Abstract
Glyburide (GLB) is a widely used oral sulfonylurea for the treatment of gestational diabetes. Therapeutic use of GLB is often complicated by a substantial inter-individual variability in the pharmacokinetics and pharmacodynamics of the drug in human populations, which might be caused by inter-individual variations in factors such as GLB metabolism. Therefore, there has been a continued interest in identifying human cytochrome P450 (CYP) isoforms that play a major role in the metabolism of GLB. However, contrasting data are available in the present literature in this regard. In the present study, we systematically investigated the contributions of various human CYP isoforms (CYP3A4, CYP3A5, CYP2C8, CYP2C9, and CYP2C19) to in vitro metabolism of GLB. GLB depletion and metabolite formation in human liver microsomes were most significantly inhibited by the CYP3A inhibitor ketoconazole compared with the inhibitors of other CYP isoforms. Furthermore, multiple correlation analysis between GLB depletion and individual CYP activities was performed, demonstrating a significant correlation between GLB depletion and the CYP3A probe activity in 16 individual human liver microsomal preparations, but not between GLB depletion and the CYP2C19, CYP2C8, or CYP2C9 probe activity. By using recombinant supersomes overexpressing individual human CYP isoforms, we found that GLB could be depleted by all the enzymes tested; however, the intrinsic clearance (Vmax/Km) of CYP3A4 for GLB depletion was 4 – 17 times greater than that of other CYP isoforms. These results confirm that human CYP3A4 is the major enzyme invovled in the in vitro metabolism of GLB.
Keywords: Glyburide, human CYP enzymes, CYP3A4, CYP2C9, in vitro metabolism
Introduction
Glyburide (GLB) is a second generation oral sulfonylurea used for the treatment of Type 2 diabetes mellitus with a potent and prolonged hypoglycemic effect (1). GLB is also commonly used to treat gestational diabetes mellitus (2–4). Two major metabolites of GLB, 4-trans-hydroxyglyburide (M1) and 3-cis-hydroxyglyburide (M2b) (Supplement 1), which account for approximately half of all the metabolites formed in vitro by human liver microsomes (5), are also pharmacologically active (6). In addition, GLB has been shown to accumulate in pancreatic beta cells after long-term use (7). These factors may contribute to the prolonged action of the drug and hypoglycemia in some cases. Glucose control and optimal GLB dosage are often hindered by a substantial inter-individual variability in the pharmacokinetics and pharmacodynamics of the drug in human populations. The inter-individual variability might be caused by the inter-individual variations in factors such as metabolism and elimination of the drug (8–12). Given the fact that GLB is extensively metabolized in the liver, the identification of major hepatic CYP enzymes involved in GLB metabolism will have significant clinical implications for rational design of optimal dosing regimen and for understanding potential drug-drug interactions.
Therefore, there has been a continued interest in identifying human CYP enzymes that play a major role in the metabolism of GLB. However, contrasting data are available in the present literature in this regard. CYP2C9 is a highly polymorphic metabolic enzyme. The CYP2C9 variants, CYP2C9*3/*3 homozygote or CYP2C9*1/*3 heterozygote, have been shown to exhibit lower catalytic activities than the wild-type CYP2C9*1/*1 (13). Kirchheriner et al. showed that the total oral clearance of GLB in the CYP2C9*3/*3 subjects (n = 3) was approximately 40% of that in the CYP2C9*1/*1 subjects (n = 4) (14). Niemi et al. reported that the area under the plasma concentration-time curve (plasma AUC) of GLB in individuals heterozygous for the CYP2C9*3 allele (n = 2) was 280% of the respective value in the CYP2C9*1/*1 subjects (n = 5) (15). Yin et al. also demonstrated that the oral plasma AUC of GLB in the CYP2C9*1/*3 subjects (n = 6) of the Chinese population was increased by approximately 100% as compared with that in the CYP2C9*1/*1 subjects (n = 12) (16). These clinical studies appear to suggest that CYP2C9 contributes significantly to GLB metabolism in vivo. However, one in vitro metabolism study with human liver microsomes revealed that CYP3A played a major role in the metabolism of GLB with no contribution from CYP2C9 (17). Our recent clinical study showed that the apparent oral clearance of GLB in pregnant women with gestational diabetes (n = 6 for CYP2C9*1/*2 and CYP2C9*1/*3 combined) was independent of the CYP2C9 genotype (4).
To verify the previous observations about in vitro metabolism of GLB by human CYP enzymes, in the present study, we systematically investigated the contributions of various human CYP isoforms (CYP3A4, CYP3A5, CYP2C8, CYP2C9, and CYP2C19) to in vitro metabolism of GLB. Several complementary experimental approaches were employed, including inhibition of GLB metabolism in human liver microsomes by selective CYP isoform inhibitors, the correlation between GLB depletion and the probe activities of individual CYP enzymes assessed using probe substrates in 16 individual human liver microsomal preparations, and the determination of kinetic parameters (Km and Vmax) of individually expressed human recombinant CYP isoforms for GLB depletion. Results of these studies suggest that human CYP3A4 is the major enzyme in the metabolism of GLB. When this study was nearly completed, another in vitro metabolism study for GLB came out and showed that CYP3A4 played a predominant role in GLB metabolism with a more than 50% contribution to the formation of total metabolites, wherease CYP2C9 had approximately 30% contribution (18).
Materials and Methods
Materials
Glyburide (GLB), glipizide (GLP), 8-methoxypsoralen (MOP), triethylenethiophosphoramide (TTEPA), omeprazole (OMP), sulfaphenazole (SUL), quinidine (QUI), ketoconazole (KTZ), testosterone, 6β–hydroxytestosterone, tolbutamide, 4-methylhydroxytobultamide, amodiaquine, desethylamodiaquine, and midazolam were purchased from Sigma (St. Louis, MO). Montelukast (MTL) was purchased from Sequoia Research Products (Pangbourne, UK). 5-hydroxyomeprazole was a gift from Astra/Zeneca. All the commercially available reagents and solvents were of either analytical or HPLC grade. Supersomes™ prepared from baculovirus-infected insect cells expressing human CYP3A4, CYP3A5, CYP2C8, CYP2C19, CYP2C9*1/*1, CYP2C9*2/*2, or CYP2C9*3/*3 isoform were purchased from GENTEST (Woburn, MA). M1 and M2b were synthesized (19).
Preparation of human liver microsomes
Sixteen individual human liver tissue samples from Caucasian donors (HL-102, 103, 108, 130, 132, 143, 144, 145, 146, 149, 155, 157, 160, 166, 167, 169) obtained from the University of Washington School of Pharmacy Human Liver Tissue Bank (Seattle, WA) were used for the correlation studies (see below). Of those human liver tissues, 3 individual human liver tissue samples (HL-143, 155, 166) were used for chemical inhibition studies (see below). The collection and use of human liver tissues for research were approved by the University of Washington Investigational Review Board. Except for HL-132 (CYP2C9*1/*2), HL-144 (CYP2C9*1/*3), HL-157 (CYP2C9*1/*3), HL-160 (CYP2C9*1/*2), and HL-169 (CYP2C9*2/*2), all other subjects carried wild-type CYP2C9*1/*1 (unpublished data). Human liver microsomes were prepared as previously described (20). Briefly, human liver tissue was thawed in 10 mM potassium phosphate buffer (pH 7.4) containing 0.9% NaCl. Microsomes were prepared by homogenizing the thawed tissue in 5 volumes of 10 mM potassium phosphate buffer (pH 7.4) containing 0.25 M sucrose in a Waring blender, followed by five strokes with a glass Teflon homogenizer. All the thawing and homogenization were done on ice. The homogenate was centrifuged at 12,000 × g for 25 min and the supernatant were further centrifuged at 110,000 × g for 70 min. Microsomal protein was carefully separated from the glycogen pellet, resuspended in 10 mM potassium phosphate buffer (pH 7.4) containing 1 mM EDTA and 1.15% KCl, and again centrifuged at 110,000 × g for 70 min. The washed pellet was resuspended in either 50 mM potassium phosphate buffer (pH 7.4) or 100 mM Tris/HCl (pH 7.4) and stored at −80°C until use. Microsomal protein concentrations were determined by the Pierce BCA protein assay kit (Rockford, IL) using bovine serum albumin as standard.
