Abstract
Objectives
To determine the association between embB mutations and drug resistance, and to further investigate the mechanism of embB mutations involved in the development of ethambutol and multidrug resistance in Mycobacterium tuberculosis.
Methods
One hundred and thirty-eight multidrug-resistant clinical M. tuberculosis isolates, including 86 ethambutol-resistant and 52 ethambutol-susceptible strains, were analysed to characterize mutations within the entire coding region of the embB gene. Moreover, a two-step genotyping was performed to identify the genetic lineage.
Results
In total, 27 embB mutation types were detected in 19 distinct codons. Though a strong association was observed between embB mutations and ethambutol resistance, 19.2% of embB306 mutants and 11.5% of embB406 or embB497 mutants were ethambutol susceptible. Among 39 ethambutol-resistant strains without embB306 mutations, 51.3% harboured mutations at codons 406 or 497. Particularly, three pairs of isolates with identical embB mutations and genotyping features were identified with variant ethambutol susceptibility. Among 77 isoniazid, rifampicin, streptomycin and ethambutol quadruple drug-resistant isolates, 89.6% carried embB mutations and 83.1% could be identified by detecting 10 embB mutations.
Conclusions
Our results suggest embB mutations alone are not sufficient for the development of full resistance to ethambutol in M. tuberculosis and mutations other than embB are also needed. Our study confirms the importance of mutations at embB406 and embB497 as hotspots, in addition to embB306, for detecting ethambutol resistance. Ten selected mutations of embB, covered by a short PCR product, can be used as candidate markers for the prediction of quadruple resistance to isoniazid, rifampicin, streptomycin and ethambutol.
Keywords: genetic background, MIRU-VNTR, embB306, embB497, ethambutol
Introduction
Multidrug-resistant (MDR) tuberculosis (TB) has an estimated 4.8% prevalence worldwide and poses a serious threat to global public health.1 In China, the significantly high (9.3%) prevalence of MDR-TB makes the prevention and control of tuberculosis especially challenging.2 The rapid and reliable detection of drug resistance is critical for optimizing treatment regimens and for preventing the spread of tuberculosis.
Ethambutol, which is an essential first-line drug in tuberculosis treatment, plays an important role in the chemotherapy of drug-resistant TB.3 Ethambutol inhibits mycobacterial arabinosyl transferases encoded by the embCAB operon, which includes three genes (embC, embA and embB). Amino acid substitutions encoded by embB are observed in non-tuberculous mycobacteria with intrinsic resistance to ethambutol.4 Exchanging wild-type embB306, embB497 and embB406 with mutant codons increases the ethambutol minimal inhibition concentrations (MICs) of Mycobacterium tuberculosis.5–7 Mutations at embB320 and embB324,5,7 as well as mutations at embB397, embB445, embB1024 and embC13, have also been found to be associated with ethambutol resistance.5
The most commonly detected point mutation in ethambutol-resistant clinical strains of M. tuberculosis is in the embB gene at codon 306, which occurs in 30%–69% of ethambutol-resistant clinical strains.4,8–11 Initial studies indicated that the embB306 mutations were only observed in ethambutol-resistant strains and led to the proposal that the embB306 locus be considered as a diagnostic marker for ethambutol resistance.10 However, the detection of embB306 from ethambutol-susceptible clinical isolates questions the validity of this assertion.12–15 Mokrousov et al.14 first described this phenomenon as a genuine discrepancy between genotypic and phenotypic tests, and noticed that embB306 mutations in ethambutol-susceptible isolates were limited to the isolates already resistant to other drugs. Based on a study of 1020 clinical isolates, Hazbon et al.12 suggested that the embB306 mutation is associated with broad drug resistance rather than ethambutol resistance per se. Shen et al.15 also proposed the embB306 locus as a candidate marker for the detection of MDR and extensively drug-resistant M. tuberculosis isolates. Although the association between embB306 mutation and ethambutol resistance or broad drug resistance has been observed in several groups' studies with both clinical or laboratorial isolates,5,6,9,16,17 the exact role embB306 mutations play in the development of ethambutol resistance and multidrug resistance in M. tuberculosis is not fully understood. Mutations in embB other than embB306 were also detected in these studies, but the contribution of such mutations to the development of ethambutol resistance is similarly not clear. It is believed that variant genetic alterations that accumulate in epidemic M. tuberculosis lead to the development of drug resistance,18–20 including ethambutol resistance,5 but not much evidence has been obtained from studies of clinical isolates.
