Abstract
The Salvador–Warts–Hippo (SWH) pathway is a key controller of tissue growth in both flies and mammals, and deregulation of pathway activity contributes to tumour formation. The SWH pathway regulates cell growth, proliferation and apoptosis by restricting activity of the Yorkie transcriptional co-activator protein. The proteins that function together with Yorkie to drive transcription and tissue growth are beginning to be revealed and include the Scalloped (Sd), Teashirt (Tsh) and Homothorax (Hth) transcription factors. In this study, we define Wbp2 as a promoter of Yorkie-dependent growth of Drosophila melanogaster tissues. Mammalian WBP2 was previously identified as a protein that interacts with the mammalian Yorkie homologue, Yes-associated protein. WBP2 has been shown to enhance steroid hormone-dependent transcription in cultured cells but its in vivo function has remained obscure. We show that D. melanogaster Wbp2 interacts with Yorkie in a WW domain- and PY motif-dependent manner and that Wbp2 can enhance Yorkie's transcriptional co-activator properties. In vivo, Wbp2 is required for growth of the D. melanogaster wing, and reduction of Wbp2 expression suppresses overgrowth of tissues that lack the warts growth-suppressive gene. Collectively, these studies define an important role for Wbp2 as a downstream component of the SWH tissue growth-control pathway.
Keywords: Drosophila melanogaster, apoptosis, proliferation, Yorkie, Wbp2, Hippo pathway
The Salvador–Warts–Hippo (SWH) pathway is an important mediator of tissue growth that was first discovered in Drosophila melanogaster,1 and was subsequently shown to have a similar function in mammals.2, 3 Altered regulation of SWH pathway activity has been observed in both human and mouse tumours.4, 5 The SWH pathway can be broken down into three major classes of proteins: upstream regulators; the core kinase cassette; and downstream transcriptional regulatory proteins. Upstream regulators consist of three main branches: the Fat-Dachsous (Ft-Ds) branch, the Kibra-Expanded-Merlin (KEM) complex and the apical basal polarity proteins.6 Upstream regulatory branches crosstalk with each other and influence tissue growth by activating the core kinase cassette, which consists of the serine/threonine kinases, Hippo (Hpo)7, 8, 9, 10, 11 and Warts (Wts), and the adaptor proteins, Salvador (Sav)1, 12 and Mob as tumour suppressor (Mats).13 The core kinase cassette limits tissue growth by repressing activity of the Yorkie (Yki) transcriptional co-activator protein (Yes-associated protein (YAP) and TAZ/WWTR1 (transcriptional co-activator with PDZ-binding motif/WW domain containing transcription regulator 1) in mammals).14 Yki drives a growth-regulatory transcriptional programme by regulating transcription factors that include Scalloped (Sd),15, 16, 17 Homothorax (Hth) and Teashirt (Tsh).18 Yki and YAP bind via a region of ∼100 amino acids in their N-termini to Sd and TEAD1-4, respectively.15, 19 Additional transcription factors have been shown to be activated by YAP including pEBP2α, ErbB-4 and p73, and these factors bind to the WW domains of YAP via PPxY (PY) motifs.20
To determine whether transcriptional regulatory proteins that bind to the Yki/YAP WW domains are required for their ability to promote cell transformation and tissue growth, we and others assessed the activity of WW domain mutant versions of Yki/YAP.21, 22, 23 Interestingly, we found that the WW domains conveyed context-specific regulatory function to YAP; in the human breast epithelial MCF10A cell line, WW domains mediated inhibition of YAP's ability to induce cell transformation,21, 23 but in NIH-3T3 fibroblasts, YAP lacking functional WW domains displayed lower activity.23 In addition, the Yki WW domains were absolutely required for Yki to promote both normal, and excessive, tissue growth.22, 23 This suggested that proteins that bind the Yki/YAP WW domains are required for the ability of Yki, YAP and TAZ to promote transcription.
To identify such proteins we performed large-scale immunoprecipitations of TAZ coupled with mass-spectrometry.24 This analysis revealed a specific interaction with WW domain-binding protein-2 (WBP2). Interestingly, WBP2 was one of the first identified YAP-interacting proteins25 and was previously shown to cooperate with YAP to increase ligand-dependent transactivation of oestrogen and progesterone receptors.26 However, WBP2 function has never before been investigated in vivo. In this study, we show that the D. melanogaster Wbp2 homologue, CG11009, binds to Yki in a WW domain- and PY motif-dependent manner and can potentiate Yki's transcriptional co-activator function. We show that Wbp2 is required for growth of the D. melanogaster wing and for overgrowth of wts mutant tissues, thus defining Wbp2 as an important regulator of Yki-dependent tissue growth in vivo.
