Abstract
The simplicity of BCR-ABL ‘oncogene addiction' characterizing leukemia contrasts with the complexity of solid tumors where multiple ‘core pathways', including receptor tyrosine kinases (RTKs) and p53, are often altered. This discrepancy illustrates the limited success of RTK antagonists in solid tumor treatment compared with the impact of Imatinib in BCR-ABL-dependent leukemia. Here, we identified c-Abl as a signaling node interconnecting Met-RTK and p53 core pathways, and showed that its inhibition impairs Met-dependent tumorigenesis. Met ensures cell survival through a new path in which c-Abl and p38-MAPK are employed to elicit p53 phosphorylation on Ser392 and Mdm2 upregulation. We found a clinical correlation between activated Met, phospho-p53, and Mdm2 levels in human tumors, supporting the role of this path in tumorigenesis. Our findings introduce the concept that RTK-driven tumors may be therapeutically treated by hitting signaling nodes interconnecting core pathways. Moreover, they underline the importance of evaluating the relevance of c-Abl antagonists for combined therapies, based on the tumor signaling signature.
Keywords: RTK signaling, Met/HGF, c-Abl, p53, oncogenic signals
It is well accepted that cancer is caused by genetic alterations of oncogenes and tumor suppressor genes, and several signaling pathways through which these genes contributing to cancer act, have been uncovered. Recently, genome-wide profiling studies have revealed a remarkable heterogeneity of mutations among different patients affected by the same type of cancer. These studies have evidenced alteration of ‘core pathways', which correspond to groups of proteins known to act together to form a signaling cascade and behave as a group of equivalence in which one mutation influences cell fate towards oncogenesis.1, 2 However, it remains unclear whether genetic alterations of one core pathway functionally deregulate the others through the action of interconnecting nodes. Understanding this issue might offer the possibility of targeting these nodes, in addition to mutated signals within the core.
Receptor tyrosine kinase (RTK) signaling is one core pathway frequently altered in cancer. Over recent years, therapeutic approaches based on compounds selectively targeting oncogenic RTKs, to which cells are dependent, have been developed.3 However, the success of these strategies has been limited since inhibition of the ‘primary RTK addiction' triggers a selective pressure to acquire resistance through RTK switching.4 As RTKs share several effectors that participate in the oncogenic process and in drug response,5 an alternative strategy would rely on the identification of druggable signals required for RTK-triggered tumorigenesis.
In contrast to the genetic complexity of solid tumors, most chronic myeloid leukemias (CML) and a subset of acute lymphocytic leukemia (ALL) are caused by the oncoprotein BCR-ABL, to which cells are addicted for the execution of the oncogenic program.6 The pivotal role of BCR-ABL in human leukemia has allowed the development of effective therapies using c-Abl-specific inhibitors such as Imatinib (Gleevec, Basel, Switzerland).7 Although c-Abl activation by RTKs contributes to distinct biological responses,8, 9 c-Abl oncogenic forms have been rarely identified in solid tumors.10, 11 The still obscure role of c-Abl in this context is paralleled by the failure of using Abl antagonists for the treatment of solid tumors.10, 11 It remains to be clarified whether a subset of tumors, identifiable by specific genetic or molecular signatures, would be highly sensitive to Abl inhibitors.
In this study, we asked whether c-Abl is aberrantly instructed and integrated in signaling mechanisms triggered by oncogenic RTKs by exploring c-Abl requirement in Met-triggered tumorigenesis. The Met-RTK and its ligand HGF have essential functions during embryogenesis and regenerative processes by regulating cell migration, survival, proliferation, and differentiation, similar to Met functions during the oncogenic processes,4, 12, 13, 14, 15, 16, 17 and effective Met antagonists have been identified.4, 18, 19 Moreover, Met confers resistance to ErbB antagonists, as cancer cells use Met to maintain the RTK-driven oncogenic pathways active.20, 21, 22 Here, we show that genetic and pharmacological knockdown of c-Abl impairs Met-triggered solid tumor formation in vivo. By investigating the molecular mechanism involved, we identified a novel signaling path by which c-Abl interconnects RTKs and p53 core pathways. In particular, we show that c-Abl activation by Met triggers p53 phosphorylation on Ser392, which in turn drives the transcriptional upregulation of Mdm2 and protection from cell death. Finally, we uncovered a clinical correlation between aberrant Met activation, wild-type p53 phosphorylation on Ser392, and Mdm2 expression in human hepatocellular carcinoma (HCC). Our findings elucidate a molecular mechanism by which c-Abl participates in the oncogenic program driven by RTKs and may disclose a novel signaling signature to identify patients whose treatment may benefit from c-Abl inhibition.
Results
c-Abl is required for survival and anchorage-independent growth of cancer cells dependent on oncogenic Met
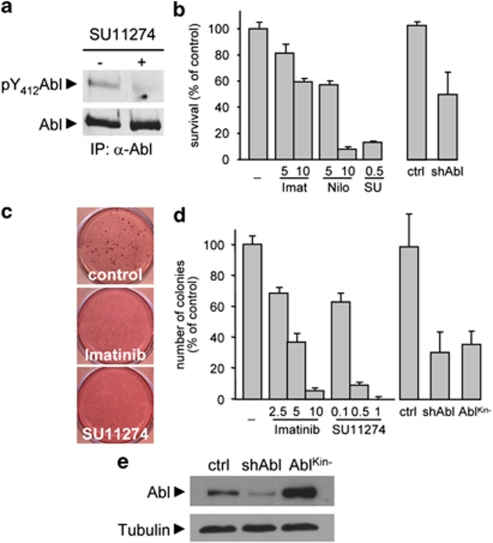
To study whether c-Abl is a signaling node in oncogenic processes triggered by RTKs, we used GTL-16 human gastric carcinoma cells, in which constitutively activated Met leads to Met dependency for cell survival, anchorage-independent growth, and in vivo tumorigenesis.23 We found that constitutive c-Abl phosphorylation on Tyr412 was dependent on Met activity in GTL-16 cells (Figure 1a). Met-triggered survival of GTL-16 cells was significantly reduced by c-Abl antagonists, in a dose-dependent manner (Figure 1b). c-Abl requirement downstream of Met for cell survival was further confirmed by using shRNA interference approach (Figures 1b and e), and found in other cancer cell lines. In particular, c-Abl phosphorylation on Tyr412 was triggered by HGF in human HepG2 HCC cells (Supplementary Figure S1a) and c-Abl inhibition impaired HGF-induced HepG2 cell survival (Supplementary Figure S1b). Imatinib and Nilotinib also inhibit PDGFR and Kit, in addition to c-Abl,7 but we excluded them as primary targets as they were not expressed in all cell types used in our studies (Supplementary Figures S1c and d). We next evaluated in GTL-16 cells whether c-Abl was required for Met-triggered anchorage-independent growth, which is a hallmark of oncogenic transformation. c-Abl inhibition, either pharmacologically, through shRNA interference, or by using a kinase dead form (AblKin−),24 severely affected Met-triggered anchorage-independent growth in a dose-dependent manner (Figures 1c–e), indicating that c-Abl is required to execute the oncogenic transformation in cancer cells dependent on oncogenic Met.
