Abstract
Ancient autophagy pathways are emerging as key defense modules in host eukaryotic cells against microbial pathogens. Apart from actively eliminating intracellular intruders, autophagy is also responsible for cell survival, for example by reducing the deleterious effects of endoplasmic reticulum stress. At the same time, autophagy can contribute to cellular suicide. The concurrent engagement of autophagy in these processes during infection may sometimes mask its contribution to differing pro-survival and pro-death decisions. The importance of autophagy in innate immunity in mammals is well documented, but how autophagy contributes to plant innate immunity and cell death is not that clear. A few research reports have appeared recently to shed light on the roles of autophagy in plant–pathogen interactions and in disease-associated host cell death. We present a first attempt to reconcile the results of this research.
Keywords: autophagy, ATG genes, innate immunity, plants
Autophagy mediates the degradation of bulk proteins and is also involved in the clearance of damaged organelles, insoluble protein aggregates and lipids.1, 2, 3 Autophagic digestion and recycling can occur as a survival mechanism to maintain cellular homeostasis and to respond to environmental stresses, such as nutrient depletion or pathogen attack, but may also function as a mediator and/or mechanism of programmed cell death.4, 5, 6, 7, 8 Several subtypes of autophagy are described, but macroautophagy (hereafter termed autophagy) is the most extensively studied9 and will be the only form described here. The process is characterized by the formation of large, double-membrane vesicles called autophagosomes. These structures arise from expanding single membranes (termed phagophores), which enclose cytoplasmic material and organelles for degradation. Completed autophagosomes fuse with the vacuole/lysosome to release the inner single-membrane vesicle, called the autophagic body, into the lumen for hydrolytic degradation and recycling.2, 10
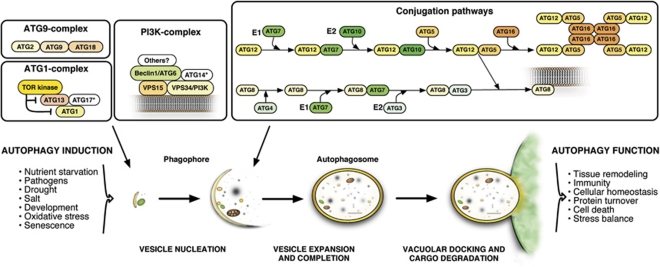
The mechanism of autophagy is conserved in yeast, plants and metazoans, and involves the action of canonical autophagy related genes (ATG) that synthesize and coordinate membrane rearrangements to allow cellular catabolism.1, 2 The core sets of ATG genes seem to be present in all eukaryotes and to be essential for the autophagy pathway (Figure 1). For instance, induction of autophagy requires the negative regulator target of rapamycin (TOR) kinase and the ATG1 kinase complex, which control the activity of the phosphatidylinositol 3-kinase complex containing, for example, ATG6/Beclin1.11 Initiation and completion of autophagosome formation involves two ubiquitin-like conjugation systems to produce ATG12-ATG5 and ATG8-phosphatidylethanolamine (ATG8-PE) conjugates. ATG8-PE conjugation involves the cysteine proteinase ATG4 and the E1-like protein ATG7, and lipidated ATG8 is linked to and translocated with autophagosomes to the vacuole.12 Therefore, conversion from soluble to lipid bound ATG8, as well as subcellular localization of green fluorescent protein (GFP)-fused protein, have been used to monitor temporal dynamics and spatial regulation of autophagy.13 Finally, recycling and retrieval of autophagy proteins require the ATG9 complex, containing ATG2, ATG9 and ATG18.2, 10
Figure 1.
The autophagy pathway in plants. Upon induction by environmental and developmental stimuli, macroautophagy starts by nucleation and expansion of a pre-autophagosomal membrane, the phagophore, which engulfs cytoplasmic material destined for degradation. Autophagosomes are then transported to and dock with the vacuole, which leads to release of the inner single-membrane vesicle, the autophagic body, into the vacuolar lumen and hydrolytic breakdown of the enclosed cargo. Autophagy induction and vesicle nucleation require the action of the TOR kinase and ATG1-complex, which activates the class III PI3K complex containing ATG6/Beclin1 and possibly other yet unknown interactors. Additionally, as known from other eukaryotes, the ATG9 complex is required for recycling and retrieval of autophagy proteins, but this function awaits verification in plants. Membrane elongation and autophagosome formation require the action of two-ubiquitin-like conjugation systems, which modify two ubiquitin-like molecules, ATG5 and ATG8, to mediate association with the phagophore and its subsequent folding and expansion. *ATG proteins not identified in plants yet
A number of excellent reviews provide more details about the molecular mechanisms of autophagy and the individual components required for autophagic complexes and processes7, 14, 15, 16, 17 (see also Figure 1). In this review, we focus on the role of autophagy in programmed cell death and innate immune responses, with special emphasis on the plant hypersensitive response associated with disease resistance.