Results
Wbp2 is broadly expressed in larval D. melanogaster tissues, and is both nuclear and cytoplasmic
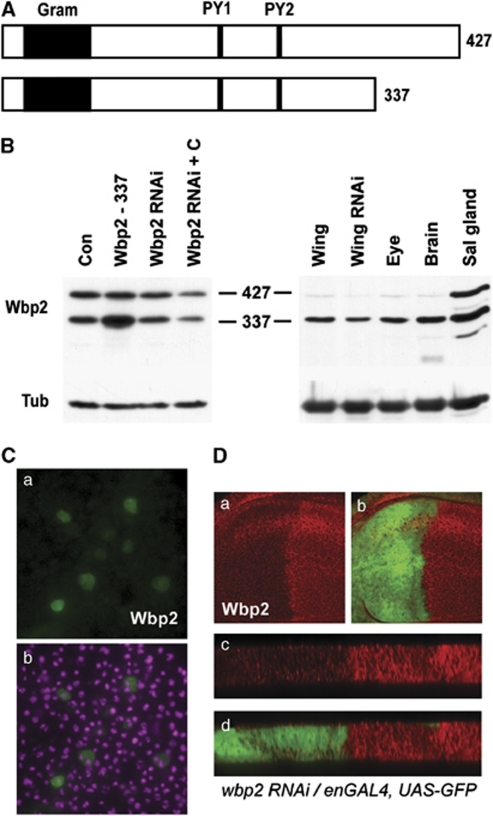
To identify candidate transcriptional regulatory proteins that interact with the WW domains of Yki and its mammalian homologues, YAP and TAZ, we performed large-scale immunoprecipitations with TAZ in cultured cells.24 A major interaction partner of TAZ was WBP2, homologues of which are found in many different species including D. melanogaster, which has a sole WBP2 homologue, CG11009 (hereafter referred to as Wbp2). The wbp2 gene is predicted to encode for two polypeptides of 337 and 427 amino acids, each of which contains a GRAM (glucosyltransferases, Rab-like GTPase activators and myotubularins) domain that has no defined function and two evolutionarily conserved P-P-X-Y (PY) motifs (Figure 1A). Using an antibody raised against recombinant Wbp2, we detected two protein isoforms of 39 and 49 kDa in S2 cells, matching the molecular mass of the two predicted Wbp2 isoforms (Figure 1B). Both isoforms were observed in several third instar larval tissues; eye and wing imaginal discs and brains expressed predominantly the 39-kDa isoform, whereas salivary glands expressed both isoforms equally (Figure 1B). Therefore, we chose to study the short Wbp2 isoform in the context of SWH pathway activity and growth control. To determine the subcellular localisation of Wbp2, we expressed an epitope-tagged version of the short Wbp2 isoform in S2 cells and found it to be present at similar amounts in both the cytoplasm and nucleus (Figure 1C).
Figure 1.
Wbp2 is broadly expressed in larval tissues and is both nuclear and cytoplasmic. (A) Schematic representation of proteins encoded by the wbp2 gene. Two protein products are predicted (337 and 427 amino-acids long) that each contain a GRAM domain and two evolutionarily conserved PY motifs. (B) Wbp2 protein expression assessed by immunoblotting in S2 cells and third instar larval tissues. S2 cells were transfected with vector control (Con), short Wbp2 isoform (Wbp2 – 337) or treated with dsRNA for Wbp2 in the presence or absence of Cellfectin. Tissues assessed were wings, eyes, brains and salivary glands. Wings expressing the wbp2-RNAi 1 transgene under the control of the 32B-Gal4 driver (Wing RNAi) were also assessed. (C) Subcellular localisation of Wbp2 (green) in S2 cells transfected with a Myc-tagged version of the short Wbp2 isoform. Nuclei are marked by DAPI (magenta). (D) The level of Wbp2 knockdown caused by expression of wbp2-RNAi 1 was assessed in vivo. Planar (a and b) and cross (c and d) sections of Wbp2 protein expression (red) in third instar larval wing discs expressing wbp2-RNAi 1 under the control of the en-GAL4 driver. Reduced Wbp2 expression was observed specifically in the posterior compartment of the wing disc, labelled with GFP (green)
We also assessed whether the Wbp2 protein could be depleted by RNA interference (RNAi) in cultured cells and in vivo. We achieved very poor knockdown of Wbp2 in S2 cells using standard methodology (∼10% reduction of Wbp2 protein), and ∼50% depletion when dsRNA delivery was aided by Cellfectin (Figure 1B). We achieved ∼50% protein depletion in larval wing imaginal discs using the 32B-Gal4 driver, which is expressed throughout the developing wing (Figure 1B). In both cultured cells and in vivo, we used two independent RNAi constructs, and observed similar results showing that Wbp2 protein is relatively resistant to RNAi-mediated depletion.
To provide confirmation for both our Wbp2 antibody and the effectiveness of two wbp2-RNAi transgenes, we assessed Wbp2 protein expression in developing wing imaginal discs that expressed wbp2-RNAi transgenes under control of the engrailed-Gal4 driver. Using either wbp2-RNAi line, we found Wbp2 expression to be lower in the engrailed expression domain of the wing disc than in the control anterior compartment, although residual protein was still present (Figures 1Da–d and data not shown). This confirmed that these wbp2-RNAi transgenes could successfully knock down Wbp2 protein, albeit partially, and that the antibody we generated is specific for Wbp2 protein isoforms.
Wbp2 and Yorkie interact in a WW domain- and PY motif-dependent manner
WBP2 was the first protein that was found to interact with murine YAP, and its discovery enabled the identification of the PY motif as the favoured ligand for WW domains.25 To determine whether the D. melanogaster WBP2 homologue, CG11009 (hereafter referred to as Wbp2, could interact with Yki we performed immunoprecipitation analysis using D. melanogaster S2 cell lysates. Wbp2 bound specifically to wild-type Yki, but not to Yki that lacked functional WW domains (Yki-WW1*2*)23 (Figure 2a).
Figure 2.
