Abstract
Objectives
Tn916-like elements are one of the most common types of integrative and conjugative element (ICE). In this study we aimed to determine whether novel accessory genes, i.e. genes whose products are not involved in mobility or regulation, were present on a Tn916-like element (Tn6087) isolated from Streptococcus oralis from the human oral cavity.
Methods
A minocycline-resistant isolate was analysed using restriction fragment length polymorphism (RFLP) analysis on amplicons derived from Tn916 and DNA sequencing to determine whether there were genetic differences in Tn6087 compared with Tn916. Mutational analysis was used to determine whether the novel accessory gene found was responsible for an observed extra phenotype.
Results
A novel Tn916-like element, Tn6087, is described that encodes both antibiotic and antiseptic resistance. The antiseptic resistance protein is encoded by a novel small multidrug resistance gene, designated qrg, that was shown to encode resistance to cetyltrimethylammonium bromide (CTAB), also known as cetrimide bromide.
Conclusions
This is the first Tn916-like element described that confers both antibiotic and antiseptic resistance, suggesting that selection of either antibiotic or antiseptic resistance will also select for the other and further highlights the need for prudent use of both types of compound.
Keywords: ICEs, CTAB, tetracycline resistance, IS1216, Tn916, conjugative transposon
Introduction
The Tn916/Tn1545 family is the most widespread of the conjugative transposons. Tn916 was first discovered in the late 1970s when tetracycline resistance was transferred from a resistant to a susceptible strain of Enterococcus faecalis in the absence of a detectable plasmid.1 Subsequently a variety of Tn916-like elements have been described2,3 with elements containing different recombinases and resistance genes. These elements comprise four functional modules: conjugation, regulation, recombination and accessory genes, the latter are often antibiotic resistance genes.3 Most Tn916-like elements possess the tetracycline resistance gene tet(M), located within the regulatory module, and some contain macrolide resistance genes erm(B) (e.g. Tn1545, Tn6002 and Tn6003)4,5 and mef(A) (e.g. Tn2009 and Tn2017),6,7 as well as kanamycin (aphA-3) (Tn1545)5 and mercury (mer) (Tn6009)8 resistance genes. Tn916-like elements have been found in >30 bacterial genera and are common among the human oral commensal bacteria, which act as a reservoir for resistance genes.9
The aim of the current study was to determine whether a Tn916-like element found in a minocycline-resistant oral isolate, Streptococcus oralis F.MI.5, contained any additional accessory resistance genes. An element, designated Tn6087, which contains tet(M) and a quaternary ammonium compound (QAC) resistance gene (qrg) was identified. Qrg is a protein that confers resistance to cetyltrimethylammonium bromide (CTAB), a cationic antiseptic belonging to the QACs. The co-localization of antiseptic and antibiotic resistance has important clinical ramifications, as the selection of one will lead to the selection of the other. Linkage between antibiotic and antiseptic resistance has been described previously in some Staphylococcus species;10,11 however, this is the first described member of the Tn916 family of elements encoding antiseptic resistance.
Materials and methods
Sampling, bacterial strains and plasmids
Saliva was obtained from 10 healthy volunteers who had not received antibiotics in the previous 3 months. Ethics approval for the project was obtained from the University College London (UCL) Ethics Committee (project no. 1364/001). Pooled saliva was cultured anaerobically on fastidious anaerobe agar (Oxoid) and at 5% CO2 on Columbia blood agar (Oxoid) at 37°C. All media contained 2 mg/L minocycline and were incubated for 48 h. Bacterial strains and plasmids used in this study are listed in Table S1 (available as Supplementary data at JAC Online).
Molecular biology
DNA was extracted using the ArchivePure DNA Yeast and Gram Positive Bacteria Kit (5Prime) according to manufacturer's instructions. Mutanolysin at a final concentration of 15 μg/mL and lysozyme at a final concentration of 3 μg/mL were added at the cell lysis step of the protocol. DNA was resuspended in molecular-grade water (Sigma-Aldrich) and stored at −20°C.