Chemical inhibition of GLB depletion and metabolite formation in human liver microsomes
To identify CYP isoforms in human liver microsomes that can metabolize GLB, the well-established and selective inhibitors of these CYP enzymes were incubated, one at a time, with GLB in reaction mixtures. GLB metabolism was determined by measuring depletion of the drug. The following compounds 8-methoxypsoralen (MOP), triethylenethiophosphoramide (TTEPA), sulfaphenazole (SUL), quinidine (QUI), omeprazole (OMP), ketoconazole (KTZ), and montelukast (MTL) have been shown to selectively inhibit CYP2A6, CYP2B6, CYP2C9, CYP2D6, CYP2C19, CYP3A, and CYP2C8, respectively, and were used in the GLB depletion study (21–25). KTZ is a potent and well-studied inhibitor of CYP3A with a Ki value of 0.05 μM (24) and therefore is often used in vitro and in vivo as a CYP3A inhibitor. However, KTZ is only a selective CYP3A inhibitor that could also inhibit other CYP enzymes such as CYP2C9, but to a significantly lesser extent (26). SUL is perhaps the most potent and selective inhibitor of CYP2C9 with a Ki value of 0.12 μM (23, 27). OMP has been shown to be a more selective inhibitor for CYP2C19 (Ki = 3 μM) over CYP3A4 (Ki = 44 μM) (21). The concentrations of MOP, TTEPA, SUL, QUI, OMP, KTZ, and MTL used were 5 μM, 20 μM, 2.2 μM, 0.45 μM, 27 μM, 1 μM, and 4 μM, respectively. The concentrations were selected so that inhibition of each target CYP isoform was estimated to be greater than or equal to 90% as previously described (21–25). These inhibitors and GLB were dissolved in methanol, and the final concentration of methanol in reaction mixtures was <1% (v/v). No effects of the solvent on GLB depletion were observed at this concentration. All the experiments were performed at GLB concentrations lower than the apparent Km values of these enzymes (see below).
The reaction mixtures (a final volume of 0.2 ml) contained 100 mM sodium phosphate buffer (pH 7.4), human liver microsomes (0.5 mg of protein per ml), 5 mM MgCl2, and GLB at a final concentration of 1.25 nM, 0.125 μM or 1.25 μM. The reactions were carried out by incubation of the reaction mixtures at 37° C in a shaking water bath. After pre-warming for 5 min, reactions were initiated by adding a NADPH-regenerating system (1 mM NADP+, 10 mM glucose-6-phosphate, and 1 unit/ml glucose-6-phosphate dehydrogenase). In the negative controls, only the NADPH regenerating system or 1% (v/v) methanol, but without human liver microsome, was added. Reactions were stopped at 0 or 60 min by adding 2 ml of the mixed organic solvent (n-hexane/methylene chloride at a 1:1 ratio, v/v). Each sample was then acidified by the addition of 20 μl of 2 M HCl and 20 μl of a stock solution containing 2 μM GLP (internal standard). The samples were briefly vortexed at room temperature. The upper organic phase was transferred to a disposable clean glass tube for each sample and dried under N2. The dried residue from each sample was reconstituted in 100 μl of the solvent (methanol/H2O at a 30:70 ratio with 0.5 mM ammonium formate). After gentle vortexing, 15 μl of each reconstituted sample was injected for HPLC/MS analysis. In this chemical study, we found that GLB depletion was linear over time up to 60 min. Three different human liver microsome preparations were used in this study to enable statistical analysis. The three HLMs used were wild-type for CYP2C9 to eliminate possible effects of CYP2C9 mutations on GLB metabolism. The effects of inhibitors on metabolite formation were determined based on the peak area ratios of the metabolites (M1, M2b, M4, and M5) to the internal standard. Except for M1 and M2b, the standards of M4 and M5 were not available to us. The standards of M1 and M2b were used to identify the respective metabolites formed. With knowing the elution orders and the pattern of formation of the metabolites under the HPLC assay conditions we used (see below), the peaks for M4 and M5 can be identified as previously described (28). The peak area ratios of the metabolites with no inhibitors added were set as 100%. The formation of the metabolites (M1, M2b, M4, and M5) in the presence of inhibitors was then calculated as percentage of those with no inhibitors added. In the control experiments with only the inhibitors added, no peaks of the metabolites were detected.
Correlation between GLB depletion and the probe activity of individual CYP isoforms in human liver microsomes
To further confirm the results obtained from the above chemical inhibition study, we determined whether GLB depletion by human liver microsomes is correlated with the probe activity of individual CYP isoforms assessed using a probe substrate. First, GLB depletion was determined in a panel of liver microsomes prepared from 16 human subjects at a GLB concentration of 0.3 μM as described above. In separate experiments, to determine the probe activities of individual CYP isoforms, a simplified cocktail assay was conducted to measure the metabolite formation of probe substrates as follows (29). Two cocktails were used. Briefly, cocktail A containing 100 μM tolbutamide for CYP2C9 and 75 μM testosterone for CYP3A or cocktail B containing 4 μM amodiaquine for CYP2C8 and 20 μM omeprazole for CYP 2C19 was incubated with human liver microsomes, one at a time, at 37°C for 60 min in a final volume of 0.2 ml. All the compounds were dissolved in methanol and the final concentration of the solvent was less than 1% (v/v). The reactions were carried out by incubation of the reaction mixtures at 37° C in a shaking water bath. After pre-warming for 5 min, reactions were initiated by adding a NADPH-regenerating system (1 mM NADP+, 10 mM glucose-6-phosphate, and 1 unit/ml glucose-6-phosphate dehydrogenase). In the negative controls, only the NADPH regenerating system or 1% (v/v) methanol, but without human liver microsome, was added. Reactions were stopped at 60 min by adding 0.2 ml of ice-cold acetonitrile. The metabolite formation, namely testosterone 6β-hydroxylation by CYP3A, tolbutamide 4-methylhydroxylation by CYP2C9, omeprazole 5-hydroxylation by CYP2C19, or amodiaquine desethylation by CYP2C8, was determined by HPLC/MS as described below. GLB depletion was then plotted against the formation of CYP-specific metabolites. Correlation coefficients (r2) between GLB depletion and the formation of metabolites (as a measure of probe activities of individual CYP enzymes) were determined by a linear regression using the GraphPad software (GraphPad Software, Inc., San Diego, CA). Multiple correlation analysis was performed using the STATA software (StataCorp LP, College Station, TX).
Determination of kinetic parameters of recombinant human CYP isoforms for GLB depletion
To further compare the contribution of individual human CYP isoforms to GLB metabolism, we also determined kinetic parameters of individually expressed human CYP isoforms for GLB depletion. Briefly, the reaction mixtures (a final volume of 0.2 ml) contained 100 mM sodium phosphate buffer (pH 7.4), supersomes containing individually expressed human recombinant CYP isoform (CYP3A4 at 5 pmol/ml, CYP3A5 at 30 pmol/ml, CYP2C8 at 40 pmol/ml, CYP2C19 at 30 pmol/ml, CYP2C9*1/*1 at 40 pmol/ml, CYP2C9*2/*2 at 40 pmol/ml, or CYP2C9*3/*3 at 40 pmol/ml), 5 mM MgCl2, and GLB at various concentrations (0.05 – 15 μM). GLB was dissolved in methanol and the final concentration of methanol was 1% (v/v). Reactions were performed by incubation at 37° C in a shaking water bath. After pre-warming for 5 min, reactions were initiated by adding a NADPH-regenerating system (1 mM NADP+, 10 mM glucose-6-phosphate, and 1 unit/ml glucose-6-phosphate dehydrogenase). In the negative controls, only the NADPH regenerating system or 1% methanol, but without supersomes, was added. Reactions were stopped at 0, 10, 20, 30, or 60 min by the addition of 2 ml of the solvent (n-hexane:methylene chloride at a 1:1 ratio, v/v). Each sample was then acidified by adding 20 μl of 2 M HCl and 20 μl of a 2 μM glipibzide stock in acetonitrile (internal standard). The samples were briefly vortexed at room temperature. The upper organic phase was transferred to a disposable clean glass tube for each sample and dried under N2. The dried residue for each sample was reconstituted in 100 μl of the solvent (methanol: H2O at a 30:70 ratio with 0.5 mM ammonium formate). After gentle vortexing, 15 μl of each reconstituted sample was injected for HPLC/MS analysis.