Yorkie and Wbp2 physically interact in a WW domain- and PY motif-dependent manner. (a) HA-tagged wild-type and mutant Yki proteins were assessed for their ability to bind to Myc-tagged Wbp2. Immunoprecipitations were performed with rabbit anti-HA antibody and immunoblotting was carried out with mouse anti-Myc to detect Myc-tagged Wbp2 and rat anti-HA to detect HA-Yki. (b) Myc-tagged wild-type and PY motif-mutant Wbp2 proteins were assessed for their ability to bind wild-type HA-Yki. Molecular mass markers (in kDa) and immunoglobulin (Ig) bands are indicated
Human WBP2 possesses three PY motifs, two of which are highly conserved in different eukaryotes, whereas a third PY motif is less conserved and present in some species, for example, humans, and not in others, for example Caenorhabditis elegans. Therefore, we focused on the two highly conserved Wbp2 PY motifs and determined whether they were essential for interaction with Yki. Each PY motif was mutated by converting the conserved Y to A, which renders PY motifs incapable of binding to WW domains.27, 28 Mutation of either Wbp2 PY motif largely prevented interaction with Yki, with only residual binding detected. A similar result was observed when both PY motifs were mutated (Figure 2b). Collectively, these studies show that the Yki WW domains are essential for interaction with Wbp2, that each Wbp2 PY motif can engage in interactions with Yki, and that both PY motifs of Wbp2 are required for optimal binding to Yki.
Wbp2 enhances Yorkie's transcriptional co-activator function
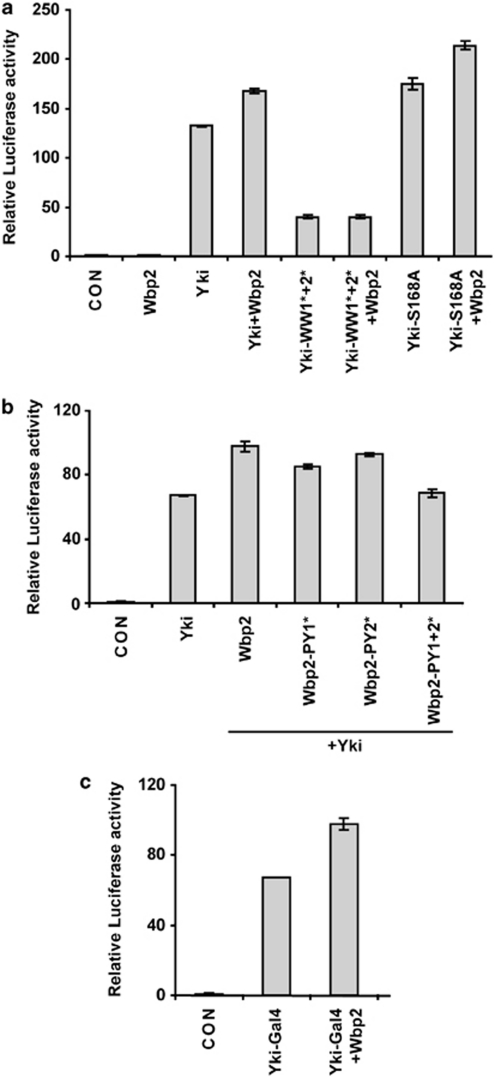
Previously, WBP2 was found to cooperate with YAP to promote ligand-dependent transactivation potential of progesterone and oestrogen receptors.26 Given that Wbp2 interacts with WW domains of Yki, which are required to promote Yki-dependent gene transcription and tissue growth, we hypothesised that Wbp2 would promote Yki's ability to activate transcription factors. To test this hypothesis, initially we determined whether Wbp2 could activate Yki in two independent luciferase-based transcription assays.
Yki-driven tissue overgrowth is dependent on the Sd transcription factor,15, 16, 17 and therefore, we investigated Wbp2's ability to regulate Yki activity in a Sd-dependent luciferase assay. The addition of wild-type Yki to S2 cells increased Sd activity by ∼130-fold, whereas in the absence of Yki, Wbp2 had no affect on Sd activity (Figure 3a). When Wbp2 was co-transfected with Yki, Sd's activity was increased a further 27% to ∼170-fold, showing that indeed Wbp2 can potentiate Yki's transcriptional co-activator function. To determine whether Yki WW domains were required for Wbp2 to potentiate Yki activity, we assayed the effect of Wbp2 expression on Yki-WW1*2*. Yki-WW1*2* increased Sd activity only ∼40-fold over control and therefore was 70% less potent than wild-type Yki at activating Sd, consistent with previous observations.23 Wbp2 was incapable of promoting the activity of Yki-WW1*2*, providing further evidence that Wbp2 activates Sd by interacting with Yki in a WW domain-dependent manner (Figure 3a).
Figure 3.