Autophagy in Plants
Much has been learned about the requirement for specific ATG genes in the model plant Arabidopsis. Loss-of-function mutations in ATG genes such as ATG7 and ATG5 implicate autophagy as a central player in cellular homeostasis.18, 19 Processing and delivery of ATG8 to the vacuole under nitrogen-starved condition requires the cysteine protease ATG4 and the ATG12-ATG5 conjugate,20, 21 and atg5, atg7, atg10, as well as atg12a/b double mutants are hypersensitive to both nitrogen and carbon starvation.21, 22, 23 Thus, both autophagic-related conjugation pathways seem to be required for autophagy in plants and, as in yeast and other models, the process is required to recycle nutrients during starvation.
Several reports have documented the roles of autophagy in plant development and under stress conditions. During senescence of Arabidopsis leaves kept in darkness (a form of carbon starvation for photosynthetic autotrophs), autophagy seems to be responsible for degradation of the chloroplasts,24 and root development also becomes impaired in different atg mutants during nitrogen starvation.18, 20 Perhaps not surprisingly, autophagy functions in the removal of oxidized proteins during oxidative stress in Arabidopsis,25 and downregulation of ATG18a using interference RNA (RNAi) renders plants more sensitive to salt and drought stress.26 Collectively, these reports demonstrate that autophagy affects plants in many aspects of their life cycle.
In contrast to autophagy mechanisms in yeast and mammals, information about the signaling pathways triggering the induction of plant autophagy in response to developmental, nutritional and environmental cues is largely lacking. Only recently, direct genetic evidence has been provided that the TOR kinase is a negative regulator of autophagy in higher plants.27 Although knockout of the single TOR gene in Arabidopsis proved to be embryo-lethal,28, 29 knockdown by RNAi resulted in constitutive autophagy under non-stressed conditions in an ATG18-dependent fashion.27 In addition, Tap46, the regulatory subunit of protein phosphatase 2A, was recently identified as a downstream effector of the TOR signaling pathway. Depletion of Tap46 reproduced the signature phenotypes of TOR inactivation, including autophagy induction.30
Autophagy in Immunity
As autophagy has the ability to eliminate unwanted cellular structures, it is not surprising that this complex and evolutionary ancient pathway also evolved to combat unwanted intracellular microbes. That autophagy could contribute to cellular clearance of microbes was evident already in the 1980s,31 but it was first a decade later that the molecular tools to study autophagy in immune responses became available. More recently, autophagy has been shown in a number of cases to contribute to defenses against microbial invasion. For example, autophagy defends mammalian cells against invading Streptococcus.32 In contrast to wild type cells, Streptococcus survives and multiplies in ATG5 deficient cells, suggesting that the autophagic machinery is engaged to actively kill the bacteria. These data were supported by micrographs of Streptococci trapped inside autophagosomal structures and these are absent in ATG5 deficient cells.32 Likewise, induction of autophagy suppressed intracellular survival of Mycobacterium tuberculosis in macrophages. In this case, the bacteria are also trapped inside autophagosomal-like structures positive for Beclin1.33 Since these reports, a number of excellent reviews have discussed other studies documenting autophagy as an innate defense mechanism for controlling intracellular pathogens in mammals.34, 35, 36
In the evolutionary arms race between pathogens and their hosts, some pathogens have also developed mechanisms to avoid or even exploit this defense mechanism to survive and establish infection. Shigella bacteria can escape autophagy by secreting effectors by means of the type III secretion system. However, mutant bacteria lacking specific effectors become trapped by autophagy during multiplication within host cells and fail to establish infection.37 Interestingly, autophagosomal-like structures in human cells provide membranous supports for poliovirus RNA replication and, in cells in which autophagy is inhibited via drugs or RNAi against a collection of ATG genes, poliovirus yield is diminished.38
More recently, increased autophagic activity has been linked directly to pathogen surveillance systems in different organisms. For example, mammalian Toll-like receptors (TLRs) detect microbial-associated molecular patterns (MAMP) and induce defense responses upon ligand detection.39 Accordingly, stimulation of TLR7 in macrophages leads to increased autophagic activity and elimination of M. tuberculosis in an ATG5-dependent manner.40 Another example includes the pathogen receptor CD46 that binds Streptococci and triggers autophagy.41 Thus, these two examples provide evidence that pathogen recognition in different animal systems is directly linked to higher levels of autophagic activity.
