Summary
Many anti-infectives inhibit the synthesis of bacterial proteins, but none selectively inhibits their degradation. Most anti-infectives kill replicating pathogens, but few preferentially kill pathogens that have been forced into a non-replicating state by conditions in the host. To explore these alternative approaches we sought selective inhibitors of the proteasome of Mycobacterium tuberculosis (Mtb). Given that proteasome structure is extensively conserved, it is not surprising that inhibitors of all chemical classes tested have blocked both eukaryotic and prokaryotic proteasomes, and no inhibitor has proved substantially more potent on proteasomes of pathogens than of their hosts. Here we show that certain oxathiazol-2-ones kill non-replicating Mtb and act as selective suicide-substrate inhibitors of the Mtb proteasome by cyclo-carbonylating its active site threonine. Major conformational changes protect the inhibitor-enzyme intermediate from hydrolysis, allowing formation of an oxazolidin-2-one and preventing regeneration of active protease. Residues outside the active site whose H-bonds stabilize the critical loop before and after it moves are extensively non-conserved. This may account for the ability of oxathiazol-2-ones to inhibit the mycobacterial proteasome potently and irreversibly while largely sparing the human homolog.
Proteasomes are structurally conserved from archaea to eukaryotes and are essential in eukaryotes1. The cytotoxicity of proteasome inhibitors has been exploited for cancer therapy2 and suggested for the treatment of infections by eukaryotic pathogens, such as plasmodia and trypanosomes3. Unfortunately, the inherent toxicity of proteasome inhibitors is a drawback in the treatment of curable infections. Inhibitors that act with comparable potency on human proteasomes and proteasomes of infectious agents have not entered clinical practice as anti-infectives.
The only known bacterial pathogens with proteasomes are mycobacteria4. The tuberculosis pandemic has been declared a global health emergency as Mtb’s growing resistance to antibiotics5 coincides with the spread of risk factors such as HIV/AIDS and diabetes6,7. Proteasomes degrade proteins that serve in signaling during adaptation; have become irreparably oxidized; or are scavenged during starvation. Mtb’s proteasome is required for degradation of certain proteins8,9, for Mtb to survive nitroxidative stress in vitro4 and for Mtb to persist in mice10. The latter observation validates the Mtb proteasome as a drug target. However, all known proteasome inhibitors tested have inhibited mammalian proteasomes more potently than those of Mtb, including peptidyl epoxyketones, peptidyl aldehydes, γ-lactam-β-lactones such as salinosporamide A (NPI0052)11,12 and the peptidyl boronate bortezomib (Velcade®), which is in clinical use2. An effort to exploit the substrate preferences of the Mtb proteasome led to bortezomib analogues with varying amino acids at P1, the most selective of which inhibited the Mtb proteasome only 8-fold more potently than a mammalian proteasome13.
Nonetheless, biochemical and structural differences between proteasomes from Mtb and mammals encouraged us to seek species-selective proteasome inhibitors. In eukaryotic proteasomes the 7 types of β subunits forming the two heteroheptameric inner rings of the core particle14 include three proteases with distinct specificities for oligopeptides. The Mtb proteasome has one type of β subunit15,16, but it is active against diverse benzyloxycarbonyl-capped tripeptides15,16, perhaps because the side chains lining the active site of the Mtb proteasome have physicochemical properties that are a composite of those contributing to the 3 distinct active sites in eukaryotes15,16. Until now, residues so distant from the active site as to play no role in binding of substrate were not considered germane to the species selectivity of inhibitors, including those listed above, that react with the catalytic hydroxyl of the N-terminal threonine (Thr1).
