Abstract
In yeast, activation of ATG1/ATG13 kinase complex initiates autophagy. This mechanism of autophagy initiation is conserved, as unc-51-like kinase 1 (ULK1) and unc-51-like kinase 2 (ULK2) are two mammalian functional homologues of ATG1 and form similar complex with mammalian ATG13. Here, we report that both ULK1 and ULK2 are transcriptional targets of tumor suppressor p53. In response to DNA damage, ULK1 and ULK2 are upregulated by p53. The upregulation of ULK1 (ULK2)/ATG13 complex by p53 is necessary for the sustained autophagy activity induced by DNA damage. In this context, elevated autophagy contributes to subsequent cell death. These findings suggest that ULK1 and ULK2 may mediate part of tumor suppression activity in mammalian cells and contribute to the efficacy of genotoxic chemotherapeutic drugs.
Keywords: autophagy, p53, DNA damage, Ulk1
Autophagy is an intracellular self-digestive process and plays a critical role in maintaining energy homeostasis. During autophagy, a double-membrane vesicle, termed autophagosome, forms around a portion of cytosol and/or damaged organelles. The autophagosome then fuses with lysosome, and the sequestered cargos are degraded inside the acidic compartment to provide cells necessary nutrient supply.
Autophagy has been extensively studied in yeast Saccharomyces cerevisiae and dozens of autophagy-related proteins (ATGs) have been discovered.1 These ATG proteins can be classified into several functional groups, one of which is autophagy-initiating complex composed of ATG1 and ATG13, as well as ATG17, ATG29 and ATG31 under certain circumstances.1 ATG1 is a serine/threonine protein kinase. In yeast, autophagy initiation is triggered by nutrient starvation or inhibition of TOR (target of rapamycin) kinase, resulting in dephosphorylation of ATG13, association between ATG13 and ATG1, and subsequent full activation of ATG1's kinase activity.2 This process is largely conserved from yeast to human.3 ATG1 has at least two mammalian functional homologues named unc-51-like kinase 1 (ULK1)4, 5 and unc-51-like kinase 2 (ULK2),6 both of which form complex with mammalian ATG13 protein. In mammals, ULK1 and ULK2 have been shown to be necessary for the proper autophagy induction and contribute to various developmental, physiological and pathological processes.7
P53 is a well-known tumor suppressor protein. It functions as a tumor suppressor partly through transcriptionally regulating the expression of genes involved in cellular senescence and apoptosis. It has also been reported to induce autophagy by inhibiting mammalian TOR (mTOR) signaling through elevating AMPK-β1, TSC2, PTEN8 and Sesn1/2,9 all of which are negative regulators of mTOR. In addition, p53 can also trigger autophagy through activating the transcription of DRAM, a novel lysosomal protein.10
Autophagy has been implicated in various cell death phenotypes, yet the true nature of these events remains elusive, and it can be linked to both death-promoting11, 12 and -inhibiting activities.13 Here in this study, we report that the two human ATG1 homologues, ULK1 and ULK2, are transcriptional targets of p53. Their upregulation by p53 leads to sustained autophagy in response to DNA damage and contributes to subsequent cell death.
Results
Sublethal DNA damage causes sustained increase of autophagy
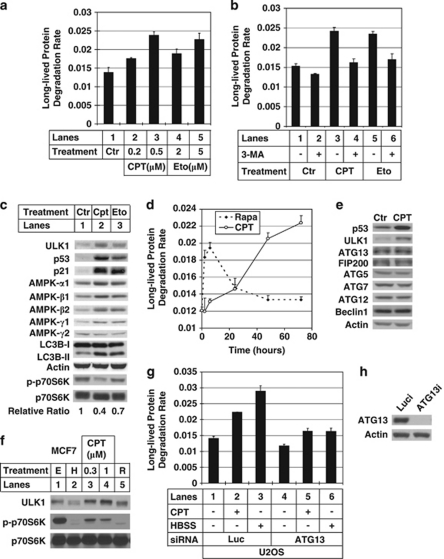
DNA topoisomerase inhibitors such as camptothecin (CPT) and etoposide (Eto) can trigger DNA damage, resulting in cell cycle arrest, apoptosis and autophagy.11 In human osteosarcoma U2OS cells, treatment with low doses of CPT (less than 1 μM) or Eto (less than 15 μM) only induced cell cycle arrest, but not apoptosis (Supplementary Figure 1). These sublethal conditions were then used in the following studies.