Figure 1.
c-Abl is constitutively phosphorylated in GTL-16 cells overexpressing Met, and required for survival and anchorage-independent growth. (a) Constitutive activation of Abl is impaired in GTL-16 cells exposed to the Met inhibitor SU11274 for 24 h. Abl activation was revealed by immunoblotting with anti-phospho-Y412-Abl antibodies that selectively recognize active Abl. (b) Survival of GTL-16 cells is impaired either by Met (SU11274: SU) or by c-Abl (Imatinib: Imat or Nilotinib: Nilo) inhibitors, in a dose-dependent manner (μM). Similarly, cell survival is affected when GTL-16 are transfected with Abl shRNA interference. GTL-16 cells were serum starved for 24 h and then incubated in the presence of the indicated inhibitors for 48 h. Viability was assessed with the Cell Titer Glo Luminescent Assay. (c and d) Anchorage-independent growth of GTL-16 cells is significantly reduced in the presence of Imatinib or SU11274, when cells are transfected with Abl shRNA, or with the Abl kinase dead form, AblKin−. Mean±S.D. The inhibitors were added every 3 days. (e) c-Abl protein levels in GTL-16 cells transfected with Abl shRNA interference or with AblKin−, compared with controls. A scramble shRNA was used as control (ctrl)
Inhibition of c-Abl interferes with Met-triggered tumor growth in vivo
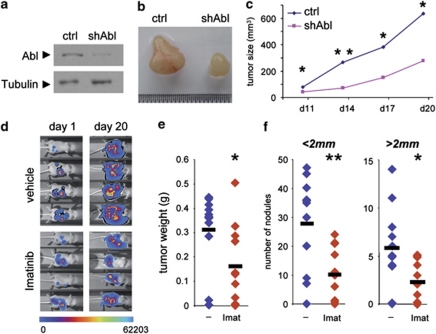
We next evaluated the requirement of c-Abl in Met-triggered tumor growth in vivo by depleting c-Abl using shRNA plasmids (Figure 2a). We found that tumor growth caused by subcutaneous injection of HepG2shAbl cells was significantly reduced compared with that induced by HepG2 control cells (Figures 2b and c), which tumorigenesis has been demonstrated to be dependent on Met.25 Similarly, we observed that c-Abl antagonists restrained Met-triggered tumor growth in vivo by following mice injected intraperitoneally with GTL-16 cells engineered for in vivo non-invasive bioluminescence imaging (Figure 2d). Imatinib treatment led to a reduction of tumor weight by 49%, and of nodule number by 64% (size<2 mm) and 61% (size>2 mm) (Figures 2e and f). Taken together, these findings provide the first in vivo demonstration that c-Abl, when aberrantly instructed by oncogenic RTKs such as Met, is required for solid tumor growth.
Figure 2.
Inhibition of c-Abl signaling interferes with Met-triggered tumor growth in vivo. (a) c-Abl protein levels in HepG2shAbl cells compared with HepG2shScramble (ctrl). (b) HepG2shScramble (ctrl; left) or HepG2shAbl (shAbl; right) cells were subcutaneously injected in nude mice and tumor growth was monitored. Pictures representative of tumors after dissection are shown. (c) Quantification of tumor growth. (d) Stable luciferase-expressing GTL-16luc cells were injected intraperitoneally in nude mice and tumor growth was monitored in mice daily treated with Imatinib or vehicle. Pictures representative of mice groups were taken with NightOwl apparatus after intraperitoneal injection of Luciferin. (e and f) Imatinib (Imat) treatment reduces tumor weight (e) and nodule numbers (f) in nude mice injected with GTL-16 cells. *P<0.05; **P<0.01; Student's t-tests; mean±S.D.
c-Abl interconnects RTK and p53 core pathways by regulating Mdm2 gene expression
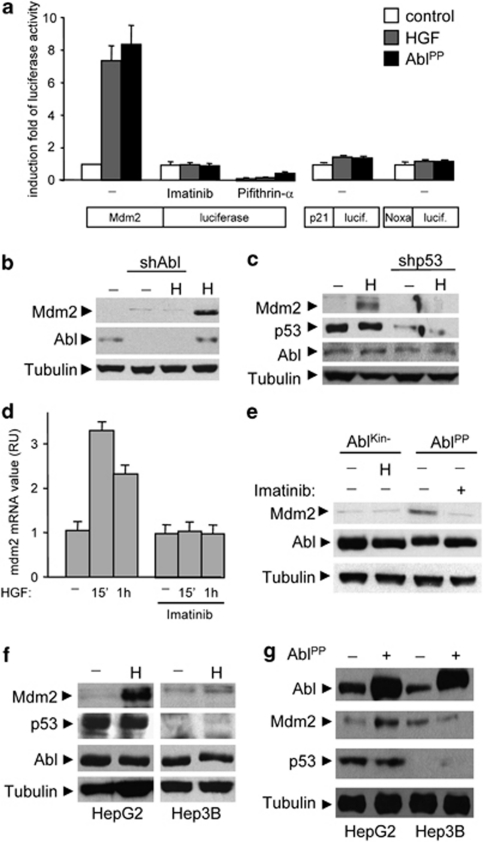
The observed major effect of c-Abl inhibition on Met-dependent cancer cells led us to hypothesize that c-Abl might act as molecular relay connecting core pathways. One of the most frequently altered core pathway evidenced by cancer genomic studies is centered on the tumor suppressor p53. We therefore asked whether c-Abl inhibition would affect the p53 core pathway by monitoring, as a readout of p53 transcriptional activity, expression of target genes such as Mdm2, cyclin-dependent kinase inhibitor p21WAF1, or Noxa.26, 27 For this purpose, HepG2 cells (expressing the transcriptionally competent wild-type p53) were transfected with the luciferase reporter gene controlled by Mdm2, p21WAF1, or Noxa promoters, which are involved in cell survival, cell-cycle arrest, and apoptosis, respectively. Luciferase activity was measured in cells either treated with HGF or co-transfected with AblPP active form.28 We found that both HGF treatment and AblPP expression selectively enhanced luciferase activity controlled by Mdm2, but not by p21WAF1 or Noxa promoters (Figure 3a; Supplementary Figure S2a). The selectivity of Met-Abl axis to upregulate Mdm2, but not p21 or Noxa, was further confirmed by western blot studies (Supplementary Figure S2b). Importantly, the activity of the Mdm2-luciferase reporter was impaired in the presence of both c-Abl and p53 inhibitors (Figure 3a).