Identification of mycobactericidal, non-cytotoxic proteasome inhibitors
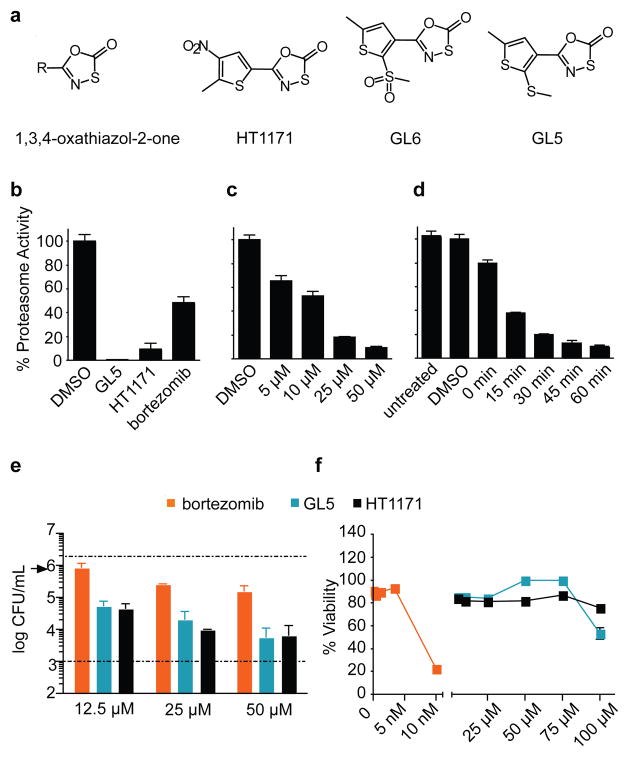
We expressed Mtb proteasome from the genes encoding proteasome components A (for α chain) and B (for β chain) (PrcBA) with and without the N-terminal octapeptide deleted from the α chains. The latter “open-gate” mutant (Mtb20SOG) is thought to mimic a physiologic mechanism for gate opening and has a higher specific activity than the wild type without a change in substrate preference13,15,16. By following Mtb20SOG’s hydrolysis of Suc-LLVY-7-amido-4-methylcoumarin (AMC), we screened 20,000 compounds (Supplementary Information) and identified two inhibitors, 5-(5-methyl-2-(methylthio)thiophen-3-yl)-1,3,4-oxathiazol-2-one (GL5) and 5-(2-methyl-3-nitrothiophen-2-yl)-1,3,4-oxathiazol-2-one (HT1171), as well as an inactive congener, 5-(5-methyl-2-(methylsulfonyl)thiophen-3-yl)-1,3,4-oxathiazol-2-one (GL6) (Fig. 1a).
Figure 1. Oxathiazol-2-ones inhibit mycobacterial proteasomes and kill non-replicating Mtb.
(a) Compound structures. (b) Inhibition of proteasomes in intact BCG (OD580nm 0.6 - 1) by GL5, HT1171 or bortezomib (each 50 μM) after 4 hours. Proteasome activity in lysates was tested with Ac-YQW-AMC (50 μM) as substrate. (c) Concentration-response for GL5 after 1 hour exposure as in (b). Control, DMSO (<1% vol/vol) alone. (d) Time-course for effect of GL5 (50 μM) as in (b) except that washing began at the times indicated. “Untreated” and “DMSO” cells were handled in the same manner but received nothing or DMSO at time 0 and were lysed at 60 min. “T0” cells were treated and washed immediately. (e) Killing of Mtb Erdman in Sauton’s medium with time 0 addition of 50 μM DETA-NO, a nitric oxide donor (t1/2, ~20 h), mimicking the nitroxidative stress that limits Mtb’s replication in mice23. Upper dashed line, CFU/ml after exposure to DMSO control. Lower dashed line, limit of detection. Arrow, initial inoculum. (f) Monkey kidney epithelial cells (Vero76) were incubated with compounds for 4 days before MTS assay for viability; microscopy gave concordant results. Data are means ± SD of triplicates in one of at least two experiments. Some error bars fall within the symbols.
Like bortezomib, oxathiazol-2-ones were able to cross Mtb’s cell wall, insofar as GL5 and HT1171 inhibited proteasome activity upon treatment of Mycobacterium bovis var. BCG (Fig. 1b). At 50 μM, GL5 and HT1171 inhibited ~90% of mycobacterial proteasome activity, while bortezomib (50μM) inhibited 52%. Exposure of BCG to 25 μM GL5 for 4 hours led to >80% reduction of proteasome activity (Fig. 1c), while as little as ~15 min exposure to GL5 at 50 μM was sufficient to reduce activity by ~50% (Fig. 1d).
Moreover, GL5 killed BCG alone and in synergy with sub-bacteriostatic fluxes of nitric oxide arising from the decomposition of 2, 2-(hydroxynitrosohydrazino)-bis-ethanamine (DETA-NO) (Fig. S1). GL5 and HT1171 also dose-dependently killed 1.5-2.5 log10 Mtb over 4 days in synergy with sufficient nitric oxide to induce a pathophysiologically relevant state of bacterial non-replication17 (Fig. 1e). Bortezomib was less mycobactericidal (Fig. 1e), and it was toxic to monkey epithelial cells (Fig. 1f) and human macrophages (Fig. S2). In contrast, GL5 and HT1171 showed no apparent toxicity to mammalian cells (Fig. 1f; Fig S2) at concentrations up to 75 μM, 3000-fold greater than those at which bortezomib destroyed the epithelial cells. The oxathiazol-2-ones exerted no antibacterial activity against Mycobacterium avium-intracellulare, Staphylococcus aureus, Salmonella enterica var. Typhimurium or Pseudomonas aeruginosa (data not shown). Although some oxathiazol-2-ones were reported to react with thiols18, those studied here did not inhibit the thiol-dependent cathepsin B (data not shown). Moreover, 11 of 23 oxathiazol-2-ones tested were <5% reactive with glutathione; the others reacted to a limited degree (Table S1). Thus, at a functional level, the oxathiazol-2-ones tested here appear to be relatively selective and nontoxic, although they may have additional targets.