HPLC/MS analysis
GLB concentrations and formation of GLB metabolites were determined using a validated HPLC/MS assay as previously described (30, 31). Briefly, HPLC/MS was performed on a Waters 2690 HPLC equipped with an auto-sampler and interfaced with a Micromass Platform LCZ 4000 single Quadrupole Mass Spectrometer (Waters, Milford, MA). Separation of GLB, its metabolites, and GLP was achieved using an Agilent Zorbax XDB-C8 analytical column (2.1 × 50 mm, 5 μm) equipped with a Phenomenex Security Guard C18 guard column (2.0 × 4 mm) with gradient elution. The mobile phase was composed of methanol and water at pH 6, both containing 0.5 mM ammonium formate. The flow rate was set at 0.25 ml/min. At time 0, the mobile phase was 30% (v/v) methanol and 70% (v/v) water. The concentration of methanol was increased linearly to 45% (v/v) over 1 min, held for 3 min, then increased linearly to 75% (v/v) over 1.5 min, held for 1 min, then further increased linearly to 90% (v/v) over 10 s, held for 3 min, and finally returned to the initial composition of 30% (v/v) methanol and 70% (v/v) water. The column was pre-equilibrated for 5.2 min, and the total run time was 15 min. The following parameters were set for optimum detection sensitivity: cone voltage 15 V, capillary voltage 3,500 V, desolvation temperature 350° C, and desolvation gas flow rate 10.6 L/min. Mass spectrometric data were collected between 2 and 15 min. At all other times, column elute was diverted to waste. Selected reaction monitoring was used to simultaneously monitor for analyte and internal standard. The following selected channels were used for the detection of analytes: 493.9 m/z for GLB, 532.0 m/z for GLP, and 446 m/z for internal standard.
The concentrations of metabolites formed from selective CYP probes (6β-hydroxy-testosterone, 4-methylhydroxy-tolbutamide, 5-hydroxyl-omeprazole, and desethyl-amodiaquine) were determined using a validated HPLC/MS assay as previously described (29) with minor modifications. Briefly, HPLC/MS was performed on a Waters 2690 HPLC equipped with an auto-sampler and interfaced with a Micromass Platform LCZ 4000 single Quadrupole Mass Spectrometer (Waters, Milford, MA). Separation of cocktail A (6β-hydroxy-testosterone, 4-methylhydroxy-tolbutamide, and midazolam as an internal standard) or cocktail B (5-hydroxy-omeprazole, desethyl-amodiaquine, and midazolam as an internal standard), was achieved using an Agilent Zorbax XDB-C8 analytical column (2.1 × 50 mm, 5 μm) equipped with a Phenomenex Security Guard C18 guard column (2.0 × 4 mm) with gradient elution. The mobile phase was composed of methanol and water containing 0.1% formic acid. The flow rate was set at 0.25 ml/min. At time 0, the mobile phase was 10% (v/v) methanol and 90% (v/v) water. The concentration of methanol was increased linearly to 30% (v/v) over 4 min, held for 2 min, then increased linearly to 75% (v/v) over 3 min, held for 4 min, and finally returned to the initial composition of 10% (v/v) methanol and 90% (v/v) water in 10 s. The column was pre-equilibrated for 4 min before each injection. The following parameters were set for optimum detection sensitivity: fragmentation voltage 25 V, capillary voltage 3,500 V, desolvation temperature 350°C, and desolvation gas flow rate 9.9 L/min. Mass spectrometric data were collected between 1 and 13 min. At all other times, column elute was diverted to waste. Selected reaction monitoring was used to simultaneously monitor analytes and internal standard. The following selected channels were used for the detection of analytes: cocktail A: 287.0 m/z for 4-methylhydroxy-tolbutamide, 305.0 m/z for 6β-hydroxy-testosterone, and 326.0 m/z for internal standard; cocktail B: 328.0 m/z for desethyl-amodiaquine, 362.0 m/z for 5-hydroxyl-omeprazole, and 326.0 m/z for internal standard.
Estimation of kinetic parameters for GLB depletion by recombinant CYP enzymes
In the GLB depletion experiments with recombinant human CYP isoforms, the ratios of GLB to GLP (internal standard) peak height were determined and normalized to the values obtained at time 0. Such ratios represent the amounts of GLB that remained undepleted. The profile of the log percentage of GLB remaining over time at each GLB concentration was used to determine the substrate depletion rate constant (kdep) according to the first-order decay function. We found that for all the recombinant human CYP isoforms examined, the log percentage of GLB remaining was linear over time up to 30 min. The Km values for GLB depletion were estimated by plotting the kdep values versus the GLB concentrations based on the following equation:
where [S] is the GLB concentration, kdep([S] ≈ 0) is the theoretical maximum depletion rate constant as substrate concentration approaches zero, and Km is the Michaelis-Menten constant (32). Finally, Clint was calculated based on the equation below:
where CYPconc is the concentration of the CYP enzyme added in the final volume of the reaction mixture. Vmax was calculated by multiplying Clint by the respective Km value.
Measurement of unbound fractions of GLB in supersome incubations
To evaluate the effect of protein binding of GLB on kinetic parameters, protein binding of GLB in supersome incubations was determined by ultracentrifugation as previously described with modifications (33). Experiments were performed using a Thermo Fisher Scientific SORVALL Discovery M150SE ultracentrifuge with a S100-AT3 rotor. Unbound fractions of GLB in supersome incubations were determined in triplicates. The GLB concentration range tested was 0.1 – 15 μM. Except for the NADPH-regenerating system, all other components in supersome incubations as described above were mixed. Two hundred μl of the mixtures were transferred to centrifuge tubes. After 5 min pre-incubation at 37° C, the mixtures were centrifuged at 400,000 × g for 140 min at 37° C. Twenty μl was then collected from each ultracentrifugation supernatant. To each collected ultracentrifugation supernatant, 2 ml of a mixed solvent (n-hexane/methylene chloride at a 1:1 ratio, v/v), 32 ng of GLP (internal standard in acetonitrile), and 20 μl of acetonitrile were added and mixed. The upper organic phase was transferred to a disposable clean glass tube and dried under N2. The dried residue from each sample was reconstituted in 100 μl of the solvent (methanol/H2O at a 30:70 ratio with 0.5 mM ammonium formate). After gentle vortexing, 15 μl of each reconstituted sample was injected for HPLC/MS analysis as described. The unbound fractions of GLB were calculated as the ratios of the free concentrations in ultracentrifugation supernatants to the total concentrations of GLB added which were also determined by HPLC/MS.
Statistical analysis
All data are presented as means ± S.D. from at least three determinations. Differences between two groups of data were compared using the two-sample Student’s t-test. Differences with p values < 0.05 were considered statistically significant.