The sequences of all primers used in the study are shown in Table S2 (available as Supplementary data at JAC Online). Standard PCR was carried out using BioMix™ Red (Bioline). Long PCR (using O1F and CT25R; CTn9824F and PW01R spanning almost the entire Tn916) was carried out with TaKaRa Ex Taq™ (Lonza) according to the manufacturer's instructions.
Single specific primer (ssp)-PCR was performed in order to find the target sequence for the insertion of Tn6087. Genomic DNA was digested with HindIII and EcoRI and ligated into HindIII/EcoRI digested and dephosphorylated pUC19. A primer specific to the left end or right end (LEO, REO, respectively) of the transposon and/or M13F or M13R were used to amplify the end of the transposon and the flanking sequence. LEO and REO were used in a PCR to amplify the joint of the excised Tn6087 in circular form. DNA sequencing was carried out at the UCL Wolfson Institute DNA sequencing facility.
Southern blot hybridization was performed on HincII- and HindIII-digested genomic DNA of the strain containing Tn6087 using a probe derived from the Tn916 int gene (U09422, 16 704–17 725 bp) and the IS1216 sequence (HQ663849, 9139–9857 bp and 11 456–12 174 bp) from within Tn6087.
Genotypic characterization of the novel transposon
The entire Tn6087 element was sequenced using primers O1F, CTn670F, CTn1320F, CTn1960F, CT14F, CT14-19 W1F, CTn4028F, CTn4470F, CT14-19 W3F, CTn5109F, CT20F, CTn6352F, CTn6990F, O7F, CTn8286F, CTn9824F, CTn10462F, CTn10593R, CTn11122F, O11F, CTn12353F, RT1-4 W2F, CTn13562F, CTn14122F, CTn14749F, CTn15401F, CTn16068F, CTn16707F, PW01F and PW01R resulting in at least triple coverage. The transposon name was designated by the transposon registry (http://www.ucl.ac.uk/eastman/tn/).12
Primers (W1F, Tn6087 IS1R, Tn6087 IS2F, W1R) were used in PCR to investigate if the excision of the IS1216 elements could be detected in S. oralis F.MI.5. Primers SMR-01 and SMR-02 were used to detect qrg in metagenomic DNA isolated from pooled saliva and faecal samples taken from Norway, Finland, France and Italy.13 Each sample (either saliva or faecal) from each country is a pooled sample derived from 20 healthy individuals.
Mutation of qrg
A construct with identical sequence (∼550 bp upstream of qrg and 400 bp downstream) to the regions flanking qrg, but with the chloramphenicol resistance gene cat(P) (derived from the plasmid pMTL900)14 inserted in its place, was used in an allelic replacement experiment to knock out qrg. The construct was cloned into pGEM T-Easy vector, linearized and transformed into S. oralis F.MI.5. For transformation, the recipient strain was grown to exponential phase; at this point 1 mg of the construct was mixed with the cells and the mix was incubated for 3 h at 37°C prior to spreading on selective plates. Chloramphenicol-resistant (10 mg/L) colonies were then selected for further analysis. One strain with the correct mutation was designated S. oralis F.MI.5Δqrg. For complementation, a construct containing qrg and flanking regions was inserted into the BamHI site of the shuttle vector pVA83815 and transformed into F.MI.5Δqrg. The complemented strain was selected on chloramphenicol (10 mg/L) and CTAB (32 mg/L). The complemented strain was designated S. oralis F.MI.5Δqrg pVA838::qrg.
Transfer studies
Mating experiments were performed using S. oralis F.MI.5 as a donor and both Enterococcus faecalis JH2-2 and Streptococcus pneumoniae DP1004 as recipients using two previously described methods.16,17 As these experiments yielded no transconjugants, transformation experiments were performed. S. oralis F.MI.5 genomic DNA (10 μg) was added to minocycline-susceptible Streptococcus australis FRStet12 (Table S1) while competent during the exponential growth phase. Transformants were selected on media containing 4 mg/L minocycline.
