU population from HapMap Project release 28, and P-value of allele frequency difference between cases and HapMap individuals.
For SFRP4, eight SNPs were detected (Table I). Six of these eight SNPs were known, located in 5′ upstream (g.−565G>T and g.−256C>T), exon 3 (g.567G>A), exon 4 (g.786C>T) and exon 6 (g.958C>A and g.1019G>A), respectively. The other two (g.−415G>A and g.−163G>A) were novel SNPs located in 5′ upstream and 5′ UTR. The G to A transition at 1019 causes a non-synonymous change of the amino acid sequence, p.R340K. Estimated allele frequencies of the 15 families varied from 2 to 60%. Population-based allele frequencies of the six known SNPs in the 1000G database varied from 3 to 63%, with frequencies found in the HapMap database for four of six SNPs varying from 24 to 58% (Table I).
Two variants were found for HOXA10: g.−351C>T in one, and g.1151G>A in three, of the 47 samples with estimated minor allele frequencies (MAFs) of 2% for both (Table I).
g.−351C>T is a novel mutation, not present in the latest version of the 1000G dataset or in HapMap. For g.1151G>A (rs34957925), the population-based frequency in 1000G was 4% (5% in HapMap). The three g.1151G>A samples were from members of the same family; the g.−351C>T sample came from a different family. g.1151G>A is a known SNP and is located near the end of the exon 2 coding region, but this transition does not cause an amino acid change. g.−351C>T is located in the 5′ UTR and is a novel mutation, so the effect caused by this polymorphism is still not known.
Discussion
In our previous whole genome linkage study in families with three or more affected individuals we reported significant linkage to chromosome 7. The best-fitting parametric models suggested that the signal was most likely due to one or more rare variants in a 6.4 Mb region with near-Mendelian inheritance patterns (Zondervan et al., 2007). This implied the presence of one or more causal mutations occurring with high frequency in cases from these families, but with rarity in the general population [similar therefore to the BRCA1 and BRCA2 variants in breast cancer (Miki et al., 1994; Wooster et al., 1995)]. Recently published GWAS studies (Uno et al., 2010; Painter et al., 2011) did not provide evidence of a significant signal in the linkage region; significant association was found for a SNP located 20 Mb away, but this was too distant to be responsible for the linkage signal on the basis of local linkage disequilibrium patterns. However—given their population-based design—GWAS have little or no power to detect associations with rare variants, but rather are designed to investigate variations that are relatively common in general population (MAFs > 5%). They are therefore complementary to family-based approaches such as presented here, which aim to detect variants responsible for familial aggregation patterns due to variants that are otherwise rare in the general population.
In the current study, we selected three promising endometriosis candidate genes (INHBA, SFPR4 and HOXA10) located within or close to the 1.5 LOD interval of the linkage signal, and screened for variants across coding regions and upstream regulatory sequences using Sanger sequencing. Several variants were identified, including two novel variants in SFRP4 and one in HOXA10. However, most of the variants were rare in the pedigree cases, as well as population-based individuals from HapMap and 1000G; they were therefore unlikely to explain, individually or cumulatively, the linkage signal. Those variants that were common among cases had similarly high allele frequencies in HapMap and 1000G.