Figure 3.
c-Abl acts on p53 to regulate Mdm2, but not p21 or Noxa, expression. (a) HepG2 cells were transfected with plasmid carrying the luciferase reporter gene controlled by the Mdm2, p21WAF1 (p21), or Noxa promoter alone or along with AblPP constructs. After 24 h, cells were treated or not with HGF for 24 h, and luciferase activity was assayed on cell lysates. (b) HepG2 cells stably transfected with Abl shRNA or control plasmid were serum starved, then stimulated with HGF, and Mdm2 protein levels were analyzed by immunoblotting. (c) HGF-induced Mdm2 upregulation is impaired in HepG2 cells transfected with p53 shRNA plasmid. (d) Imatinib impairs HGF-induced upregulation of Mdm2 mRNA levels in HepG2 cells. Cells were serum starved for 3 h and then stimulated with HGF in the presence or not of Imatinib (10 μM). Total RNA was extracted at the indicated times with Trizol and reverse transcription was performed. Levels of Mdm2 mRNA were measured by quantitative real-time PCR (RU, relative unit). (e) HepG2 cells transiently transfected with dominant negative (AblKin−) or constitutive active (AblPP) forms of c-Abl were serum starved, stimulated with HGF as in (c) and Mdm2 protein levels were analyzed by immunoblotting. (f) Upregulation of Mdm2 by HGF does not occur in p53-null Hep3B cells. (g) Constitutive active AblPP does not upregulate Mdm2 levels in the absence of p53
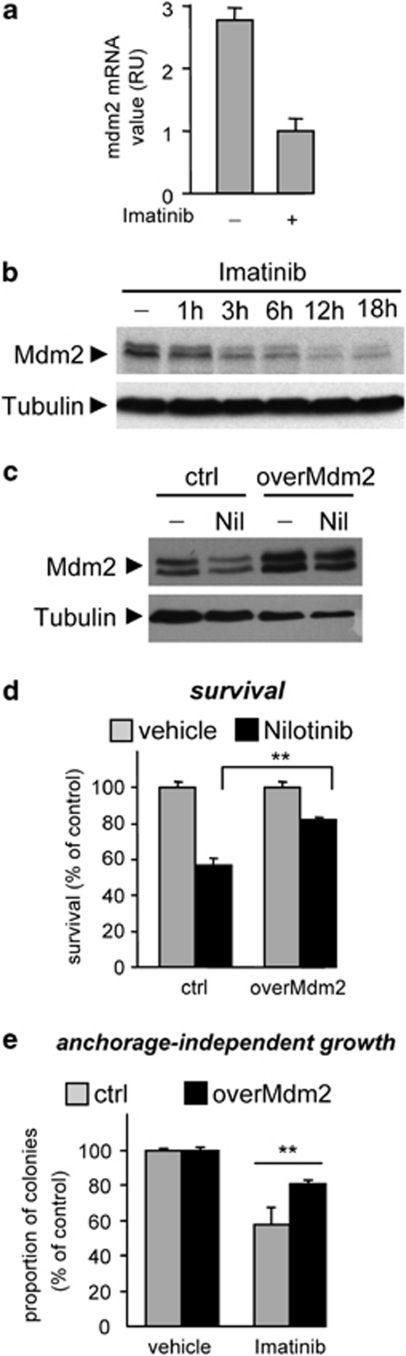
Mdm2 overexpression is frequently observed in several tumors and contributes to tumorigenesis through p53-dependent and -independent mechanisms.29, 30 Through a series of experimental settings, we evaluated Mdm2 regulation by the Met-Abl-p53 axis. Pharmacological impairment or shRNA interference of either c-Abl or p53 prevented Mdm2 upregulation in HepG2 cells exposed to HGF stimulation (Figures 3b–e; Supplementary Figure S2c). Conversely, the AblPP constitutively active form led to an increase of Mdm2 levels even in the absence of HGF stimulation (Figure 3e). Moreover, HGF or Abl-PP failed to induce Mdm2 upregulation in p53-deficient Hep3B cells (Figures 3f and g). We found also high Mdm2 levels in GTL-16 cells (expressing the transcriptionally competent wild-type p53), which were drastically reduced when either c-Abl or p53 was impaired (Figures 4a and b; Supplementary Figures S2d and e).
Figure 4.
Upregulation of Mdm2 expression by the Abl-p53 axis in Met-dependent cells, which in turn ensures cell survival and tumorigenesis. (a and b) High levels of Mdm2 mRNA (a) and protein (b) are significantly reduced in GTL-16 cells exposed to Imatinib (5 μM) for 24 h. Levels of Mdm2 mRNA were measured by quantitative real-time PCR (RU, relative unit). Total protein extracts were analyzed by immunoblotting. (c) High Mdm2 protein levels are present in GTL-16overMdm2 cells compared with parental cells. Mdm2 levels were drastically reduced by Nilotinib (5 μM) in control cells, but not in GTL-16overMdm2 cells. (d and e) Overexpression of Mdm2 significantly restores survival (d) and anchorage-independent growth (e) of GTL-16 cells exposed to c-Abl inhibitors (5 μM). **P<0.01
We next ascertained the relative contribution of Mdm2 regulation by c-Abl downstream of oncogenic Met by assessing the sensitivity of Mdm2 overexpressing GTL-16 cells (GTL-16overMdm2) to c-Abl antagonists. Mdm2 levels were drastically reduced by c-Abl inhibitors in control cells, but not in GTL-16overMdm2 cells (Figure 4c). When cells were exposed to c-Abl inhibitors, we observed an ∼30% higher survival and tumorigenesis capacity of GTL-16overMdm2 compared with controls (Figures 4d and e). In contrast, no changes were observed in the absence of c-Abl inhibitors (Figures 4d and e), indicating that the enhanced survival and tumorigenic properties correlated with c-Abl signaling. The enhanced survival properties were accompanied by impaired caspase activation in GTL-16overMdm2 cells exposed to c-Abl antagonists compared with controls (Supplementary Figure S3a and b). Altogether, these findings show that c-Abl signaling interconnects the Met-RTK to p53, leading to Mdm2 upregulation and survival of cancer cells.
c-Abl regulates p53 phosphorylation on Ser392, ensuring a qualitative readout of p53 transcriptional outcomes
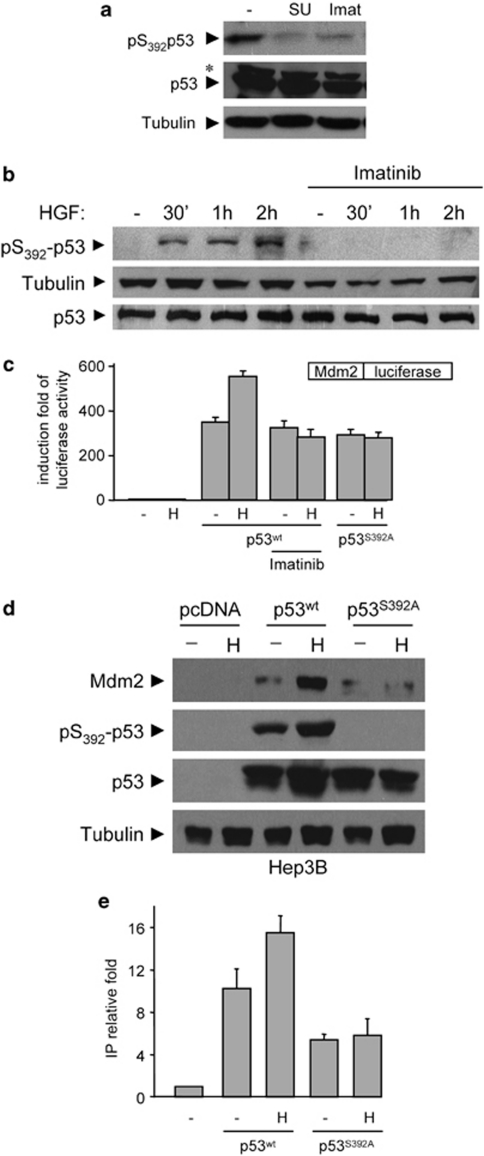
We next investigated how c-Abl acted on p53 to regulate Mdm2 transcription in cells exposed to survival stimuli such as those emanating from Met. As p53 functions are highly modulated by post-translational modifications,26, 31 we evaluated whether c-Abl inhibition would affect p53 phosphorylation. Basal low levels of p53 phosphorylation on Ser15 and Ser46 residues in the N-terminal region, which are usually phosphorylated in response to stress stimuli, were found in GTL-16 cells. No significant changes were observed in the presence of c-Abl antagonists (Supplementary Figure S4a). In contrast, high levels of p53 phosphorylation on Ser392 were found in GTL-16 cells, which were impaired by c-Abl antagonists (Figure 5a; Supplementary Figure S4a). Consistently, c-Abl-dependent p53 phosphorylation on Ser392 was also found in HepG2 cells exposed to HGF (Figure 5b).