MICs
MICs were investigated according to the BSAC guidelines (http://www.bsac.org.uk/) using tetracycline, minocycline, CTAB, acriflavine, Acridine Orange and ethidium bromide. These were carried out using S. oralis F.MI.5, S. australis FRStet12 and S. australis FRStet12::Tn6087. Iso-sensitest agar containing 5% horse blood was used and plates were incubated at 37°C for 24 h. Antibiotics and antiseptics were used at concentrations ranging from 0 to 128 mg/L. All cultures were used at a 1/10 dilution of the 0.5 McFarland standard. MICs for CTAB were also investigated for S. oralis F.MI.5Δqrg and F.MI.5Δqrg pVA838s::qrg.
Results
Isolation of Tn6087 and identification of qrg
Minocycline-resistant S. oralis F.MI.5 was isolated from pooled saliva. Partial 16S rRNA gene sequencing showed that the strain had 99.7% identity to S. oralis strain VS2971S (AY944234).
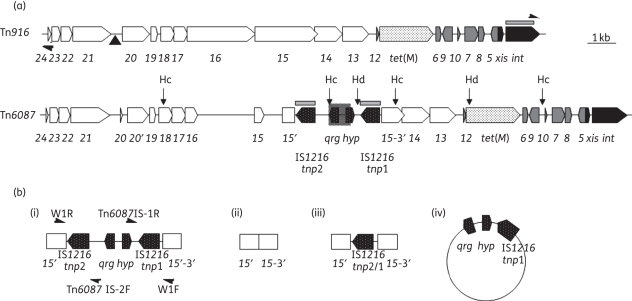
To identify the gene responsible for minocycline resistance, degenerate primers (RPP-F and RPP-R) (Table S2), which amplify ribosome protection genes, were used to amplify DNA from S. oralis F.MI.5. Sequencing of the PCR products showed that tet(M) was present, and further PCR showed that it was located on a Tn916-like element. Two long PCRs spanning almost the entire length of Tn916 then indicated that the conjugation module was ∼3 kb longer than that from wild-type Tn916 in Bacillus subtilis BS34A (Tables S1 and S3; available as Supplementary data at JAC Online). The entire element was subsequently sequenced, and designated Tn6087 (Figure 1). Excluding the insertion in the conjugation region, Tn6087 is 99.03% identical to Tn916 (U09422) at the nucleotide level. The base pair differences result in some changes to the open reading frames (ORFs) of the element. Orf24 contains a stop codon at 263 bp, resulting in a 16 amino acid truncation in the polypeptide. Also, orf20 contains a premature stop codon at 2906 bp, resulting in a 314 amino acid truncation; orf16 contains a stop codon at 5621 bp, resulting in a 673 amino acid truncation; and orf15 contains a stop codon at 7997 bp, resulting in truncated protein of 119 amino acids. The Tn6087 tet(M) is 99.7% identical to tet(M) from the E. faecalis Tn916-like conjugative transposon (DQ223248), and is 93.85% identical to tet(M) of Tn916 (U09422).
Figure 1.
(a) A schematic representation of Tn916 and Tn6087 showing the functional modules (conjugation in white; regulation in light grey; the accessory gene in white with black dots; and recombination in dark grey). The position of the primers LEO (at the left end) and REO (at the right end) are shown on Tn916. A black triangle indicates the position of the origin of transfer, oriT, of Tn916. The IS1216 flanked insertion of Tn6087 is shown in black with white dots. The shaded region between the IS1216 sequences represents the sequence similar to S. gallolyticus strain UCN34 (FN597254). HindIII and HincII (denoted Hd and Hc) restriction sites are shown; the int and IS1216 probes are shown as grey rectangles appearing above the relevant genes. (b) The four forms of the qrg gene and IS1216 insertion obtained using PCR analysis of S. oralis F.MI.5 with the primers shown in Figure 1b(i). (i) The entire 3124 bp insertion within orf15 (primers W1R, Tn6087 IS-2F, Tn6087 IS-1R, W1F are shown from left to right); (ii) the entire insertion excised from orf15; (iii) only a chimeric form of the two IS1216 sequences remaining; and (iv) a circular molecule consisting of the qrg gene, the hypothetical protein and the IS1216 1 sequence.