Therefore, to further investigate the mechanism of embB mutations in the development of drug resistance and to evaluate the association between embB mutations and drug resistance, including ethambutol, multidrug and broad drug resistance, a relatively large population of MDR-TB isolated from Henan province, China was examined. In the present study, we characterized the mutations of the embB complete coding sequence and documented the variable number tandem repeat of mycobacterial interspersed repetitive units (MIRU-VNTR) genotypes of this MDR-TB population, to further analyse the mechanism underlying the development of ethambutol drug resistance in clinical M. tuberculosis isolates.
Materials and methods
M. tuberculosis clinical strains
One hundred and fifty MDR-TB strains were collected by sequentially screening 1605 clinical M. tuberculosis strains isolated from patients from Henan province in 2007–09. Meanwhile, 22 pan-susceptible strains were collected from the same location to be used as controls in this study.
Drug susceptibility testing (DST)
DST to four first-line antituberculosis drugs was performed in the Tuberculosis Reference Laboratory at Henan Provincial Centers for Disease Control and Prevention, China. The Löwenstein–Jensen (LJ) proportion method, recommended by WHO/International Union Against Tuberculosis and Lung Disease (IUATLD), was used to perform DST with the following critical drug concentrations: 0.2 mg/L isoniazid; 40.0 mg/L rifampicin; 2.0 mg/L ethambutol; and 4.0 mg/L streptomycin.21,22
RD105 deletion-targeted multiplex PCR (DTM-PCR) and MIRU-VNTR genotyping
DTM-PCR was performed to identify the Beijing family strains.23 A China-specified MIRU-VNTR genotyping method (VNTR-7) was performed on all MDR-TB isolates with seven VNTR loci in this study, and additional nine VNTR loci (VNTR-9) was applied to the isolates with identical first seven VNTR loci.24,25 Samples with more than one band in the PCR product on any VNTR locus were considered as mixed strains and excluded from the studied population. The VNTR genotyping data, transformed into a distance matrix on the web site MIRU-VNTRplus (http://www.miru-vntrplus.org) by default setting, were treated as categorical variables and the phylogenetic analysis of the distance data was conducted using MEGA version 4.26,27
PCR amplification, sequencing and data analysis
The full-length embB gene coding region of the studied strains was amplified with three overlapped fragments. Chromosomal DNA was extracted using the boiling method.28 Phusion® Hot Start High-Fidelity DNA Polymerase (Finnzymes, Finland), an ultrahigh-fidelity DNA polymerase, was used for the amplification. The primers synthesized by Sangon Biochemical for DNA amplification were: embB1-1 (5′-TCGACGATCGCCACGTACCT-3′) and embB1-2 (5′-CAGCAGCAGCCAGCA CACTA-3′); embB2-1 (5′-TATTCGGCTT CTGCTCTGG-3′) and embB2-2 (5′-CACACCGTAGCTGGAGACAT-3′); and embB3-1 (5′-GTTCCTGGC GGCGTTATTCT-3′) and embB3-2 (5′-AGCCTG ACGCTATGGACCAA-3′). The sequencing primers were embB(S)1 (5′-CGTCCTTGCCTTGCTTGGGT-3′) and embB(S)3 (5′-GCGTGGTATCTCCTGCCTAAG-3′); other sequencing primers were the same as embB1-2, embB2-1, embB2-2 and embB3-1. PCR products were sequenced by Sinogenomax Co. Ltd. Sequence data were assembled by Seqman pro (version 7.1, DNAstar Lasergene), and mutations were determined by comparing with the H37Rv sequence of embB from Tuberculist (http://genolist.pasteur.fr/TubercuList/) and the GenBank database (http://www.ncbi.nih.gov/gene). The frequency calculation and association analysis were performed using SPSS for Windows® (version 10.0, SPSS, Inc., USA).
Results and discussion
General profile of genotyping, drug resistance and embB mutations
Twelve isolates (8%) of the primary study population, which were identified as a mixture of different individual MDR-TB strains by a two-step genotyping method of VNTR-7 and VNTR-9, were not included in the following analysis. In total, 138 MDR isolates included in the study showed 116 unique and 11 clustered genotyping patterns based on VNTR-7 genotyping (Figure 1a). Most of the MDR isolates (95%, 131/138) were identified as Beijing family M. tuberculosis strains by DTM-PCR. Among the 138 clinical MDR-TB isolates, 9 presented no additional resistance (isoniazid/rifampicin resistant), 43 were isoniazid/rifampicin/streptomycin resistant, 9 were isoniazid/rifampicin/ethambutol resistant and 77 were isoniazid/rifampicin/ethambutol/streptomycin resistant. There were 86 ethambutol-resistant and 52 ethambutol-susceptible isolates.