Figure 5.
c-Abl signaling by Met leads to p53 phosphorylation on Ser392, which is required for Mdm2 upregulation. (a) High phosphorylation levels of p53 on Ser392 are found in GTL-16 cells, but not when cells are exposed to Met (SU11274: 5 μM) or Abl (Imatinib: 10 μM) inhibitors. The asterisk indicates not specific band. (b) HGF treatment leads to c-Abl-dependent p53 phosphorylation on Ser392 in HepG2 cells. Cells were serum starved for 24 h and then stimulated with HGF in the presence or in the absence of Imatinib (10μM). (c) Ser392 on p53 is required for Mdm2 transcriptional regulation by HGF. p53-deficient Hep3B cells were transfected with Mdm2-promoter-luciferase plasmid along with the indicated p53 constructs, and cells were treated as in Figure 3a. (d) HGF upregulates p53 phosphorylation and Mdm2 levels in Hep3B cells transfected with p53wt, but not with p53S392A. Hep3B cells were transfected with the indicated p53 constructs and treated as in (c). (e) Chromatin immunoprecipitation assay of exogenous p53 bound to Mdm2 promoter using anti-p53 antibody. p53-null Hep3B cells were transfected with empty vector, p53wt, or p53S392A mutant, as indicated. Cells were treated 24 h after transfection with or without HGF for 20 min. Q-RT-PCR of immunoprecipitated DNA was normalized to sample input (no chromatin immunoprecipitated signal was obtained in the immunoprecipitated samples without antibody) and values displayed are relative to sample transfected with an empty vector
It has been shown that phosphorylation on Ser392 leads to a p53 conformational change that enhances DNA binding and transcriptional activity.31 We therefore ascertained whether p53 phosphorylation on Ser392 was required for Met-Abl-triggered Mdm2 transcriptional regulation by reconstituting either p53wt or p53S392A mutant in p53-deficient Hep3B cells. By following luciferase activity, we found that p53S392A failed to trigger Mdm2-promoter induction in response to HGF, in contrast to p53wt (Figure 5c; Supplementary Figure S4b). Enhanced luciferase activity by HGF in Hep3B cells transfected with p53wt was blocked in the presence of c-Abl antagonists (Figure 5c). Furthermore, HGF induced p53 phosphorylation on Ser392 and upregulation of endogenous Mdm2 levels in Hep3B cells transfected with p53wt, but not with p53S392A (Figure 5d), thus demonstrating the requirement of p53 phosphorylation on Ser392 for Mdm2 transcriptional regulation by the Met-Abl axis.
We next assayed whether p53 phosphorylation on Ser392 enhances its DNA binding by performing chromatin immunoprecipitation studies in Hep3B cells transfected either with p53wt or p53S392A. We found higher basal levels of Mdm2 promoter bound to p53wt compared with the p53S392A mutant (Figure 5e). Moreover, HGF increased the levels of Mdm2 promoter bound to p53wt, but not to p53S392A (Figure 5e). Altogether, these findings indicate that c-Abl signaling by the Met-RTK regulates distinct p53 transcriptional outcomes through its phosphorylation on Ser392.
p38-MAPK links Met-Abl to p53 phosphorylation on Ser392 and Mdm2 upregulation
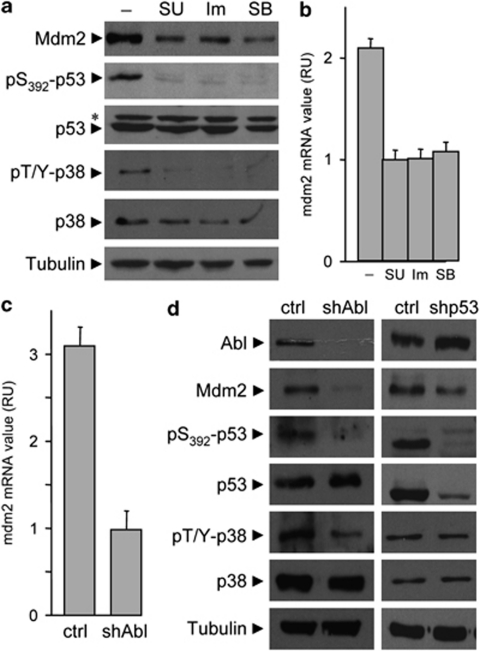
We next investigated how c-Abl tyrosine kinase triggered p53 phosphorylation on Ser392. It has been reported that p38-MAPKs may regulate the phosphorylation of p53 on Ser392 in nasopharyngeal carcinogenesis and in cells exposed to UV treatment.26, 31 We previously reported that HGF stimulation leads to transient p38-MAPK phosphorylation in embryonic cells.32 We therefore analyzed whether p38-MAPK signaling was required downstream of Met-Abl for p53 phosphorylation and Mdm2 expression. We found that phosphorylation of p38-MAPK was dependent on activated Met and c-Abl in GTL-16 cells (Figure 6a). Inhibition of p38-MAPK interfered with p53 phosphorylation on Ser392 and Mdm2 levels in GTL-16 cells (Figures 6a and b). Moreover, c-Abl shRNA interference significantly reduced levels of Mdm2, p53 phosphorylation on Ser392, and p38 phosphorylation (Figures 6c and d). Finally, low levels of Mdm2 and p53 phosphorylation on Ser392, but not of p38 phosphorylation, were observed in GTL-16 transfected with p53 shRNA interference (Figure 6d), indicating that p53 phosphorylation is epistatic to p38 function. The requirement of p38-MAPK downstream of activated c-Abl was further conformed through a series of experimental setting in HepG2 (Supplementary Figures S5a–c). Altogether, these findings show that p38-MAPK signaling links Met-Abl activation to phospho-Ser392-p53 and Mdm2 upregulation.
Figure 6.