Autophagy and Cell Death
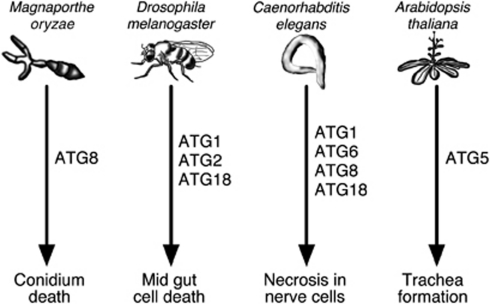
Apart from being required to tolerate nutrient deprivation and other stresses, autophagy also represents a cell death pathway conserved across the eukaryotic kingdom. In 2004, Yu et al.42 reported the requirement for ATG7 and Beclin1 in certain types of cell death in mammalian cell cultures and provided a primary example of autophagic cell death. Since then, numerous reports have argued for or against autophagy as a cell death mechanism, but evidence in favor of autophagic cell death has recently emerged in various genetic models. For example, the conidium of the rice blast fungus Magnaporthe grisea undergoes autophagic cell death to establish an infection and, accordingly, M. oryzae atg8 null-mutants are unable to infect plants.43 In Drosophila, physiological cell death of the salivary gland requires the action of ATG genes44 and autophagy is essential for midgut cell death as well.45 In C. elegans, necrotic breakdown of neurons is autophagy-dependent and necrotic cell death is accompanied by elevated autophagic activity.46 A recent report documented that cell death in the formation of tracheary elements in Arabidopsis is inhibited in atg5 null-mutants and stimulated by increased levels of autophagy.47 Such examples indicate that autophagy effectuates cell death pathways critical for many aspects of eukaryotic development (Figure 2).
Figure 2.
Examples of cell death that depend on autophagy components in different eukaryotic models. In the rice blast fungus Magnaporthe oryzae, autophagic cell death is required for degradation of conidia and thus fungal pathogenicity, which can be inhibited by knockout of ATG8.43 In Drosophila melanogaster, knockouts of ATG1, ATG2 and ATG18 demonstrated the requirement of autophagy for mid gut cell death.45 In Caenorhabiditis elegans, autophagosome formation is required for necrotic cell death of neurons, which has been shown to be inhibited by knockout of ATG1 or RNAi knockdown of ATG6, ATG8 and ATG18.46 Finally, in Arabidopsis thaliana, development of tracheary elements depends on autophagic cell death, which is inhibited in atg5 knockout mutants47
Beclin1 provides a primary example of a molecular connection between autophagy and apoptosis, an important pathway to cellular destruction in metazoans. Beclin1 is a haplo-insufficient tumor suppressor in mice48 involved in the initial formation of autophagosomes. Beclin1 also forms complexes with the anti-apoptotic protein Bcl-2 in mammalian cells49 and loss of Beclin1 in C. elegans triggers apoptotic cell death.50 In Arabidopsis, Beclin1 is essential for pollen germination, a feature not associated with other ATG-null mutants,51 but knockdown of Beclin1 through antisense or viral-induced gene silencing leads to premature chlorosis and cell death in both Arabidopsis and Nicotiana benthamiana.52, 53 It is therefore tempting to speculate that, like Beclin1 in metazoans, plant Beclin1 could also represent a molecular link between autophagy and another cell death route. ATG5 represents an additional molecular link between autophagy and apoptosis, because calpain-mediated cleavage of ATG5 promotes apoptosis through mitochondrial cytochrome C release and caspase activation.54 Recently, a conjugate between ATG12 and ATG3 was shown to affect mitochondrial homeostasis and sensitize cells to apoptosis in a context completely separated from the established roles of the ATG proteins in the autophagic pathway.55 Collectively, these findings imply that specific ATG genes can act as molecular switches and have autophagy-independent functions in homeostatic processes and cell death.