Wbp2 promotes Yorkie's transcriptional co-activator properties. (a and b) Luciferase assays measuring Sd-dependent transcriptional activity in S2 cells expressing the indicated plasmids. The effect of wild-type Wbp2 on wild-type Yki, WW domain mutant-Yki (Yki-WW1*2*) or Yki-S168A was assessed in a. The ability of wild-type Wbp2 or Wbp2 carrying mutations in PY motif 1, PY motif 2 or both PY motifs, to enhance Yki's ability to activate Sd was assessed in b. (c) Luciferase assay in S2 cells expressing the indicated plasmids, measuring Wbp2's ability to activate Yki-Gal4. Error bars in a–c represent S.D., n=3
Mammalian studies have shown that WBP2 complexes with DNA from gene promoters and that WBP2 possesses intrinsic transactivation function and can enhance the transactivation potential of the progesterone and oestrogen receptors together with YAP.25, 26 These findings suggest that WBP2 assembles into a transcriptional co-activator complex with YAP/Yki that influences the activity of cognate transcription factors. An alternate possibility is that WBP2 promotes YAP/Yki activity by interfering with SWH pathway-mediated repression of YAP/Yki. In an initial attempt to discern between these two possibilities, we investigated whether Wbp2 could also activate a version of Yki that lacks the major Wts-phosphorylation site, S168 (Yki-S168A). We reasoned that if Wbp2 activated Yki by abrogating SWH pathway-mediated repression, then Wbp2 would not activate Yki-S168A. When incubated in the presence of Yki-S168A, Sd activity was increased 175-fold and in the presence of Wbp2 and Yki, Sd was activated a further 22% (to ∼215-fold) (Figure 3a). Recently, Wts was shown to inhibit Yki by phosphorylation at two additional residues, S111 and S250.22 To rule out the possibility that Wbp2 could affect Wts-dependent phosphorylation of these sites, we repeated our luciferase assays with a version of Yki where S111, S168 and S250 were mutated to A (Yki-3SA), thus rendering it impervious to the Wts kinase. Wbp2 still retained the ability to strongly enhance the activity of Yki-3SA, thus ruling out this possibility (Supplementary Figure S1). These data suggest that Wbp2 does not activate Yki by influencing phosphorylation-mediated inhibition of Yki by the SWH pathway.
To further interrogate the role of WW domain–PY motif interactions between Yki and Wbp2, we performed luciferase assays using wild-type Yki and Wbp2 with various PY motif mutations. Wbp2 lacking either PY motif 1 or PY motif 2 activated Yki with slightly lower potency than wild-type Wbp2, whereas mutation of both conserved Wbp2 PY motifs completely abolished its ability to enhance Yki activity (Figure 3b). These results are consistent with our immunoprecipitation data showing that both Wbp2 PY motifs are required for interaction with Yki. Wbp2's ability to activate Yki was not restricted to Yki-dependent activation of the Sd transcription factor; in an independent assay measuring transactivation potential of a Yki-Gal4 fusion protein, Wbp2 enhanced Yki-Gal4 activity by 46% (Figure 3c).
Wbp2 is required for growth of the D. melanogaster wing
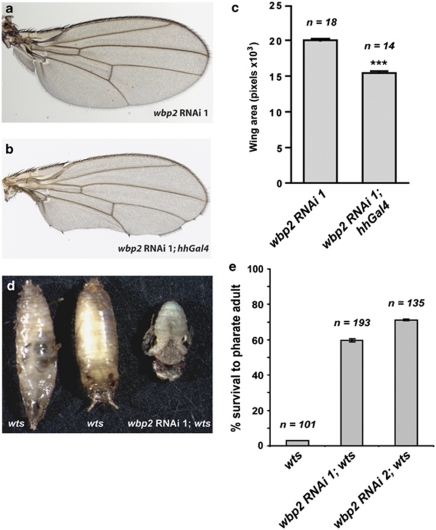
On the basis of our findings that Wbp2 interacted with and activated Yki, we reasoned that like Yki, Wbp2 would be required for tissue growth. To investigate this hypothesis, we expressed two independent transgenic RNAi constructs targeted against different regions of wbp2 in the posterior compartment of the wing using hedgehog-GAL4 (hh-GAL4). At 29°C, both wbp2-RNAi transgenes produced wings with scalloped edges that were reduced in size (Figure 4b and data not shown) compared with wbp2-RNAi flies lacking hh-GAL4 (Figure 4a). Quantification of the posterior compartment indicated that wbp2-RNAi 1; hh-GAL4 wings were 23% smaller than negative control flies (Figure 4c). Collectively, these data show that Wbp2 is required for wing growth during normal development.
Figure 4.
Wbp2 depletion restricts wing growth and partially rescues lethality caused by deficiency of the warts gene. (a and b) RNAi-mediated depletion of wbp2 in the posterior compartment of the wing (using hh-GAL4) results in reduced wing size (b) compared with wbp2-RNAi control wings that lack the hh-GAL4 driver (a). Flies were reared at 29°C to increase GAL4/UAS expression. (c) Quantification of the effect of Wbp2 knockdown in the hh-GAL4 expression domain in adult wings. wbp2-RNAi 1; hh-GAL4 wings showed a 23% reduction in the posterior compartment of the adult wing (right column) compared with wbp2-RNAi 1 flies that lacked the hh-GAL4 driver (left column). The posterior compartment of the indicated genotypes was measured (in pixels) and compared using an unpaired Student's t-test. Values represent the Mean±S.E.M., ***P<0.0001. (d) The effect of Wbp2 knockdown on lethality caused by the generation of wtsX1 mutant eye discs was assessed. Lethal pupae harbouring wtsX1 mutant eye discs generated using eyFLP/MARCM are shown on the left, and a live adult generated using eyFLP/MARCM harbouring wtsX1 mutant eyes that also expressed wbp2-RNAi 1 is shown on the right. (e) Rescue of animals with wtsX1 mutant eye tissues from late larval lethality to the pharate adult stage of development by two independent wbp2-RNAi lines. Percentage of pupae of the indicated genotypes that developed to pharate adults is shown, with error bars representing S.E.M.. In c and e the number of animals (n) assessed for each genotype is indicated
Wbp2 is required for overgrowth of eye tissue lacking warts
Next, to determine whether Wbp2 is required for tissue overgrowth arising from repression of SWH pathway signalling, we depleted Wbp2 expression in wts-deficient tissue. Clones of tissue harbouring a null allele of wts, wtsX1, were generated in the developing D. melanogaster eye using an eyFlp/MARCM system.29 These tissues overgrew dramatically and caused lethality at late third instar larval or early pupal stages of development. Strikingly, when wbp2-RNAi 1 was expressed in wtsX1-deficient tissues using eyFlp/MARCM, we observed a significant increase in survival of flies with the majority reaching late pupal development and forming pharate adults, with a small number of flies emerging as fully developed adults (Figures 4d and e). These observations were phenocopied using a second independent wbp2 RNAi line (wbp2-RNAi 2), providing strong evidence that the observed phenotype was not due to off-target effects (Figure 4e).