A 3124 bp composite transposon-like structure was found inserted in orf15 at 9099 bp, which contained ORFs predicted to encode a small multidrug resistance protein and a hypothetical protein, flanked by IS1216 elements (Figure 1). The two IS1216 elements were 99.38% identical to each other at the nucleotide level. Approximately half (812 bp) of the region between the two IS1216 sequences, including the small multidrug resistance gene qrg (Figure 1), showed 95% identity to genomic sequence from a Streptococcus gallolyticus strain UCN34 (FN597254).
Southern blots probed with Tn916 int and IS1216 confirmed that S. oralis F.MI.5 contained only one copy of the int gene and two IS1216 elements, suggesting that the strain possessed a single copy of Tn6087 (results not shown).
MICs of minocycline, tetracycline, CTAB, acriflavine, Acridine Orange and ethidium bromide were carried out on S. oralis F.MI.5, S. australis FRStet12 and S. australis FRStet12::Tn6087. The results showed that the presence of Tn6087 increased resistance to minocycline, tetracycline and CTAB (16-fold, 32-fold and 16-fold, respectively; Table 1). A 2-fold increase in resistance to acriflavine was also detected; however, there was no difference seen in resistance to Acridine Orange or ethidium bromide. A 16-fold increase in resistance to CTAB suggested that Qrg, encoded for by Tn6087, provided resistance to some QACs. To prove this, qrg was replaced by the chloramphenicol resistance gene cat(P) in S. oralis F.MI.5, generating S. oralis F.MI.5Δqrg. MICs for CTAB were performed on these strains and on a complemented F.MI.5Δqrg designated S. oralis F.MI.5Δqrg pVA838::qrg. The results showed a 4-fold decrease in CTAB resistance in F.MI.5Δqrg and an increase of resistance to CTAB in F.MI.5Δqrg pVA838::qrg (Table 1), confirming that qrg was responsible for CTAB resistance.
Table 1.
MICs of antibiotics and QACs (mg/L)
| TET | MIN | EB | AO | AF | CTAB | |
|---|---|---|---|---|---|---|
| S. oralis F.MI.5 Tn6087 | 32 | 8 | 8 | 8 | 4 | 64 |
| S. oralis F.MI.5 Tn6087Δqrg | 32 | 8 | 8 | 8 | 4 | 16 |
| S. oralis F.MI.5 Tn6087Δqrg pVA838::qrg | 32 | 8 | 8 | 8 | 4 | 32 |
| S. australis FRStet12 | 4 | 0.25 | 8 | 16 | 4 | 4 |
| S. australis FRStet12::Tn6087 | 128 | 4 | 8 | 16 | 8 | 64 |
TET, tetracycline; MIN, minocycline, EB, ethidium bromide; AO, Acridine Orange; AF, acriflavine; CTAB, cetyltrimethylammonium bromide.
To investigate if qrg is present in other populations, PCR analysis of oral and faecal metagenomic DNA taken from healthy volunteers from four European countries was performed. All samples (eight samples in total; pooled saliva and pooled faecal samples from four countries) contained amplicons of the expected size. Subsequent sequence analysis of the PCR products amplified from saliva samples were shown to have an identity of at least 97.81% to qrg from Tn6087.
