Abstract
The reverse transcriptase telomerase is a ribonucleoprotein complex that adds telomeric repeats to chromosome ends, using a sequence within its endogenous RNA component as a template. Although templating domains of telomerase RNA have been studied in detail, little is known about the roles of the remaining residues, particularly in yeast. We examined the functions of nontemplate telomerase residues in the telomerase RNA of budding yeast Kluyveromyces lactis. Although approximately half of the RNA residues were dispensable for function, four specific regions were essential for telomerase action in vivo. We analyzed the effects of mutating these regions on in vivo function, in vitro telomerase activity, and telomerase RNP assembly. Deletion of two regions resulted in synthesis of stable RNAs that appeared unable to assemble into a stable RNP. Mutating a region near the 5′ end of the RNA allowed RNP assembly but abolished enzymatic activity. Mutations in another specific small region of the RNA led to an inactive telomerase RNP with an altered RNA conformation.
Keywords: Telomerase, RNA, ribonucleoprotein, telomere, yeast, K. lactis
Ribonucleoprotein (RNP) complexes play central roles in essential cellular processes that include translation, rRNA, pre-mRNA and pre-tRNA processing, and telomere maintenance. In the ribosome and the spliceosome, large RNA molecules form a structural framework for the assembly of proteins and provide critical active site residues. The reverse transcriptase (RT) telomerase is also an RNP complex (for review, see Blackburn 1992, 1998), which uses a small part of an intrinsic RNA component as a template for telomeric DNA synthesis. Telomerase copies this template to synthesize new telomeric repeat units onto the termini of eukaryotic chromosomes. Telomeric DNA consists of a tandem array of repeat units whose length and number are often tightly controlled. The inability of conventional DNA polymerases to replicate the very ends of chromosomes would cause progressive loss of DNA sequences from the ends (Watson 1972; Olovnikov 1973). Telomerase action compensates for this loss and is thus required for long-term chromosomal maintenance.
Telomerase activity, purified partially from ciliate, human, and yeast extracts, has been characterized in detail in vitro (Morin 1989; Greider 1991; Lee and Blackburn 1993; Cohn and Blackburn 1995). In vitro enzyme action requires binding to a telomeric oligonucleotide primer and dNTP substrates, alignment of the primer 3′ end to the templating domain of telomerase RNA, and its extension by incorporation of nucleotide triphosphates complementary to the templating residues. Yeast telomerases typically synthesize only one repeat unit in vitro and stay bound to the product after completion of DNA synthesis (Prescott and Blackburn 1997a; Fulton and Blackburn 1998). The core catalytic components of the enzyme are telomerase RNA (TER) and a protein subunit containing conserved RT motifs (Weinrich et al. 1997; Beattie et al. 1998). Conserved aspartate residues in these motifs are required for catalysis (Counter et al. 1997; Harrington et al. 1997b; Lingner et al. 1997b; Weinrich et al. 1997). Additional proteins are associated with the core catalytic unit and may alter enzymatic properties or mediate the interaction of the enzyme with telomeres (Collins et al. 1995; Steiner et al. 1996; Harrington et al. 1997a; Nakayama et al. 1997; Gandhi and Collins 1998).
Telomerase RNA sizes vary widely among eukaryotes, from ∼150 to 200 nucleotides in ciliates (Greider and Blackburn 1989; Shippen-Lentz and Blackburn 1990) and ∼400 to 450 nucleotides in mammals (Blasco et al. 1995; Feng et al. 1995) to 1200–1300 nucleotides in the yeasts Saccharomyces cerevisiae and Kluyveromyces lactis (Singer and Gottschling 1994; McEachern and Blackburn 1995). This size variability is accompanied by a lack of overall primary sequence conservation among each group of organisms, although core secondary structure is conserved among widely divergent ciliate RNAs (Romero and Blackburn 1991; Lingner et al. 1994; McCormick-Graham and Romero 1995). The templating domain has been the most well-studied portion of telomerase RNA. The consequences of telomerase RNA template mutations have been studied in vivo and in vitro in ciliates (for review, see Greider 1996 and references therein) and yeasts (Prescott and Blackburn 1997b) and examined minimally in mammals (Marusic et al. 1997). Most of these mutations were faithfully copied into telomeric DNA by the enzyme in vivo and in vitro. Strikingly, several of these mutations affected enzymatic properties such as processivity, fidelity, and proper termination of synthesis; in one case eliminating telomerase activity in vivo and in vitro (Gilley et al. 1995; Gilley and Blackburn 1996; Prescott and Blackburn 1997a,b). Hence, the template region of the RNA must contribute to active site functions. Less is known about the roles of the nontemplate regions of telomerase RNA, and in this regard the large size of the yeast telomerase RNAs is intriguing. An exciting possibility is that the RNA component of telomerase may contribute to the catalytic properties of the enzyme. In addition, telomerase RNA may provide a matrix for the assembly of the telomerase complex.
Experiments with ciliate telomerase RNAs in vivo have revealed the importance of nontemplate residues (Bhattacharyya and Blackburn 1997). When the nontemplate portion of the Tetrahymena telomerase RNA is replaced with the corresponding region from another ciliate species, Glaucoma, and the chimeric RNA is expressed in Tetrahymena, it assembles into an only partially functional enzyme with an aberrant cleavage activity. In vitro studies by reconstitution of enzyme activity from in vitro synthesized telomerase RNA and partially purified cell extracts have shown that mutations in some regions outside the template domain of Tetrahymena telomerase RNA abolish or significantly reduce reconstituted activity (Autexier and Greider 1998). Similarly a 30-nucleotide nontemplate region of the human telomerase RNA is necessary for reconstitution of enzyme activity (Autexier et al. 1996). However, these studies did not distinguish whether these sequences are required for enzyme activity itself or for telomerase RNP assembly.
Here we report a detailed examination of the role of nontemplate residues in the telomerase RNA (TER1) of the budding yeast K. lactis. Each of the 12 K. lactis telomeres contains an average of 15 tandem 25-bp repeats, with the range being 10–20 repeats. TER1 RNA contains a 30-nucleotide template region that is complementary to 1 telomeric repeat units (McEachern and Blackburn 1995), and an in vitro telomerase activity in K. lactis has been described recently (Fulton and Blackburn 1998). We report that although approximately half of the TER1 RNA sequences are dispensable, several small sequences are critical for enzyme function in vivo and in vitro. Two small regions are essential for activity, although mutations in them still allow telomerase RNP assembly. Interestingly, mutating one of these regions abolishes detectable telomerase activity both in vivo and in vitro and significantly changes the conformation of the mutant telomerase RNA as well as the telomerase RNP complex. This is the first demonstration that in yeast telomerase RNAs, residues far from the template play essential roles in the telomerase RNP and its activity both in vitro and in vivo.
telomeric repeat units (McEachern and Blackburn 1995), and an in vitro telomerase activity in K. lactis has been described recently (Fulton and Blackburn 1998). We report that although approximately half of the TER1 RNA sequences are dispensable, several small sequences are critical for enzyme function in vivo and in vitro. Two small regions are essential for activity, although mutations in them still allow telomerase RNP assembly. Interestingly, mutating one of these regions abolishes detectable telomerase activity both in vivo and in vitro and significantly changes the conformation of the mutant telomerase RNA as well as the telomerase RNP complex. This is the first demonstration that in yeast telomerase RNAs, residues far from the template play essential roles in the telomerase RNP and its activity both in vitro and in vivo.
Results
A BclI template mutation provides a marker for assaying in vivo functions of TER1 RNA
To investigate the roles of telomerase RNA sequences outside the template, we created a series of TER1 deletion or substitution mutants and assayed their function in vivo. Function was assessed in a K. lactis strain deleted for the entire TER1 gene (strain 7BΔTER1). As described previously for a TER1 allele containing a 300-bp deletion including the templating domain (McEachern and Blackburn 1995), telomeric repeats in strain 7BΔTER1 were progressively lost from the chromosome ends (Fig. 1A, lanes 5,6). Upon further passaging, the cells grew very poorly, forming small senescent colonies with extremely rough edges and critically short telomeric fragments. The occasional healthy, smooth survivor colonies contained cells that had regenerated long tracts of telomeric DNA by an alternative RAD52-dependent recombination pathway (McEachern and Blackburn 1996; Fig. 1A, lanes 7,8). As described previously, such survivor colonies with elongated telomeric DNA continue to be prone to telomeric shortening and additional episodes of growth senescence (McEachern and Blackburn 1996). When the 7BΔTER1 strain was transformed with a centromeric plasmid expressing the wild-type TER1 gene (pTERWT), wild-type length telomeres were regained. These telomeres were stably maintained after numerous serial passages (Fig. 1A, lanes 9,10), each passage representing ∼25 cell doublings.