Selective inhibition of mycobacterial proteasomes
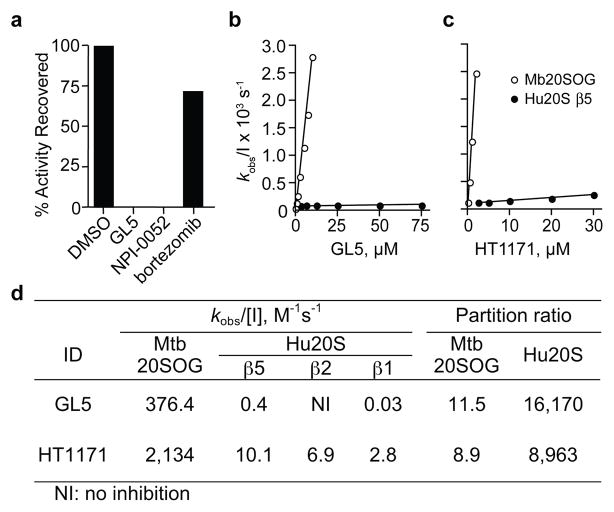
The different impact of oxathiazol-2-ones on Mtb and mammalian cells prompted us to ask if these compounds differentially inhibit isolated mycobacterial and human proteasomes. In dialysis (Fig. 2a) and kinetic studies (Fig. 2b, c), oxathiazol-2-ones inhibited Mtb proteasomes irreversibly (Fig. 2a), while inhibition of human proteasome β1, β2 and β5 sites was so minimal (Fig. 2b, c; Table S1) as to preclude definition of a mode of inhibition. After establishing that oxathiazol-2-ones spontaneously hydrolyze in tissue culture medium to the corresponding amide with t1/2’s ranging from 7 to 180 minutes (Table S2), we used partition ratios (the ratios of rate constants for catalysis and inactivation) to assess their relative potency19. By this measure, GL5 and HT1171 were >1000-fold more effective against Mtb proteasomes than human proteasomes (Fig. 2d; Fig. S3a, b). Inhibition was competitive with the substrate benzyloxycarbonyl-valylleucylarginyl-AMC (Z-VLR-AMC) (Fig. S3c, d), suggesting that the inhibitor binds at or near the active site, and was time-dependent (Fig. S4). Potencies of all 21 oxathiazol-2-ones tested against wild type Mtb proteasome correlated closely (r2 = 0.82) with their potencies against the open-gate form (Fig. S5). Oxathiazol-2-one-treated Mtb proteasomes lost the ability to degrade not only oligopeptides, but also a protein substrate, β-casein (Fig. S6). In contrast, oxathiazol-2-ones were inactive or very weak inhibitors of trypsin (IC50 >50 μM), cathepsin B (IC50 >50 μM), matrix metalloproteinase-2 (IC50 >100 μM) and mycobacterial blaC-encoded®-lactamase, a serine protease-like enzyme (IC50 >100 μM). Although GL5 reversibly inhibited α-chymotrypsin with Ki 64 nM, anti-chymotryptic potency was minimal in 13 other oxathiazol-2-ones tested (Table S4). Table S1 summarizes biochemical and mycobactericidal properties of 24 oxathiazol-2-ones, including 21 we synthesized as described in Supplemental Information.
Figure 2. Kinetic analysis of inactivation of Mtb 20SOG and human proteasomes (Hu20S) by oxathiazol-2-ones.
(a) Mtb20SOG (0.23 nM) was pre-treated with DMSO, GL5 (5 μM), salinosporamide A (NPI-0052, 10 nM), or bortezomib (2.7 μM) for 3 hours and assayed, then dialyzed against assay buffer overnight and assayed again. (b), (c) Plots of kobs as function of inhibitor concentration for GL5 (b) and HT1171 (c). Values for kobs, derived from the data in Fig S4, were plotted against inhibitor concentration [I]. (d) Kinetic parameters and partition ratios of GL5 and HT1171, determined as in Figs. S3 and S4. Kobs/[I] is second-order rate constant of inactivation. Partition ratios for human proteasomes refer to the β5 subunit. Standard errors were <10%.