Tn6087 could be transferred by transformation to S. australis FRStet12
Excision and circularization of Tn6087 was detected using PCR for the joint of the circular form in strain S. oralis F.MI.5. However, no transconjugants could be isolated following mating experiments with both E. faecalis JH2-2 and S. pneumoniae DP1004 as recipients (numbers of donor cells varied from 1.6 × 107 to 1 × 1010 and numbers of recipient cells varied from 1.5 × 107 to 2.6 × 1010). Tn6087 was transferred by transformation using S. oralis F.MI.5 genomic DNA and the minocycline-susceptible recipient S. australis FRStet12 at a frequency of ∼1.2 × 10−8 transformants/μg of DNA. A minocycline-resistant transformant was isolated and PCR analysis confirmed that the transformant possessed the entire Tn6087 element. Sequencing of the sod(A) gene confirmed transfer, as the sequence in the recipient was identical to that of the transformant but different from that of the donor (99.32% identity). The presence of a circular intermediate of Tn6087 was confirmed in the transformant by performing a PCR with primers LEO and REO (Table S2). The target sites of Tn6087 were investigated using ssp-PCR in the transformant and were identical to S. oralis F.MI.5 (Figure 2).
Figure 2.
(a) The Tn6087 target site within S. oralis F.MI.5. (b) The left end and (c) right end of Tn6087 are shown in bold. (d) The joint of the circular intermediate of Tn6087 is shown. The transposon's coupling sequences are shown in bold italics. As can be seen in (d), the coupling sequence at the joint of the circular form is a heteroduplex.
The region containing IS1216 undergoes genetic rearrangement
In order to investigate the plasticity of the insertion within orf15 of Tn6087, a series of PCRs were carried out to detect recombination and/or transposition products using S. oralis F.MI.5 genomic DNA. PCR analysis showed at least two different rearrangements were present in the region of the transposon containing IS1216 (Figure 1b). The first was a complete deletion of the IS elements and the DNA between them [Figure 1b(ii)]. In the second, a single chimeric copy of the IS element remained [Figure 1b(iii)], and in addition a circular form of the element containing one IS element was detected [Figure 1b(iv)]. It is likely that the sequences seen in Figure 1[b(iii) and b(iv)] arose by separate recombination events between the directly repeated IS elements. One reaction deletes the DNA between the ISs, resulting in one chimeric copy of IS1216 within Tn6087, and the other reaction results in a circular molecule containing a copy of IS1216 and qrg, as observed.
Discussion
This is the first reported Tn916-like element that encodes both antibiotic and antiseptic resistance to tetracyclines and CTAB, respectively. Plasmids encoding both antiseptic and antibiotic resistance have been described previously.10,11,18,19 The gene combinations found on plasmids have usually been qacA/B/C and blaZ in Staphylococcus sp., and these are often associated with IS elements such as IS256 and IS257.10,20,21 In one case a plasmid-located qacB was found adjacent to the incomplete and non-functional β-lactamase-encoding transposon Tn552.10 Additionally, a putative qacF was found on a transposon present on a plasmid belonging to the IncP-1β family.18
IS1216 elements have been associated with transposons, e.g. Tn1546, which encodes resistance to vancomycin,22,23 and the partially sequenced Tn5397-like erm(B) containing transposon Tn1116.24 Additionally, a composite transposon present on a plasmid comprising a benzalkonium chloride (a QAC) resistance cassette (ORFs bcrABC) flanked by IS1216 elements was recently reported.25 In another example, IS1216 elements have been involved in the evolution of the multiresistant plasmid pK214.26 This 30 kb plasmid encodes resistance to tetracyclines, macrolides, chloramphenicol and streptomycin and contains three copies of IS1216. In this case the IS1216 sequences seem to have been involved in the insertion of a set of Staphylococcus-derived genes including a chloramphenicol acetyltransferase and a streptomycin adenyltransferase, as well as the insertion of a group of Tn916-like genes including tet(S) and spanning part of the regulation module.