Autophagy in Plant Immunity and Hypersensitive Cell Death
Plants rely on a multilayered innate immune system to prevent pathogen invasion and proliferation. Pathogen recognition can occur on the cell surface by pattern recognition receptors that detect MAMPs and induce immune responses such as cell wall thickening and production of antimicrobial proteins.56 However, diverse pathogens deliver a variety of virulence determinants, commonly referred to as effectors, into plant cells to evade or suppress MAMP-triggered immunity and to manipulate the host machinery for their own benefit.57 In turn, plants have evolved another layer of defense to recognize effector modifications of host target proteins via host surveillance proteins (resistance (R) proteins). These R-mediated defenses often include a localized programmed cell death reaction known as the hypersensitive response (HR) to limit pathogen spread.58
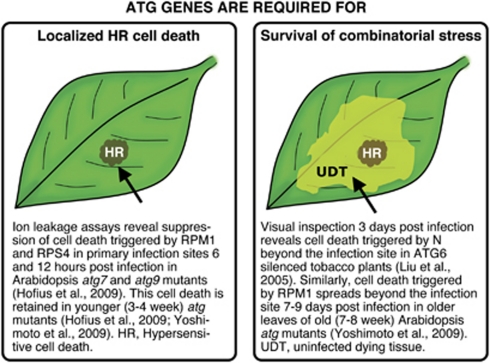
Several examples of the involvement of autophagy in plant immunity and hypersensitive-related cell death have emerged in the past few years. However, there remains some doubt and apparent contradictions concerning the function(s) of autophagy as a pro-survival or pro-death pathway. In 2005, Liu et al.,52 linked the activation of autophagy to infection in plants and nicely demonstrated that autophagy contributes to resistance. In addition, they also presented evidence that autophagy was required to restrict the spread of plant hypersensitive cell death, thus functioning as a pro-survival pathway. Activation of the N-resistance gene in N. benthamiana by the p50 helicase protein of tobacco mosaic virus (TMV) during infection or upon transient expression triggered hypersensitive cell death, and a few days after local infection, dead cell patches became visible in non-infected tissues on plants silenced for Beclin1.52 A similar approach in Arabidopsis supported the observations in N. benthamiana, because activation of hypersensitive cell death via the R gene RPM1 upon infection with bacteria also led to macroscopic cell death beyond the infection site in plants silenced for Beclin1, starting roughly 5 days post-infection.53 In this context, it should be noted that cell death triggered by RPM1 is executed rapidly and ends after roughly 6–8 h.59 This raises the question of whether cell death emerging several days later is directly connected to uncontrolled HR cell death. Nevertheless, the data led to the hypothesis that autophagy prevents unrestricted HR cell death by yet unknown mechanisms and thus functions as a pro-survival pathway in plant–pathogen interactions.60, 61
More recently, a pro-death function of autophagy during hypersensitive cell death was reported.62 Here, autophagic activity in the infected tissue accompanied the onset of cell death execution triggered by some, but not all types of R proteins. In addition, in cases in which autophagy was induced, cell death was suppressed in local infected tissues in different atg mutants. Strongest suppression was found for cell death conditioned by the R proteins RPS4 and RPP1 that signal through the signaling component Enhanced Disease Susceptibility1 (EDS1).62 HR cell death triggered by RPM1 was also significantly suppressed in atg mutants, but in this case suppression was most prominent in combination with cathepsin inhibitors.62 Cathepsins contribute to HR triggered by Phytophthora infestans in N. benthamiana HR,63 suggesting that both autophagy and cathepsins are engaged in RPM1-triggered hypersensitive cell death.62 In this context, Hatsugai et al.64 recently reported that the 20S proteasome subunit PBA1 has caspase-3-like activity and contributes to cell death triggered by RPM1. This further supported the view that RPM1 recruits autophagic mechanisms for suicidal cell death, in parallel with various other execution pathways.65
How can these apparent discrepancies about the role of autophagy in HR cell death be explained? First, it is important to emphasize differences in the experimental systems. Liu et al.52 primarily investigated plants visually and concluded that collapse of tissues next to infection sites a few days after local infections is caused by uncontrolled HR. Patel and Dinesh-Kumar53 performed similar assays with similar observations in Arabidopsis. In contrast, Hofius et al.62 examined cell death in the actual infection site, using a widely accepted electrolyte leakage assay59 during the first hours after bacterial infection, as well as trypan blue staining of leaves infected with avirulent Hyaloperonospora arabidopsidis and bacteria. So, the apparently different conclusions come from studies of non-infected tissue, days after infection versus infected tissue during HR execution (summarized in Figure 3). In addition, the link between direct activation of autophagy in R gene-triggered immunity is supported by recent discoveries in mammals. Here, an R protein homolog, the cytosolic NOD1 receptor that provides a surveillance system for the detection of intracellular pathogens, recruits the autophagic protein ATG16L to the plasma membrane at the site of bacterial entry.66
Figure 3.