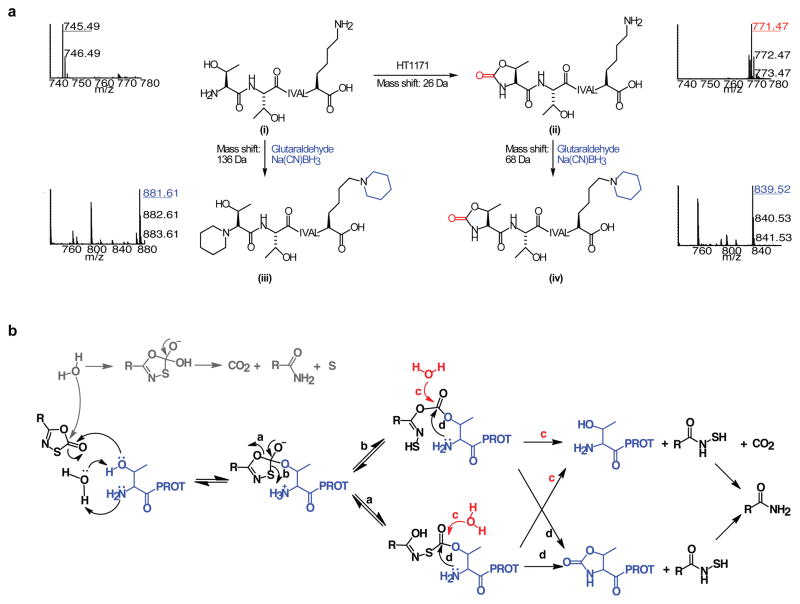
Competitive, irreversible, mechanism-based inhibition suggested that the oxathiazol-2-ones inactivate the Mtb proteasome by covalent attack on the active site Thr1. To test this, we trypsinized Mtb proteasomes that had been treated or not with HT1171. Results were identical for both wild type and open-gate forms. LC-MS/MS identified peptides representing 98% of the β subunit. Only one peptide ion was unique to the Mtb proteasome β subunit treated with HT1171 (Fig. 3a). Its mass was 26 Da higher than that of the N-terminal heptapeptide (TTIVALK) identified only in the untreated samples (Figs. 3a and S7a, b), suggesting the addition of a carbonyl at the expense of two hydrogen atoms (i→ii in Fig. 3a). MS/MS analysis of this modified peptide (Fig. S7a) indicated that the modification was on one of the first two residues. To determine if the N-terminal residue was modified, the tryptic peptides from untreated and treated Mtb proteasome β subunits were subjected to reductive glutaraldehydation, which modifies amino groups of proteins and peptides20. Subjecting the trypsinized oligopeptides to reductive glutaraldehydation without prior exposure to oxathiazol-2-one increased the mass of the N-terminal heptapeptide by 136.13 Da, consistent with modification of two primary amines, those of Thr1 and Lys7 (Fig. 3a [i→iii] and Fig. S7c). In contrast, applying the same procedure to proteasomes that had been pre-treated with HT1171 increased the mass of the same peptide by only 68.06 Da (Fig. 3a [ii→iv] and Fig. S7d), indicating that one primary amine was no longer available for reductive alkylation. MS/MS analysis (Fig. S7d) of this mono-alkylated peptide showed that only the lysine was modified, confirming that the N-terminal Thr was modified upon HT1171 treatment. The mass spectrometric results were the same when HT1171 was replaced by GL5 or GL3 and when the proteasome was wild type or open-gate (not shown). These findings support the mechanism of inhibition in Fig. 3b: the attack of the oxathiazol-2-one by the OH of Thr1 forms a carbonated or carbonothioated enzyme intermediate on Thr1 that can either undergo hydrolysis to reactivate the enzyme, or donate a carbonyl to Thr1 that links its α-amino and γ-hydroxyl groups to form an oxazolidin-2-one, a chemically stable moiety21, consonant with the ability of oxathiazol-2-ones to cyclocarbonylate 1,2-aminoalcohols22.
Figure 3. LC-MS/MS identification of the modified N-terminus of the Mtb proteasome treated with oxathiazol-2-ones.