Proteins with long half-life are generally degraded in lysosomes through autophagy. Rate of long-lived protein degradation (LLPD) can therefore be used to monitor autophagy activity.14 As shown in Figure 1a, LLPD assay indicates that both CPT and Eto treatments upregulated autophagy in a dose-dependent manner. 3-Methyladenine (3-MA), an autophagy inhibitor, attenuated the increase of LLPD rate induced by CPT/Eto (Figure 1b, compare lanes 4, 6 with lane 3, 5, respectively).
Figure 1.
Autophagy induction by sub-lethal DNA damage. (a) U2OS cells were treated with different concentrations of camptothecin (CPT, lanes 2 and 3) or etoposide (Eto, lanes 4 and 5) for 2 days before doing the long-lived protein degradation (LLPD) assay as described in Materials and Methods. Y axis is the LLPD rate after 2 h. (b) U2OS cells were treated with either 0.5 μM CPT (lanes 3 and 4) or 5 μM Eto (lanes 5 and 6) for 2 days. On the day of measuring LLPD rate, 5 mM of 3-methyladenine (3-MA) was used to pretreat cells for 1 h (lanes 2, 4 and 6) before doing the LLPD assay. (c) U2OS cells were treated with either 0.5 μM CPT (lane 2) or 5 μM Eto (lane 3) for 2 days. The cells were collected and lysed to run western blot. The blot were probed with the antibody against unc-51-like kinase 1 (ULK1), p53, p21, T-389 phosphorylated form of p70S6K (p-p70S6K), p70S6K, AMPK-α1, -β1, -β2, -γ1, -γ2, LC3B and actin. To measure the ratio of p-p70S6K over p70S6K, the blot was developed using enhanced chemiluminescence (ECL) reagent and the band intensities were measured using Kodak Image Station 4000R pro. The relative ratio for the control is defined as 1. (d) U2OS cells were treated with 1 μM rapamycin or 0.5 μM CPT for 2, 6, 24, 48 and 72 h, after which LLPD rates were measured for each sample and plotted against treatment time. (e) U2OS cells were treated with 1 μM CPT for 1 day. The cells were then collected and lysed to do western blot. The blots were probed with the antibodies against p53, ULK1, ATG13, FIP200, ATG5, ATG7, ATG12, beclin 1 and actin. (f) MCF7 cells were untreated (E, lane 1) or treated with either Hank's buffered salt solution (HBSS) for 2 h (H, lane 2), 0.3 μM CPT for 1 day (lane 3), 1 μM CPT for 1 day (lane 4) and 1 μM rapamycin for 6 h (R, lane 5). The cells were then collected and lysed to do western blot. The blots were probed with the antibodies against ULK1, p-p70S6K and p70S6K. (g) U2OS cells were transfected with either luciferase control small interfering (si)RNA (lanes 1–3) or ATG13 siRNA (lanes 4–6) as described in Materials and Methods. After 3 days of siRNA transfection, they were treated with (lanes 2 and 5) or without (lanes 1 and 4) 0.5 μM CPT for 2 days and measured the LLPD rates. HBSS treatments (lanes 3 and 6) were used as controls. (h) The same samples as in (g) were used to do western blot to confirm the knockdown efficiency of ATG13. These data are presented as mean+S.D. and are representative of at least two independent experiments
Microtubule-associated protein light chain 3B (LC3B, or LC3) is the mammalian functional homologue of yeast Atg8 protein. Cytoplasmic LC3B (LC3B-I) is translocated to autophagosomes during autophagy and converted to LC3B-II by proteolysis and lipidation at its C terminus.15 The conversion of LC3B-I to -II is a classical marker for autophagy induction. As shown in Figure 1c, LC3B-II levels were increased after CPT or Eto treatment.