To assess the effect of reducing Wbp2 protein on the growth of wts tissues, we observed the overall size and differentiation of developing eye imaginal discs. Control third instar larval eye imaginal discs generated with the parent FRT82B chromosome and eyFlp/MARCM showed a roughly equivalent proportion of GFP-positive clones to control unlabelled clones. F-Actin was observed in an ordered pattern posterior to the morphogenetic furrow (MF) corresponding to presumptive ommatidial clusters containing differentiating photoreceptor cells. The MF was observed as a concentration of F-Actin, caused by cells in this region of the developing eye undergoing apical constriction (Figures 5a–a″). When wts clones were generated using eyFlp/MARCM, eye discs substantially overgrew with wts clones occupying almost the entire eye disc (Figure 5b′). In addition, third instar larval wts mutant eye discs displayed strong differentiation defects; very few presumptive ommatidial clusters were observed as determined by F-Actin morphology and there was no obvious sign of the MF, consistent with previous studies30 (Figures 5b–b″). When Wbp2 was knocked down in wts eye tissues using eyFlp/MARCM and either wbp2-RNAi line, we observed a reduction in eye disc size compared with wts mosaic eye discs, a decrease in the amount of wts tissue (GFP positive) relative to control tissue, and almost complete restoration of differentiation of the developing eye, as assessed by F-Actin morphology (Figures 5c–c″).
Figure 5.
Wbp2 depletion partially reverts overgrowth, differentiation defects and ectopic CycE expression of eye tissue lacking the warts gene. (a–c″) The effect of Wbp2 knockdown on the overgrowth of wtsX1 mutant tissue was analysed in third instar larval eye discs with GFP-positive eyFLP/MARCM clones of FRT82B (a–a″), wtsX1 (b–b″) and wbp2-RNAi 1; wtsX1 (c–c″). Discs were stained for F-Actin (red in a, b and c). Control and mutant clones express GFP (green in a′, b′ and c′) and are merged with F-Actin staining images in a″, b″ and c″. Inset images in a, b and c are high magnification views of F-Actin. Arrowheads in a and c indicate the morphogenetic furrow, which was undetectable in b. Knockdown of Wbp2 in wtsX1 clones resulted in reduced overgrowth, a partial return to wild-type F-Actin staining and an increased amount of GFP-negative wild-type tissue. (d–e″) CycE expression (red in d and e) and the merged images (d″ and e″) were assessed in third instar larval eye discs with genotypes wtsX1 (d–d″) or wbp2-RNAi 1; wtsX1 (e–e″). In d–e″ arrowheads indicate the morphogenetic furrow, whereas arrows in d–d″ indicate a large wtsX1 clone with elevated CycE. Scale bars represent 50 μM in a–e″. (f–h) The specificity of Wbp2's role as a regulator of tissue growth was determined by assessing the effect of Wbp2 knockdown on tissue lacking the Tsc1 gene. Eyes from the following adult female flies are shown: (f) eyFLP/MARCM control containing wild-type clones; (g) eyFLP/MARCM generated clones of Tsc1Q87X; (h) eyFLP/MARCM generated clones of UAS-wbp2 RNAi 1; Tsc1Q87X. Expression of UAS-wbp2 RNAi did not suppress eye overgrowth caused by Tsc1 deficiency. Anterior is to the right in all images
Similar data were observed when expression of the neuronal marker, ELAV, was assessed in wts eye tissue, with or without expression of wbp2-RNAi (Supplementary Figure S2). Differentiation was largely defective in third instar larval eye discs harbouring wts clones, however, disruption to differentiation was variable. For example some wts mosaic discs displayed very few ELAV-positive cells, whereas other discs contained regions of eye displaying some signs of differentiation, as detected by ELAV expression (Supplementary Figure S2). Consistently, however, we observed a more regular ELAV-staining pattern in wts discs expressing wbp2-RNAi, showing that eye differentiation was largely restored (Supplementary Figure S2). These data are consistent with Wbp2 having an essential role in enhancing Yki's ability to promote tissue growth.
To further investigate this, we determined whether Wbp2 knockdown could reduce ectopic expression of the SWH pathway-target protein, Cyclin E (CycE). In the developing third instar larval eye disc, elevated CycE posterior to the second mitotic wave (SMW) is a characteristic feature of diminished SWH pathway activity or of overexpression of Yki.1, 14 Consistent with previous reports, CycE was elevated in wts tissue posterior to the SMW1 (Figures 5d–d″). However, when Wbp2 was reduced by RNAi in wts clones, CycE expression posterior to the SMW returned to baseline levels suggesting that Yki's ability to drive high levels of CycE expression was disabled (Figures 5e–e″). Importantly, the ability of Wbp2 knockdown to revert tissue overgrowth was specific to the SWH pathway, as tissue overgrowth caused by deficiency of Tsc1, an inhibitor of the target of rapamycin (TOR) pathway, was not affected when Wpb2 was knocked down in Tsc1Q87X mutant eyFlp/MARCM clones (Figures 5f–h).