Figure 1.
Deletion of TER1 results in telomere shortening, which is complemented by TER1–BclI. Southern blots of yeast genomic DNA showing telomeric profiles from wild-type strain 7B520, ΔTER1, or derivatives thereof. DNAs are digested with EcoRI (A) or in pairs of digestions with EcoRI (R) and EcoRI plus BclI (R/B) (B,C) and probed with radiolabeled oligonucleotide probes for telomeric repeats as indicated. The wild-type (WT) probe (oligonucleotide Klac 1–25) hybridizes to wild-type and mutant repeats. The BclI probe (oligonucleotide KTelBcl) hybridizes only to BclI-marked repeats. The same filter was probed first with the BclI probe (C) and subsequently with the wild-type probe (B). DNA markers in kilobases are indicated to the left of each panel. All 12 telomeres are visualized. The band around 1 kb contains seven telomeres and the band just above 1.6 kb contains two telomeres. The telomeric fragment marked with an asterisk in B contains a BclI restriction site between the subtelomeric EcoRI site and the telomere end. The arrowheads demonstrate examples of telomeric fragment shortening with BclI digestion. Each passage represents ∼25 cell doublings.
To differentiate between telomeric repeats added by telomerase containing the mutant RNAs and the preexisting wild-type repeats, we marked the template of the TER1 RNA with a mutation: a single base change in the template (A458G, TER1–BclI allele), resulting in the creation of a BclI restriction site (M. McEachern and E.H. Blackburn, unpubl.). Telomeric repeats synthesized by TER1–BclI incorporate the single base change without otherwise altering the telomeric repeat sequence (D. Hager and M. McEachern, pers. comm.). Unlike previously described mutations in the TER1 template region (McEachern and Blackburn 1995; Krauskopf and Blackburn 1996), the TER1–BclI mutation is phenotypically silent and does not detectably affect telomere function and telomere length regulation. The 7BΔTER1 cells transformed with a centromeric plasmid expressing the TER1–BclI gene (pTERBclI) regained stably maintained wild-type length telomeres, with no change in telomere length and no apparent colony or cellular phenotype observed even after multiple serial cell passages (Fig. 1A, lane 11; data not shown). The BclI mutation was incorporated into newly synthesized telomeric repeats at the telomere termini, as described previously for other K. lactis and S. cerevisiae telomerase RNA template mutants (McEachern and Blackburn 1995; Prescott and Blackburn 1997b). The BclI mutant repeats were detected by hybridization of EcoRI-digested genomic DNA to a radio-labeled oligonucleotide probe specific for the BclI repeat (Fig. 1C, lanes 1,3). The hybridizing bands were lost when the DNA was digested secondarily with BclI restriction enzyme (Fig. 1C, lanes 2,4). As seen by hybridization with a wild-type telomeric probe, this second digestion with BclI resulted in the expected shortening of EcoRI telomeric fragments (Fig. 1B, lanes 2,4, arrowheads), demonstrating that the BclI sites were at the ends of the telomeres, and were incorporated onto every chromosome. When 7BΔTER1 yeast cells were cotransformed with two plasmids, one expressing wild-type TER1 and one expressing TER1–BclI, we estimated that equal numbers of wild-type and BclI marked repeats were incorporated onto telomeres (data not shown).
Various nontemplate mutations were created in the TER1–BclI allele (Fig. 2). We anticipated that both telomere length and the extent of incorporation of BclI-marked telomeric repeats would provide an accurate readout of varying degrees of function of the mutant alleles in vivo. Test plasmids with a HIS3 marker, containing mutant TER1–BclI genes, unmutagenized TER1–BclI, or no TER1 gene, were transformed into K. lactis strain 7BΔTER1 expressing the URA3 marked plasmid pTERWT. Subsequently, pTERWT was selected against by plating cells on medium containing 5-fluoro-orotic acid (5-FOA). We compared telomere length and incorporation of BclI-marked repeats onto telomeres both before and after loss of the pTERWT plasmid. Thus, the function of each mutant RNA in vivo was examined both when it was coexpressed with wild-type telomerase RNA and when it was expressed alone.
Figure 2.
A schematic diagram of the TER1 RNA. The template region is contained within residues 435 and 464. The boxed areas represent regions removed or substituted in mutant alleles.
In control experiments, when the HIS3 plasmid contained no TER1 gene, loss of the pTERWT plasmid resulted in the expected shortening of telomeres followed by telomere lengthening by the recombination pathway (Fig. 3A, lanes 1–6), and no BclI repeat incorporation (Fig. 3B, lanes 1–6). With the unmutagenized TER1–BclI gene, telomere length remained unchanged before and after transformation and after 5-FOA selection (Fig. 3A, lanes 7,9; data not shown). BclI repeats were incorporated at the earliest times observable after transformation (∼25 cell doublings). Similarly, we determined the extent to which each deletion mutant was able to provide TER1 function in vivo. The deletion mutants assayed are shown schematically in Figure 2 and the in vivo results obtained are summarized in Table 1.
Figure 3.
Telomeric profile of genomic DNA from the ΔTER1 strain transformed with control pHIS3 and pTERBclI plasmids (A,B) or fully functional TER1 deletion mutants (C,D). Each pair of lanes contains DNA digested with EcoRI (R) and EcoRI plus BclI (R/B) enzymes, respectively. Each passage represents ∼25 doublings after the loss of the pTERWT plasmid. The DNAs in C and D were prepared from cells after five passages. The same filters were probed first with the BclI probe (B,D) and subsequently with the wild-type probe (A,C). The asterisk in B is a nonspecific band.
Table 1.
Summary of TER1 mutant alleles assayed in vivo
|
TER1 allele
|
BclI repeat incorporation in presence of pTERWT
|
BClI repeat incorporation after loss of pTERWT
|
Telomere length after loss of pTERWT
|
Telomerase function in vivo
|
|---|---|---|---|---|
| Δter1 | N/A | N/A | short, followed by lengthening in survivor cells (S) | − |
| TER1–BclIa | + | ++ | wild type | ++ (fully functional) |
| ter1-Δ80–120b | + | ++ | wild type | ++ |
| ter1-Δ125–204 | + | ++ | marginally shorter than wild type | ++ |
| ter1-Δ275–335 | + | ++ | marginally shorter than wild type | ++ |
| ter1-Δ360–420 | + | ++ | wild type | ++ |
| ter1-Δ697–714 | + | ++ | wild type | ++ |
| ter1-Δ718–731 | + | ++ | wild type | ++ |
| ter1-Δ810–818 | + | ++ | wild type | ++ |
| ter1-Δ825–850 | + | ++ | wild type | ++ |
| ter1-Δ77–424 | − | + | ∼100–150 bp shorter than wild type | + (partially functional) |
| ter1-Δ1077–1229 | − | + | ∼200 bp shorter than wild type | + |
| ter1-ΔCombc | − | + | ∼200 bp shorter than wild type | + |
| GAL1–TER1d | + | ++ | wild type | ++ |
| GAL1–ter1-Δ77–424d | + | ++ | wild type | ++ |
| ter1-Δ20–60 | − | − | S | − |
| ter1-Δ493–580 | − | − | S | − |
| ter1-Δ630–730 | − | − | S | − |
| ter1-Δ914–1031 | − | − | S | − |
| ter1-Δ917–945 | + | ++ | wild type | ++ |
| ter1-Δ967–1000 | − | − | S | − |
| ter1-Δ1021–1033 | + | ++ | wild type | ++ |
| ter1-sb980–987loopeh | − | − | S | − |
| ter1-Δ bulgefh | − | − | S | − |
| ter1-bulgetostemgh | − | − | S | − |
TER1 nucleotide A458 is changed to G. All subsequent ter1 mutant alleles also contain this mutation.