Although the precise aetiology of endometriosis remains unclear, its development must, according to the implantation theory, involve several important mechanisms such as adhesion, angiogenesis and the invasion of endometrial-like tissue into organs in the peritoneal cavity. Genes that regulate the fate of cells, such as those which help ectopic endometrial reflux survive clearance by immune responses, as well as apoptosis, intercellular adhesion, angiogenesis and cell proliferation have therefore been suggested as plausible candidate. Many genes with such functions are reported to be differentially expressed in ectopic and eutopic endometrial tissue in women with endometriosis compared with unaffected controls (Ueda et al., 2002; Matarese et al., 2003; Osteen et al., 2003; Braun et al., 2007; Kim et al., 2007), but it is uncertain whether these findings are causal or secondary to the disease, and whether they are robust to replication. Studies investigating the influence of genetic polymorphisms on disease risk, focusing on specific candidate genes because of a biological hypothesis, aim to identify causal mechanisms. However, as biological pathways of interest contain many potential candidate genes, the effect sizes of genetic variants on endometriosis are known to be modest and studies in endometriosis have often been too small to have sufficient power of association detection (Zondervan et al., 2002). In addition, the results from candidate gene studies are rarely replicated or consistent (Montgomery et al., 2008). Theoretically selecting candidate genes based on hypothesis-free, positional evidence—such as from linkage or GWAS—has a much greater prior probability of success, as the number of potential genes of interest is greatly reduced.
The three genes screened here are putative candidates because they are located in a region that shows significant linkage to endometriosis and they have been found to play roles in endometrial development and function. All three genes are expressed in human endometrium and their levels are altered in women with endometriosis (Abu-Jawdeh et al., 1999; Gui et al., 1999; Matsuzaki et al., 2004). INHBA is a multifunctional molecule that is involved in cell proliferation, differentiation and apoptosis in the form of homo- or hetero-dimers (Chen et al., 2002); SFRP4 is also involved in the regulation of cytoskeletal rearrangements, apoptosis and proliferation by antagonising the WNT signalling pathway (Abu-Jawdeh et al., 1999), and HOXA10 is a member of the HOX gene family, which is highly conserved in evolution and a principal regulator of tissue differentiation in the embryo, playing a very important role in endometrial development and receptivity.
In this study, several variants were identified in SFRP4 and HOXA10, including one non-synonymous mutation in exon 6 of SFRPR (g.1019G>A, causing a change in amino acid sequence p.R340K) and three novel variants in coding or UTRs (two in SFRP4 and one in HOXA10). More analyses would be required to study what effects these changes have (if any). However, all the variants in the three candidate genes were found with insufficient frequency among the cases sequenced to account for the linkage signal on chromosome 7, or were found to be as common in reference population samples. It should be noted that this does not mean that these genes should be entirely disregarded as candidates. First, sequencing was limited to the coding, and part of the regulatory, regions as the most likely location of rare variants responsible for the linkage signal. However, meaningful variants that alter the expression levels of these genes might also exist in regulatory elements located in regions other than those screened, such as introns, 3′ UTR and downstream regions and 5′ upstream regulatory regions. These regulatory elements were not included in the present study because their position was not known. A sequencing analysis of HOXA10 comparing endometriosis cases and controls was previously conducted by Wu et al. (2008). Although they also did not identify a significant association between the number of genetic alterations in HOXA10 coding regions and endometriosis, they found that—among endometriosis cases—the presence of such variants appeared significantly associated with rAFS stage and serum CA-125 concentration (Wu et al., 2008). In addition to alterations in the DNA sequence itself, differential expression levels of the candidate genes might be caused by nucleotide modification, for example methylation. Of interest, therefore, is the report that methylation in the promoter and intron 1 of HOXA10 might be associated with an increased risk of endometriosis (Wu et al., 2005). However, as such differential methylation patterns tend to arise as a consequence of environmental exposures, they would be unlikely to manifest themselves as significant linkage, which could only be caused by the presence of inherited DNA mutations.
All the samples sequenced were derived from affected members in those families that contributed most to the linkage signal on chromosome 7. No unaffected controls were screened; instead, variants were compared with the frequency of the CEU population in HapMap or 1000G where data were available. Such a strategy was adopted because it was the most cost-efficient and powerful method to identify variants, including InDels and/or malfunction mutations, which are highly prevalent in the endometriosis cases, but rare in the general population.