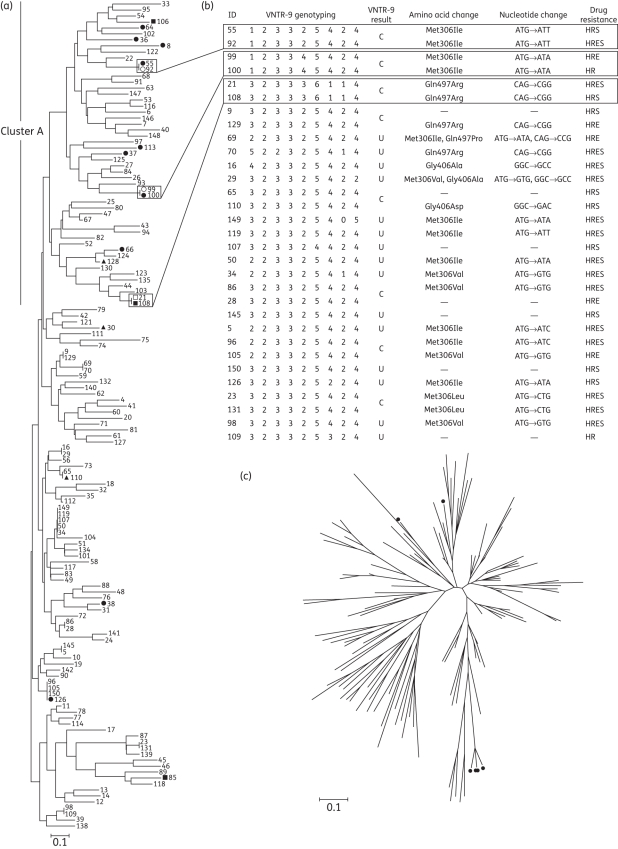
Figure 1.
Phylogenetic analysis of MDR-TB isolates, and characteristics of VNTR-9 genotyping, embB mutations and drug resistance patterns of isolates with the clustered VNTR-7 genotypes. (a) Phylogenetic map generated by the neighbour-joining (NJ) method, based on VNTR-7 genotyping data of 138 MDR-TB isolates. Numbers indicate strain ID. Filled circles indicate ethambutol-susceptible embB306 mutants, while open circles indicate ethambutol-resistant counterparts. Filled squares indicate ethambutol-susceptible embB497 mutants, while open squares indicate ethambutol-resistant embB497 counterparts. Filled triangles indicate ethambutol-susceptible embB406 mutants. (b) VNTR-9 genotypes, embB mutations and drug resistance patterns of isolates showing the clustered VNTR-7 genotypes. Blocks highlight three pairs of isolates with identical VNTR-16 genotypes and embB mutations, but opposite ethambutol resistance. C indicates isolates with identical VNTR-9 genotyping patterns. U indicates isolates with unique VNTR-9 genotyping patterns. H, isoniazid; R, rifampicin; E, ethambutol; S, streptomycin. (c) Radial NJ phylogeny based on VNTR-7 genotyping data of MDR-TB isolates, which shows the genetic background of six EmbB Met306Leu mutants. Filled circles represent strains with a Met→Leu amino acid substitution.
One hundred and thirty-eight MDR-TB strains were screened for embB mutations. A total of 119 embB mutations representing 27 mutations types were detected in 19 distinct codons, including 4 synonymous mutation types at codons 304, 424, 534 and 539 (Table 1). Ninety-four mutations were detected in 86 ethambutol-resistant MDR-TB strains. Twenty-five mutations were detected in 52 ethambutol-susceptible MDR-TB strains. Of the 138 MDR-TB isolates studied, 38 (27.5%) showed wild-type embB sequences while 100 (72.5%) showed mutated sequences (Table 2).
Table 1.