(a) Mass spectra of tryptic N-terminal heptapeptides from Mtb proteasomes that were untreated (i), HT1171-treated (ii), treated with glutaraldehyde/Na(CN)BH3 after trypsin digestion (iii), or treated with glutaraldehyde/Na(CN)BH3 after HT1171 treatment and trypsin digestion (iv). All ions were confirmed by MS/MS analysis (Fig. S7a–d). Reaction equations illustrate proposed modification of active site Thr1 by oxathiazol-2-one (i
 ii), and modification of primary amino groups at Thr1 and Lys7 with glutaraldehyde and Na(CN)BH3 (i
ii), and modification of primary amino groups at Thr1 and Lys7 with glutaraldehyde and Na(CN)BH3 (i
 iii and ii
iii and ii
 iv). (b) Proposed mechanism of proteasome inactivation by oxathiazol-2-one. Paths marked by a, b, d lead to irreversible inhibition. In paths marked by c, hydrolysis of the inhibitor-enzyme intermediate allows the proteasome to degrade the oxathiazol-2-one without losing activity.
iv). (b) Proposed mechanism of proteasome inactivation by oxathiazol-2-one. Paths marked by a, b, d lead to irreversible inhibition. In paths marked by c, hydrolysis of the inhibitor-enzyme intermediate allows the proteasome to degrade the oxathiazol-2-one without losing activity.
Structural basis of species selectivity
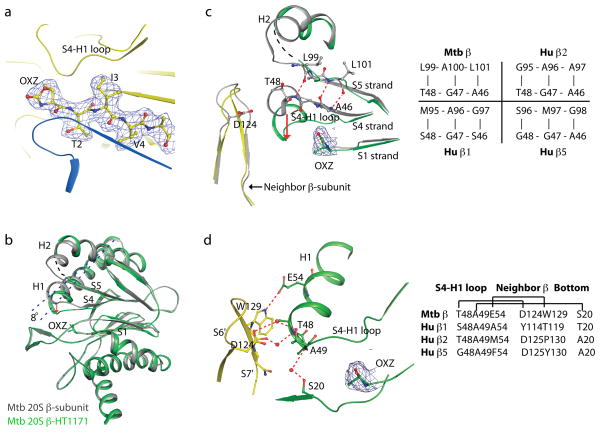
To determine the basis for species selectivity, we solved four crystal structures: wild type Mtb proteasome following exposure to GL1 at 2.4 Å resolution and to HT1171 at 2.5 Å resolution, and the open-gate variant (20SOG) alone at 2.5 Å resolution or following exposure to HT1171 at 2.9 Å resolution. N-terminal octa-peptide deletion in the α-subunit of 20SOG did not alter the overall structure of the Mtb proteasome (Fig. S8a). Furthermore, wild type and open-gate proteasomes underwent the same conformational changes (described below) upon inhibitor treatment (Fig. S8b). The three structures (Figs. 4, S8, S9; Table S5) each confirmed that oxathiazol-2-ones cyclo-carbonylate Thr1. Not only the oxazolidin-2-one ring, but also its protruding methyl group and carbonyl oxygen, were resolved in the electron density (Fig. 4). The oxazolidin-2-one ring is stabilized by an H-bond network involving Ala180, Ser141, Asn24 of the neighboring β-subunit, and a water molecule in the substrate cavity (Fig. S9). Use of HT1171 and GL1 in the crystallographic studies and HT1171, GL5 and GL3 in the mass spectroscopic studies brought to 4 the number of oxathiazol-2-ones for which the same suicide-substrate inhibition mechanism was confirmed.
Figure 4. Crystal structure of the full-length Mtb 20S proteasome after exposure to HT1171 reveals cyclo-carbonylation of active site Thr1 and conformational changes in the β-subunit.