(Δa–b) A deletion of nucleotides a through b.
The ΔComb deletion is a combination of the Δ697–714, Δ718–731, Δ750–785, Δ810–818, and Δ825–850 deletions.
In the GAL1–ter1 alleles nucleotides 1–20 of TER1 are replaced with GAL1 RNA leader sequences.
TER1 nucleotides 980–987 are substituted with TTCG.
TER1 nucleotides 971–975 and 992–996 are deleted.
TER1 nucleotides 992–996 are replaced with TGAGG.
Loop, bulge, and stem refer to putative secondary structural elements.
Several regions of the TER1 RNA are fully dispensable for telomerase activity in vivo
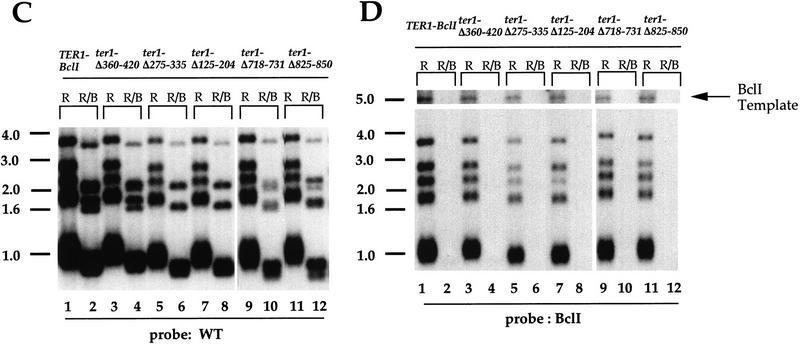
Several small deletions of telomerase RNA provided wild-type levels of TER1 function in vivo. The deletions between the 5′ end of the RNA and the template (ter1-Δ80–120, ter1-Δ125–204, ter1-Δ275–335, and ter1-Δ360–420) were almost indistinguishable from the TER1–BclI gene in terms of incorporation of BclI-marked telomeric repeats and the length of telomeres (Table 1; examples are shown in Fig 3C,D). With the ter1-Δ125–204 and ter1-Δ275–335 mutants, telomeres shortened slightly (<50 bp; Fig. 3C,D, lanes 5,7). However, the amount of BclI incorporation in these mutants both in the presence and absence of pTERWT was comparable to TER1–BclI (Table 1). We suggest that telomerase function is only slightly reduced in these two mutants. Four small deletions in the central part of the RNA downstream of the template (ter1-Δ697–714, ter1-Δ718–731, ter1-Δ810–818, and ter1-Δ825–850) also had telomere lengths and BclI repeat incorporation indistinguishable from TER1–BclI (Table 1; examples are shown in Fig. 3C,D). BclI repeats were also incorporated when each of these mutant RNAs was coexpressed with wild-type TER1 RNA (Table 1). On the basis of these criteria, these deleted regions of TER1 RNA are dispensable for its function in vivo.
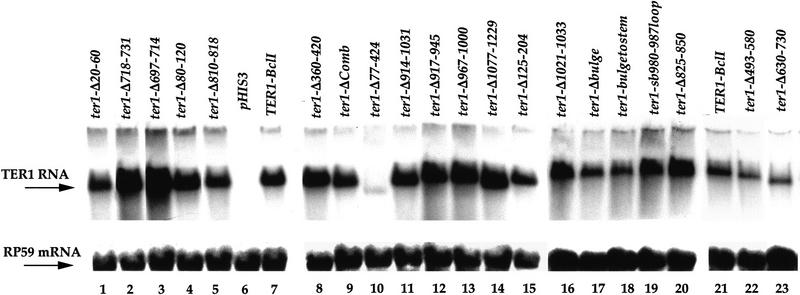
We also tested, by Northern blotting analysis, whether the deletions affected the steady-state levels of these mutant RNAs. Total RNA was extracted from the 7BΔTER1 strain expressing mutant TER1 alleles or the TER1–BclI control and hybridized to a TER1 probe (Fig. 4). A second probe for an unrelated mRNA, ribosomal protein 59 (RP59), was used as a control for RNA loading in each lane. As expected, no detectable TER1 RNA was found in the 7BΔTER1 strain transformed with HIS3 plasmid only (Fig. 4; lane 6). The level of each of the essentially fully functional mutant RNAs was comparable to the level of TER1–BclI RNA. Two of these mutant RNAs (ter1-Δ697–714 and ter1-Δ718–731) were in fact present at twofold higher levels than the TER1–BclI RNA. Therefore, these deletions do not decrease the synthesis or stability of TER1 RNA.
Figure 4.
Steady-state levels of wild-type and mutant TER1 RNAs. Northern blot of RNA extracted from the ΔTER1 strain expressing pHIS3, TER1–BclI or deletion mutants of TER1 and probed with a TER1 DNA probe (top) or a probe for RP59 mRNA (bottom).
Deleting three large domains of TER1 results in partial telomerase function
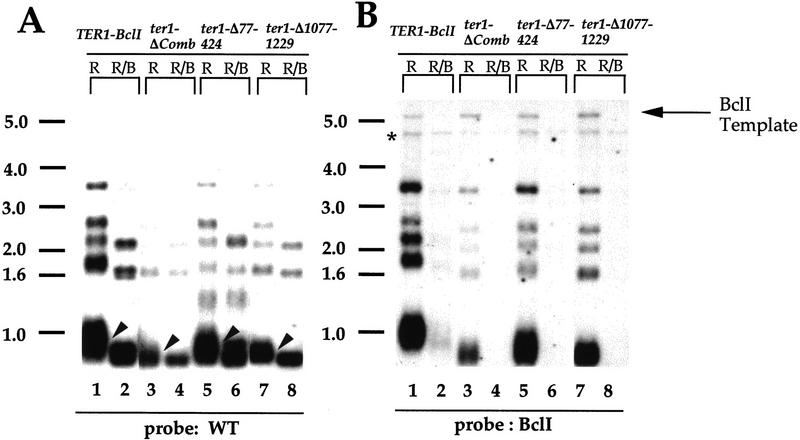
Three larger deletions that remove substantial portions of the RNA were tested for in vivo telomerase action: ter1-Δ77–424, a 347-nucleotide deletion between the 5′ end of TER1 RNA and the template; ter1-ΔComb, a combination of five small deletions in the central part of the RNA removing a total of 129 nucleotides; and ter1-Δ1077–1229, a 154-nucleotide deletion near the 3′ end of the RNA (Fig. 2; Table 1).
All three mutant RNAs functioned in vivo by the criterion of incorporation of BclI mutant repeats (Fig. 5A,B). However, fewer BclI-marked repeats were incorporated into the ends of the telomeres than in the TER1–BclI control, particularly in the case of the ter1-ΔComb mutant (Fig. 5B, cf. lanes 1, 3, 5, and 7). Furthermore, digestion of telomeres with BclI restriction enzyme resulted in less shortening than in TER1–BclI cells, indicating the presence of fewer BclI repeats at the ends (Fig. 5A, arrowheads). Thus, we categorized these mutant RNAs as partially functional (Table 1). The telomeric fragments in these strains were also shorter than wild-type by ∼200 nucleotides (i.e., equivalent to eight 25-bp telomeric repeats) for the ter1-ΔComb and ter1-Δ1077–1229 mutants, and ∼100–150 nucleotides (i.e., four to six 25-bp telomeric repeats) for the ter1-Δ77–424 mutant. Interestingly, in all these cases the shortened telomeres were maintained stably at their new length, with no further change in telomere length occurring for up to 250 generations (data not shown).
Figure 5.
BclI repeat incorporation and telomere shortening in partially functional TER1 mutants. Telomeric profile of DNAs prepared from the fifth passage of the ΔTER1 strain expressing TER1–BclI or partially functional TER1 alleles alone (A,B) or together with the pTERWT plasmid (C). Each pair of lanes contains DNA digested with EcoRI (R) and EcoRI plus BclI (R/B) enzymes, respectively. The same filter was probed first with the BclI probe (B) and subsequently with the WT probe (A). The asterisk in B and C is a nonspecific band.