Results
Chemical inhibition of GLB metabolism by human liver microsomes
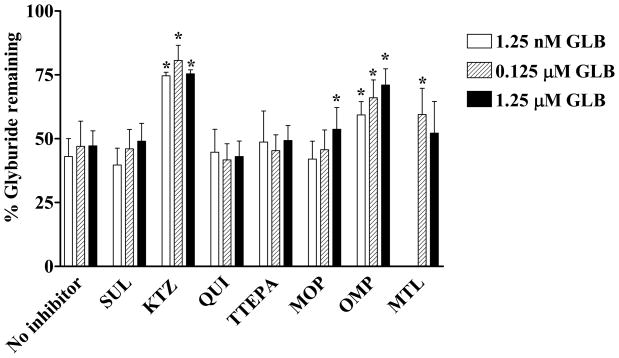
Inhibition of GLB metabolism in human liver microsomes at three different GLB concentrations (1.25 nM, 0.125 μM, and 1.25 μM) was carried out in the presence and absence of selective inhibitors of respective human CYP isoforms. Figure 1 shows the effects of various human CYP inhibitors on GLB depletion. The amount of GLB in samples in the absence of the NADPH regenerating system was set as 100%. When no inhibitor was added, approximately 40 – 50% of GLB remained undepleted. The addition of 1 μM ketoconazole, a relatively selective inhibitor for CYP3A, significantly increased the amount of remaining GLB to 75 – 85%, indicating that inhibition of CYP3A-mediated GLB depletion occurred. The addition of 27 μM omperazole, a selective inhibitor of CYP2C19, and 4 μM montelukast, a selective inhibitor of CYP2C8, also significantly increased the amount of remaining GLB to approximately 60 – 70% and 50 – 60%, respectively. The addition of 5 μM 8-methoxypsoralen, a selective inhibitor of CYP2A6, slightly but significantly increased the amount of remaining GLB only when GLB at 1.25 μM was incubated. The addition of other inhibitors had no significant effects on GLB depletion. Overall, these data suggest that CYP3A4 contributed approximately 50% to the total GLB depletion, wherease CYP2C19 and CYP2C8 together contributed 50%. Hence, CYP3A, CYP2C19, and CYP2C8 all appear to contribute to in vitro GLB metabolism in human liver microsomes, with CYP3A being the most effective enzyme.
Figure 1. Effects of human CYP isoform inhibitors on glyburide depletion in human liver microsomes.
GLB depletion was measured in the absence or presence of selective inhibitors of human CYP isoforms as described. SUL, KTZ, QUI, TTEPA, MOP, OMP, and MTL are relatively selective inhibitors for CYP2C9, CYP3A, CYP2D6, CYP2B6, CYP2A6, CYP2C19, and CYP2C8, respectively. The concentrations of SUL, KTZ, QUI, TTEPA, MOP, OMP, and MTL used were 2.2 μM, 1.0 μM, 0.45 μM, 20 μM, 5.0 μM, 27 μM, and 4.0 μM, respectively. Data shown are means ± S.D. of three independent experiments using three individual human liver microsomal preparations. The amounts of GLB remained undepleted in the negative controls with no NADPH regenerating system added were set as 100%. *Indicates the statistically significant differences with p values < 0.05 as compared with the 100% controls by the Student’s t-test.
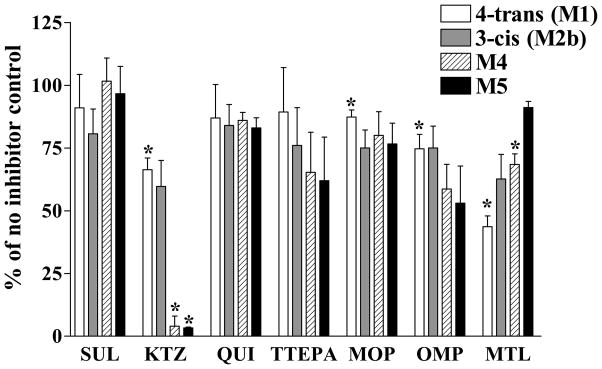
In the same experiments, the effects of these CYP inhibitors on the formation of GLB metabolites were also evaluated (Figure 2). The relative formation of the metabolites in reaction mixtures with no inhibitor added was set as 100%. The addition of ketoconazole significantly decreased the formation of the M1, M4, and M5 metabolites to approximately 60%, 5%, and 4% of the controls, respectively, with a particularly strong inhibition of the formation of M4 and M5. The addition of montelukast also significantly inhibited the formation of M1 and M4 to approximately 50% and 65% of the controls, respectively. The addition of omeprazole and 8-methoxypsoralen showed approximately 15 – 25% of significant inhibition for the formation of M1. Sulfaphenazole, quinidine, and thioTEPA had no significant effects on the formation of any of the four metabolites. Overall, these results are consistent with the data of chemical inhibition shown in Figure 1.
Figure 2. Effects of human CYP isoform inhibitors on the formation of glyburide metabolites in human liver microsomes.
Four metabolites of GLB were detected in the absence or presence of the CYP inhibitors as described. SUL, KTZ, QUI, TTEPA, MOP, OMP, and MTL are relatively selective inhibitors for CYP2C9, CYP3A, CYP2D6, CYP2B6, CYP2A6, CYP2C19, and CYP2C8, respectively. The concentrations of these inhibitors used were the same as described in Figure 1. The GLB concentration used in the experiments was 1.25 μM. The amount of metabolites formed in the reaction mixtures without any inhibitor added was set as 100%. Shown are means ± S.D. of three independent experiments with three individual human liver microsomal preparations. *Indicates the statistically significant differences with p values < 0.05 as compared with the 100% controls by the Student’s t-test.
Correlation between GLB depletion and individual CYP activities assessed using probe substrates
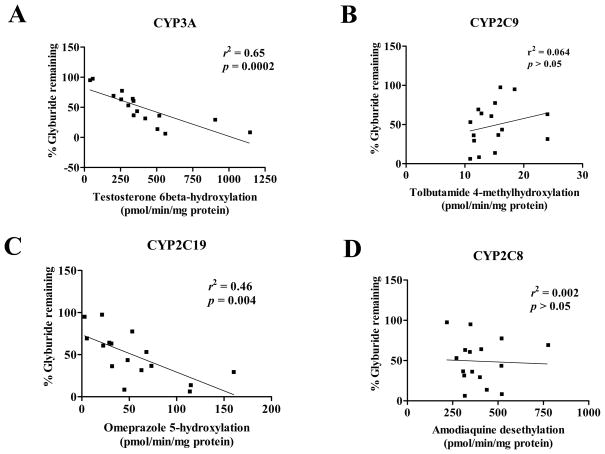
Results of the chemical inhibition study suggest that CYP3A appears to be a major player in the in vitro metabolism of GLB. If this is correct, we would expect to see a positive correlation between GLB depletion and the specific activity of CYP3A assessed using its probe substrate. Therefore, we performed the correlation study using human liver microsomes isolated from 16 Caucasian subjects. Single time-point determinations of testosterone-6β-hydroxylation, omeprazole-5-hydroxylation, amodiaquine-desethylation, and tolbutamide-4-hydroxlation were used as the marker activities of CYP3A, CYP2C19, CYP2C8, and CYP2C9, respectively, and we assumed that these activities were all within a linear range. As shown in Figure 3, there was a significant correlation between GLB depletion and testosterone 6β-hydroxylation for CYP3A (r2 = 0.65, p < 0.0001). GLB depletion also significantly correlated with omeprazole-5-hydroxylation for CYP2C19, but with a lower correlation coefficient (r2 = 0.46, p < 0.005). GLB depletion did not correlate with tolbutamide 4-methylhydroxylation nor amodiaquine-desethylation. Furthermore, multiple correlation regression analysis between GLB depletion and individual CYP isoform activities indicated that only CYP3A, but not CYP2C19, CYP2C8, and CYP2C9, significantly contributed to the overall correlation. We noted that there were 5 CYP2C9 variants among the 16 human liver tissue samples used. Although we did not find a significant difference in the selective CYP2C9 probe activity assessed using tolbutamide 4-methylhydroxylation between the wild-type and variant CYP2C9 groups, the trend was as expected with the variant CYP2C9 group (5 CYP2C9 variants combined) showing lower probe activities than the wild-type group (data not shown). GLB depletion did not correlate with tolbutamide 4-methylhydroxylation. The liver microsomal preparations from the two CYP2C9*1/*3 subjects that exhibited particularly poor CYP2C9 probe activities showed high GLB depletion and were found to have high CYP3A probe activities (data not shown). Overall, this correlation study again suggests that CYP3A is the major enzyme in in vitro metabolism of GLB in human liver microsomes.