(a) 2Fo-Fc electron density map contoured at 1.2 σ and superimposed on the crystal structure of HT1171-treated proteasome at the active site in the β-subunit. Map was calculated by omitting oxazolidin-2-one and the N-terminal four amino acids from the crystal structure. (b) Superposition of HT1171-modifiedβ-subunit in green with native β-subunit in gray (PDB 2FHG). OXZ labels oxazolidin-2-one ring on Thr1. The conformational changes can be approximately described by an 8° tilt of H1 (dashed line) and a downward shift of the S4-H1 loop (red arrow). (c) Active site structure of HT1171-modified β-subunit in green, in comparison with native β-subunit structure in gray. In the native structure, A46G47T48 is part of the S4 strand forming a β-sheet with the S5 strand. In the HT1171-treated structure, A46G47T48 loses contacts with S5 and converts to the S4-H1 loop. Panel at right specifies amino acid pairs that form S4–S5 β-sheet interaction in this region in Mtb β chain prior to oxathiazol-2-one treatment and compares these with corresponding sequences in the proteolytically active human proteasome β chains. (d) Active site structure of HT1171-treated Mtb 20S oriented to view H bonds stabilizing new position of S4-H1 loop in the Mtb β chain after oxathiazol-2-one treatment. S4-H1 loop as upper surface of the constricted substrate-binding pocket is stabilized by an H-bond between Glu54 and Trp129 of the neighboring βsubunit and by 3 pairs of water-mediated H-bonds with the neighboring β-subunit and with Ser20 at the lower substrate-binding surface of the same subunit. Panel at right compares Mtb’s H-bonding residues with the human counterparts. The 2Fo-Fc electron density map contoured at 1.2 σ is superimposed at the oxazolidin-2-one ring site in (c) and (d). Thr1 modification and β-subunit conformational changes upon exposure to GL1 or HT1171 in the wild type and the open gate Mtb 20S are virtually the same.
Surprisingly, the substrate-binding pocket of the Mtb proteasome underwent a major conformational change upon cyclo-carbonylation of Thr1 by HT1171 or GL1. Such a change is unprecedented among the dozens of crystal structures of proteasomes in complex with inhibitors11. An ~8° downward tilt of the H1 helix in the β-subunit moved the N-terminal end of H1 (Ala49-Phe55) downward by as much as 4.2 Å (Fig. 4b). The H1 shift brought with it a 3-amino-acid stretch (Ala46-Thr48) of the S4 β-strand, converting this segment into a short loop (S4-H1). As a consequence, another short loop (Met95Gln96Gly97) between H2 and S5 lost stabilizing contacts with the shifted components and became disordered, as illustrated by a black dashed curve in Fig. 4b. The S4-H1 loop region comprises the upper surface of the substrate-binding pocket11,15. Its downward shift constricted the pocket to the point that it could not accommodate a peptide substrate, affording an additional mechanism of inhibition over and above incorporation of the active site hydroxyl into an oxazolidin-2-one.
In the native Mtb β-subunit, the three S4-H1 loop amino acids Ala46, Gly47 and Thr48 are in βstrand configuration and form a β-sheet interaction with Leu101, Ala100 and Leu99, respectively, in the S5 β-strand (Fig. 4c). None of these residue pairs is conserved in the human proteasome β5 subunit and only one of them (G47-A96) in the human β1 and β2 subunits. One direct H-bond and three pairs of water-mediated H-bonds stabilized the new position of the S4-H1 loop (Fig. 4d): between Glu54 and Trp129 of neighboring β-subunit (3.0 Å); between Thr48, a water, and Asp124 in the S6-S7 loop of neighboring β-subunit (2.8 Å and 3.1 Å, respectively); between Ala50, a water, and the carbonyl O of Trp129 in the short S7 β-strand of the neighboring β-subunit (3.0 Å and 3.1 Å, respectively); and between Ala 49, a water, and Ser20 in the same β-subunit (3.2 Å and 3.1 Å, respectively). This last pair of H-bonds cross-linked the upper and the lower substrate-binding surfaces, sealing the entrance of the substrate pocket. The residue pairs involved in the post-shift H bonds are not well conserved in the human proteasome (Fig. 4d). We speculate that the upper substrate-binding surfaces (S4-H1 loops) in the 3 catalytic β-subunits of the human proteasome might have difficulty breaking off from their corresponding β-sheet cores upon initial modification of Thr1 by oxathiazol-2-ones.
Thus, selectivity of oxathiazol-2-ones appears to be imparted in 3 ways: by the presence of a 1,2-aminoalcohol at the active site of the target, which, among enzymes, is likely to be limited to the N-terminal Thr hydrolase family; by the ability of the inhibitor to bind rapidly (before its spontaneous decay) and precisely adjacent to the 1,2-aminoalcohol; and the degree to which the amino group of the 1,2-aminoalcohol has better access to the inhibitor’s carbonyl (now attached to the alcohol) than water has. The protein landscape near the 1,2-aminoalcohol can thus determine species selectivity, both in how it binds the R group on the oxathiazol-2-one and in the conformations it adopts, which may either permit or limit access of water to the intermediate formed during reaction of the oxathiazol-2-one with the active site. Detailed understanding the sequence of steps by which oxathiazol-2-ones cause a major conformational shift in the substrate-binding domain could guide design of the next generation of inhibitors selective for the Mtb proteasome over the human proteasome.