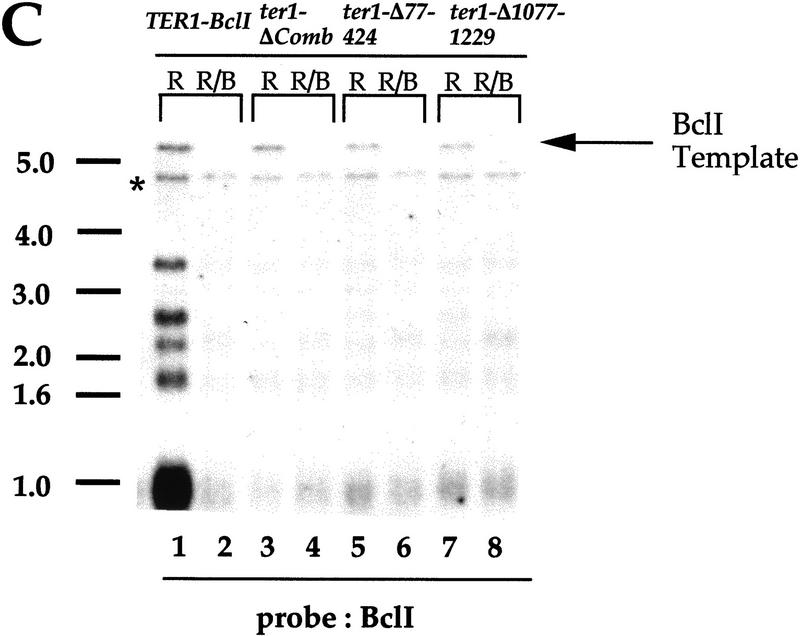
When these mutant genes were coexpressed with wild-type TER1 and assayed for BclI repeat incorporation, no BclI-specific hybridization was obtained (Fig. 5C). Because we were able to detect the BclI template of the mutant gene with the BclI-specific probe, we conclude that the probe can detect approximately one to three repeats per telomere. Hence, in contrast to the essentially fully functional mutants, these deleted RNAs, although functional when expressed alone, were unable to act at telomeres to cause incorporation of telomeric repeats in the presence of the wild-type TER1 RNA.
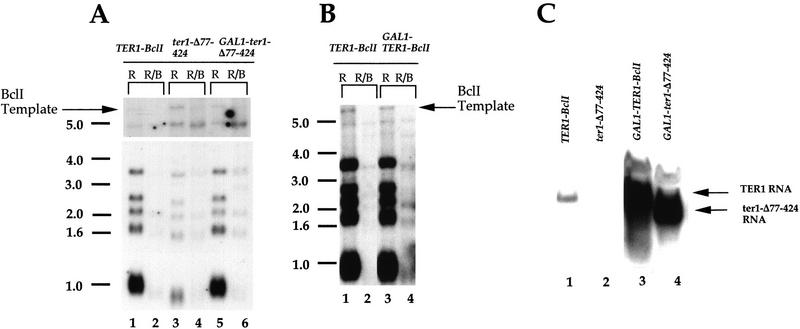
We tested whether the decreased telomerase function of these partially functional mutants reflected decreased steady-state RNA levels. Levels of the ter1-Δ1077–1229 and ter1-ΔComb RNAs were comparable to the wild-type TER1–BclI RNA (Fig. 4, lanes 7,9,14). Thus, the Δ1077–1229 and ΔComb deletions do not affect the synthesis and stability of the mutant RNAs. In contrast, the ter1-Δ77–424 mutant RNA was markedly less abundant than TER1–BclI (Fig. 4, lane 10). In Northern blots using a probe that is completely complementary to both RNAs, the ter1-Δ77–424 RNA was found to be approximately eightfold less abundant than TER1–BclI (Fig. 6C; data not shown).
Figure 6.
Overexpression of a partially functional TER1 mutant restores telomere length. (A,B) Telomeric profile of DNAs prepared from the fifth passage of the ΔTER1 strain expressing TER1 alleles on plasmids as indicated. The cells were maintained in medium containing galactose as the carbon source. Each pair of lanes contains DNA digested with EcoRI (R) and EcoRI plus BclI (R/B) enzymes, respectively. The blots are probed with the BclI probe. (C) Northern blot of RNA prepared from the ΔTER1 strain expressing TER1 alleles on plasmids. The cells were grown in galactose-containing medium. The blot is probed with radiolabeled TER1 DNA. Equal amounts of RNA were loaded in each lane.
Overexpression of a partially functional RNA restores telomere length
We tested directly whether the lower steady-state levels of ter1-Δ77–424 mutant RNA caused the decreased BclI repeat incorporation and shortened telomeres. The ter1-Δ77–424 gene was placed under the control of the inducible GAL1 promoter resulting in ∼50-fold higher levels of the induced GAL1-ter1-Δ77–424 RNA (Fig. 6C). This restored both the extent of BclI repeat incorporation in telomeric ends and the length of the telomeric fragments to wild-type levels (Fig. 6A). In contrast, overexpression of the wild-type TER1–BclI RNA from the GAL1 promoter (Fig. 6C, cf. lanes 1 and 3) caused no change in the length of telomeric fragments or BclI repeat incorporation (Fig. 6B). Thus, increasing the expression of the poorly expressed or unstable ter1-Δ77–424 RNA restores full in vivo function to this telomerase RNA. Hence, we conclude that the decreased BclI repeat incorporation by the ter1-Δ77–424 mutant is attributable to the lower steady-state level of the mutant RNA.
Deletion mutants identify four regions of TER1 outside the template that are crucial for telomerase function
Deletions of four regions of the RNA (ter1-Δ20–60, ter1-Δ493–580, ter1-Δ630–730, and ter1-Δ914–1031) comprising 40, 87, 100, and 117 residues, respectively, abolished detectable telomerase action in vivo. BclI repeats were not incorporated in the telomeres of cells expressing these RNAs, even up to 100 generations after the loss of the pTERWT plasmid (Fig. 7). We detected clear hybridization of the BclI-specific probe to the BclI template of the mutant genes, which is estimated to be present at one to three copies per cell (Fig. 7A). This hybridization signal was specific because unlike nonspecific hybridization, it was completely lost upon digestion of genomic DNA with BclI enzyme. Thus, our inability to detect any specific hybridization to telomeric DNA indicated that BclI repeats are not incorporated into the telomeres in these strains. Furthermore, the telomeres in the mutant cells exhibited the typical behavior of telomeres in the absence of telomerase activity; they shortened to a critical minimal length, and then in the population of surviving cells were generally lengthened, presumably by recombinational events (Fig. 7B). Consistent with a lack of telomerase activity, the colony phenotype of these cells was identical to the ΔTER1 strain in which TER1 is deleted completely.
Figure 7.
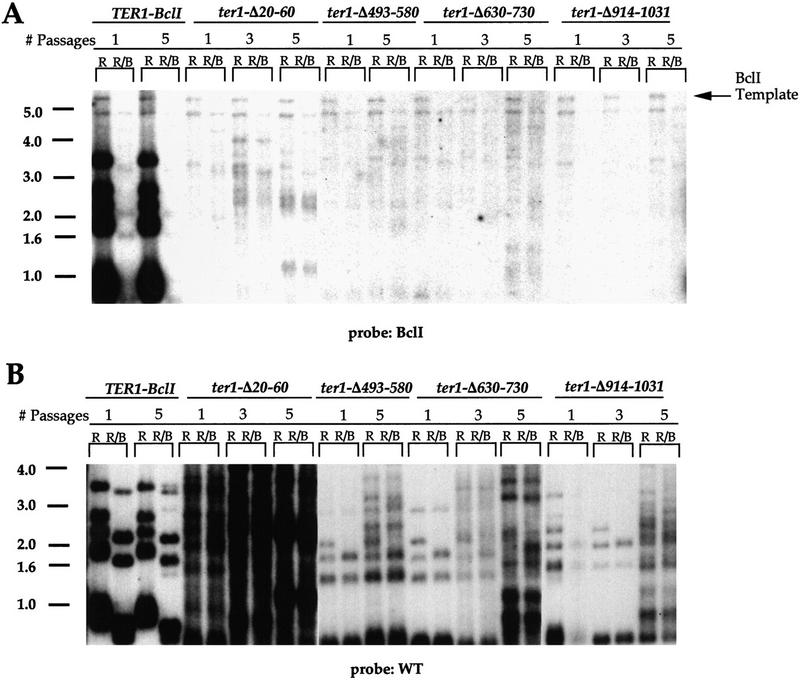
BclI repeats are not incorporated in telomeres of nonfunctional TER1 mutants. Telomeric profiles of DNAs prepared from the ΔTER1 strain expressing TER1–BclI or TER1 deletion alleles, as indicated. Each pair of lanes contains DNA digested with EcoRI (R) and EcoRI plus BclI (R/B) enzymes, respectively. Each passage represents ∼25 doublings after the loss of the pTERWT plasmid. The same filter was probed first with the BclI probe (A) and subsequently with the wild-type (WT) probe (B).