Wbp2 protein expression and subcellular localisation is not modulated by reduced SWH pathway activity
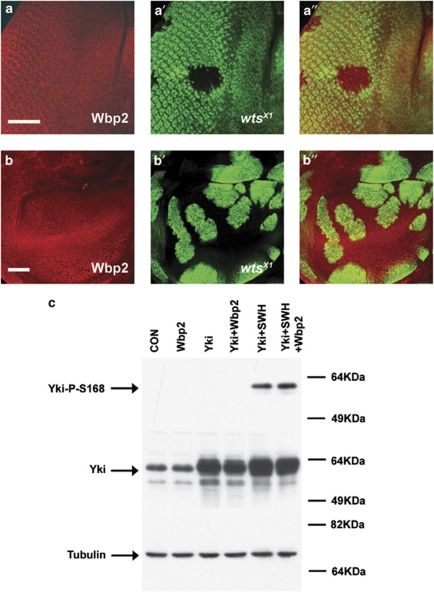
In developing eye and wing imaginal discs, Yki accumulates in the nucleus of cells when SWH pathway activity is compromised.3, 22 Therefore, we considered the possibility that Wbp2 protein levels and localisation might be influenced by SWH pathway activity. However, analysis of endogenous Wbp2 protein in wtsX1 mosaic eye and wing imaginal discs showed no obvious change in Wbp2 localisation in wtsX1 tissue compared with wild-type tissue, suggesting that Wbp2 expression and subcellular localisation are not influenced by SWH pathway activity (Figures 6a–b″).
Figure 6.
Wbp2 subcellular localisation is not altered by the SWH pathway and Wbp2 does not interfere with SWH pathway-mediated phosphorylation of Yorkie. (a and b) Expression and localisation of Wbp2 in tissue lacking wtsX1 were assessed in third instar larval discs. Wbp2 protein expression (red in a and b) is shown in mosaic eye discs with GFP-negative hsFLP wtsX1 clones (a–a″) and wing discs with GFP-positive hsFLP/MARCM wtsX1 clones (b–b″). Protein and GFP expression images are merged in a″ and b″. Scale bars represent 50 μM. (c) The effect of Wbp2 on SWH-induced Yki phosphorylation was assessed by immunoblotting protein extracts derived from S2 cells transfected with control vector (CON); Wbp2, Yki, Yki and Wbp2; Yki, Sav, Wts and Hpo; or Yki, Wbp2, Sav, Wts and Hpo. Lysates were probed with antibodies against phospho-Yki-S168, total Yki and Tubulin
Wbp2 does not affect Warts-mediated phosphorylation of Yorkie
Several transcription factors have been shown to control tissue growth in conjunction with Yki but how these proteins regulate transcription is still unclear.15, 16, 17, 18 What is better defined is the mechanism by which Yki activity is regulated by the SWH pathway; Wts/LATS1/2-mediated phosphorylation of Yki/YAP results in 14-3-3-dependent cytoplasmic sequestration of Yki/YAP, thus preventing Yki/YAP activity in the nucleus.3 To investigate whether Wbp2 affected Yki by influencing SWH pathway-mediated repression, we determined whether Wbp2 influenced Wts-dependent phosphorylation of Yki in S2 cells using anti-phospho-Yki-S168 antibody. As shown in Figure 6c, co-transfection of Yki with Sav, Wts and Hpo caused a substantial increase in Yki phosphorylation. However, co-transfection of Wbp2 with Yki, Sav, Wts and Hpo had no effect on Yki phosphorylation (Figure 6c). These results suggest that Wbp2 does not promote Yki activity by alleviating SWH pathway-mediated repression of Yki. This conclusion is entirely consistent with our luciferase assay results in Figure 2a and Supplementary Figure S1 that showed that Wbp2 could activate wild-type Yki, as well as versions of Yki that are resistant to SWH pathway-mediated repression, with similar potency.
Discussion
Previously, we and others found that the Yki/YAP WW domains were required for these proteins to drive tissue growth or transformation of at least some cell lines, respectively, suggesting that these domains interact with proteins that are necessary for the transcriptional co-activator function of Yki/YAP.22, 23 One such candidate protein was Wbp2, which was previously shown to bind the WW domains of YAP and influence its ability to promote transcription.25, 26 We found that Wbp2 enhances Yki's intrinsic transcriptional co-activator activity and that the ability of Wbp2 to bind and activate Yki was dependent on Yki's WW domains and Wbp2's PY motifs (Figures 2 and 3). Thus, these data are consistent with previous observations that defined a key positive regulatory role of the WW domains of Yki.
Consistent with the idea that Wbp2 promotes the activity of Yki, we found that RNAi-mediated depletion of Wbp2 compromised growth of the D. melanogaster wing. In addition, Wbp2 depletion partially reverted tissue overgrowth and organism lethality caused by wts deficiency, as well as ectopic expression of the downstream SWH pathway target, CycE (Figures 4 and 5). Collectively, these data provide strong in vivo evidence that Wbp2 is an important mediator of SWH pathway-dependent tissue growth. Further investigation of tissues that lack Wbp2 altogether will enable us to determine whether Wbp2 is obligate for Yki function in all settings, or whether it controls Yki activity in tissue- or temporally restricted manners, akin to the transcription factors Sd, Hth and Tsh.