Figure 3. Correlation between glyburide depletion and the activity of CYP3A, CYP2C19, CYP2C9, and CYP2C8 assessed by selective probe substrates.
GLB depletion and the CYP activities were determined by incubation for 60 min as described. Testosterone 6β-hydroxylation, omeprazole 5-hydroxylation, tolbutamide 4-methylhydrolation, or amodiaquine desethylation was analyzed as the marker activity of CYP3A, CYP2C19, CYP2C9, or CYP2C8, respectively. Shown are means of duplicated experiments with 16 individual human liver microsomal preparations. The correlation coefficients (r2) and p values of statistical analysis are indicated for each CYP isoform.
GLB depletion by recombinant human CYP isoforms
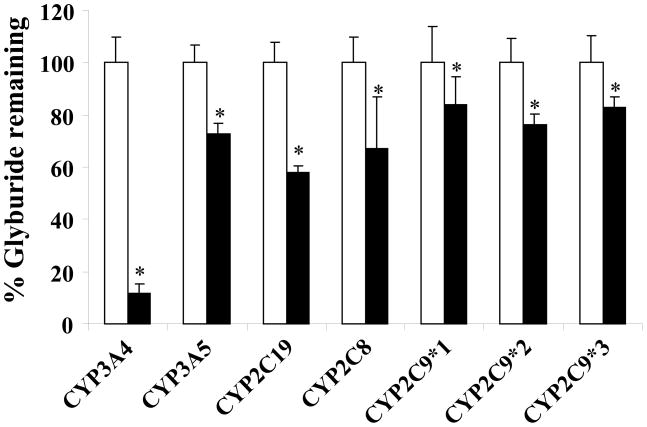
Since there are two adult CYP3A isoforms, CYP3A4 and CYP3A5, which share a very similar substrate specificity, to further establish which human CYP enzymes play a major role in GLB metabolism, we performed the GLB depletion study with individually expressed recombinant human CYP enzymes. In this study, we tested CYP3A4, CYP3A5, CYP2C8, CYP2C9, and CYP2C19 because only these human CYP isoforms were implicated in metabolizing GLB in the chemical inhibition study. We also examined the activities of two common CYP2C9 variants, CYP2C9*2/*2 and CYP2C9*3/*3. As shown in Figure 4, CYP3A4, CYP3A5, CYP2C19, CYP2C8, CYP2C9*1/*1, CYP2C9*2/*2, and CYP2C9*3/*3 were all capable of metabolizing GLB. Approximately 90% of GLB was depleted by CYP3A4 under the experimental conditions used, which was the most extensive among all the CYP enzymes tested. Other CYP enzymes significantly depleted approximately 20 – 40% of GLB. With respect to the extent of GLB depletion, the rank order was CYP3A4 > CYP2C19 > CYP2C8 and CYP3A5 > CYP2C9*1/*1, CYP2C9*2/*2, and CYP2C9*3/*3.
Figure 4. Glyburide depletion by individually expressed recombinant human CYP isoforms.
The amounts of CYP enzymes as shown in “Materials and Methods” were used to get sufficient depletion of GLB. The amounts of GLB remained undepleted in the negative controls (open bars) with no NADPH regenerating system added were set as 100%. Shown are means ± S.D. of three independent experiments. *Indicates the statistically significant differences with p values < 0.05 as compared with the 100% controls by the Students’ t-test.
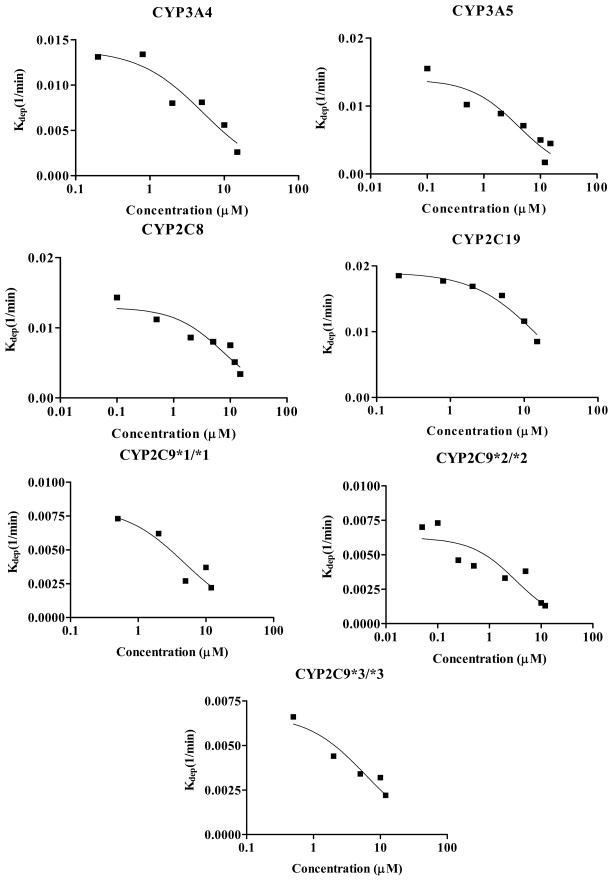
Kinetic parameters of recombinant CYP enzymes for GLB depletion
We next determined the kinetic parameters (Km and Vmax) and Clint of individual recombinant human CYP isoforms for GLB depletion by measuring the depletion rate constant (kdep) at various GLB concentrations. Such kinetic measurements are illustrated in Figure 5. The kinetic parameters obtained are shown in Table 1. Except for CYP2C19, the Km values of other CYP enzymes were comparable and at a low μM range. However, the Clint value of CYP3A4 was 4 17 times greater than that of other CYP isoforms. The rank order of the Clint values was CYP3A4 ≫ CYP2C19 and CYP3A5 > CYP2C8 > CYP2C9*1/*1, CYP2C9*2/*2, and CYP2C9*3/*3. Since CYP3A4 is the most abundant CYP isoform in the human liver, these kinetic data suggest that CYP3A4 is likely the major metabolic enzyme for GLB in vivo. Also, it appeared that there was no effect of CYP2C9 genotype on GLB depletion in vitro.
Figure 5. Determination of Km and kdep([s] ≈ 0) values of CYP enzymes for glyburide depletion.
Shown are determinations of Km and kdep([s] ≈ 0) for human CYP enzyme-mediated GLB depletion. GLB at six different concentrations (0.5, 1, 2.5, 5, 10, and 15 μM) was incubated with supersomes expressing individual human CYP isoforms (CYP3A4, CYP3A5, CYP2C8, CYP2C19, CYP2C9*1/*1, CYP2C9*2/*2, and CYP2C9*3/*3). Shown are means of triplicated experiments. Experimental details were described in “Materials and Methods”.
Table 1.
Kinetic parameters of individually expressed recombinant human CYP isoforms for glyburide depletion.
| CYP isoform | Km (μM) | Vmax (nmol/min/nmol CYP) | Clint (ml/min/nmol CYP) |
|---|---|---|---|
| CYP3A4 | 5.2 ± 1.8 | 14.4 ± 5.1 | 2.78 ± 0.26 |
| CYP3A5 | 4.2 ± 1.5 | 1.9 ± 0.73 | 0.46 ± 0.05 |
| CYP2C19 | 15.1 ± 2.3 | 9.6 ± 1.5 | 0.63 ± 0.02 |
| CYP2C8 | 7.7 ± 2.3 | 2.5 ± 0.8 | 0.32 ± 0.03 |
| CYP2C9*1/*1 | 4.7 ± 2.2 | 1.0 ± 0.5 | 0.20 ± 0.03 |
| CYP2C9*2/*2 | 3.2 ± 1.6 | 0.5 ± 0.3 | 0.16 ± 0.02 |
| CYP2C9*3/*3 | 6.0 ± 1.9 | 1.0 ± 0.3 | 0.17 ± 0.02 |
Km, Vmax, and Clint were determined using recombinant supersomes expressing individual human CYP isoforms as described in “Materials and Methods”. Such kinetic measurements are presented in Figure 5.