Both the LLPD and the LC3B-II conversion assays confirm the authenticity of autophagy induced by DNA damage. As the LLPD rate more quantitatively reflects the flux of autophagy, we used it as our primary assay in later experiments. In agreement with previous reports,8, 11 expression level of human ULK1 and AMPK-β1/2 were upregulated after DNA damage (Figure 1c). AMPK-α1 and -γ1 were also upregulated by DNA damage, whereas AMPK-γ2 was not. Activity of mTOR was inhibited after treatment, as reflected by the decrease of p70S6K phosphorylation at threonine 389 (Figure 1c), a classical marker for mTOR activity.16 Compared with Eto treatment, CPT induced a stronger response, an observation that prompted us to use CPT treatment in the following studies.
Time-course analysis of the LLPD rate after CPT treatment was performed and rapamycin treatment was used as the positive control. CPT treatment resulted in sustained increase of autophagy activity up to 3 days. In comparison, the maximum autophagy induced by rapamycin was achieved during the first few hours after treatment, and the LLPD rate actually declined back to the basal level after rapamycin treatment for 2 days (Figure 1d).
Interestingly, only the protein level of ULK1 was increased after CPT treatment, whereas other core autophagy components including ATG13, FIP200, ATG5, ATG7, ATG12 and beclin 1 (mammalian ATG6) stayed unchanged (Figure 1e). The increase of ULK1 after DNA damage was also observed in human breast cancer MCF7 cells (Figure 1f).
As ULK1/ULK2, ATG13 and FIP200 form an autophagy-initiating kinase complex,17, 18, 19 we next tested whether other component besides ULK1/ULK2 in this complex contributed to DNA damage-induced autophagy. Consistently, knock down of ATG13 attenuated CPT/Eto-induced autophagy (Figure 1g, compare lane 2 with lane 5; Supplementary Figure 2A, compare lane 3 with lane 4). As reported previously,19 removal of ATG13 resulted in destabilization of ULK1 as well (Supplementary Figure 3, lanes 3–6, 8 and 10). These data indicate that ULK1/ATG13 autophagy-initiating complex is involved in the DNA damage response.
Upregulation of ULK1 and autophagy by DNA damage is dependent on p53
It has been reported that DNA damage induces autophagy through p53.10 P53 does so by transcriptionally elevating AMPK-β1/2, TSC2, PTEN,8 DRAM10 or Sesn1/2.9 To our knowledge, there is no report showing that p53 can directly regulate any core component of autophagy machinery (ATG proteins).
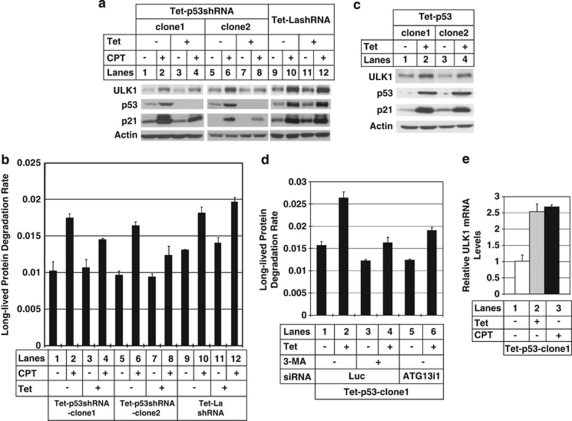
DNA damage reagent like Eto has been shown to elevate ULK1 protein level, although it remains unclear as to whether it is through p53 or not.11 We noticed that upregulation of ULK1 by CPT treatment was attenuated by knocking down p53 in U2OS cells (Figure 2a, compare lane 2 with lane 4, and lane 6 with lane 8). Consistently, the LLPD induced by DNA damage was also attenuated (Figure 2b with CPT treatment and Supplementary Figure 2B with Eto treatment). As a control, knockdown of La, an autophagy-unrelated protein, did not affect either ULK1 protein level (Figure 2a) or autophagy activity (Figure 2b).