Inhibition of p38α and p38β interferes with p53 phosphorylation on Ser392 and Mdm2 upregulation by the Met-Abl axis. (a and b) Basal levels of p38-MAPK phosphorylation in GTL-16 cells requires intact Met and c-Abl signaling. Inhibition of p38α and p38β by SB202190 (SB: 10 μM) affects p53 phosphorylation and upregulation of Mdm2 protein (a) and mRNA (b; P<0.01; Student's t-tests), similar to Met (SU) and c-Abl (Im) inhibitors. The asterisk indicates not specific band. (c and d) Transfection of GTL-16 cells with Abl shRNA interference leads to reduced levels of Mdm2 mRNA (c) as well as Mdm2 protein, p53 and p38 phosphorylation (d), while transfection of GTL-16 cells with p53 shRNA reduced levels of Mdm2 protein, but does not affect p38 phosphorylation (d)
Clinical correlation of phospho-Met, wild-type phospho-Ser392-p53, and Mdm2 levels
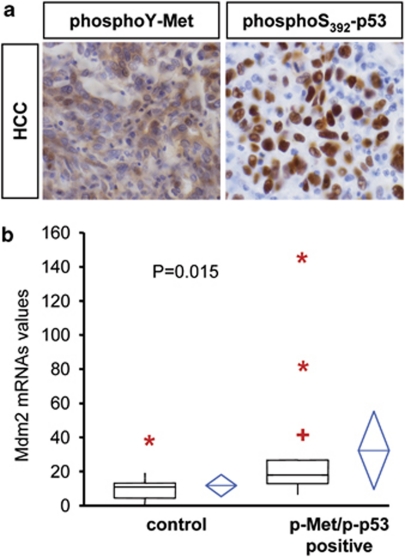
The identification of a novel mechanism by which oncogenic Met regulates Mdm2 through Abl-p53 led us to determine whether there was a clinical correlation between oncogenic Met, phospho-Ser392-p53, and Mdm2 levels in human tumors. We examined a total of 69 patient samples by applying a tumor array screening of human HCCs, where it has been reported that Met contributes to oncogenesis.16, 33 We found that 35 samples (∼50%) were positive for phospho-Met staining and 24 samples (∼35%) for nuclear phospho-Ser392-p53 staining. Notably, 20/69 HCCs (∼29%) showed coincidental immunoreactivity for both antigens. We next evaluated the p53 status in 20 double phospho-Met and phospho-Ser392-p53 positive tumors, and found that p53 gene was mutated in only 6 HCCs (3 in exon 5, 3 in exon 7). Thus, 20/24 tumors positive for phospho-Ser392-p53 were also positive for phospho-Met, indicating that p53 phosphorylation was preferentially associated with the Met oncogenic state (Figure 7a; Supplementary Table 1).
Figure 7.
Mdm2 levels correlate with phosphorylation of Met and wild-type p53 on Ser392 in human hepatocellular carcinomas (HCCs). (a) Immunohistochemistry of phospho-Met (left) and phospho-p53 on Ser392 in HCC. (b) Comparison of Mdm2 mRNAs levels between HCC samples grouped as phospho-Met and phospho-p53 double positive or not double positive (control). Each plot graphically shows the central location and scatter/dispersion of the values of each group: the line series in the rectangular shape box indicate the median value of the data and the end of vertical lines indicates the minimum and the maximum data value. ‘Plus' (+) indicate possible outliers between 1.5 and 3 interquartile range, while ‘asterisk' (*) indicate possible outliers >3 interquartile range. The mean and its confidence interval are shown in the diamond shape box. P-value was calculated according to independent samples t-test (n=27)
We next evaluated Mdm2 levels in double phospho-Met/phospho-Ser392-p53 positive HCCs by performing Q-RT-PCR and found a significant Mdm2 upregulation compared with controls (Figure 7b; Supplementary Table 1). Double-positive cases were not associated with other clinical-pathological parameters (age, sex, etiology) nor with Mdm2 gene amplification, evaluated by FISH assay. A similar correlation between phospho-Met and wild-type phospho-Ser392-p53 was also observed in 6/10 non-small-cell lung cancers. Thus, oncogenic Met and wild-type p53 phosphorylation on Ser392 together with elevated Mdm2 levels in HCCs strongly supports a role for Met-Abl-p53-Mdm2 path in subpopulations of human cancers.
Discussion
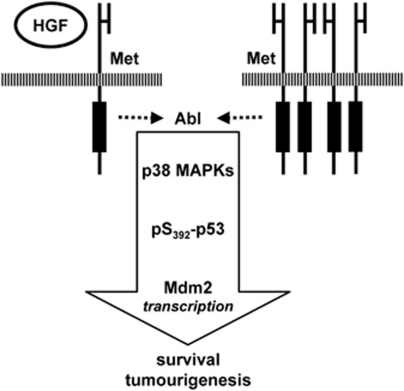
In this work, we showed that inhibition of c-Abl interferes with the Met-driven oncogenic program in vivo. Activated c-Abl signals through p38-MAPK to phosphorylate p53 on Ser392 in cancer cells with an oncogenic Met. As a result, we observed a differential transcriptional modulation of p53 target genes, leading to the selective upregulation of the Mdm2 survival gene (Figure 8). Notably, Mdm2 overexpression decreased the sensitivity of cancer cells to c-Abl antagonists. Our findings unveil the existence of a previously unrecognized mechanism by which RTKs modulate cell survival through c-Abl.
Figure 8.
Integration of c-Abl-p38-p53-Mdm2 signaling in Met-triggered cell survival and tumorigenesis. Activated Met, following its overexpression or HGF stimulation, acts on c-Abl to trigger cell survival and tumorigenesis. Mechanistically, c-Abl leads to p53 phosphorylation on Ser392 by p38-MAPK, leading to Mdm2 upregulation. The functional consequence of this mechanism is genetically and pharmacologically demonstrated. The arrow indicates a relationship between the identified components in the pathway. Additional signaling intermediates and partners remain to be identified
Aberrant activation of RTKs such as Met and ErbBs is associated with the evolution of several solid tumors, and their inhibition is deeply explored for molecular therapies.4, 20, 21, 22 However, this strategy may not be fully effective since cells exposed to an inhibitor of the ‘primary' activated kinase soon become resistant by a mechanism described as RTK switching.4, 20, 21, 22 Our findings provide the first in vivo evidence that inhibition of c-Abl hampers the oncogenic program triggered by the activation of RTKs such as Met. The findings also suggest an alternative strategy to counteract oncogene dependency by targeting downstream signaling nodes, such as c-Abl, shared by several RTKs,8, 9, 10, 11 which may therefore overcome resistance associated to RTK switching. Notably, protein-network-based strategies have revealed that some proteins, although not mutated, have important roles in interconnecting key signals in oncogenic processes.34 Even though c-Abl genetic alterations are rarely found in solid tumors,11 the identification of c-Abl as a mandatory passage in RTK-triggered oncogenesis designates it as a promising therapeutic target.
p53 transcriptional regulation depends on multiple factors such as the strength and reversibility of the stress, and the cellular context.26, 27, 35 The p53 tumor suppressor function can be converted into tumor promoting by a few mutations found in tumors.31, 35 Recent studies have shown that, rather than being a simple tumor suppressor gene, p53 is integrated in instructive signals to orchestrate developmental cell fate.36, 37 Interestingly, it has been shown that while p53 acetylation is required for its transactivation activity on several p53 target genes, the Mdm2 promoter is regulated by p53 even in the absence of this post-translational regulation, suggesting that Mdm2 transcription by p53 may involve specific pathways.38 We identified Ser392 phosphorylation as a mechanism to specifically tune the expression of genes like Mdm2 and regulate cell survival. In contrast, Met does not modulate the phosphorylation of other Ser residues regulating p53 levels, which would be incompatible with tumorigenesis. Notably, in the cancer cell systems we have used, the increase Mdm2 levels do not impact significantly the basal levels of p53, which are, and remain, low compared with those found in cells exposed to agents triggering apoptosis, such as adriamycin. It has been shown that Abl activation may also trigger Mdm2 phosphorylation, which in turn impairs the inhibition of p53 by Mdm2.39 Thus, c-Abl appears to modulate Mdm2 at multiple levels, and the signaling outcomes appear to be distinct from the well-known feedback loop on p53 regulation. Interestingly, our findings raise the provocative idea that oncogenic RTKs instruct wild-type p53 to participate in tumor evolution. Our results agree with those of recent studies aimed at uncovering the molecular mechanisms through which wild-type p53 activity is tuned in tumorigenesis.40 Future work will clarify to which extent Ser392 phosphorylation confers selectivity towards specific DNA sequences (and consequently target genes), which mechanisms are triggered by Mdm2 to impair cell death, and whether other signaling components are recruited within this path at different levels.