Discussion
In the present study, we systematically investigated the contributions of various human CYP isoforms to in vitro metabolism of GLB. There is a single nucleotide polymorphism linkage between CYP2C8*3 and CYP2C9*2, in which 96% of the Swedish subjects carrying CYP2C8*3 also possessed the CYP2C9*2 allele and 85% of the subjects with CYP2C9*2 also carried the CYP2C8*3 allele (34). We therefore included CYP2C8 in this study. Three GLB concentrations were used in the chemical inhibition study for two reasons. First, 1.25 nM and 0.125 μM are within the range of the free and total therapeutic plasma concentrations of GLB, respectively (35), and a peak plasma concentration of approximately 1.25 μM could be achieved after a single oral does of 20 mg GLB (36). Thus, the results obtained would have direct clinical implications. Second, the use of different GLB concentrations may provide us with information regarding whether the relative contributions of CYP isoforms to GLB metabolism are dependent on substrate concentration. The chemical inhibition study revealed that CYP3A, CYP2C19, and CYP2C8 were able to catalyze GLB metabolism with the rank order in catalytic activity of CYP3A > CYP2C19 > CYP2C8 (Figures 1 and 2). The activities of these enzymes were independent of GLB concentrations used, suggesting that these enzymes were not saturated at 1.25 μM, which is consistent with the Km values determined using recombinant human CYP isoforms (Table 1). We noted that the contribution of CYP3A to total GLB metabolism in human liver microsomes was approximately 50% as estimated from the chemical inhibition study. Next, multiple correlation regression anaysis revealed that CYP3A was the only CYP enzyme that showed significant correlation between GLB depletion and its marker activity in 16 individual human liver microsomal preparations. Hence, both the chemical inhibition and correlation studies support the notion that CYP3A plays a major role in the in vitro metabolism of GLB, although other CYP isoforms particularly CYP2C19 and CYP2C8 also contribute to GLB metabolism. Similar conclusions were obtained in the previous study using commercially available human liver microsomes (17). The contribution of CYP2C19 to GLB metabolism in vivo is expected to be low as this enzyme accounts for only a small fraction (~ 4%) of the total CYP content in human liver. A recent studyshowed by chemical and antibody inhibition analysis that, although CYP2C8 and CYP2C9 also substantially contributed to GLB metabolism in human liver microsomes, CYP3A contributed more than 50% (18). Hence, like what we have shown in this study, their data also indicate that CYP3A plays a greater role (>50%) in GLB metabolism than any other CYP isoforms. A major difference between this study and the work by Zharikova et al (18) is that we could not show the contribution of CYP2C9 to GLB metabolism to any significant extent by chemical inhibition of GLB metabolism in human liver microsomes. Nevertheless, our data are comparable to those reported by Naritomi et al (17). The reason for the above difference is not clear. We noted that the GLB concentrations used in chemical inhibition by Zharikova et al (18) were approximately the reported apparent Km values for GLB metabolism (5 – 11 μM) (5), which were much higher than the GLB concentrations used in this study. The concentration of the CYP2C9 inhibitor SUL used by Zharikova et al. was also much greater than that of this study (40 μM versus 2.2 μM). These differences in experimental conditions might affect the results. It also might be possible that the human liver tissues used by Zharikova et al. express higher levels of CYP2C9 than the human liver tissues used in this study and hence exhibited higher CYP2C9 metabolic activities for GLB. In addition to chemical inhibition, Zharikova et al (18) also used antibodies against individual CYP isoforms and showed that the antibodies against CYP2C9 inhibited the formation of GLB metabolites, supporting the contribution of CYP2C9 to the in vitro metabolism of GLB in human liver microsomes.
To further differentiate the contribution of CYP3A isoforms (CYP3A4 and CYP3A5) to GLB metabolism which has not been established in previous studies (17, 18), we performed GLB depletion with supersomes expressing individual human CYP isoforms. Since CYP2C9 polymorphism was shown to markedly affect the pharmacokinetics of GLB in humans (14–16), we included CYP2C9*1/*1, CYP2C9*2/*2, and CYP2C9*3/*3 to confirm their roles in GLB metabolism in vitro. We noted that the CYP enzymes tested were all able to catalyze GLB depletion; however, GLB depletion by CYP3A4 was the greatest (Figure 4). Except for CYP2C19, the Km values of other CYP enzymes were all within a low μM range (3 – 7 μM) (Table 1). The intrinsic Clint of CYP3A4 was much greater than that of any other CYP enzyme including CYP3A5. Thus, for the first time, we have shown that it is CYP3A4, but not CYP3A5, that plays a major role in in vitro metabolism of GLB, a conclusion that cannot be obtained in studies using human liver microsomes. The Km values obtained in this study are generally in agreement with those reported previously for CYP3A4, CYP2C8, and CYP2C19 except that others have reported an approximately 5-time lower Km value for CYP2C9*1/*1 (18). This difference could be caused by the different methods used in kinetic measurements (GLB depletion of this study versus metabolite formation of the study by Zharikova et al. (18)). Zharikova et al estimated the relative contributions of individual CYP enzymes to GLB metabolism using the kinetic parameters they obtained and the contribution of CYP2C9 was approximately 30% (18). If the kinetic parameters obtained in this study were used for the estimation, the contribution of CYP2C9 would be much less. Thus, our results of the kinetic measurements support the notion that GLB could be metabolized by CYP2C9, but to a much lesser extent than by CYP3A4. The Km or Clint values obtained in this study were not corrected for GLB protein binding. This was because we found that the unbound fractions of GLB were not significantly different in supersome incubations of different CYP isoforms and were independent of GLB concentrations in the range of 0.1 – 15 μM (data not shown). Hence, the comparison of the Km or Clint values between different CYP enzymes would not be affected no matter whether unbound or total concentrations of GLB were used for the calculations.
Taken together, we have further confirmed previous observations that human CYP3A4 is the major enzyme responsible for GLB metabolism in vitro (17, 18). Since CYP3A4 is the most abundant CYP isoform in human liver which accounts for approximately 50% of the total CYP content, it is reasonable to assume that CYP3A4 is likely the major metabolizer of GLB in vivo. In supersome incubations, we found that the metabolite M5 was predominantly formed by CYP3A4 and CYP3A5 (data not shown). If CYP3A4 is the major contributor to the metabolism of GLB in vivo, M5 should also be the major metabolite formed in vivo. Indeed, in the human liver microsome incubations, we noted that M5 was the major metabolite formed because the peak areas of M1, M2b, M3 and M4 were approximately 56, 43, 16, and 8% of that of M5, respectively (data not shown). Whether M5 is pharmacologically active is currently not known. It should be noted that the expression levels of CYP enzymes in human liver vary substantially, depending on the race, gender, or age. Moreover, many CYP enzymes are inducible by dietary constituents, drug intake, or other factors. Therefore, the contributions of CYP isoforms to GLB metabolism in vivo are expected to be highly variable. However, such an inter-individual variation does not override the general conclusion that CYP3A4 is a major player in GLB metabolism. Several clinical studies have shown that CYP2C9 polymorphism markedly affects the pharmacokinetics of GLB in vivo in humans (14–16). However, the data of the present study and those reported by others (17, 18) all support the notion that CYP3A, but not CYP2C9, plays a predominant role in GLB metabolism. Moreover, the activities of three CYP2C9 variants in GLB depletion are indistinguishable (Figure 4 and Table 1). At the present, we have no explanation for this discrepancy between in vitro and in vivo studies. Based on the Km values determined, none of the CYP isoforms is likely saturated at therapeutically relevant GLB concentrations in vivo. Hence, the effect of CYP2C9 polymorphism on the pharmacokinetics of GLB is unlikely attributable to a possible difference in GLB concentration. In contrast to previous clinical studies, we recently showed that the apparent oral clearance of GLB in pregnant women with gestational diabetes was independent of CYP2C9 genotype (4). Since CYP2C8 can also metabolize GLB in vitro (Figure 1 and Table 1) and CYP2C8 has been shown to have a single nucleotide polymorphism linkage with CYP2C9, this may partially explain the previous clinical observations with respect to the role of CYP2C9 in in vivo metabolism of GLB. More studies are needed to resolve this issue in future work. It would be important to know if there is a correlation between the clearance of GLB and in vivo marker activities of CYP3A4 in human subjects. Contributions of drug transporters such as P-glycoprotein, the breast cancer resistance protein (BCRP/ABCG2), and the organic anion transporter polypeptide 2B1(OATP2B1) and their linkage with CYP2C9 variants should also be considered in future work.