In addition to the novel architecture, Tn6087 also represents the first proven QAC resistance gene identified in oral bacteria. The oral environment is home to a rich and complex community of bacteria that have ample opportunity to exchange genes by multiple mechanisms.9 It was not possible to show transfer by conjugation to the proven recipient strains E. faecalis JH2-2 or S. pneumoniae DP1004; however, this is likely to be due to the insertion of the IS1216-like element containing qrg into the region thought to be involved in conjugation, as well as the truncation of a number of other conjugation genes. It is also possible that Tn6087 might be able to transfer by conjugation if the conjugation functions lost are provided in trans by another mobile element. However, the element could transfer by transformation, a mechanism of genetic exchange likely to be important in the oral cavity due to the large number of naturally competent streptococci. The qrg gene was shown by PCR to be present in metagenomic oral and faecal DNA isolated from other European countries, demonstrating that the gene is likely to be distributed throughout Europe.
This work has resulted in the detection of a novel small multidrug resistance gene, qrg, that encodes resistance to CTAB. It is located on a composite transposon consisting of two IS1216 elements flanking qrg, inserted in a Tn916-like element capable of transfer by transformation to another streptococcal strain.
Funding
This research was funded by the Commission of the European Communities, specifically the Infectious Diseases research domain of the Health theme of the 7th Framework Programme, contract 241446, ‘The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiotas at various body sites’ and the 5th Framework research and technological development (RTD) program ‘Quality of Life and Management of Living Resources’, QLK2-CT-2002-00843, ‘Antimicrobial resistance transfer from and between Gram-positive bacteria of the digestive tract and consequences for virulence’.
Transparency declarations
None to declare.
Supplementary data
Tables S1–S3 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org).
Acknowledgements
We would like to thank Dr Philip Warburton and Miss Azmiza Jasni (University College London) and Dr Francesco Santoro (University of Siena) for bacterial strains and experimental advice.
References
- 1.Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of conjugal transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flannagan SE, Zitzow LA, Su YA, et al. Nucleotide-sequence of the 18-Kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid. 1994;32:350–4. doi: 10.1006/plas.1994.1077. doi:10.1006/plas.1994.1077. [DOI] [PubMed] [Google Scholar]
- 3.Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–8. doi: 10.1016/j.tim.2009.03.002. doi:10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Warburton PJ, Palmer RM, Munson MA, et al. Demonstration of in vivo transfer of doxycycline resistance mediated by a novel transposon. J Antimicrob Chemother. 2007;60:973–80. doi: 10.1093/jac/dkm331. doi:10.1093/jac/dkm331. [DOI] [PubMed] [Google Scholar]
- 5.Cochetti I, Tili E, Mingoia M, et al. erm(B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother. 2008;52:1285–90. doi: 10.1128/AAC.01457-07. doi:10.1128/AAC.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Grosso M, Camilli R, Libisch B, et al. New composite genetic element of the Tn916 family with dual macrolide resistance genes in a Streptococcus pneumoniae isolate belonging to clonal complex 271. Antimicrob Agents Chemother. 2009;53:1293–4. doi: 10.1128/AAC.01066-08. doi:10.1128/AAC.01066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Grosso M, d'Abusco AS, Iannelli F, et al. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2004;48:2037–42. doi: 10.1128/AAC.48.6.2037-2042.2004. doi:10.1128/AAC.48.6.2037-2042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soge OO, Beck NK, White TM, et al. A novel transposon, Tn6009, composed of a Tn916 element linked with a Staphylococcus aureus mer operon. J Antimicrob Chemoth. 2008;62:674–80. doi: 10.1093/jac/dkn255. doi:10.1093/jac/dkn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther. 2010;8:1441–50. doi: 10.1586/eri.10.106. doi:10.1586/eri.10.106. [DOI] [PubMed] [Google Scholar]