These mutant RNAs were expressed and stably maintained in cells at levels comparable to the TER1–BclI RNA, as judged by Northern blotting analysis (see Fig. 4, lanes 1,11,22,23). Thus, RNA residues in each of these four deleted regions (see Fig. 2) are essential for an aspect of telomerase activity other than the synthesis and stability of the RNA.
Small mutations in a domain of TER1 RNA abolish telomerase function
We examined in more detail the region of the TER1 RNA between residues 914 and 1031. This part of the RNA was predicted to form a discrete structural domain, on the basis of a computer folding program (see Materials and Methods) and phylogenetic comparisons of TER1 RNA sequences of four different Kluyveromyces species (Y. Tzfati and E.H. Blackburn, unpubl.). This computer-predicted RNA secondary structure contains three putative stable stem-loops and was used to guide construction of three small deletion mutants (ter1-Δ917–945, ter1-Δ967–1000, and ter1-Δ1021–1033; Fig. 8A). The steady-state levels of all three mutant RNAs were similar to TER1–BclI RNA (see Fig. 4, lanes 12,13,16). The ter1-Δ917–945 and ter1-Δ1021–1033 mutants incorporated BclI repeats into telomeres and were fully functional in vivo (Fig. 8B,C, lanes 1,2,5,6). In contrast, the ter1-Δ967–1000 mutant RNA was completely nonfunctional in vivo; there was no incorporation of BclI repeats into telomeres (Fig. 8B,C, lanes 3,4), and the telomeres behaved similarly to those in strain 7BΔTER1. Hence, RNA sequences critical for telomerase function lie between nucleotides 967 and 1000 of TER1.
Figure 8.
Mutations in a small region of TER1 abolish telomerase function in vivo. (A) Predicted secondary structure of the TER1 RNA between residues 901 and 1054. (B,C) Telomeric profiles of DNAs prepared from the 10th passage of the ΔTER1 strain expressing TER1 deletion alleles. Each pair of lanes contains DNA digested with EcoRI (R) and EcoRI plus BclI (R/B) enzymes, respectively. The same filter was probed first with the BclI probe (C) and subsequently with the wild-type (WT) probe (B).
We then constructed three smaller deletion and substitution mutants between residues 967 and 1000 to further narrow down the crucial residues. The first, ter1–sb980–987loop, substitutes the proposed 8-base ‘loop’ of this stem–loop region (residues 980–987) with a stable tetraloop sequence (UUCG). The second mutant, ter1–Δbulge, deleted the putative ‘bulge/internal loop’ in the stem region (deletion of residues 971–975 and 992–996). The third mutant, ter1–bulgetostem, was designed to convert the putative bulge to a stem and was a substitution of residues 992–996, making these positions complementary to residues 971–975 (see Table 1).
All three mutants were completely nonfunctional in vivo. They showed no BclI repeat incorporation for up to 10 passages (Fig. 8B, lanes 7–12), and telomeres shortened for the first two to three passages and then lengthened as in strain 7BΔTER1 (Fig. 8C, lanes 7–12; data not shown). The levels of the ter1–Δbulge and ter1–bulgetostem mutant RNAs were ∼30% lower than the control TER1–BclI RNA (Fig. 4, lanes 7,17,18). Thus, residues 971–975 and 992–996 are important for both telomerase action and stability or expression. The level of the ter1–sb980–987loop RNA (Fig. 4, lane 19) was identical to the TER1–BclI control. Thus, altering residues 980–987 of the RNA specifically abolished telomerase action in vivo.
The TER1 RNA mutants lacking function in vivo have undetectable telomerase activity in vitro
We tested two possibilities for the lack of telomerase action in vivo for the nonfunctional TER1 mutant RNAs: (1) a catalytically inactive telomerase enzyme and (2) defective interactions between a catalytically active enzyme and other components of the telomere maintenance pathway that prevent telomerase from adding telomeric repeats to telomeres. The latter, for example, is true for the S. cerevisiae est1, est3, and est4 mutants, in which telomerase is active in vitro (Cohn and Blackburn 1995; Lingner et al. 1997a). Four of the nonfunctional TER1 mutants, ter1-Δ20–60, ter1-Δ493–580, ter1-Δ630–730, and ter1–sb980–987loop, were tested directly for telomerase activity in vitro.
An in vitro telomerase assay for K. lactis cell extracts has been developed recently (Fulton and Blackburn 1998). This activity extends telomeric oligonucleotides by the incorporation of deoxynucleotides (one of which is radio-labeled) and can undergo one round of telomeric repeat synthesis. Control wild-type TER1 or TER1–BclI cell extracts extended a telomeric DNA oligonucleotide primer (Fig. 9A, lanes 1,3), producing the characteristic pattern of mid-template and near-terminal products described previously for wild-type telomerase reactions with this primer (Fulton and Blackburn 1998). The activity was sensitive to RNase A (Fig. 9A, lanes 2,4). No significant difference in activity was found between the extracts containing TER1 and TER1–BclI RNA under the conditions used, confirming that the BclI mutation does not obviously affect the enzymatic properties of telomerase.
Figure 9.
(A,B) Nonfunctional TER1 mutants contain undetectable telomerase activity in vitro. DEAE fractions of extracts from strains expressing TER1 alleles were assayed for K. lactis telomerase activity in vitro with primer KL13(12). The sequence of the TER1 templating domain and the predicted alignment of the primer are shown. The boxed template residue corresponds to the site of the A → G mutation in the TER1–BclI strains examined. Terminal transferase-labeled KL13(12) primer is shown in M lanes, and the positions of the +1 products are marked correspondingly. (A) Telomerase reactions with DEAE fractionated extracts were carried out with all four dNTPs. RNase pretreatment (lanes 2,4,6) consisted of incubation of extracts with 10 μg/ml RNase A at 25°C for 5 min. Mid-template products are denoted with brackets, and near-terminal products are marked with arrowheads. A background ladder of RNase A insensitive bands was detected in lanes 5 and 6 and is most likely caused by contaminating polymerases in the fractions assayed. The asterisk marks a nontelomerase generated background band described previously (Fulton and Blackburn 1998). (B) Reactions with DEAE fractionated extracts from TER1 (lane 1), TER1–Bcl (lane 2), ter1–sb980–987loop (lane 3), ter1-Δ630–730 (lane 4), ter1-Δ20–60 (lane 5), and ter1-Δ493–580 (lane 6) strains were carried out as in A but with ddTTP substituted for dTTP. (C,D) Profiles of TER1 RNA-containing complexes in wild-type and mutant cell extracts. DEAE fractions of extracts from strains expressing TER1 alleles were fractionated on nondenaturing gels and probed with a radio-labeled TER1 DNA probe. The gel in C was run for 13 hr, whereas the gel in D was run for 10 hr.