Although high levels of phospho-Ser392-p53 have been observed in tumors with mutant p53 and have been correlated with malignancy,31 this phosphorylation in human tumors with wild-type p53 has been neglected so far. Our HCC array studies revealed that p53 was wild type in 70% of double-positive phospho-Met/phospho-Ser392-p53, pointing to the path we have identified as a novel putative mechanism to modulate wild-type p53 function in cancer. These studies deserve further investigation to uncover the relationship between phospho-Ser392-p53 and deregulated RTK in solid tumors, and to clarify whether this correlation can be used as a signaling signature to identify those tumors that may be sensitive to c-Abl antagonists.
In summary, we introduce c-Abl as a signaling node interconnecting RTK and p53 core pathways, and provide clinical correlations between oncogenic Met, wild-type phospho-Ser392-p53, and Mdm2 levels in human tumors. Our findings point to c-Abl as a novel Achille's heel in RTK-dependent tumors with wild-type p53. Abl antagonists, while highly effective in some leukemias, have met little success when tested in patients affected by solid tumors. This failure, however, may be related to the molecular heterogeneity of the treated tumors. Here, we have identified a molecular signature that may help to discriminate which tumors could be sensitive to c-Abl inhibitors. Thus, the promise of c-Abl antagonists for combined treatment of solid tumors, either with traditional or with RTK-targeted agents, should deserve further evaluation according to tumor signaling signature.
Materials and Methods
Antibodies and reagents
Antibodies used were anti-tubulin (Sigma, St. Louis, MO, USA), anti-Kit, anti-phospho-Y1234−1235-Met, T180/Y182-p38 and S392-p53 (Cell Signaling, Danvers, MA, USA), anti-MDM2 (Oncogene Science, San Diego, CA, USA), anti-Met, anti-p53 (FL-393), and anti-PDGFR-α (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), mouse anti-Abl (monoclonal antibody Ab3; Calbiochem-Merck, Darmstadt, Germany), anti-phospho-Y412-Abl (ab4717; Abcam, Cambridge, MA, USA). HGF (R&D Systems, Minneapolis, MN, USA) was used at a concentration of 50 ng/ml. The pharmacological inhibitors used were Imatinib and Nilotinib (Abl inhibitors, kindly provided by E Buchdunger and P Manley, Novartis Pharma AG, Basel, Switzerland), JLK1214 (p53 inhibitor, kindly provided by JL Kraus, IBDML),41 Pifithrin-α (p53 inhibitor), SU11274 (Met inhibitor), K252a (RTK inhibitor), z-DVED-fmk (Caspase inhibitor; Calbiochem). The concentrations of each inhibitor are the following; Imatinib (from 5 to 10 μM), Nilotinib (from 1 to 10 μM), SU11274 (from 0.1 to 5 μM), JLK1214 (10 μM), Pifithrin-α (from 1 to 10 μM), K252a (750 nM), SB202190 (from 1 to 10 μM), z-DVED-fmk (10 μM). No toxic effects were observed at the concentrations used. Actinomycin D (Sigma) was used at 0.2 μg/ml.
DNA constructs
p53-HA-S392A was generated from pCDNA3-p53-HA using the QuickChange site-directed mutagenesis kit (Stratagene, Santa Clara, CA, USA), according to the manufacturer's instructions. pcDNA.AblKin− and pcDNA.AblPP have been previously described.24, 28 psuper-shAbl and psuper-shp53 plasmids were generated according to the siRNA sequence previously described. Luciferase constructs were generated using mdm2, p21WAF1 or Noxa promoters. pCMV-h-mdm2 plasmid was kindly provided by K Vousden.
Cell cultures
GTL-16, HepG2 cells with wild-type p53,42 and p53-deficient Hep3B cells43 were grown in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with penicillin–streptomycin and 10% fetal calf serum. GTL-16luc, GTL-16overMdm2, and GTL-16shp53 cells were obtained by stable transfection of parental GTL-16 cells with the corresponding constructs, using lipofectamin reagent, according to the manufacturer's instructions. Clones were selected using geneticin (for GTL-16luc and GTL-16overMdm2; 0.5 mg/ml, Invitrogen) and puromycin (for GTL-16shp53; 2 μg/ml, Invitrogen). Expression of luciferase in resistant clones was confirmed by assaying luciferase activity in cell lysates. Expression levels of Mdm2 and p53 in GTL-16overMdm2 and GTL-16shp53 cells were analyzed by western blots. For GTL-16luc, a pool of six positive clones was confirmed to behave like the parental cells in soft agar growth assays and was subsequently used for in vivo studies. For GTL-16overMdm2 and GTL-16shp53, pools of resistant clones were used for biochemical and biological assays.
Biochemistry and quantitative real-time PCR
For biochemical studies, cell extracts, immunoprecipitations, and western blots were performed as described previously.14, 15, 44 For quantitative real-time PCR, total RNA was extracted with Trizol (Invitrogen), according to the manufacturer's instructions. Reverse transcription was performed with Superscript Reverse Transciptase II (Invitrogen), according to the manufacturer's instructions. Levels of Mdm2 mRNA were measured by quantitative real-time PCR (qRT-PCR), based on TaqMan methodology, using the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA), as previously described.45 Briefly, custom-designed TaqMan primers and probes (Applied Biosystems) specific for Mdm2 were used. Results are expressed as relative units of target mRNA, referred to a sample called calibrator, chosen to represent 1 × expression of the target gene. The calibrator used was the lowest value in the tissue collection under study. All analyzed samples express n-fold mRNA relative to the calibrator. Each sample mRNA was normalized relative to the β-actin mRNA. Pre-designed TaqMan primers and probe (Applied Biosystems) for the housekeeping gene β-actin were used. Each sample was treated with deoxyribonuclease (DNase) I amplification grade (Invitrogen), tested before and after DNase treatment.
Survival and anchorage-independent growth
For survival assays, GTL-16 cells were seeded in serum-free medium; inhibitors were applied 24 h later. HepG2 cells were seeded in medium containing 10% serum, starved for 24 h, and then inhibitors were applied. Viability was assessed with the Cell Titer Glo Luminescent Assay (Promega, Madison, WI, USA). Survival assays and anchorage-independent growth assays were performed as previously described.14, 15, 18 Data on biological assays are representative of three independent experiments performed in duplicate.