Clarification of studies on autophagic cell death in plant immunity. HR, hypersensitive response; UDP, uninfected, dying tissue
In any event, these reports also seem to underscore the importance of the autophagic machinery in limiting pathogen infection in plants. Liu et al.52and co-workers found increased titers of avirulent TMV in infected tissues of Beclin1 silenced N. benthamiana plants, and Patel and Dinesh-Kumar53 observed increased susceptibility towards virulent strains of P. syringae in Beclin1 antisense Arabidopsis plants. Similarly, Hofius et al.62 observed increased growth of both virulent P. syringae and Hyaloperonospora arabidopsidis in different Arabidopsis atg mutants. Together, all these findings demonstrate that autophagy can function in plant innate immunity.
Another important contribution comes from Yoshimoto et al.67 Primarily scoring macroscopic lesions or visible death in leaves, the authors found no difference in RPM1-triggered cell death beyond the initial infection site in younger atg mutants. However, in older atg mutants such as atg5, they observed lesions in non-infected tissues 6–9 days after infection. Interestingly, these effects were suppressed by removal of the phytohormone salicylic acid (SA) and by mutations in non expressor of pr genes (npr1).67 The authors thus proposed that autophagy negatively regulates cell death by controlling NPR1-dependent SA signaling, although it is unclear why there is a difference between young and old atg mutants.
Autophagy-deficient mutants lack the autophagic machinery to remove accumulating cellular ‘garbage', and in contrast to younger or newly emerged leaves, older atg mutant leaves contain higher levels of metabolites, disrupted organelles and oxidized proteins.24, 25 Such accumulated cellular debris may well disrupt homeostasis, leading to pleiotropic effects including accumulation of danger-associated molecular patterns (DAMPs)68 triggering SA accumulation, and subsequent production of secreted pathogenesis-related (PR) proteins accompanied by ER stress. Autophagy is required to dampen the deleterious effects caused by ER stress and it is well described in other models that atg mutants die upon increased ER stress.69 Thus, accumulated ER stress would be expected to increase the susceptibility of older atg mutants to additional stresses. Interestingly, npr1 has reduced ER stress and expression of PR genes, and mutants like bip2 die upon SA-analog treatment.70 This would explain why npr1 (and SA-deficient mutants) rescue older atg mutants; even in uninfected tissues, DAMP signals and ER stress-related effects are reduced due to reduced expression of defense genes in npr1/atg5 double mutants. Again, Figure 3 attempts to summarize some of the most important observation done by the different groups.
Concluding Remarks
Many questions remain to be addressed on the roles of autophagy in plants. Because autophagy is required for cellular homeostasis, more specific autophagic functions in plant–microbe interactions and immunity are hard to unravel. For example, are plant atg mutants, in contrast to those of other organisms, able to cope with prolonged ER stress and, if not, what is the outcome? Similarly, it may be problematic to use older atg mutants plants, because of pleiotropic effects caused by lifelong accumulation of the kind of cellular ‘garbage' normally removed by autophagy. In addition, we need to be circumspect in the selection of autophagic mutants amenable for specific studies. For example, the strong chlorotic phenotype of Beclin1 antisense plants suggests that Beclin1 may also be involved in other cell death programs, as is now apparent in metazoans. Moreover, the available collection of atg mutants needs to be further explored to analyze the role of autophagy in MAMP- and effector-triggered immune responses of various host-pathogen systems. Similarly, autophagy components and mechanisms might be specifically targeted by pathogen effector proteins to either suppress defense responses or to promote pathogenicity, for example, of necrotrophic pathogens. Finally, diverse microbial life styles and the changing ‘rules of engagement' in the evolutionary arms race indicate that autophagy may be co-opted for various purposes by hosts and microbes alike. Therefore, it is possible that host-derived, perimicrobial membranes71 are actually autophagic in origin. If so, this could represent an armistice to enable symbiosis for the benefit of both microbes and plants.