Evidence for copying of the template domain of TER1 RNA came from termination of synthesis with the incorporation of dideoxy-TTP (dd-TTP) at the expected position along the template (Fig. 9B, lanes 1,2). Such termination reactions provided a sensitive assay for telomerase activity in this system. Telomerase activity was undetectable in extracts prepared from cells expressing the four nonfunctional mutant RNAs, when synthesis was examined in the presence of all four dNTPs (Fig. 9A, lanes 5,6; data not shown) or with ddTTP substituted for dTTP (Fig. 9B, lanes 3–6). No activity was detected with these mutant extracts in assays using different concentrations of oligonucleotide primer, or primers aligning to different regions of the TER1 template, or under conditions where the incorporation of even a single templated nucleotide would be detected (data not shown). The mutant extracts did not contain trans-acting factors that inhibited telomerase activity, because mixing equal amounts of mutant and wild-type extracts did not inhibit the activity of the wild-type extract (data not shown). Because telomerase activity was detected in the TER1–BclI extract when it was diluted up to fivefold (data not shown), we estimated that the mutant extracts contained <20% of wild-type telomerase activity. The in vivo lack of function suggests that activity may in fact be abolished completely.
These results showed that the ter1-Δ20–60, ter1-Δ493–580, ter1-Δ630–730, and ter1–sb980–987loop mutants are defective in core enzymatic properties of telomerase, although it is possible that they may also be defective for other aspects of telomerase function. Any one or more of the following steps could be affected: proper RNA folding and/or assembly of a functional telomerase RNP complex, binding of core telomerase to accessory proteins or to the oligonucleotide primer and/or nucleotide triphosphate substrates, and polymerization.
Mutations in the TER1 971–996 region result in a major conformational change in the RNA but allow assembly into an RNP complex
We examined telomerase RNA and RNP complexes by native gel electrophoresis (Fig. 9C; see Materials and Methods). Fractions from TER1 or TER1–BclI cell extracts contained a single major RNP complex (Fig. 9C, lanes 1,3). The mobility of the hybridizing species increased with pretreatment with proteinase K or phenol/chloroform extraction, indicating that the complex contains at least one protein component (Fig. 9C, lanes 2,4; data not shown). As expected, the RNP complex was also sensitive to RNase A treatment (data not shown). For the nonfunctional mutants ter1-Δ493–580 and ter1-Δ630–730, a single major hybridizing species was observed, whose mobility remained unchanged with proteinase K digestion or phenol extraction (Fig. 9C, lanes 7–10; data not shown). Hence these two mutants appear to be defective in telomerase RNP assembly, which we propose accounts for their failure to exhibit telomerase activity in vivo or in vitro.
The mobilities of the TER1-hybridizing species of the nonfunctional ter1-Δ20–60 mutant, before and after proteinase K treatment, were identical to those of TER1 and TER1–BclI cell extracts (Fig. 9C, lanes 11,12). Thus, although subtle changes may not be detected by mobility in these gels, the ter1-Δ20–60 mutant does not appear to be impaired for stable telomerase RNP assembly or have a greatly changed RNA conformation. Because telomerase in this mutant was nonfunctional in vitro, we suggest that the ter1-Δ20–60 mutation affects an aspect of enzymatic activity rather than RNP assembly.
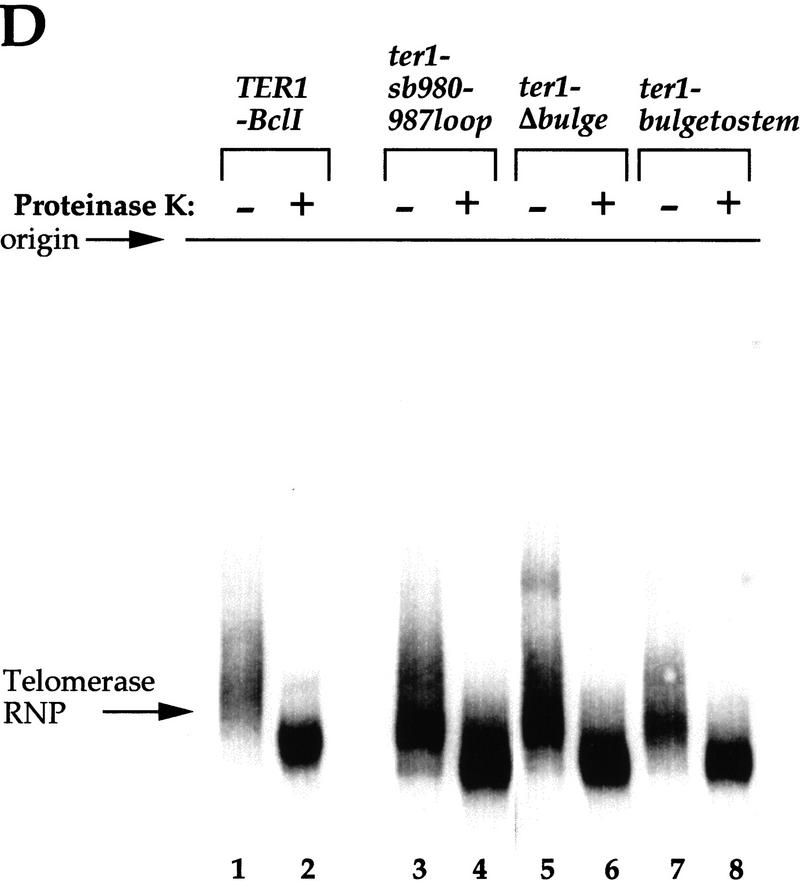
The ter1–sb980–987loop mutant contained a single RNP complex (Fig. 9C, lane 5), which migrated more slowly than its proteinase K-treated counterpart (Fig. 9C, lane 6). Strikingly, both this RNP complex and the proteinase K-treated species migrated significantly faster than their TER1 or TER1–BclI counterparts (Fig. 9C, cf. lanes 3 with lanes 5 and 4 with 6). This fast migration cannot be accounted for by the removal of only four nucleotides in this RNA mutant. Hence, we conclude that the ter1–sb980–987loop RNA has a different conformation from wild-type, even although it is still assembled with at least some proteins. The protein composition of this enzymatically inactive mutant complex may be similar to that of the wild-type complex, as proteinase K treatment caused approximately the same degree of change in mobility for both types of complexes.
Results similar to those with the ter1–sb980–987loop mutant were found with the ter1–Δbulge and ter1–bulgetostem mutants (Fig. 9D, cf. lanes 3–8). Extracts from these strains also contained undetectable levels of telomerase activity in vitro (data not shown). Therefore, for each of these three mutants, both the mutant RNA itself and its RNP complex differed in conformation from wild type.
Discussion
Here we have determined the effects of mutations in K. lactis yeast telomerase RNA on the properties of telomerase in vivo and in vitro. A silent mutation marking the template of the mutant RNAs created a sensitive assay for the ability of mutant telomerases to add the marked telomeric repeats in vivo. By using TER1 RNAs in a yeast strain completely deleted for wild-type TER1, any telomerase activity observed in vivo and in vitro was attributable only to the mutant RNA. By also examining the function of mutant TER1 RNAs when they were coexpressed in cells with wild-type RNA, we tested the ability of mutant RNAs to compete with wild-type RNA for assembly with telomerase proteins, or other factors involved in the biogenesis and function of the telomerase complex. Conversely, by such coexpression we would also be able to detect cooperativity between mutant and wild-type RNAs, as was found for the 476GUG mutant of S. cerevisiae telomerase RNA (Prescott and Blackburn 1997b).
Small regions of TER1 are critical for enzyme activity
Four specific TER1 sequences several hundred residues from the template were critical for telomerase function in vivo and in vitro. This is the first example of specific nontemplate telomerase RNA residues that are essential for enzyme activity both in vivo and in vitro.
With wild-type enzyme, we observed a single RNP complex containing TER1 RNA. Because this complex is functional, it must contain the RT protein subunit and may also contain other components such a homolog of Est1p. Two of the essential TER1 RNA regions, defined by residues 493–580 and 630–730, map to the central part of the RNA (Fig. 2). Deletion of these regions result in a lack of telomerase RNP assembly; the simplest interpretation of our results is that these mutant RNAs are unable to assemble with the RT protein subunit. S. cerevisiae telomerase functions as a dimer with two RNAs (Prescott and Blackburn 1997a) suggesting that the K. lactis enzyme may be similar. Therefore, another possibility is that the mutant RNAs do not achieve proper secondary, tertiary, or quaternary structural conformation, and this prevents correct RNP assembly.