In vivo tumorigenesis assays
Mice were kept at the IBDML animal facilities and all experiments were performed in accordance with institutional guidelines. Xenografts of GTL-16luc cells were established by intraperitoneal (i.p.) injection of cells (106) in nude mice (S/SOPF SWISS NU/NU; Charles River, Wilmington, MA, USA). Mice were then treated daily with Imatinib (i.p. 50 mg/kg, according to the guidelines provided by Novartis Pharma AG) or vehicle. For in vivo bioluminescence imaging, after i.p. injection of luciferin (3 mg/mouse), mice were anesthetized and placed in the NightOwl LB981 (Berthold Technologies, Bad Wildbad, Germany) under continuous anesthetization (1.5–3% isoflurane in O2). First, a black and white photographic image was acquired using 100 ms exposure. Next, the luminescent image was acquired using a 2-min photon integration period with background subtraction (pixel binning 8 × 8). Analysis was performed using Berthold Technologies software. Mice were then killed after 20 days of treatment. Tumor nodules present in the peritoneal cavity were isolated and quantified according to their diameter and their total weight. Three independent tumorigenesis assays were performed (eight mice per group were used). For tumorigenesis assays with HepG2pSuper and HepG2shAbl cells, nude mice were inoculated subcutaneously into the flank/leg region with control pSuper or shAbl mutant cells. Tumor size was externally monitored every 3 days.
ChIP studies
Formaldehyde cross-linking and ChIP were performed as described previously.46 p53-null Hep3B cells (0.5 × 108–1 × 108) were transfected with 3 μg of p53wt or p53S392A expression vector, or with an empty vector pCDNA3 using lipofectamine 2000 reagent (Invitrogen). Cells were treated 24 h after transfection with or without HGF (50 ng/ml) for 20 min, washed with phosphate-buffered saline, and incubated for 10 min with 1% formaldehyde. After the reaction was quenched with 0.1 M glycine, the cells were sonicated into chromatin fragments with an average length of 500–800 bp. Immunoprecipitation was performed with protein G-Sepharose (Amersham Biosciences Corp., Uppsala, Sweden) and 3–5 μg of anti-p53 antibody (Ab7; Oncogene Science). The chromatin solution was precleared by incubation with protein G-Sepharose for 2 h at 4°C, divided into aliquots, and incubated with the antibodies overnight at 4°C. Before use, protein G-Sepharose was blocked twice at 4°C with salmon sperm DNA (1 μg/μl) that had been sheared to a 500-bp length and BSA (1 μg/μl) for 2 h and overnight, respectively. Cross-linking was reversed by incubation of the eluate for 12 h at 65°C with 200 mM NaCl. Finally, samples were treated with proteinase K (Invitrogen) and DNA was purified by extraction with phenol/chloroform and precipitated with isopropyl alcohol plus glycogen. The resulting DNA was analyzed by real-time PCR using following primers. Mdm2 promoter Forward: 5′-GCAGGTTGACTCAGCTTTTCCTCT-3′ Mdm2 promoter Reverse: 5′-GTGGTTACAGCCCCATCAGTAGGTA-3′.
p53 mutations analysis
Only p-p53 nuclear immunoreactive cases were analyzed for p53 exons 5–8 mutations using SSCP analysis (single-strand conformational polymorphism analysis). Briefly, PCR amplification of exons 5, 6, 7, and 8 were done in 50 μl reaction volumes with a final concentration of 0.2 mmol/l deoxynucleotide triphosphates (Finnzymes), 0.2 μmol/l of each primer (Sigma), and 1 unit of Taq polymerase (Finnzymes). PCR products, denaturated in a buffer containing 100% formamide (Sigma), were run at room temperature on 15% acrylamide-bisacrylamide gel (Sigma) for 1 h at 100 V, and 4 h at 300 V. DNA containing the wild-type p53 (human placental DNA) was always included as negative control. Silver staining was done using PlusOne DNA Silver Staining kit (Amersham Biosciences Corp.), according to the manufacturer's instructions.
Tumor series under study and Immunohistochemistry
Sixty-nine cases of HCC resected at the Istituto Clinico Humanitas from 2004 to 2006 were analyzed. Clinicopathologial features are reported in Supplementary Table 1. From the paraffin tissue of the single most representative tumor sample, four punches were obtained and collected on a tissue microarray (TMA). Standard 2 μm-thick sections of TMA were submitted to antigen retrieval (30 min at 98°C in W-CAP TEC Buffer pH 8, Bio-Optica, Milano, Italy) and then incubated (1 h) with mouse monoclonal p-p53 (1 : 100; Novocastra Laboratories, New Castle upon Tyne, UK) or rabbit polyclonal p-Met (Tyr1230-1234-1235; 1 : 50; Assay Designs, Ann Arbor, MI, USA). The Immpress reagent was used as detection system (Vector Laboratories, Burlingame, CA, USA). Staining was obtained using 3,3′diaminobenzidine as chromogen followed by hematoxylin counterstaining. Positive (colorectal cancer) and negative (primary antibody omitted) controls were used. The nuclear p-p53 and the cytoplasmic p-Met immunoreactivity were evaluated as follows: negative – no or very weak/equivocal staining; positive – unequivocal staining. Total RNA purification from paraffin-embedded HCC samples was performed using Absolutely RNA FFPE Kit (Stratagene). Levels of Mdm2 RNA were measured by quantitative real-time PCR.
Statistical analysis
Quantification of tumor growth in mice was analyzed by Student's t-tests. The relationship between Mdm2 mRNA levels and double-positive sample for phospho-Met and phospho-S392-p53 was analyzed by a two-tailed Student's t-test. A P<0.05 was considered as significant. Statistical analysis was carried out using the Analyse-it software for Microsoft Excel (Analyse-it Software, Ltd, Leeds, UK, version 1.62).
Acknowledgments
We are particularly grateful to C Ponzetto, G Superti-Furga, G Cesareni, A Porras, F Helmbacher, R Dono, and all the members of our laboratories for helpful discussions and comments. We thank A Moumen for initial studies on p53 requirement for Mdm2 transcriptional regulation by Met; L Leserman and M Buferne for luciferase plasmids and for valuable help with in vivo bioluminescence imaging; S Soddu and F Moretti for p21Luc, Noxaluc, Mdm2, and p53 constructs, for helpful discussion, and comments; JC Bourdon for Mdm2Luc constructs; MG diBari for the shAbl construct; A Totaro for helping with point mutagenesis; C Ponzetto and T Crepaldi for GTL-16 cells; P Dubreuil for TF1 cells and anti-Kit antibodies; Novartis Pharma AG, Basel (Switzerland) for Imatinib and Nilotinib; and staff at the IBDML and CIML animal house facilities. This work was funded by the INCa, ARC, FRM, AFM, FdF, Fondation Bettencourt-Schueller to FM, and by the Telethon Foundation, AIRC, AICR (07-0461) to DB. AF was supported by INCa and ARC fellowships; FC by AIRC fellowship; VS was supported by Fondazione Santa Lucia and by a FIRC fellowship.