In conclusion, sequencing of coding regions in three candidate genes, INHBA, SFRP4 and HOXA10, selected from a region of significant linkage on chromosome 7, did not reveal any mutation(s) responsible for the signal. Given that only coding regions, 5′ UTR and part of the upstream regions were screened, the three genes cannot be entirely excluded as positional candidates, although their involvement is unlikely. In addition, other candidate genes in the region need to be explored in the future.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors’ roles
J.L.: design, experiments, analysis and interpretation, drafting article and final approval. L.Z.: experiments and final approval. S.H.K.: conception and design, interpretation, revising for critical content and final approval. K.T.Z.: conception and design, interpretation, revising for critical content and final approval.
Funding
The current study was partly supported by funding from the National Institutes of Health (R01HD050537). S.H.K. is supported by the NIHR Biomedical Research Centre Programme, Oxford. K.T.Z. is supported by the Wellcome Trust Research Career Development Fellowship (WT085235/Z/08/Z).
Supplementary Material
Acknowledgements
We thank Dr Grant Montgomery from Queensland Institute of Medical Research, Australia, for commenting on the paper.
References
- Abu-Jawdeh G, Comella N, Tomita Y, Brown LF, Tognazzi K, Sokol SY, Kocher O. Differential expression of frpHE: a novel human stromal protein of the secreted frizzled gene family, during the endometrial cycle and malignancy. Lab Invest. 1999;79:439–447. [PubMed] [Google Scholar]
- Braun DP, Ding J, Shaheen F, Willey JC, Rana N, Dmowski WP. Quantitative expression of apoptosis-regulating genes in endometrium from women with and without endometriosis. Fertil Steril. 2007;87:263–268. doi: 10.1016/j.fertnstert.2006.06.026. doi:10.1016/j.fertnstert.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Browne H, Taylor H. HOXA10 expression in ectopic endometrial tissue. Fertil Steril. 2006;85:1386–1390. doi: 10.1016/j.fertnstert.2005.10.072. doi:10.1016/j.fertnstert.2005.10.072. [DOI] [PubMed] [Google Scholar]
- Chen YG, Lui HM, Lin SL, Lee JM, Ying SY. Regulation of cell proliferation, apoptosis, and carcinogenesis by activin. Exp Biol Med (Maywood) 2002;227:75–87. doi: 10.1177/153537020222700201. [DOI] [PubMed] [Google Scholar]
- Florio P, Severi FM, Luisi S, Ciarmela P, Calonaci G, Cobellis L, Petraglia F. Endometrial expression and secretion of activin A, but not follistatin, increase in the secretory phase of the menstrual cycle. J Soc Gynecol Investig. 2003;10:237–243. doi: 10.1016/s1071-5576(03)00045-5. doi:10.1016/S1071-5576(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Fujita M, Ogawa S, Fukuoka H, Tsukui T, Nemoto N, Tsutsumi O, Ouchi Y, Inoue S. Differential expression of secreted frizzled-related protein 4 in decidual cells during pregnancy. J Mol Endocrinol. 2002;28:213–223. doi: 10.1677/jme.0.0280213. doi:10.1677/jme.0.0280213. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. doi:10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- Gui Y, Zhang J, Yuan L, Lessey BA. Regulation of HOXA-10 and its expression in normal and abnormal endometrium. Mol Hum Reprod. 1999;5:866–873. doi: 10.1093/molehr/5.9.866. doi:10.1093/molehr/5.9.866. [DOI] [PubMed] [Google Scholar]