Discussion
Non-replicating Mtb displays “phenotypic tolerance,” that is, relative resistance to conventional anti-infectives, imposing the need to treat tuberculosis longer than almost any other infectious disease. Prolonged treatment leads to interruption of therapy and emergence of hereditable drug resistance. Hence agents are needed that can kill Mtb when its replication is halted by conditions encountered in the host, such as those imposed by inducible nitric oxide synthase23. Oxathiazol-2-ones identified here phenocopy genetic deletion of the Mtb proteasome10 by killing Mtb rendered non-replicative by exposure to sub-lethal nitric oxide. Along with thioxothiazolidines that inhibit Mtb’s dihydrolipoamide acyltransferase17, oxathiazol-2-ones are only the second class of compounds, to our knowledge, that are selectively bactericidal for a non-replicating pathogen.
Certain oxathiazol-2-ones are selective in other ways as well. They inhibit the Mtb proteasome but not the human proteasome N-terminal threonine β1, β2 or β5 proteases, cysteine proteases, serine proteases, or metalloproteases. They do not kill monkey epithelial cells, human macrophages or any bacteria we tested other than mycobacteria. The oxathiazol-2-ones react with proteasomes at their active site. However, residues distant from the active site, with which oxathiazol-2-ones do not interact, appear to impart species selectivity.
Inhibition of protein synthesis at the stages of transcription (e.g., by rifamycins) or translation (e.g., by aminoglycosides and capreomycin) are among the best validated antibiotic strategies24. It may prove synergistic to interfere at the same time with bacterial protein degradation. The ability of a brief exposure to oxathiazol-2-ones to inhibit Mtb proteasomes permanently is a potential advantage. Mtb may have difficulty replacing irreversibly inactivated proteasomes, not only when Mtb’s protein synthesis is markedly diminished in the non-replicative state25, but also when its protein synthesis is impaired by antibiotics.
Much of biology can be viewed as the interplay of two principles, conservation and diversity. The conservation of proteasomes across vast evolutionary distances is striking. The present work demonstrates that functionally exploitable diversity exists between the proteasomes of Mtb and its obligate human host.
Methods Summary
High throughput screening was performed in 384-well plates with 33 μM test compounds. To test inhibition of proteasomes in intact mycobacteria, treated cells were centrifuged, washed twice and lysed mechanically, and aliquots (10 μg protein) of the supernates assayed with Suc-LLVY-AMC (50 μM). Killing of DETANO-treated, non-replicating Mtb Erdman was tested in Sauton’s medium, pH 7.4. After 4 days, CFU were plated for enumeration 3 weeks later. Monkey kidney cells were cultured in Dulbecco’s Eagle’s medium with 10% fetal bovine serum for cytotoxicity assessment after 48 hours by tetrazolium reduction and microscopy. Recombinant Mtb proteasomes were purified as reported16. Human proteasomes were activated with PA28 (Boston Biochem). Kinetic studies were conducted in a Molecular Devices fluorescent plate reader with AMC-derivatized peptide substrates according to which proteasomal protease was being studied (Mtb or human β1, β2 or β5). Values of kobs were derived from the fit of data to equation (1)26 in Prism (GraphPad Software, Inc. La Jolla, CA).
| (1) |
LC-MS/MS analysis used Thermo LTQ Orbitrap and Applied Biosystems QSTAR mass spectrometers. MALDI-TOF analysis was performed on a PerSeptive DE-STR instrument. Crystallization of the Mtb proteasome, diffraction data collection and structure solution and refinement are included along with other methodologic details in the Methods section that appears online in the full-text HTML and PDF versions of this article.
Supplementary Material
Acknowledgments
S. Eswaramoorthy (BNL) helped with crystallography software, C. Lipinski (Melior Discovery, Waterford CT) and C. Walsh and M. Fischbach (Harvard Medical School) proposed reaction mechanisms, C. Karan (High Throughput Screening Facility, The Rockefeller University and Weill Cornell Medical College) assisted with screening. S. Ehrt and S. Gandotra (Weill Cornell Medical College) performed some bactericidal assays, and C. Tsu and L. Dick (Millennium Pharmaceuticals, Cambridge, MA) donated a fluorimeter. Supported by NIH PO1-AI056293, NIH R01AI070285 and the Milstein Program in Chemical Biology of Infectious Diseases. X-ray diffraction data were collected at beamline X6A, X25, and X29 in the National Synchrotron Light Source, a facility supported by US DOE and NIH. The Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature and includes details of chemical synthesis and characterization, Figs S1 to S9 and Tables S1 to S5.