The precise mechanism by which Wbp2 activates Yki is still unclear. We considered the possibility that Wbp2 activates Yki by interfering with SWH pathway-mediated repression of Yki. However, two points of data refuted this notion: (1) Wbp2 overexpression had no effect on the ability of overexpression of Sav, Wts and Hpo to induce Yki phosphorylation at S168, an event that inhibits Yki's activity; and (2) Wbp2 activated both wild-type Yki and Yki mutant proteins that were impervious to SWH pathway-mediated repression, with similar potency (Figures 3 and 6). In overexpression studies in cultured cells, Wts, Ex and Hpo can interact with Yki and repress its activity in a phosphorylation-independent mechanism.31, 32 We found that Wbp2 depletion reduced overgrowth of tissues harbouring amorphic alleles of wts, showing that Wbp2 exerts an effect epistatically to Wts. Therefore, Wbp2 does not activate Yki by interfering with Wts-mediated Yki repression, whether it is dependent or independent of phosphorylation. Further studies are required to determine whether Wbp2 can interfere with the ability of Ex or Hpo to repress Yki in a phosphorylation-independent manner. However, we believe a more likely mechanism for Wbp2 function is that it directly promotes the transcription co-activator function of Yki. This notion is supported by the fact that Wbp2 was observed in the nucleus and by the following mammalian studies: YAP and WBP2 can complex with DNA, presumably as part of higher order protein complexes as YAP at least can not bind DNA directly;26, 33 WBP2 and YAP possess intrinsic gene transactivation potential and can exert an effect cooperatively to promote transactivation of oestrogen and progesterone receptors.26, 34 Clearly, however, further experimentation is required to delineate the precise mechanism by which Wbp2 cooperates with Yki/YAP to control transcription and tissue growth.
An interesting facet of this study is that it further emphasises the importance of WW domain–PY motif interactions in the SWH pathway. Previously identified PY motif-containing proteins in this pathway include Wts, Ex and Hpo, whereas Sav, Yki and Kibra all have WW domains.20 The reiterated use of this ligand-domain pair in the SWH pathway is unique among signalling pathways and raises the possibility that other as yet unidentified WW domain- or PY motif-containing proteins form part of the SWH tissue growth-control pathway.20 From a human disease perspective, evidence supporting an important role for deregulation of SWH pathway activity in the genesis of human cancer is mounting.4, 5 Given that reduction of Wbp2 expression partially rescued overgrowth and lethality associated with hyperactivated Yki protein in wts-deficient tissues, WBP2 constitutes a potential therapeutic target in human tumours that are driven by hyperactivation of the Yki homologues, YAP and TAZ.
Materials and Methods
D. melanogaster stocks
Wbp2 knockdown was performed using two different RNAi lines: UAS-wbp2-RNAi 1 (KK) and UAS-wbp2-RNAi 2 (GD), both from the Vienna Drosophila RNAi Center.35 eyFlp and hsFlp MARCM clones and loss-of-function clones were generated with the following stocks: w; FRT82B wtsX1,36 w; FRT82B Tsc1Q87X,37 eyFlp, UAS-GFP; tub-GAL4 FRT82B tub-GAL80, hsFlp, UAS-GFP; tub-GAL4 FRT82B tub-GAL80,29 and y w hsFlp; FRT82B ubi-GFP. Heat shocks for 20 min at 37°C were performed 3 days after egg deposition for experiments using hsFlp. Other stocks used were engrailed-Gal4, hedgehog-Gal4, 32B-Gal4, UAS-GFP and w; FRT82B.
Quantification of wing size
Wings from male adult flies reared at 29°C were dissected, fixed in xylene (BDH, Poole, UK) and mounted in Canada balsam (Sigma-Aldrich, St. Louis, MO, USA). Wings were imaged on an Olympus BX51 microscope (Olympus, Tokyo, Japan) at 4 × magnification using Advanced Spot software (version 4.7, Diagnostic Instruments, Sterling Heights, MI, USA). The posterior compartment size of each wing was quantified using the Measurement function of Adobe Photoshop (CS3), with pixel length and logical length both set at 1. The mean and S.E.M. values of wing area were determined with GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA). For statistical analysis, genotypes were compared with an unpaired Student's t-test using GraphPad Prism. P values <0.05 were considered significant.
Immunofluorescence
Primary antibodies were specific for Wbp2 (this study), CycE (H Richardson),38 Anti-Myc (9E10, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and ELAV (Developmental Studies Hybridoma Bank). Anti-rat and anti-mouse secondary antibodies were from Molecular Probes (Eugene, OR, USA). Tritc-Phalloidin and 4,6-diamidino-2-phenylindole (DAPI) were from Sigma-Aldrich. Tissues were fixed in 4% paraformaldehyde and incubated with antibodies overnight at 4 °C in phosphate-buffered saline with 10% goat serum and 0.1% Triton X-100 as in Bennett and Harvey.39 S2 cells were fixed in 4% paraformaldehyde and incubated with anti-Myc at 4 °C for 1 h in phosphate-buffered saline with 10% goat serum and 0.2% Triton X-100.
Expression plasmids
A wbp2 cDNA encoding the 337-amino-acids isoform was amplified from clone PM01318 (Drosophila Genomics Resource Center via the Australian Drosophila Research Support Facility) using Myc-tagged primers and cloned into pAc5.1 to generate pAc-Myc-Wbp2.
Forward: 5′-CCGAATTCATGGAACAAAAACTCATCTCAGAAGAGGATCTGTCGGTTAATACGGCTCACG-3′
Reverse: 5′-GGCTCGAGTTACATTCCAGGGGGCAAG-3′.