- Kennedy S, Mardon H, Barlow D. Familial endometriosis. J Assist Reprod Genet. 1995;12:32–34. doi: 10.1007/BF02214126. doi:10.1007/BF02214126. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Taylor HS, Lu Z, Ladhani O, Hastings JM, Jackson KS, Wu Y, Guo SW, Fazleabas AT. Altered expression of HOXA10 in endometriosis: potential role in decidualization. Mol Hum Reprod. 2007;13:323–332. doi: 10.1093/molehr/gam005. doi:10.1093/molehr/gam005. [DOI] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Amer J Hum Genetics. 2001;69(supplement):504.. [Google Scholar]
- Lee GH, Kim SH, Choi YM, Suh CS, Kim JG, Moon SY. Estrogen receptor beta gene +1730 G/A polymorphism in women with endometriosis. Fertil Steril. 2007;88:785–788. doi: 10.1016/j.fertnstert.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Matalliotakis I, Arici A, Cakmak H, Goumenou A, Koumantakis G, Mahutte N. Familial aggregation of endometriosis in the Yale Series. Arch Gynecol Obstet. 2008;278:507–511. doi: 10.1007/s00404-008-0644-1. [DOI] [PubMed] [Google Scholar]
- Matarese G, De Placido G, Nikas Y, Alviggi C. Pathogenesis of endometriosis: natural immunity dysfunction or autoimmune disease? Trends Mol Med. 2003;9:223–228. doi: 10.1016/s1471-4914(03)00051-0. doi:10.1016/S1471-4914(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Matsuzaki S, Canis M, Vaurs-Barriere C, Pouly JL, Boespflug-Tanguy O, Penault-Llorca F, Dechelotte P, Dastugue B, Okamura K, Mage G. DNA microarray analysis of gene expression profiles in deep endometriosis using laser capture microdissection. Mol Hum Reprod. 2004;10:19–728. doi: 10.1093/molehr/gah097. doi:10.1093/humupd/dmh004. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. doi:10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Moen MH, Magnus P. The familial risk of endometriosis. Acta Obstet Gynecol Scand. 1993;72:560–564. doi: 10.3109/00016349309058164. doi:10.3109/00016349309058164. [DOI] [PubMed] [Google Scholar]
- Montgomery GW, Nyholt DR, Zhao ZZ, Treloar SA, Painter JN, Missmer SA, Kennedy SH, Zondervan KT. The search for genes contributing to endometriosis risk. Hum Reprod Update. 2008;14:447–457. doi: 10.1093/humupd/dmn016. doi:10.1093/humupd/dmn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteen KG, Yeaman GR, Bruner-Tran KL. Matrix metalloproteinases and endometriosis. Semin Reprod Med. 2003;21:155–164. doi: 10.1055/s-2003-41322. doi:10.1055/s-2003-41322. [DOI] [PubMed] [Google Scholar]
- Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43:51–54. doi: 10.1038/ng.731. doi:10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis FM, Di Blasio AM, Florio P, Ambrosini G, Di Loreto C, Petraglia F. Evidence for local production of inhibin A and activin A in patients with ovarian endometriosis. Fertil Steril. 2001;75:367–373. doi: 10.1016/s0015-0282(00)01720-9. doi:10.1016/S0015-0282(00)01720-9. [DOI] [PubMed] [Google Scholar]
- Rombauts L, Donoghue J, Cann L, Jones RL, Healy DL. Activin-A secretion is increased in the eutopic endometrium from women with endometriosis. Aust N Z J Obstet Gynaecol. 2006;46:148–153. doi: 10.1111/j.1479-828X.2006.00546.x. doi:10.1111/j.1479-828X.2006.00546.x. [DOI] [PubMed] [Google Scholar]
- Sampson JA. Peritoneal endometriosis due to menstrual dissemination of endometrial tissue into the peritoneal cavity. Am J Obstet Gyneco. 1927;14:422–469. [Google Scholar]
- Seli E, Arici A. Endometriosis: interaction of immune and endocrine systems. Semin Reprod Med. 2003;21:135–144. doi: 10.1055/s-2003-41320. doi:10.1055/s-2003-41320. [DOI] [PubMed] [Google Scholar]
- Simpson JL, Elias S, Malinak LR, Buttram VC., Jr Heritable aspects of endometriosis. I. Genetic studies. Am J Obstet Gynecol. 1980;137:327–331. doi: 10.1016/0002-9378(80)90917-5. [DOI] [PubMed] [Google Scholar]