- 10.Sidhu MS, Heir E, Sorum H, et al. Genetic linkage between resistance to quaternary ammonium compounds and beta-lactam antibiotics in food-related Staphylococcus spp. Microb Drug Resist. 2001;7:363–71. doi: 10.1089/10766290152773374. doi:10.1089/10766290152773374. [DOI] [PubMed] [Google Scholar]
- 11.Bjorland J, Steinum T, Kvitle B, et al. Widespread distribution of disinfectant resistance genes among staphylococci of bovine and caprine origin in Norway. J Clin Microbiol. 2005;43:4363–8. doi: 10.1128/JCM.43.9.4363-4368.2005. doi:10.1128/JCM.43.9.4363-4368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts AP, Chandler M, Courvalin P, et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60:167–73. doi: 10.1016/j.plasmid.2008.08.001. doi:10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seville LA, Patterson AJ, Scott KP, et al. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb Drug Resist. 2009;15:159–66. doi: 10.1089/mdr.2009.0916. doi:10.1089/mdr.2009.0916. [DOI] [PubMed] [Google Scholar]
- 14.Roberts AP, Hennequin C, Elmore M, et al. Development of an integrative vector for the expression of antisense RNA in Clostridium difficile. J Microbiol Methods. 2003;55:617–24. doi: 10.1016/s0167-7012(03)00200-8. doi:10.1016/S0167-7012(03)00200-8. [DOI] [PubMed] [Google Scholar]
- 15.Macrina FL, Tobian JA, Jones KR, et al. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982;19:345–53. doi: 10.1016/0378-1119(82)90025-7. doi:10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 16.Roberts AP, Mullany P, Wilson M. Gene transfer in bacterial biofilms. Microbial Growth in Biofilms Pt A. 2001;336:60–5. doi: 10.1016/s0076-6879(01)36578-3. doi:10.1016/S0076-6879(01)36578-3. [DOI] [PubMed] [Google Scholar]
- 17.Santoro F, Oggioni MR, Pozzi G, et al. Nucleotide sequence and functional analysis of the tet(M)-carrying conjugative transposon Tn5251 of Streptococcus pneumoniae. FEMS Microbiol Lett. 2010;308:150–8. doi: 10.1111/j.1574-6968.2010.02002.x. [DOI] [PubMed] [Google Scholar]
- 18.Schluter A, Heuer H, Szczepanowski R, et al. Plasmid pB8 is closely related to the prototype IncP-1 beta plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein. Plasmid. 2005;54:135–48. doi: 10.1016/j.plasmid.2005.03.001. doi:10.1016/j.plasmid.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Bjorland J, Sunde M, Waage S. Plasmid-borne smr gene causes resistance to quaternary ammonium compounds in bovine Staphylococcus aureus. J Clin Microbiol. 2001;39:3999–4004. doi: 10.1128/JCM.39.11.3999-4004.2001. doi:10.1128/JCM.39.11.3999-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam MM, Kobayashi N, Uehara N, et al. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb Drug Resist. 2003;9:109–21. doi: 10.1089/107662903765826697. doi:10.1089/107662903765826697. [DOI] [PubMed] [Google Scholar]
- 21.Berg T, Firth N, Apisiridej S, et al. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol. 1998;180:4350–9. doi: 10.1128/jb.180.17.4350-4359.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen LB. Internal size variations in Tn1546-like elements due to the presence of IS1216V. FEMS Microbiol Lett. 1998;169:349–54. doi: 10.1111/j.1574-6968.1998.tb13339.x. doi:10.1111/j.1574-6968.1998.tb13339.x. [DOI] [PubMed] [Google Scholar]
- 23.Sletvold H, Johnsen PJ, Wikmark OG, et al. Tn1546 is part of a larger plasmid-encoded genetic unit horizontally disseminated among clonal Enterococcus faecium lineages. J Antimicrob Chemother. 2010;65:1894–906. doi: 10.1093/jac/dkq219. doi:10.1093/jac/dkq219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenciani A, Bacciaglia A, Vecchi M, et al. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob Agents Chemother. 2007;51:1209–16. doi: 10.1128/AAC.01484-06. doi:10.1128/AAC.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elhanafi D, Dutta V, Kathariou S. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998–1999 Outbreak. Appl Environ Microbiol. 2010;76:8231–8. doi: 10.1128/AEM.02056-10. doi:10.1128/AEM.02056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perreten V, Schwarz F, Cresta L, et al. Antibiotic resistance spread in food. Nature. 1997;389:801–2. doi: 10.1038/39767. doi:10.1038/39767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.