Glossary
- RTK
receptor tyrosine kinase
- CML
chronic myeloid leukemias
- ALL
acute lymphocytic leukemia
- HGF
hepatocyte growth factor
- i.p.
intraperitoneal
- HCC
human hepatocellular carcinoma
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by B Dynlacht
Supplementary Material
References
- Network TCGAR Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Zwick E, Bange J, Ullrich A. Receptor tyrosine kinases as targets for anticancer drugs. Trends Mol Med. 2002;8:17–23. doi: 10.1016/s1471-4914(01)02217-1. [DOI] [PubMed] [Google Scholar]
- Knudsen BS, Vande Woude G. Showering c-MET-dependent cancers with drugs. Curr Opin Genet Dev. 2008;18:87–96. doi: 10.1016/j.gde.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci USA. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschel O, Superti-Furga G. Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi: 10.1016/S0065-230X(04)91001-9. [DOI] [PubMed] [Google Scholar]
- Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13:2400–2411. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan D, Plattner R. Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 2006;66:5648–5655. doi: 10.1158/0008-5472.CAN-06-0734. [DOI] [PubMed] [Google Scholar]
- Lin J, Arlinghaus R. Activated c-Abl tyrosine kinase in malignant solid tumors. Oncogene. 2008;27:4385–4391. doi: 10.1038/onc.2008.86. [DOI] [PubMed] [Google Scholar]
- Sirvent A, Benistant C, Roche S. Cytoplasmic signalling by the c-Abl tyrosine kinase in normal and cancer cells. Biol Cell. 2008;100:617–631. doi: 10.1042/BC20080020. [DOI] [PubMed] [Google Scholar]
- Maina F, Casagranda F, Audero E, Simeone A, Comoglio P, Klein R, et al. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell. 1996;87:531–542. doi: 10.1016/s0092-8674(00)81372-0. [DOI] [PubMed] [Google Scholar]
- Maina F, Pante G, Helmbacher F, Andres R, Porthin A, Davies AM, et al. Coupling Met to specific pathways results in distinct developmental outcomes. Mol Cell. 2001;7:1293–1306. doi: 10.1016/s1097-2765(01)00261-1. [DOI] [PubMed] [Google Scholar]
- Moumen A, Ieraci A, Patane S, Sole C, Comella JX, Dono R, et al. Met signals hepatocyte survival by preventing Fas-triggered FLIP degradation in a PI3k-Akt-dependent manner. Hepatology. 2007;45:1210–1217. doi: 10.1002/hep.21604. [DOI] [PubMed] [Google Scholar]
- Moumen A, Patane S, Porras A, Dono R, Maina F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134:1443–1451. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genestine M, Caricati E, Fico A, Richelme S, Hassani H, Sunyach C, et al. Enhanced neuronal Met signalling levels in ALS mice delay disease onset Cell Death Dis 2011. doi: 10.1038/cddis.2011.11 [DOI] [PMC free article] [PubMed]
- Patane S, Pietrancosta N, Hassani H, Leroux V, Maigret B, Kraus JL, et al. A new Met inhibitory-scaffold identified by a focused forward chemical biological screen. Biochem Biophys Res Commun. 2008;375:184–189. doi: 10.1016/j.bbrc.2008.07.159. [DOI] [PubMed] [Google Scholar]
- Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Carraway KL, III, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2-overexpressing breast cancer cells. Cancer Res. 2008;68:1471–1477. doi: 10.1158/0008-5472.CAN-07-5962. [DOI] [PubMed] [Google Scholar]
- Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- Barila D, Mangano R, Gonfloni S, Kretzschmar J, Moro M, Bohmann D, et al. A nuclear tyrosine phosphorylation circuit: c-Jun as an activator and substrate of c-Abl and JNK. EMBO J. 2000;19:273–281. doi: 10.1093/emboj/19.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman DA, Overmeer RM, van Beusechem VW, Lamers WH, Hakvoort TB, Snijders PJ, et al. Inhibition of angiogenesis and HGF-cMET-elicited malignant processes in human hepatocellular carcinoma cells using adenoviral vector-mediated NK4 gene therapy. Cancer Gene Ther. 2005;12:954–962. doi: 10.1038/sj.cgt.7700856. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Barilà D, Superti-Furga G. An intramolecular SH3-domain interaction regulates c-Abl activity. Nat Genet. 1998;18:280–282. doi: 10.1038/ng0398-280. [DOI] [PubMed] [Google Scholar]
- Daujat S, Neel H, Piette J. MDM2: life without p53. Trends Genet. 2001;17:459–464. doi: 10.1016/s0168-9525(01)02369-1. [DOI] [PubMed] [Google Scholar]
- Ganguli G, Wasylyk B. p53-independent functions of MDM2. Mol Cancer Res. 2003;1:1027–1035. [PubMed] [Google Scholar]
- Matsumoto M, Furihata M, Ohtsuki Y. Posttranslational phosphorylation of mutant p53 protein in tumor development. Med Mol Morphol. 2006;39:79–87. doi: 10.1007/s00795-006-0320-0. [DOI] [PubMed] [Google Scholar]
- Segarra J, Balenci L, Drenth T, Maina F, Lamballe F. Combined signaling through ERK, PI3K/AKT, and RAC1/p38 is required for Met-triggered cortical neuron migration. J Biol Chem. 2006;281:4771–4778. doi: 10.1074/jbc.M508298200. [DOI] [PubMed] [Google Scholar]
- Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Piccolo S. p53 regulation orchestrates the TGF-beta response. Cell. 2008;133:767–769. doi: 10.1016/j.cell.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Lowe SW. Stem cells: the promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Z, Vogt Sionov R, Berger M, Zwang Y, Perets R, Van Etten RA, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: implications for p53 regulation. EMBO J. 2002;21:3715–3727. doi: 10.1093/emboj/cdf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- Pietrancosta N, Moumen A, Dono R, Lingor P, Planchamp V, Lamballe F, et al. Imino-tetrahydro-benzothiazole derivatives as p53 inhibitors: discovery of a highly potent in vivo inhibitor and its action mechanism. J Med Chem. 2006;49:3645–3652. doi: 10.1021/jm060318n. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Yoshida T, Hayashi K, Sekiya T, Yokota J, Hirohashi S, et al. p53 gene mutations in gastric cancer metastases and in gastric cancer cell lines derived from metastases. Cancer Res. 1991;51:5800–5805. [PubMed] [Google Scholar]
- Farshid M, Tabor E. Expression of oncogenes and tumor suppressor genes in human hepatocellular carcinoma and hepatoblastoma cell lines. J Med Virol. 1992;38:235–239. doi: 10.1002/jmv.1890380402. [DOI] [PubMed] [Google Scholar]
- Barilà D, Rufini A, Condo I, Ventura N, Dorey K, Superti-Furga G, et al. Caspase-dependent cleavage of c-Abl contributes to apoptosis. Mol Cell Biol. 2003;23:2790–2799. doi: 10.1128/MCB.23.8.2790-2799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodosmo A, Giglio S, Moretti S, Mancini F, Barbi F, Avenia N, et al. Analysis of human MDM4 variants in papillary thyroid carcinomas reveals new potential markers of cancer properties. J Mol Med. 2008;86:585–596. doi: 10.1007/s00109-008-0322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner A, Manni I, Fuschi P, Mantovani R, Guadagni F, Sacchi A, et al. Requirement for down-regulation of the CCAAT-binding activity of the NF-Y transcription factor during skeletal muscle differentiation. Mol Biol Cell. 2003;14:2706–2715. doi: 10.1091/mbc.E02-09-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.