Mutations in Wbp2 PY motif 1 (Y203 to A), PY motif 2 (Y252 to A) or both PY motifs were introduced into pAc-Myc-Wbp2 by QuickChange Mutagenesis (Stratagene, La Jolla, CA, USA). The S168A mutation was introduced into pAc-Yki-HA by QuickChange Mutagenesis (Stratagene) and WW domain mutations in pAc-Yki-HA were described previously.23
Luciferase assays
Sd-dependent luciferase assays were performed as described previously.23 D. melanogaster S2 cells were transfected with Sd-Gal4, pUAST-Luciferase17 and pGL3-HSP-Renilla (internal control) plasmids, along with control pAc5.1 plasmid or plasmids expressing wild-type or mutant Yki proteins (Yki-S168A, Yki-WW1*2* or Yki-3SA),16, 23 and/or wild-type or mutant Wbp2 proteins. Yki-Gal4 luciferase assays were performed as described in Huang et al.14 S2 cells were transfected with a control plasmid or plasmids encoding Yki and/or Wbp2. After 36 h, cells were lysed and luciferase activity was recorded using the Dual-luciferase reporter assay system (Promega, Madison, WI, USA). Results represent the mean of three experiments, whereas error bars represent S.D. (n=3).
Antibody production
wbp2 cDNA encoding the 337-amino-acid isoform was cloned into pGEX-4T-1, and full-length yki cDNA into pGEX-4T-3. GST-Wbp2 or GST-Yki proteins were induced in BL21 cells and excised from a SDS-PAGE gel. Rats were inoculated with GST-Wbp2 and rabbits with GST-Yki by IMVS Veterinary Services (Adelaide, South Australia, Australia) and serum was collected as in Milton et al.40
Immunoprecipitations and immunoblotting
S2 cells were transfected with plasmids encoding wild-type or mutant Yki and Wbp2 proteins and lysed after 48 h. Lysates were immunoprecipitated with rabbit anti-HA (Sigma-Aldrich). Immunoprecipitates were subjected to SDS-PAGE and immunoblotted with either mouse anti-Myc (9E10, Santa Cruz Biotechnology), rat anti-HA (Roche, Basel, Switzerland), rabbit anti-Yki (this study), rabbit anti-Yki-phospho-S168 (From D Pan)3 and mouse anti-Tubulin (Developmental Studies Hybridoma Bank).
Acknowledgments
We thank N Albanese for technical assistance, A Brumby, S Cohen, I Hariharan, J Jiang, L Luo, D Pan, H Richardson, A Zider, the Developmental Studies Hybridoma Bank, the Drosophila Genomics Resource Center, the Vienna Drosophila RNAi Center, the Australian Drosophila Research Support Facility (http://www.ozdros.com) and the Bloomington Stock Centre for fly stocks, plasmids and antibodies. KH is a Sylvia and Charles Viertel Senior Medical Research Fellow. This work was supported by a Project Grant from the National Health and Medical Research Council of Australia and a Discovery Grant from the Australian Research Council.
Glossary
- CycE
Cyclin E
- DAPI
4, 6-diamidino-2-phenylindole
- Ft
Fat
- Ds
Dachsous
- Ex
Expanded
- GFP
green fluorescent protein
- GRAM
glucosyltransferases, Rab-like GTPase activators and myotubularins
- hh
hedgehog
- Hpo
Hippo
- Hth
Homothorax
- KEM
Kibra-Expanded-Merlin
- Mats
Mob as tumor suppressor
- Mer
Merlin
- MF
morphogenetic furrow
- RNAi
RNA interference
- Sav
Salvador
- Sd
Scalloped
- SMW
second mitotic wave
- SWH
Salvador–Warts–Hippo
- TAZ/WWTR1
transcriptional co-activator with PDZ-binding motif/WW domain containing transcription regulator 1
- TOR
target of rapamycin
- Tsh
Teashirt
- Wbp2
WW domain-binding protein-2
- Wts
Warts
- YAP
Yes-associated protein
- Yki
Yorkie
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Kumar
Supplementary Material
References
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway – an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the hippo size control pathway. Curr Biol. 2010;20:R574–R582. doi: 10.1016/j.cub.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Humbert PO, Harvey KF. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ Oncogene 2010. doi: 10.1038/onc.2010.438 [DOI] [PMC free article] [PubMed]
- Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, Peterson S, et al. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- Dhananjayan SC, Ramamoorthy S, Khan OY, Ismail A, Sun J, Slingerland J, et al. WW domain binding protein-2, an E6-associated protein interacting protein, acts as a coactivator of estrogen and progesterone receptors. Mol Endocrinol. 2006;20:2343–2354. doi: 10.1210/me.2005-0533. [DOI] [PubMed] [Google Scholar]
- Chen HI, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J Biol Chem. 2002;277:9307–9317. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Menut L, Vaccari T, Dionne H, Hill J, Wu G, Bilder D. A mosaic genetic screen for Drosophila neoplastic tumor suppressor genes based on defective pupation. Genetics. 2007;177:1667–1677. doi: 10.1534/genetics.107.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Richardson H, O'Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development. 1995;121:3371–3379. doi: 10.1242/dev.121.10.3371. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Milton CC, Zhang X, Albanese NO, Harvey KF. Differential requirement of Salvador-Warts-Hippo pathway members for organ size control in Drosophila melanogaster. Development. 2010;137:735–743. doi: 10.1242/dev.042309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.