Mutation pattern of the embB gene in isolates with different phenotypes
| Number of mutations |
|||||||
|---|---|---|---|---|---|---|---|
| ethambutol-susceptible MDR |
ethambutol-resistant MDR |
||||||
| Locus | isoniazid/rifampicin | isoniazid/rifampicin/streptomycin | total ethambutol susceptible | isoniazid/rifampicin/ethambutol | isoniazid/rifampicin/ethambutol/streptomycin | total ethambutol resistant | pan-susceptible |
| embB306 | 3 | 7 | 10 | 4 | 43 | 47 | 0 |
| embB497 | 3 | 3 | 3 | 13 | 16 | 0 | |
| embB406 | 3 | 3 | 1 | 6 | 7 | 0 | |
| embB354 | 2 | 2 | 2 | 2 | 0 | ||
| embB534a | 1 | 1 | 3 | 8 | 11 | 1 | |
| embB304a | 0 | 1 | 1 | 0 | |||
| embB328 | 0 | 1 | 1 | 0 | |||
| embB330 | 0 | 1 | 1 | 0 | |||
| embB424a | 0 | 1 | 1 | 0 | |||
| embB439 | 0 | 1 | 1 | 0 | |||
| embB469 | 1 | 1 | 0 | 0 | |||
| embB508 | 1 | 1 | 0 | 0 | |||
| embB539a | 0 | 1 | 1 | 0 | |||
| embB627 | 0 | 1 | 2 | 3 | 0 | ||
| embB651 | 1 | 1 | 0 | 0 | |||
| embB667 | 1 | 1 | 0 | 0 | |||
| embB1000 | 0 | 1 | 1 | 0 | |||
| embB1002 | 0 | 1 | 1 | 0 | |||
| embB1024 | 2 | 2 | 0 | 0 | |||
| Total | 3 | 22 | 25 | 13 | 81 | 94 | 1 |
aSynonymous mutations.
Table 2.
Characteristics of embB mutants within the MDR-TB isolates
| Locus | Nucleotide change | Amino acid change | No. of isolates (percentage of 138 MDR isolates) |
|---|---|---|---|
| embB306 | atg→Ctg | Met→Leu | 46 (33.3) |
| atg→Gtg | Met→Val | ||
| atg→atA | Met→Ile | ||
| atg→atC | Met→Ile | ||
| atg→atT | Met→Ile | ||
| embB306 and embB534 | atg→atC and gac→gaT | Met→Ile and Asp→Asp | 6 (4.3) |
| atg→Gtg and gac→gaT | Met→Val and Asp→Asp | ||
| embB306 and embB354 | G inserts between AT and gac→gCc | frameshift and Asp→Ala | 1 (0.7) |
| embB306 and embB406 | atg→Gtg and ggc→gCc | Met→Val and Gly→Ala | 1 (0.7) |
| embB306 and embB424 | atg→Gtg and cgg→cgA | Met→Val and Arg→Arg | 1 (0.7) |
| embB306 and embB497 and embB304 | atg→Ctg and cag→cCg and ctg→Ttg | Met→Leu and Gln→Pro and Leu→Leu | 1 (0.7) |
| embB306 and embB497 and embB534 | atg→atA and cag→cCg and gac→gaT | Met→Ile and Gln→Pro and Asp→Asp | 1 (0.7) |
| embB328 | gat→Tat | Asp→Tyr | 1 (0.7) |
| embB330 | ttc→tCc | Phe→Ser | 1 (0.7) |
| embB354 | gac→gCc | Asp→Ala | 3 (2.2) |
| embB406 | ggc→Agc | Gly→Ser | 8 (5.8) |
| ggc→gAc | Gly→Asp | ||
| ggc→gCc | Gly→Ala | ||
| embB406 and embB534 and embB539 | ggc→gAc and gac→gaT and cgg→cgA | Gly→Asp and Asp→Asp and Arg→Arg | 1 (0.7) |
| embB439 | gca→Aca | Ala→Thr | 1 (0.7) |
| embB469 | cgt→cAt | Arg→His | 1 (0.7) |
| embB497 | cag→cCg | Gln→Pro | 15 (10.9) |
| cag→cGg | Gln→Arg | ||
| embB497 and embB627 | cag→cGg and T inserts between CG | Gln→Arg and frameshift | 1 (0.7) |
| embB497 and embB1024 | cag→cCg and gac→Aac | Gln→Pro and Asp→Asn | 1 (0.7) |
| embB508 | gtt→Ttt | Val→Phe | 1 (0.7) |
| embB534 | gac→gaT | Asp→Asp | 2 (1.4) |
| embB534 and embB1000 | gac→gaT and atg→aGg | Met→Arg | 1 (0.7) |
| embB534 and embB1002 | gac→gaT and cac→cGc | His→Arg | 1 (0.7) |
| embB627 | T inserts between CG | frameshift | 2 (1.4) |
| embB651 | agc→aCc | Ser→Thr | 1 (0.7) |
| embB667 | aca→aAa | Thr→Lys | 1 (0.7) |
| embB1024 | gac→Aac | Asp→Asn | 1 (0.7) |
| Mutant isolates | 100 (72.5) | ||
| Wild-type isolates | 38 (27.5) |
Overall, 16 isolates carried more than one mutation in the entire coding region of embB, including 7 isolates with two non-synonymous embB mutations (Table 2). There were three isolates that carried triple embB mutations. Thirteen isolates were detected to have double embB mutations, of which 5 isolates carried two non-synonymous mutations. Among the seven isolates carrying two non-synonymous embB mutations, six were resistant to ethambutol (isoniazid/rifampicin/ethambutol resistant or isoniazid/rifampicin/ethambutol/streptomycin resistant). Particularly, among those ethambutol-resistant isolates with multiple mutations, three isolates contained mutation combinations involving codons 306, 406 and 497 (embB306 plus embB406; embB306 plus embB497), which has not been documented in previous studies.8,10,29
Characteristics of embB306 mutation