In conclusion, in the present study, we have shown that CYP3A4 is a major contributor to in vitro metabolism of GLB. This finding is consistent with the observation that the oral clearance of GLB in pregnant women with gestational diabetes is significantly increased compared with non-pregnant controls (4). This is because we have shown that CYP3A activity is induced by pregnancy in vivo (37). The contribution of CYP3A to GLB metabolism could be more pronounced in pregnancy than in non-pregnant women owing to pregnancy-induced activity of CYP3A. Therefore, we hypothesize that the increased oral clearance of GLB in pregnancy is caused by the induction of CYP3A4, but not CYP2C9. Consistent with this hypothesis, we have shown that the CYP2C9 polymorphism does not affect the pharmacokinetics of GLB in pregnant women (4). CYP2C19 also seems unlikely to be involved because CYP2C19 activity appears to be decreased during pregnancy (38, 39). Identification of CYP3A4 as the major metabolizer of GLB may lead to valuable prediction of potential interactions with GLB and other drugs that are CYP3A4 inhibitors or inducers, and will help design more efficacious and safer pharmacotherapy for both pregnant and non-pregnant patients with diabetes mellitus receiving GLB treatment.
Supplementary Material
Acknowledgments
The authors thank Astra/Zeneca for providing us with 5-hydroxy-omeprazole. We thank Drs. Zhanglin Ni and Xiaokun Cai (Department of Pharmaceutics, the University of Washington) for technical assistance in the glyburide metabolism studies. We are grateful to Dr. Mahmoud S. Ahmed (Department of Obstetrics & Gynecology, University of Texas Medical Branch, Galveston, TX) for providing the standards of the M1 and M2b metabolites of glyburide.
Abbreviations
- CYP
cytochrome P450
- GLB
glyburide
- GLP
glipizide
- AUC
area under the concentration-time curve
- MOP
8-methoxypsoralen
- TTEPA
triethylenethiophosphoramide
- OMP
omeprazole
- SUL
sulfaphenazole
- QUI
quinidine
- KTZ
ketoconazole
- MTL
montelukast
- HPLC/MS
High performance liquid chromatography mass spectrometry
- M1
4-trans-hydroxycyclohexyl glyburide
- M2a
4-cis-hydroxycyclohexyl glyburide (axial)
- M2b
3-cis-hydroxycyclohexyl glyburide (axial)
- M3
3-trans-hydroxycyclohexyl glyburide
- M4
2-trans-hydroxycyclohexyl glyburide
- M5
an ethyl hydroxycyclohexyl glyburide
Footnotes
This work was supported by the grant U10HD047892 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) through support of the Obstetric-Fetal Pharmacology Research Unit Network grant, and the NICHD grant P50HD044404. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development. L Zhou was the recipient of the William E. Bradley Endowed Fellowship from School of Pharmacy, University of Washington.
References
- 1.Feldman JM. Glyburide: a second-generation sulfonylurea hypoglycemic agent. History, chemistry, metabolism, pharmacokinetics, clinical use and adverse effects. Pharmacotherapy. 1985;5:43–62. doi: 10.1002/j.1875-9114.1985.tb03404.x. [DOI] [PubMed] [Google Scholar]
- 2.Langer O, Conway DL, Berkus MD, Xenakis EM, Gonzales O. A comparison of glyburide and insulin in women with gestational diabetes mellitus. N Engl J Med. 2000;343:1134–8. doi: 10.1056/NEJM200010193431601. [DOI] [PubMed] [Google Scholar]
- 3.Moore TR. Glyburide for the treatment of gestational diabetes. A critical appraisal. Diabetes Care. 2007;30(Suppl 2):S209–13. doi: 10.2337/dc07-s218. [DOI] [PubMed] [Google Scholar]
- 4.Hebert MF, Ma X, Naraharisetti SB, Krudys KM, Umans JG, Hankins GD, Caritis SN, Miodovnik M, Mattison DR, Unadkat JD, Kelly EJ, Blough D, Cobelli C, Ahmed MS, Snodgrass WR, Carr DB, Easterling TR, Vicini P. Are we optimizing gestational diabetes treatment with glyburide? The pharmacologic basis for better clinical practice. Clin Pharmacol Ther. 2009;85:607–14. doi: 10.1038/clpt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zharikova OL, Ravindran S, Nanovskaya TN, Hill RA, Hankins GD, Ahmed MS. Kinetics of glyburide metabolism by hepatic and placental microsomes of human and baboon. Biochem Pharmacol. 2007;73:2012–9. doi: 10.1016/j.bcp.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Rydberg T, Jonsson A, Roder M, Melander A. Hypoglycemic activity of glyburide (glibenclamide) metabolites in humans. Diabetes Care. 1994;17:1026–30. doi: 10.2337/diacare.17.9.1026. [DOI] [PubMed] [Google Scholar]
- 7.Hellman B, Sehlin J, Taljedal IB. Glibenclamide is exceptional among hypoglycaemic sulphonylureas in accumulating progressively in beta-cell-rich pancreatic islets. Acta Endocrinol (Copenh) 1984;105:385–90. doi: 10.1530/acta.0.1050385. [DOI] [PubMed] [Google Scholar]
- 8.Sartor G, Melander A, Schersten B, Wahlin-Boll E. Serum glibenclamide in diabetic patients, and influence of food on the kinetics and effects of glibenclamide. Diabetologia. 1980;18:17–22. doi: 10.1007/BF01228296. [DOI] [PubMed] [Google Scholar]
- 9.Huupponen R, Viikari J, Saarimaa H. Chlorpropamide and glibenclamide serum concentrations in hospitalized patients. Ann Clin Res. 1982;14:119–22. [PubMed] [Google Scholar]
- 10.Matsuda A, Kuzuya T, Sugita Y, Kawashima K. Plasma levels of glibenclamide in diabetic patients during its routine clinical administration determined by a specific radioimmunoassay. Horm Metab Res. 1983;15:425–8. doi: 10.1055/s-2007-1018746. [DOI] [PubMed] [Google Scholar]
- 11.Ferner RE, Chaplin S. The relationship between the pharmacokinetics and pharmacodynamic effects of oral hypoglycaemic drugs. Clin Pharmacokinet. 1987;12:379–401. doi: 10.2165/00003088-198712060-00001. [DOI] [PubMed] [Google Scholar]
- 12.Marchetti P, Navalesi R. Pharmacokinetic-pharmacodynamic relationships of oral hypoglycaemic agents. An update. Clin Pharmacokinet. 1989;16:100–28. doi: 10.2165/00003088-198916020-00004. [DOI] [PubMed] [Google Scholar]
- 13.Cavallari LH, Limdi NA. Warfarin pharmacogenomics. Curr Opin Mol Ther. 2009;11:243–51. [PubMed] [Google Scholar]
- 14.Kirchheiner J, Brockmoller J, Meineke I, Bauer S, Rohde W, Meisel C, Roots I. Impact of CYP2C9 amino acid polymorphisms on glyburide kinetics and on the insulin and glucose response in healthy volunteers. Clin Pharmacol Ther. 2002;71:286–96. doi: 10.1067/mcp.2002.122476. [DOI] [PubMed] [Google Scholar]
- 15.Niemi M, Cascorbi I, Timm R, Kroemer HK, Neuvonen PJ, Kivisto KT. Glyburide and glimepiride pharmacokinetics in subjects with different CYP2C9 genotypes. Clin Pharmacol Ther. 2002;72:326–32. doi: 10.1067/mcp.2002.127495. [DOI] [PubMed] [Google Scholar]