Author contributions GL purified recombinant proteasome, conducted the screen, designed new oxathiazol-2ones and performed most of the assays. DL and HL purified and crystallized recombinant proteasomes and solved their structures. LPSC helped analyze kinetics. HD performed mass spectrometry. HT synthesized oxathiazol-2-ones under the supervision of JDW. GV studied human macrophages. KW conducted studies with viable Mtb. JS and TC performed kinetic, bactericidal and cytotoxicity experiments. CN organized the effort and helped design and interpret experiments. CN, GL and HL wrote the paper.
Author information Reprints and permissions information is available at www.nature.com/reprints.
Coordinates have been deposited in the Protein Data Bank under ID codes 3H6F, 3H6I, 3HFA, 3HF9.
References
- 1.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 2.Kropff M, et al. Bortezomib in combination with intermediate-dose dexamethasone and continuous low-dose oral cyclophosphamide for relapsed multiple myeloma. Br J Haematol. 2007;138:330–337. doi: 10.1111/j.1365-2141.2007.06656.x. [DOI] [PubMed] [Google Scholar]
- 3.Glenn RJ, et al. Trypanocidal effect of alpha', beta'-epoxyketones indicates that trypanosomes are particularly sensitive to inhibitors of proteasome trypsin-like activity. Int J Antimicrob Agents. 2004;24:286–289. doi: 10.1016/j.ijantimicag.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 4.Darwin KH, et al. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 5.Raviglione MC, Smith IM. XDR tuberculosis--implications for global public health. N Engl J Med. 2007;356:656–659. doi: 10.1056/NEJMp068273. [DOI] [PubMed] [Google Scholar]
- 6.Corbett EL, et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 7.Restrepo BI. Convergence of the tuberculosis and diabetes epidemics: renewal of old acquaintances. Clin Infect Dis. 2007;45:436–438. doi: 10.1086/519939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce MJ, et al. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns KE, et al. Proteasomal protein degradation in mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284:3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandotra S, et al. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 12.Lin G, et al. Unpublished results. [Google Scholar]
- 13.Lin G, et al. Distinct specificities of Mycobacterium tuberculosis and mammalian proteasomes for N-acetyl tripeptide substrates. J Biol Chem. 2008;283:34423–34431. doi: 10.1074/jbc.M805324200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 15.Hu G, et al. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Mol Microbiol. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin G, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Mol Microbiol. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 17.Bryk R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3:137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huth JR, et al. Toxicological evaluation of thiol-reactive compounds identified using a La assay to detect reactive molecules by nuclear magnetic resonance. Chem Res Toxicol. 2007;20:1752–1759. doi: 10.1021/tx700319t. [DOI] [PubMed] [Google Scholar]
- 19.Copp LA. In: Enzyme Kinetics: A Modern Approach. Marangoni AG, editor. Ch 13. Wiley; 2002. pp. 158–173. [Google Scholar]
- 20.Russo A, Chandramouli N, Zhang L, Deng HT. Reductive glutaraldehydration of amine groups for identification of protein N-termini. J Proteome Res. 2008;7:4178–4182. doi: 10.1021/pr800224v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu LQ, et al. A new entry to cascade organocatalysis: reactions of stable sulfur ylides and nitroolefins sequentially catalyzed by thiourea and DMAP. J Am Chem Soc. 2008;130:6946–6948. doi: 10.1021/ja800746q. [DOI] [PubMed] [Google Scholar]
- 22.Rajca A, Grobelny D, Witek S, Zbirovsky M. 5-Aryl-2-oxo-1,2,4-oxathiazoles as cyclocarbonylating agents for 2-aminoalcohols and 1,2-diamines. Synthesis-Stuttgart. 1983;12:1032–1033. [Google Scholar]
- 23.MacMicking JD, et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94:5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh C. Where will new antibiotics come from? Nat Rev Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 25.Hu YM, et al. Protein synthesis is shutdown in dormant Mycobacterium tuberculosis and is reversed by oxygen or heat shock. FEMS Microbiol Lett. 1998;158:139–145. doi: 10.1111/j.1574-6968.1998.tb12813.x. [DOI] [PubMed] [Google Scholar]
- 26.Copeland RA. Enzymes: a practical introduction to structure, mechanism, and data analysis. Charpter 9-10. John Wiley & Sons, Inc., Publication; 2000. pp. 305–349. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.