One of the major mutations was embB306, accounting for the highest proportion of all mutations detected in our study. Altogether, 41.3% (57/138) of MDR-TB isolates carried the embB306 mutation. The proportion of embB306 mutants among ethambutol-resistant MDR strains (54.7%, 47/86) was much higher than in the ethambutol-susceptible MDR strains (19.2%, 10/52). While the association between embB306 mutation and ethambutol resistance is statistically significant (odds ratio = 5.1, χ2 = 16.7, P < 0.0001), our data suggest that embB306 is not the sole causative mutation of ethambutol resistance, but is a sensitive candidate marker for ethambutol resistance analysis. Our finding that embB306 mutations exist in both ethambutol-resistant and -susceptible clinical M. tuberculosis isolates differs from the work of Plinke et al.,9 in which they showed no embB306 mutation in ethambutol-susceptible MDR strains, but agrees with the observations of others.12,15 One possible explanation for this inconsistency between phenotypic and genotypic testing results is that embB306 mutations confer M. tuberculosis variable ethambutol MICs and clinical strains with low to moderate levels of resistance may readily show opposite ethambutol susceptibility results using different testing methods.6,16,17,30 The occurrence of the embB306 mutations has been compared in different lineages and genotypes to identify the association between mutations and genetic structures.12,14 However, in MDR strains, it is not well addressed whether the genetic background contributes to the levels of ethambutol resistance in embB306 mutants. In this study, after locating the ethambutol-susceptible isolates with embB306 mutations into the phylogenetic map, we found that although 80% (8/10) of ethambutol-susceptible embB306 mutants niched in a major cluster (indicated as ‘cluster A’ in Figure 1a), the genetic background of these strains was diverse and failed to identify an obvious relationship of any specific genetic lineage with pheno-genotype discordance of embB306 mutants (Figure 1a). Interestingly, two pairs of embB306 mutants sharing identical VNTR-16 genotyping patterns but with different ethambutol susceptibility for each paired isolates clearly indicates that ethambutol resistance and susceptibility can exist in clinical isolates with highly similar genetic background (Figure 1a). Different types of embB306 mutation have been reported to affect the ethambutol MICs for M. tuberculosis and the results of ethambutol susceptibility testing.6,10,17 However, the pair 1 isolates (isolate 55 versus isolate 92) both carried a single embB306 mutation (ATG→ATT), of which isolate 92 was ethambutol resistant and isolate 55 was ethambutol susceptible. The pair 2 isolates (isolate 99 versus isolate 100) carried another single embB306 mutation (ATG→ATA), of which isolate 99 was ethambutol resistant while isolate 100 was ethambutol susceptible (Figure 1b). Thus, the pheno-genotype discordance of embB306 mutation and ethambutol resistance is not likely related to different types of embB306 mutation or multiple mutations of embB. In addition, this finding is unlikely related to additional antibiotic resistances, as observed between multidrug resistance and ethambutol resistance,15 because each of the paired isolates share identical first-line drug resistance patterns (isoniazid/rifampicin resistant for pair 1 and isoniazid/rifampicin/streptomycin resistant for pair 2). The inconsistency between ethambutol DST results and embB306 mutation is more likely related to other mutations occurring outside the embB gene in the genome of these clinical strains. Several previous works indicate that ethambutol resistance is a multigene mutation process that requires mutations in the embB gene and other currently unknown loci.5,16 Our findings clearly suggest that the determinant of ethambutol resistance may invoke more than one gene variation including embB gene, and that concurrent mutations incident at different sites of the bacterial genome are needed to confer overt ethambutol resistance of M. tuberculosis.
Interestingly, the ATG→CTG mutation at embB306, which was associated with a higher ethambutol MIC and conferred a growth advantage under sub-MICs of isoniazid or rifampicin,6,10 was detected in six strains with quadruple first-line drug resistance (isoniazid/rifampicin/ethambutol/streptomycin resistant) in our study (Table S1, available as Supplementary data at JAC Online) (Figure 1c).