- 16.Yin OQ, Tomlinson B, Chow MS. CYP2C9, but not CYP2C19, polymorphisms affect the pharmacokinetics and pharmacodynamics of glyburide in Chinese subjects. Clin Pharmacol Ther. 2005;78:370–7. doi: 10.1016/j.clpt.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Naritomi Y, Terashita S, Kagayama A. Identification and relative contributions of human cytochrome P450 isoforms involved in the metabolism of glibenclamide and lansoprazole: evaluation of an approach based on the in vitro substrate disappearance rate. Xenobiotica. 2004;34:415–27. doi: 10.1080/00498250410001685728. [DOI] [PubMed] [Google Scholar]
- 18.Zharikova OL, Fokina VM, Nanovskaya TN, Hill RA, Mattison DR, Hankins GD, Ahmed MS. Identification of the major human hepatic and placental enzymes responsible for the biotransformation of glyburide. Biochem Pharmacol. 2009;78:1483–90. doi: 10.1016/j.bcp.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill RA, Rudra S, Peng B, Roane DS, Bounds JK, Zhang Y, Adloo A, Lu T. Hydroxyl-substituted sulfonylureas as potent inhibitors of specific [3H]glyburide binding to rat brain synaptosomes. Bioorg Med Chem. 2003;11:2099–113. doi: 10.1016/s0968-0896(02)00606-5. [DOI] [PubMed] [Google Scholar]
- 20.Thummel KE, Kharasch ED, Podoll T, Kunze K. Human liver microsomal enflurane defluorination catalyzed by cytochrome P-450 2E1. Drug Metab Dispos. 1993;21:350–7. [PubMed] [Google Scholar]
- 21.Funck-Brentano C, Becquemont L, Lenevu A, Roux A, Jaillon P, Beaune P. Inhibition by omeprazole of proguanil metabolism: mechanism of the interaction in vitro and prediction of in vivo results from the in vitro experiments. J Pharmacol Exp Ther. 1997;280:730–8. [PubMed] [Google Scholar]
- 22.Walsky RL, Obach RS, Gaman EA, Gleeson JP, Proctor WR. Selective inhibition of human cytochrome P4502C8 by montelukast. Drug Metab Dispos. 2005;33:413–8. doi: 10.1124/dmd.104.002766. [DOI] [PubMed] [Google Scholar]
- 23.Hickman D, Wang JP, Wang Y, Unadkat JD. Evaluation of the selectivity of In vitro probes and suitability of organic solvents for the measurement of human cytochrome P450 monooxygenase activities. Drug Metab Dispos. 1998;26:207–15. [PubMed] [Google Scholar]
- 24.Wandel C, Kim RB, Guengerich FP, Wood AJ. Mibefradil is a P-glycoprotein substrate and a potent inhibitor of both P-glycoprotein and CYP3A in vitro. Drug Metab Dispos. 2000;28:895–8. [PubMed] [Google Scholar]
- 25.Rae JM, Soukhova NV, Flockhart DA, Desta Z. Triethylenethiophosphoramide is a specific inhibitor of cytochrome P450 2B6: implications for cyclophosphamide metabolism. Drug Metab Dispos. 2002;30:525–30. doi: 10.1124/dmd.30.5.525. [DOI] [PubMed] [Google Scholar]
- 26.Sai Y, Dai R, Yang TJ, Krausz KW, Gonzalez FJ, Gelboin HV, Shou M. Assessment of specificity of eight chemical inhibitors using cDNA-expressed cytochromes P450. Xenobiotica. 2000;30:327–43. doi: 10.1080/004982500237541. [DOI] [PubMed] [Google Scholar]
- 27.Baldwin SJ, Bloomer JC, Smith GJ, Ayrton AD, Clarke SE, Chenery RJ. Ketoconazole and sulphaphenazole as the respective selective inhibitors of P4503A and 2C9. Xenobiotica. 1995;25:261–70. doi: 10.3109/00498259509061850. [DOI] [PubMed] [Google Scholar]
- 28.Ravindran S, Zharikova OL, Hill RA, Nanovskaya TN, Hankins GD, Ahmed MS. Identification of glyburide metabolites formed by hepatic and placental microsomes of humans and baboons. Biochem Pharmacol. 2006;72:1730–7. doi: 10.1016/j.bcp.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 29.Dixit V, Hariparsad N, Desai P, Unadkat JD. In vitro LC-MS cocktail assays to simultaneously determine human cytochrome P450 activities. Biopharm Drug Dispos. 2007;28:257–62. doi: 10.1002/bdd.552. [DOI] [PubMed] [Google Scholar]
- 30.Zhou L, Naraharisetti SB, Wang H, Unadkat JD, Hebert MF, Mao Q. The breast cancer resistance protein (Bcrp1/Abcg2) limits fetal distribution of glyburide in the pregnant mouse: an Obstetric-Fetal Pharmacology Research Unit Network and University of Washington Specialized Center of Research Study. Mol Pharmacol. 2008;73:949–59. doi: 10.1124/mol.107.041616. [DOI] [PubMed] [Google Scholar]
- 31.Naraharisetti SB, Kirby BJ, Hebert MF, Easterling TR, Unadkat JD. Validation of a sensitive LC-MS assay for quantification of glyburide and its metabolite 4-transhydroxy glyburide in plasma and urine: an OPRU Network study. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;860:34–41. doi: 10.1016/j.jchromb.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obach RS, Reed-Hagen AE. Measurement of Michaelis constants for cytochrome P450-mediated biotransformation reactions using a substrate depletion approach. Drug Metab Dispos. 2002;30:831–7. doi: 10.1124/dmd.30.7.831. [DOI] [PubMed] [Google Scholar]
- 33.Nakai D, Kumamoto K, Sakikawa C, Kosaka T, Tokui T. Evaluation of the protein binding ratio of drugs by a micro-scale ultracentrifugation method. J Pharm Sci. 2004;93:847–54. doi: 10.1002/jps.20012. [DOI] [PubMed] [Google Scholar]
- 34.Yasar U, Lundgren S, Eliasson E, Bennet A, Wiman B, de Faire U, Rane A. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem Biophys Res Commun. 2002;299:25–8. doi: 10.1016/s0006-291x(02)02592-5. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson A, Rydberg T, Ekberg G, Hallengren B, Melander A. Slow elimination of glyburide in NIDDM subjects. Diabetes Care. 1994;17:142–5. doi: 10.2337/diacare.17.2.142. [DOI] [PubMed] [Google Scholar]
- 36.Coppack SW, Lant AF, McIntosh CS, Rodgers AV. Pharmacokinetic and pharmacodynamic studies of glibenclamide in non-insulin dependent diabetes mellitus. Br J Clin Pharmacol. 1990;29:673–84. doi: 10.1111/j.1365-2125.1990.tb03688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hebert MF, Easterling TR, Kirby B, Carr DB, Buchanan ML, Rutherford T, Thummel KE, Fishbein DP, Unadkat JD. Effects of pregnancy on CYP3A and P-glycoprotein activities as measured by disposition of midazolam and digoxin: a University of Washington specialized center of research study. Clin Pharmacol Ther. 2008;84:248–53. doi: 10.1038/clpt.2008.1. [DOI] [PubMed] [Google Scholar]
- 38.Na-Bangchang K, Manyando C, Ruengweerayut R, Kioy D, Mulenga M, Miller GB, Konsil J. The pharmacokinetics and pharmacodynamics of atovaquone and proguanil for the treatment of uncomplicated falciparum malaria in third-trimester pregnant women. Eur J Clin Pharmacol. 2005;61:573–82. doi: 10.1007/s00228-005-0969-7. [DOI] [PubMed] [Google Scholar]
- 39.McGready R, Stepniewska K, Seaton E, Cho T, Cho D, Ginsberg A, Edstein MD, Ashley E, Looareesuwan S, White NJ, Nosten F. Pregnancy and use of oral contraceptives reduces the biotransformation of proguanil to cycloguanil. Eur J Clin Pharmacol. 2003;59:553–7. doi: 10.1007/s00228-003-0651-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.