Critical nontemplate RNA residues may provide active site functions
The phenotypes of the ter1-Δ20–60 mutant and the mutations in the TER1 971–996 region create the most compelling argument for a direct role of nontemplate telomerase RNA residues in active site functions. Interestingly, recent phylogenetic comparisons of closely related Kluyveromyces species have indicated that these two regions include some of the most conserved TER1 sequences (Y. Tzfati and E.H. Blackburn, unpubl.). These mutations, which remove or substitute very small regions of the RNA, abolish detectable enzyme activity in vitro and enzyme function in vivo but are able to interact with at least some telomerase proteins. The ter1-Δ20–60 telomerase RNP particle appears similar in size and shape to the wild-type complex. Thus, this mutant is likely to be defective at a step after RNP assembly. The ter1–sb980–987loop, ter1–Δbulge, and ter1–bulgetostem mutants delete or substitute residues in the same region of TER1, resulting in mutant RNAs that migrate considerably faster than wild-type under nondenaturing electrophoretic conditions, indicative of a significantly different conformation. However, this does not prevent the mutant RNAs from interacting with proteins. Therefore, these mutated nucleotides are likely to be involved in functions other than protein binding. These could include recognition and binding of DNA or dNTP substrates, positioning of the template region in the active site, and incorporation of nucleotides. Alternatively, this RNA may also fail to form a dimeric/oligomeric telomerase. However, unlike the S. cerevisiase 476GUG template mutant (Prescott and Blackburn 1997b), the ter1–sb980–987loop, ter1–Δbulge, and ter1–bulgetostem mutants are not rescued by coexpression with wild-type TER1.
Unpaired RNA residues provide critical enzymatic functions in both the spliceosome and the ribosome (recently reviewed in Green and Noller 1997; Staley and Guthrie 1998). For example, the 530-loop region of the 16S rRNA interacts with the mRNA and EF-Tu (for review, see Powers and Noller 1994) and mutations in the conserved domain V loop of 23S rRNA affect proper use of the aminoacyl tRNA substrate (Green et al. 1997). In the spliceosome, mutations in a loop region of U5 snRNA affect exon recognition, highly conserved U6 snRNA residues are essential for catalytic activity, and extensive RNA–RNA interactions between U2 and U6 snRNA residues are also necessary for catalysis (for review, see Nilsen 1998). On the basis of these precedents and the results reported above, we suggest that the 971- to 996-region of TER1 may be involved in RNA–RNA, RNA–protein, or RNA–DNA interactions that are critical for the functioning of the telomerase active site.
More than half of TER1 is dispensable for in vivo telomerase function
Several stretches of TER1 sequences are apparently fully dispensable for telomerase action in vivo. Furthermore, large deletions of the RNA maintain partial function in vivo, suggesting peripheral roles for these regions. These results are consistent with the findings that more than half of the human telomerase RNA is not essential for in vitro-reconstituted activity (Autexier et al. 1996; Beattie et al. 1998).
Deletion mutants lacking large sections of the RNA, ter1-Δ77–424, ter1-ΔComb, and ter1-Δ1077–1229, had decreased telomerase function in vivo. This could result from decreased stability of the mutant RNA, partial impairment of enzyme activity, or defective ability of the enzyme to interact with telomeres. The reduced function of mutant ter1-Δ77–424 was attributable to the reduction in the steady-tate levels of mutant RNA. This deleted region may be involved in binding factors that stabilize newly synthesized TER1 RNA or aid in its assembly into telomerase RNP, failure of which leads to RNA degradation.
The TER1 domains defined by nucleotides 1077–1229 and the ΔComb deletion are not involved in the synthesis and stability of the RNA, as deletion of these sequences did not influence steady-state expression levels. In both cases, the mutant RNAs did not function in the presence of wild-type RNA, suggesting that they are unable to compete with wild-type RNA for the binding of one or more limiting factors. If this factor were the RT protein subunit, these mutants would be expected to demonstrate defects in telomerase RNP assembly or decreased in vitro telomerase activity. Alternatively, the mutant RNAs may be unable to bind properly proteins that mediate the interaction between telomerase and the chromosome ends. Candidates for such proteins have been identified in other species; for example, the EST1 protein in S. cerevisiae, and p80 and p95 in Tetrahymena.
The phenotypes of the partially functional TER1 mutants reveal interesting properties of telomere length control. In these mutants, telomeres are steadily maintained for at least 200 generations at a length 100–200 nucleotides shorter than wild type. Unlike previously observed short and stable mutant telomeres (McEachern and Blackburn 1995), the telomeric repeats analyzed here are phenotypically wild type and thus the binding to proteins that regulate telomere length (such as RAP1p) is expected to be unchanged. These results indicate that the decreased ability of the mutant telomerases to add repeats results in shorter telomeres, reflecting a new balance between telomere addition and loss of telomeric DNA. Thus, the mechanism for telomere length regulation can respond to a reduction in telomerase activity. Telomerase enzyme of K. lactis, like that of S. cerevisiae, is nondissociative in vitro and is proposed to be stably bound to the telomeric end after one round of repeat synthesis (Prescott and Blackburn 1997a; Fulton and Blackburn 1998). It is possible that telomerase is part of the structure that caps the ends of the chromosomes and the interaction between telomerase RNA and other components of this cap structure may be a target of telomere length regulation.
Materials and methods
Strains, plasmids, and media
Yeast and bacteria were grown as described elsewhere (Sherman et al. 1986). The wild-type K. lactis strain used in this study is 7B520 (Ura−, Trp−, His−, Ade−) (Wray et al. 1987) and all other strains were derived from this. Transformation of K. lactis was done by electroporation, identical to the method described for S. cerevisiae (Becker and Guarente 1991). A deletion allele of TER1 was constructed as follows. A 2.5-kb XhoI–EcoRI fragment containing TER1 was cloned into the URA3 marked yeast integrating vector YIP5. A 1.3-kb PmeI–SphI fragment containing TER1 was then removed from this construct and the remaining DNA was blunted with T4 DNA polymerase and circularized by ligation. This TER1 deletion clone was used to transform K. lactis strain 7B520 to Ura+ colonies. Transformants that had correctly ‘looped in’ the construct by homologous recombination were plated on medium containing 5-FOA and resulting ‘loop out’ colonies were screened for the Ura−, ΔTER combination by Southern analysis.
Centromeric plasmids that can be maintained in K. lactis were constructed as follows. A 2.0-kb BamHI–Sau3A DNA fragment containing the K. lactis CEN2 was ligated to a 2.5-kb BamHI–SalI DNA fragment containing the K. lactis ARS1B. The resulting 4.5-kb CEN–ARS cassette was cloned into the AatII restriction site of the URA3 or HIS3 containing yeast plasmids pRS306 and pRS303, to result in vectors pKL316 and pKL313, respectively. These plasmids are maintained stably in K. lactis; 0.1% of the cells per doubling lost the plasmids when maintained in nonselective medium. The copy number of the vectors was determined by Southern analysis to be an average of two copies per cell.
The 2.5-kb XhoI–EcoRI fragment of TER1 was cloned into pKl316 to result in plasmid pTERWT. A similar TER1 fragment, containing a mutation in the template region that changes nucleotide A458 to G was cloned into plasmid pKL313 to result in pTERBclI. The A458G mutation creates a BclI restriction site in the template of TER1. Mutant versions of pTERBclI were constructed by oligonucleotide mediated site-directed mutagenesis as described elsewhere (Kunkel et al. 1987). All mutant clones were confirmed by DNA sequencing of relevant fragments of the constructs.
A GAL1-TER1 allele was constructed as follows. A 1.2-kb DNA fragment containing the S. cerevisiae GAL1 promoter and CYC1 terminator was cloned into pKL313 to result in pGAL–KL313. A BamHI site was introduced in the TER1 sequence of pTERBclI at position 21 by site-directed mutagenesis. The BamHI–EcoRV fragment containing TER1 was removed from this construct and cloned into pGAL–KL313 (BamHI and SmaI digested) to result in the GAL–TER1 allele. This places nucleotide 22 of TER1 ∼50 nucleotides 3′ of the RNA start site in the GAL–TER1 construct. Thus, the GAL–TER1 construct contains the BclI template mutation and replaces the first 20 nucleotides of TER1 with 50 nucleotides of GAL1 RNA sequence. The Δ77–424 deletion mutation was introduced into this plasmid by site-directed mutagenesis as above.