Figure 2.
Unc-51-like kinase 1 (ULK1) upregulation after DNA damage is dependent on p53. (a) Three U2OS clones containing tetracycline (Tet)-responsive p53shRNA (Tet-p53shRNA-clone1 and clone2) or a nonspecific shRNA (Tet-LashRNA) were treated with (lanes 3, 4, 7, 8, 11 and 12) or without (lanes 1, 2, 5, 6, 9 and 10) 1 μg/ml Tet for 2 days to knock down p53 or La, and then treated with (lanes 2, 4, 6, 8, 10 and 12) or without (lanes 1, 3, 5, 7, 9 and 11) 0.5 μM CPT for 2 days. The cells were then collected and lysed to do western blot. The blots were probed with antibodies against ULK1, p53 and p21. Actin was used as a loading control. (b) The same cells were treated as in (a) before doing the long-lived protein degradation (LLPD) assay. (c) Two U2OS clones containing Tet-responsive p53 (Tet-p53-clone1 and -clone2) were treated with (lanes 2 and 4) or without (lanes 1 and 3) 1 μg/ml Tet for 1 day to induce the expression of p53. The cells were then lysed to do western blot to test the expression level of ULK1, p53 and p21. Actin was used as a loading control. (d) One of the clone (clone1) as in (c) was transfected with either Luciferase small interfering (si)RNA (Luc, lanes 1–4) or ATG13 siRNA (ATG13i1, lanes 5 and 6) as described in Materials and Methods. After 3 days of siRNA transfection, p53 expression was induced with 1 μg/ml of Tet (lanes 2, 4 and 6) for 1 day. 3-MA (5 mM) was used to pretreat the cells for 1 h (lanes 3 and 4) before doing the LLPD assay. (e) The same clone (clone 1) as in (d) was either induced to express p53 (lane 2) or treated with 0.5 μM CPT for 2 days (lane 3), after which total RNAs were extracted from these cells. Quantitative PCR was used to test the mRNA expression levels of ULK1. These data are presented as mean±standard deviation (S.D.) and are representative of at least two independent experiments
Ectopic expression of p53 transcriptionally upregulates ULK1 and increases autophagy
To show that ULK1 is a transcriptional target of p53, we generated stable U2OS cell lines in which p53 could be inducibly expressed upon addition of tetracycline (Tet) into the culture medium. As shown in Figures 2c and d, both ULK1 protein level and the LLPD rate were increased after p53 induction. The LLPD rate can be attenuated by either 3-MA treatment or ATG13 knockdown (Figure 2d, Supplementary Figure 4), confirming that p53 regulates autophagy at the step of ULK1/ATG13 kinase complex.
Increase of ULK1 protein level may be due to either transcriptional upregulation or post-translational modifications. To differentiate these two possibilities, we measured the mRNA level of ULK1 with quantitative PCR (Q-PCR). As shown in Figure 2e, ULK1 mRNA level was increased after p53 induction or CPT treatment in U2OS cells.
There are five potential ATG1 homologues in human, among which ULK1 and ULK2 are considered the true functional homologues of yeast ATG1 kinase.20 Interestingly, upregulation of ULK2 mRNA level was also observed, but only after CPT treatment, not after p53 ectopic expression (Supplementary Figure 5). Therefore, ectopic expression of p53 is sufficient to upregulate ULK1, but not ULK2.