Characteristics of embB497 and embB406 mutations
The major embB mutations detected in our isolates include not only embB306, but embB497 and embB406 as well. Mutations in embB497 were found in 16/86 (18.6%) ethambutol-resistant strains and 3/52 (5.8%) ethambutol-susceptible strains. Seven of 86 (8.1%) ethambutol-resistant strains and 3/52 (5.8%) ethambutol-susceptible isolates carried an embB406 mutation. However, a low frequency of mutations embB497 and embB406 (≤6%) has been reported in ethambutol-resistant strains by a limited number of studies.8,10,31 A recent study, which analysed all variations in the entire embCAB operon of ethambutol-resistant isolates without an embB306 mutation, showed that 10/34 (29.4%) non-embB306-mutated ethambutol-resistant isolates carried embB497 mutations and 8/34 (23.5%) carried embB406 mutations,29 while our results showed 14/39 (35.9%) and 6/39 (15.4%), respectively. The high proportion of embB497 and embB406 mutations that occurred in ethambutol-resistant isolates lacking an embB306 mutation in our study indicates the importance of embB497 and embB406 mutations as additional hotspots to embB306 for the rapid detection of ethambutol resistance using molecular assays, especially in Henan, China. Although embB406 mutations had been detected in ethambutol-susceptible isolates in two previous studies,13,31 embB497 mutations have not been found in ethambutol-susceptible strains until now.8,10,13,29,32 The reason may be that embB497 mutations are located out of the common region for detecting embB306 mutations and such regions were not examined by many existing studies.
Similar to the finding in embB306 mutants, discordant ethambutol susceptibility of embB497 mutants with identical genetic background was also observed. Pair 3 isolates (isolate 21 versus isolate 108) sharing an identical VNTR-16 genotype and sole embB497 mutation (CAG→CGG) showed different drug susceptibility; isolate 21 is ethambutol resistant while isolate 108 is ethambutol susceptible (Figure 1a and b). This result reveals that the contradiction between ethambutol drug susceptibility and embB497 mutation testing results is probably related to other mutations occurring outside the embB gene in the genome of embB497 mutants, and provides more evidence that the development of full ethambutol resistance may require certain mutations occurring at multiple genes and embB497 is one such mutation site.
embB mutations and broad drug resistance
We observed that embB mutations among clinical MDR-TB strains from Henan are common, with 100/138 (72.5%) of MDR-TB isolates carrying at least one mutation. To determine whether these 27 mutation types of embB are specific to drug resistance, 22 pan-susceptible clinical M. tuberculosis isolates from Henan patients were adopted as controls for the embB coding region analysis. Among 22 pan-susceptible clinical MDR-TB isolates, a single mutation at embB534 was detected in one strain, which indicates that embB534 is not a specific mutation in drug-resistant M. tuberculosis. Nonetheless, among the 12 embB534 MDR mutants, only 2 carried a single embB534 mutation; 10 of them had additional embB mutations (Table 2). Regardless of the embB534 mutation, the percentage of embB mutants among MDR-TB isolates is 71.0% (98/138), of which 58.2% (57/98) harboured embB306 mutations. Our results suggest that excluding embB534 mutations, all other embB-specific mutations in our patient population (and not simply limited to the most frequently documented embB306 mutation) may be candidate markers for the prediction of multidrug resistance and broad first-line polydrug resistance.
Existing commercial line probe assays are directed at detecting rifampicin or rifampicin and isoniazid mutations that are associated with phenotypic resistance. A rapid molecular assay that could predict polyresistance beyond MDR would be of significant utility to the diagnostic laboratory. Several studies show that the combination of a limited number of mutation sites can be applied to predict the drug resistance of M. tuberculosis.24,33 To assess the possibility of using only embB mutations to predict drug resistance, we performed trend analysis correlating any embB mutation and the number of first-line drug resistances. The statistically significant association (χ2 for trend = 26.5, P < 0.0001) between embB mutations and resistance to increasing numbers of antituberculosis reagents [33.3% (3/9) of two-drug-resistant isolates (isoniazid/rifampicin resistant) carried embB mutations, 53.8% (28/52) of three-drug-resistant isolates (isoniazid/rifampicin/streptomycin resistant or isoniazid/rifampicin/ethambutol resistant) carried embB mutations and 89.6% (69/77) of four-drug-resistant isolates (isoniazid/rifampicin/ethambutol/streptomycin resistant) carried embB mutations] strongly suggests that embB mutations might be sensitive candidate markers for the prediction of concurrent resistance to isoniazid, rifampicin, streptomycin and ethambutol in M. tuberculosis clinical isolates. Moreover, 83.1% of these four drug-resistant isolates can be detected by sequencing a short fragment (606 bp) of the embB gene, which covers 10 mutation sites at codons 306, 328, 330, 354, 406, 424, 439, 469, 497 and 508. While it is yet unproven why mutations affecting embB-encoded arabinosyl transferases may predict resistance to other drugs, this study supports the findings of others that embB306 is a predictor of multidrug resistance.12,15,24 In addition, our findings may lead to the development of a simple molecular test to rapidly identify isolates that are likely to express resistance to four first-line drugs in Henan, China, based on only a limited number of embB mutations. Further studies would be required to examine such a strategy in clinical practice and in other geographic locations around the world.