DNA analysis
Yeast genomic DNA was prepared from wild type or transformants grown in YPD or YP galactose media. Strains containing plasmids were grown in the appropriate minimal medium. The DNA was digested with restriction endonucleases, electrophoresed in 0.8% agarose gels in 1× TBE buffer, and blotted onto Hybond-N+ (Amersham) membrane. The DNA was cross-linked to the membrane using a Sratalinker 1800 (Stratagene). Hybridization of the blots to DNA probes was done according to Church and Gilbert (1984). For oligonucleotide probes, hybridization was carried out for 4 hr, whereas for random primed DNA probes hybridization was carried out overnight. The 5′ radio-labeled oligonucleotide Klac1–25 (ACGGATTTGATTAGGTATGTGGTGT) was hybridized to blots at 55°C to visualize mutant and wild-type telomeric repeats. To detect BclI-marked telomeric repeats specifically, the 5′ radiolabeled oligonucleotide KTelBcl (GATCAGGTATGTGG) was hybridized to the blots at 40°C. Two 5-min washes were carried out at 55°C for the Klac1–25 oligonucleotide and 42°C for the KTelBcl oligonucleotide, respectively. Where applicable, DNA blots were first hybridized to the KTelBcl probe, stripped by washing with 0.4 n NaOH solution and then hybridized to the Klac1–25 probe. The hybridized blots were exposed to film or PhosphorImager screens. Quantitation of bands was done using the ImageQuant program.
RNA analysis
Total RNA extracted from K. lactis strains was electrophoresed in 1.0% agarose, 1× TBE gels containing 7% formaldehyde and blotted to Hybond-N+ in 10× SSC buffer. The membrane was subjected to cross-linking and hybridized to random primed DNA probes as above. For the gels shown in Figures 5 and 9B,C the radio-labeled probe used was a random-primed mixture of two PCR fragments spanning nucleotides 26–277 and 704–1273 of TER1, whereas for Figure 6C only the TER1-704–273 PCR fragment was used. For detecting RP59 mRNA, a DNA fragment containing the entire RP59 coding region was used as the probe.
In vitro telomerase assays
Partially purified telomerase fractions were prepared as described previously (Fulton and Blackburn 1998) with the following modifications. Extracts were prepared from 3 to 4 litres of cells, at optical density (OD600) of 2.0, for each plasmid bearing strain (∼10–14 grams of cell pellet for each strain). Cells were grown in selective medium to OD600 of 2.0, then diluted four- to fivefold in YPD medium and allowed to continue dividing for about three generations. Cells were harvested and frozen in liquid nitrogen as strings of noodles. Subsequently, frozen cell noodles were homogenized into a powder with a prechilled mortar and pestle by grinding under liquid nitrogen for ∼20 min. The powdered cells were resuspended in two volumes of TMG [10 mm Tris-HCl (pH 8.0), 1.2 mm MgCl2, 10% glycerol, 0.1 mm EDTA, 0.1 mm EGTA] buffer and S100 fractions were obtained as in Cohn and Blackburn (1995). S100 supernatants were adjusted to 0.35 m NaOAc (pH 8.0) and loaded onto 2-ml DEAE–agarose columns equilibrated in 0.35 m NaOAc (pH 8.0)/TMG. Elutions were collected in 0.5 m, 0.7 m, 1 m, and 2 m NaOAc (pH 8.0)/TMG, respectively. Fractions were assayed for TER1 RNA by dot blot analysis. Peak fractions containing TER1 RNA at 0.7 m NaOAC were pooled, dialyzed in TMG, and used for further analysis. Extracts from different mutant strains were normalized for the amount of TER1 RNA by dilution with TMG. Assays for telomerase activity were carried out as previously described (Fulton and Blackburn 1998).
Analysis of RNP complexes
For observation of telomerase RNA-containing complexes, DEAE fractions were first incubated in 10 mm Tris-HCl (pH 7.4), 5 mm CaCl2 and either 1 μg/ml proteinase K or water at 37°C for 45 min. RNase pretreatment consisted of incubation of the DEAE fractions with 10 μg/ml RNase A at 25°C for 5 min before proteinase K addition. The samples were then adjusted to 50 mm Tris-acetate (pH 8.0). Heparin was added to 1 μg/ml and the samples were loaded on a 3.5% polyacrylamide (acrylamide:bisacrylamide, 76:1), 0.45% agarose gel. Electrophoresis was carried out in 50 mm Tris-acetate (pH 8.0) at 15 V/cm for 10–13 hr at 4°C. The gels were electroblotted to Hybond-N+ membrane in 0.5× TBE buffer for 2 hr at 70 V and hybridized to random-primed TER1 probes as above. Figure 9 shows samples electrophoresed in the presence of heparin. Omitting heparin caused no obvious changes in the mobility of complexes, although resolution was decreased.
Sequence analysis
RNA secondary structures were predicted using the Mfold program available at the web site http://ibc.wustl.edu/∼zuker/rna. All other DNA and RNA sequence analysis was carried out with the genetics computer group software package (version 9, University of Wisconsin, Madison).
Acknowledgments
We thank Mike McEachern for providing the TER1–BclI allele and sharing unpublished results; Yehuda Tzfati for help with RNA secondary structure analysis and sharing unpublished results; Anamitra Bhattacharyya, Simon Chan, Mike McMeachern, Yehuda Tzfati, and He Wang for critical reading of the manuscript and members of the Blackburn laboratory for helpful discussions. This work was supported by National Institutes of Health (NIH) grants GM26259 and DE11356 to E.H.B. and NIH training grant T32CA09270 to T.B.F. J.R. was supported by a post-doctoral fellowship from the Damon Runyon–Walter Winchell Cancer Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL maura@itsa.ucsf.edu; FAX (415) 476-8201.
References
- Autexier C, Greider CW. Mutational analysis of the Tetrahymena telomerase RNA: Identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 1998;26:787–795. doi: 10.1093/nar/26.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Becker DM, Guarente L. High-efficiency transformation of yeast by electroporation. In: Guthrie C, Fink GR, editors. Guide to yeast genetics and molecular biology. San Diego, CA: Academic Press; 1991. pp. 182–187. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH. A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc Natl Acad Sci. 1997;94:2823–2827. doi: 10.1073/pnas.94.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- ————— . Telomerase. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. . (In press.) [Google Scholar]
- Blasco MA, Funk W, Villeponteau B, Greider CW. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Fulton TB, Blackburn EH. Identification of Kluyveromyces lactis telomerase: Discontinuous synthesis along the thirty-nucleotide-long templating domain. Mol Cell Biol. 1998;18:4961–4970. doi: 10.1128/mcb.18.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes & Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH. Specific RNA residue interactions required for enzymatic functions of Tetrahymena telomerase. Mol Cell Biol. 1996;16:66–75. doi: 10.1128/mcb.16.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Lee MS, Blackburn EH. Altering specific telomerase RNA template residues affects active site function. Genes & Dev. 1995;9:2214–2226. doi: 10.1101/gad.9.18.2214. [DOI] [PubMed] [Google Scholar]
- Green R, Noller HF. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- Green R, Samaha RR, Noller HF. Mutations at nucleotides G2251 and U2585 of 23 S rRNA perturb the peptidyl transferase center of the ribosome. J Mol Biol. 1997;266:40–50. doi: 10.1006/jmbi.1996.0780. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomerase is processive. Mol Cell Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein [see comments] Science. 1997a;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DS, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes & Dev. 1997b;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf A, Blackburn EH. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol Cell Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lingner J, Cech TR, Hughes TR, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci. 1997a;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase [see comments] Science. 1997b;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- ————— Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes & Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: A gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. RNA–RNA interactions in nuclear pre-mRNA splicing. In: Simons R, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. pp. 279–307. [Google Scholar]
- Olovnikov AM. A theory of marginotomy. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Powers T, Noller HF. The 530 loop of 16S rRNA: A signal to EF-Tu? Trends Genet. 1994;10:27–31. doi: 10.1016/0168-9525(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes & Dev. 1997a;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes & Dev. 1997b;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase [see comments] Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Steiner BR, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. Origin of concatomeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Wray LV, Witte MM, Dickson RC, Riley MI. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: Structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]