ULK1 is upregulated at transcription level by p53 after DNA damage
To further confirm p53 is responsible for the transcriptional upregulation of ULK1/ULK2 after CPT treatment, mRNA levels of ULK1/ULK2 were compared in cells with or without p53. Q-PCR experiments were performed in either p53-inducible knockdown U2OS stable cell line or various human colon carcinoma cell lines with different p53 status, including HCT116, HCT116/p53KO (p53 knockout cells), RKO and RKO-E6 (p53-blunted). For U2OS cells, knocking down p53 attenuated mRNA increase for both ULK1 and ULK2 after CPT treatment (Supplementary Figure 6A). In colon carcinoma lines, ULK1 expression level also increased after CPT treatment and was attenuated in p53-blunted cells (Supplementary Figure 6B). However, in these colon cancer lines, ULK2 mRNA level did not change after CPT treatment, indicating that the regulation of ULK1 and ULK2 may differ in different tissues (Supplementary Figure 6B). In HCT116 p53 knockout cells, although the upregulation of ULK1 mRNA level was attenuated compared with parental cells after CPT treatment, it was still higher than the non-CPT treatment control group (Supplementary Figure 6B, comparing lane 3 with lane 4), suggesting the existence of a p53-independent pathway for upregulating ULK1 after DNA damage in this colon cancer cell line.
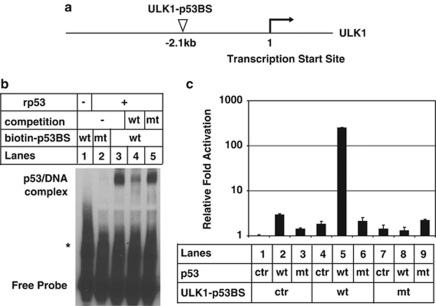
p53-responsive elements in the promoter region of ULK1 and ULK2
To confirm ULK1 and ULK2 are bona fide transcriptional targets of p53, we identified putative p53 binding sites in the promoter regions of ULK1 (Figure 3a) and ULK2 (Supplementary Figure 7A) using bioinformatics methods. To verify the authenticity of these p53 binding sites, electrophoretic mobility shift assay was performed using biotinylated oligonucleotides containing these sequences. Recombinant p53 caused the up-shift of wild-type (wt) probes (ULK1, Figure 3b, lane 3; ULK2, Supplementary Figure 7B, lane 3), but not the mutants (mts) (ULK1, Figure 3b, lane 2; ULK2, Supplementary Figure 7B, lane 2). Moreover, excessive non-biotinylated oligonucleotide containing the wt p53 site was able to compete for binding (ULK1, Figure 3b, lane 4; ULK2, Supplementary Figure 7B, lane 4), whereas the mt could not (ULK1, Figure 3b, lane 5; ULK2, Supplementary Figure 7B, lane 5), confirming the specificity of these up-shifts.
Figure 3.
Unc-51-like kinase 1 (ULK1) is a bona fide transcriptional target of p53. (a) Schematic representation of the putative p53 binding site of ULK1. (b). Electrophoretic mobility shift assay was carried out using biotinylated DNA oligos containing either the duplicated putative wild-type (wt) p53 binding sites (biotin-p53BS, lanes 1 and 3–5) or mutant (mt) p53 binding site (lane 2) in ULK1 gene as described in Materials and Methods. Wt (lane 4) or mt (lane 5) p53 binding sites of ULK1 were used for the competition assay. (The bands labeled with asterisk indicates nonspecific DNA complexes.) (c) Luciferase reporter assay was performed as described in Materials and Methods. Result for each condition was normalized to the firefly/Renilla luciferase activity ratio of empty control (lane 1). Ctr: empty pcDNA3.1 vector. Y axis is depicted as log scale. These data are representative of at least two independent experiments
We next performed a luciferase reporter assay to further verify the transcriptional regulation of p53 at the ULK1 site. Four copies of the wt or mt form of ULK1-p53 binding site (p53BS) was cloned into pGL3-promoter vector and co-transfected into U2OS cells with either wt or mt p53.21 As shown in Figure 3c, only the combination of wt p53 and wt ULK1-p53 binding sites (Figure 3c, lane 5) resulted in strong luciferase activity (over 200-fold increase), but not any other combinations. As for ULK2, as overexpression of p53 did not result in ULK2 upregulation, we did not test the activity of ULK2 p53 binding site with this assay. It is possible that after DNA damage, some other transcription factors are required to upregulate ULK2 together with p53.