In conclusion, our results demonstrated the mutation characteristics of the entire embB gene in clinical MDR-TB isolates, an undertaking not previously reported from China. Our results clearly suggest that the ethambutol resistance of M. tuberculosis depends on concurrent multiple mutations in the genome and that embB mutations alone are not sufficient for the development of full resistance to ethambutol. The three pairs of clinical isolates identified with identical VNTR genotypes and matched embB mutations but different ethambutol phenotypic susceptibility warrants further study. Using genome-wide single nucleotide polymorphism analysis of the three pairs of isolates, novel mutations contributing to ethambutol resistance are likely to be discovered.
Funding
This work was supported by China-Australia Health and HIV/AIDS Facility (CAHHF) (FA10EID23), and part supported by the National Special Key Project of China on Major Infectious Diseases (2008ZX10003-008, 2008ZX10003-005 and 2008ZX10004-303), National Natural Science Foundation of China (30970117) and Beijing Municipal of Science and Technology Key Project on Control and Prevention of Tuberculosis Disease (D0805070064801).
Transparency declarations
None to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac. oxfordjournals.org/).
References
- 1.WHO. Anti-Tuberculosis Drug Resistance in the World: Fourth Global Report. http://whqlibdoc.who.int/hq/2008/WHO_HTM_TB_2008.394_eng.pdf. (22 April 2011, date last accessed)
- 2.He GX, Zhao YL, Jiang GL, et al. Prevalence of tuberculosis drug resistance in 10 provinces of China. BMC Infect Dis. 2008;8:166. doi: 10.1186/1471-2334-8-166. doi:10.1186/1471-2334-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Thoracic Society, CDC, Infectious Diseases Society of America. Treatment of tuberculosis. MMWR Recomm Rep. 2003;52:1–77. [PubMed] [Google Scholar]
- 4.Alcaide F, Pfyffer GE, Telenti A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–3. doi: 10.1128/aac.41.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Safi H, Fleischmann RD, Peterson SN, et al. Allelic exchange and mutant selection demonstrate that common clinical embCAB gene mutations only modestly increase resistance to ethambutol in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2010;54:103–8. doi: 10.1128/AAC.01288-09. doi:10.1128/AAC.01288-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Safi H, Sayers B, Hazbon MH, et al. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob Agents Chemother. 2008;52:2027–34. doi: 10.1128/AAC.01486-07. doi:10.1128/AAC.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starks AM, Gumusboga A, Plikaytis BB, et al. Mutations at embB codon 306 are an important molecular indicator of ethambutol resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2009;53:1061–6. doi: 10.1128/AAC.01357-08. doi:10.1128/AAC.01357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad S, Jaber AA, Mokaddas E. Frequency of embB codon 306 mutations in ethambutol-susceptible and -resistant clinical Mycobacterium tuberculosis isolates in Kuwait. Tuberculosis (Edinb) 2007;87:123–9. doi: 10.1016/j.tube.2006.05.004. doi:10.1016/j.tube.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Plinke C, Rusch-Gerdes S, Niemann S. Significance of mutations in embB codon 306 for prediction of ethambutol resistance in clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2006;50:1900–2. doi: 10.1128/AAC.50.5.1900-1902.2006. doi:10.1128/AAC.50.5.1900-1902.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramaswamy SV, Amin AG, Goksel S, et al. Molecular genetic analysis of nucleotide polymorphisms associated with ethambutol resistance in human isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2000;44:326–36. doi: 10.1128/aac.44.2.326-336.2000. doi:10.1128/AAC.44.2.326-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sreevatsan S, Stockbauer KE, Pan X, et al. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–81. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazbon MH, Bobadilla del Valle M, Guerrero MI, et al. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob Agents Chemother. 2005;49:3794–802. doi: 10.1128/AAC.49.9.3794-3802.2005. doi:10.1128/AAC.49.9.3794-3802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee AS, Othman SN, Ho YM, et al. Novel mutations within the embB gene in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2004;48:4447–9. doi: 10.1128/AAC.48.11.4447-4449.2004. doi:10.1128/AAC.48.11.4447-4449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokrousov I, Otten T, Vyshnevskiy B, et al. Detection of embB306 mutations in ethambutol-susceptible clinical isolates of Mycobacterium tuberculosis from northwestern Russia: implications for genotypic resistance testing. J Clin Microbiol. 