Glossary
- ATG
autophagy related genes
- ATG8-PE
ATG8-phosphatidylethanolamine
- DAMPs
danger-associated molecular patterns
- EDS1
enhanced disease susceptibility1
- GFP
green fluorescent protein
- HR
hypersensitive response
- MAMP
microbial associated molecular patterns
- npr1
non expressor of PR genes
- PR
pathogenesis-related
- R proteins
resistance proteins
- SA
salicylic acid
- TLRs
toll-like receptors
- TMV
tobacco mosaic virus
- TOR kinase
target of rapamycin
- UDP
uninfected dying tissue
The authors declare no conflict of interest.
Footnotes
Edited by J Dangl
References
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro JA, Cuervo AM. Autophagy and lipids: tightening the knot. Semin Immunopathol. 2010;32:343–353. doi: 10.1007/s00281-010-0219-7. [DOI] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Autophagy and cell death in model organisms. Cell Death Differ. 2009;16:21–30. doi: 10.1038/cdd.2008.120. [DOI] [PubMed] [Google Scholar]
- Scarlatti F, Granata R, Meijer AJ, Codogno P. Does autophagy have a license to kill mammalian cells. Cell Death Differ. 2009;16:12–20. doi: 10.1038/cdd.2008.101. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N. Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin Cell Dev Biol. 2010;21:683–690. doi: 10.1016/j.semcdb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Espert L, Biard-Piechaczyk M, Codogno P. Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie. 2008;90:313–323. doi: 10.1016/j.biochi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:PMID: 20484971. doi: 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277:33105–33114. doi: 10.1074/jbc.M204630200. [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, et al. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010;62:483–493. doi: 10.1111/j.1365-313X.2010.04166.x. [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138:2097–2110. doi: 10.1104/pp.105.060673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178:1339–1353. doi: 10.1534/genetics.107.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, et al. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–893. doi: 10.1104/pp.108.130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS ONE. 2010;5:e11883. doi: 10.1371/journal.pone.0011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, et al. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn CS, Han JA, Lee HS, Lee S, Pai HS. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 2011;23:185–209. doi: 10.1105/tpc.110.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat Rec. 1984;208:319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Sumpter R, Jr, Levine B. Selective autophagy and viruses. Autophagy. 2011;7:PMID: 21150267. doi: 10.4161/auto.7.3.14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Levine B. Autophagy and innate immunity: triggering, targeting and tuning. Semin Cell Dev Biol. 2010;21:699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312:580–583. doi: 10.1126/science.1124550. [DOI] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara C, Syntichaki P, Tavernarakis N. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 2008;15:105–112. doi: 10.1038/sj.cdd.4402231. [DOI] [PubMed] [Google Scholar]
- Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010;64:151–164. doi: 10.1111/j.1365-313X.2010.04315.x. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143:1132–1139. doi: 10.1104/pp.106.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4:20–27. doi: 10.4161/auto.5056. [DOI] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF, III, Dangl JL. Recognition and response in the plant immune system. Annu Rev Genet. 2003;37:579–609. doi: 10.1146/annurev.genet.37.110801.142628. [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Seay M, Patel S, Dinesh-Kumar SP. Autophagy and plant innate immunity. Cell Microbiol. 2006;8:899–906. doi: 10.1111/j.1462-5822.2006.00715.x. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Takano Y, Sakai Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010;584:1350–1358. doi: 10.1016/j.febslet.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- Gilroy EM, Hein I, van der Hoorn R, Boevink PC, Venter E, McLellan H, et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 2007;52:1–13. doi: 10.1111/j.1365-313X.2007.03226.x. [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, et al. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009;23:2496–2506. doi: 10.1101/gad.1825209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar K, Dong X. A kiss of death--proteasome-mediated membrane fusion and programmed cell death in plant defense against bacterial infection. Genes Dev. 2009;23:2449–2454. doi: 10.1101/gad.1861609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, et al. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21:2914–2927. doi: 10.1105/tpc.109.068635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B, et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J Biol Chem. 2010;285:13471–13479. doi: 10.1074/jbc.M109.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- Ivanov S, Fedorova E, Bisseling T. Intracellular plant microbe associations: secretory pathways and the formation of perimicrobial compartments. Curr Opin Plant Biol. 2010;13:372–377. doi: 10.1016/j.pbi.2010.04.005. [DOI] [PubMed] [Google Scholar]