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. doi:10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Geirsson RT, Steinthorsdottir V, Jonsson H, Manolescu A, Kong A, Ingadottir G, Gulcher J, Stefansson K. Genetic factors contribute to the risk of developing endometriosis. Hum Reprod. 2002;17:555–559. doi: 10.1093/humrep/17.3.555. doi:10.1093/humrep/17.3.555. [DOI] [PubMed] [Google Scholar]
- Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14:1328–1331. doi: 10.1093/humrep/14.5.1328. doi:10.1093/humrep/14.5.1328. [DOI] [PubMed] [Google Scholar]
- Treloar SA, O'Connor DT, O'Connor VM, Martin NG. Genetic influences on endometriosis in an Australian twin sample. Fertil Steril. 1999;71:701–710. doi: 10.1016/s0015-0282(98)00540-8. doi:10.1016/S0015-0282(98)00540-8. [DOI] [PubMed] [Google Scholar]
- Treloar SA, Wicks J, Nyholt DR, Montgomery GW, Bahlo M, Smith V, Dawson G, Mackay IJ, Weeks DE, Bennett ST, et al. Genomewide linkage study in 1,176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet. 2005;77:365–376. doi: 10.1086/432960. doi:10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulac S, Nayak NR, Kao LC, Van Waes M, Huang J, Lobo S, Germeyer A, Lessey BA, Taylor RN, Suchanek E, et al. Identification, characterization, and regulation of the canonical Wnt signaling pathway in human endometrium. J Clin Endocrinol Metab. 2003;88:3860–3866. doi: 10.1210/jc.2003-030494. doi:10.1210/jc.2003-030494. [DOI] [PubMed] [Google Scholar]
- Ueda M, Yamashita Y, Takehara M, Terai Y, Kumagai K, Ueki K, Kanda K, Yamaguchi H, Akise D, Hung YC, et al. Survivin gene expression in endometriosis. J Clin Endocrinol Metab. 2002;87:3452–3459. doi: 10.1210/jcem.87.7.8682. doi:10.1210/jc.87.7.3452. [DOI] [PubMed] [Google Scholar]
- Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K, Nakamura Y.A. genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. 2010;42:707–710. doi: 10.1038/ng.612. doi:10.1038/ng.612. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. doi:10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, Wang D, Li R, Yi X, Zhang H, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature. 2005;438:981–987. doi: 10.1038/nature04225. doi:10.1038/nature04225. [DOI] [PubMed] [Google Scholar]
- Wu HH, Wang NM, Lin CY, Tsai HD. Genetic alterations of HOXA10 and their effect on the severity of endometriosis in a Taiwanese population. Reprod Biomed Online. 2008;16:416–424. doi: 10.1016/s1472-6483(10)60604-9. doi:10.1016/S1472-6483(10)60604-9. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR, Kennedy SH. What makes a good case-control study? Design issues for complex traits such as endometriosis. Hum Reprod. 2002;17:1415–1423. doi: 10.1093/humrep/17.6.1415. doi:10.1093/humrep/17.6.1415. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Weeks DE, Colman R, Cardon LR, Hadfield R, Schleffler J, Trainor AG, Coe CL, Kemnitz JW, Kennedy SH. Familial aggregation of endometriosis in a large pedigree of rhesus macaques. Hum Reprod. 2004;19:448–455. doi: 10.1093/humrep/deh052. doi:10.1093/humrep/deh052. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Treloar SA, Lin J, Weeks DE, Nyholt DR, Mangion J, Mackay IJ, Cardon LR, Martin NG, Kennedy SH, et al. Significant evidence of one or more susceptibility loci for endometriosis with near-Mendelian inheritance on chromosome 7p13–15. Hum Reprod. 2007;22:717–728. doi: 10.1093/humrep/del446. doi:10.1093/humrep/del446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.