2002;40:3810–3. doi: 10.1128/JCM.40.10.3810-3813.2002. doi:10.1128/JCM.40.10.3810-3813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, Shen GM, Wu J, et al. Association between embB codon 306 mutations and drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51:2618–20. doi: 10.1128/AAC.01516-06. doi:10.1128/AAC.01516-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perdigao J, Macedo R, Ribeiro A, et al. Genetic characterisation of the ethambutol resistance-determining region in Mycobacterium tuberculosis: prevalence and significance of embB306 mutations. Int J Antimicrob Agents. 2009;33:334–8. doi: 10.1016/j.ijantimicag.2008.09.021. doi:10.1016/j.ijantimicag.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Plinke C, Cox HS, Kalon S, et al. Tuberculosis ethambutol resistance: concordance between phenotypic and genotypic test results. Tuberculosis (Edinb) 2009;89:448–52. doi: 10.1016/j.tube.2009.09.001. doi:10.1016/j.tube.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–66. [PubMed] [Google Scholar]
- 19.Cohen T, Sommers B, Murray M. The effect of drug resistance on the fitness of Mycobacterium tuberculosis. Lancet Infect Dis. 2003;3:13–21. doi: 10.1016/s1473-3099(03)00483-3. doi:10.1016/S1473-3099(03)00483-3. [DOI] [PubMed] [Google Scholar]
- 20.Zaczek A, Brzostek A, Augustynowicz-Kopec E, et al. Genetic evaluation of relationship between mutations in rpoB and resistance of Mycobacterium tuberculosis to rifampin. BMC Microbiol. 2009;9:10. doi: 10.1186/1471-2180-9-10. doi:10.1186/1471-2180-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canetti G, Froman S, Grosset J, et al. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ. 1963;29:565–78. [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes: Approved Standard M24-A. Wayne, PA, USA: NCCLS; 2003. [PubMed] [Google Scholar]
- 23.Chen J, Tsolaki AG, Shen X, et al. Deletion-targeted multiplex PCR (DTM-PCR) for identification of Beijing/W genotypes of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2007;87:446–9. doi: 10.1016/j.tube.2007.05.014. doi:10.1016/j.tube.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Luo T, Zhao M, Li X, et al. Selection of mutations to detect MDR TB in Shanghai, China. Antimicrob Agents Chemother. 2009;54:1075–81. doi: 10.1128/AAC.00964-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Chen J, Shen X, et al. Highly polymorphic variable-number tandem repeats loci for differentiating Beijing genotype strains of Mycobacterium tuberculosis in Shanghai, China. FEMS Microbiol Lett. 2008;282:22–31. doi: 10.1111/j.1574-6968.2008.01081.x. doi:10.1111/j.1574-6968.2008.01081.x. [DOI] [PubMed] [Google Scholar]
- 26.Allix-Beguec C, Harmsen D, Weniger T, et al. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–9. doi: 10.1128/JCM.00540-08. doi:10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Dudley J, Nei M, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. doi:10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.van Soolingen D, Hermans PW, de Haas PE, et al. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–86. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plinke C, Cox HS, Zarkua N, et al. embCAB sequence variation among ethambutol-resistant Mycobacterium tuberculosis isolates without embB306 mutation. J Antimicrob Chemother. 2010;65:1359–67. doi: 10.1093/jac/dkq120. doi:10.1093/jac/dkq120. [DOI] [PubMed] [Google Scholar]
- 30.Johnson R, Jordaan AM, Pretorius L, et al. Ethambutol resistance testing by mutation detection. Int J Tuberc Lung Dis. 2006;10:68–73. [PubMed] [Google Scholar]
- 31.Ramaswamy SV, Dou SJ, Rendon A, et al. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J Med Microbiol. 2004;53:107–13. doi: 10.1099/jmm.0.05343-0. doi:10.1099/jmm.0.05343-0. [DOI] [PubMed] [Google Scholar]
- 32.Parsons LM, Salfinger M, Clobridge A, et al. Phenotypic and molecular characterization of Mycobacterium tuberculosis isolates resistant to both isoniazid and ethambutol. Antimicrob Agents Chemother. 2005;49:2218–25. doi: 10.1128/AAC.49.6.2218-2225.2005. doi:10.1128/AAC.49.6.2218-2225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Rie A, Warren R, Mshanga I, et al. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J Clin Microbiol. 2001;39:636–41. doi: 10.1128/JCM.39.2.636-641.2001. doi:10.1128/JCM.39.2.636-641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.