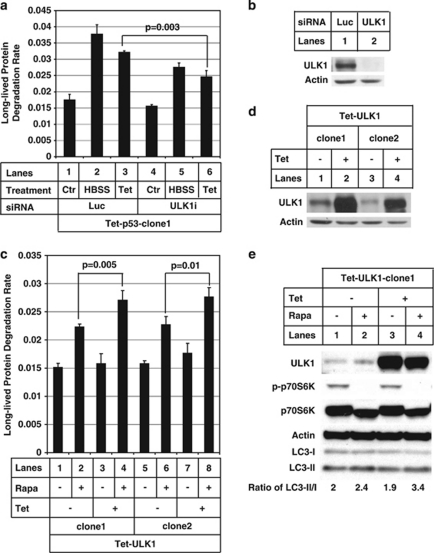
ULK1 knockdown attenuates p53 ectopic expression-induced autophagy
We subsequently focused on ULK1 to study autophagy using gain-of-function and loss-of-function assays as ULK2 upregulation by p53 remains complex in different cell lines under different treatment conditions (Supplementary Figure 5 and 6B). As shown in Figure 4a, knocking down ULK1 was sufficient to attenuate autophagy induced by p53 ectopic expression. To eliminate the possibility of off-target RNAi effect, the specificity of ULK1 siRNA was confirmed with rescue experiments using cDNA bearing silent mutations at the shRNA targeting site (Supplementary Figure 8A, compare lane 4 with lane 8). Total nutrient and serum starvation by switching the culture medium to Hank's buffered salt solution was used here as a positive control for autophagy induction (Figure 4a, lanes 2 and 5). As CPT treatment caused upregulation of both ULK1 and ULK2 in U2OS cells, it is then difficult to observe attenuation of autophagy by just knocking down ULK1. As an indirect evidence, ATG13 knockdown attenuated autophagy induced by CPT treatment (Figure 1g, compare lane 2 with 5), suggesting a putative involvement of ULK2 and/or other uncharacterized human ATG1 homologue22 in DNA-damage-induced autophagy.
Figure 4.
Unc-51-like kinase 1 (ULK1) is required for p53-dependent autophagy. (a) The p53-inducible expression cell line (tetracycline (Tet)-p53-clone1) as in Figure 2 (d) was transfected with either luciferase small interfering (si)RNA (lanes 1–3) or ULK1 siRNA (lanes 4–6) as described in Materials and Methods. After 4 days of siRNA transfection, p53 was induced to be expressed (lanes 3 and 6) for 1 day with 1 μg/ml of Tet to do the long-lived protein degradation (LLPD) assay. Hank's buffered salt solution (HBSS) treatment (lanes 2 and 5) were used as positive controls. (b) The same samples in (a) were used to do western blot to test the knockdown efficiency of ULK1. Actin was used as a loading control. (c) Two U2OS clones containing Tet-responsive ULK1 (Tet-ULK1-clone1 and -clone2) were treated with 1 μg/ml of Tet for 1 day (lanes 3, 4, 7 and 8). Then, 1 μM of rapamycin was used to treat the cells for 6 h (lanes 2, 4, 6 and 8) before doing the LLPD assay as described in Materials and Methods. (d) The samples from (c) were used to do western blot to test the expression level of ULK1. Actin was used as a loading control. (e) The Tet-ULK1-clone1 cell line was treated as in (c) and the samples were collected to do western blot to test the expression level of ULK1, p-p70S6K (T-389), p70S6K, actin and LC3-I and -II. To measure the ratio of LC3-II over LC3-I, the blot was developed with enhanced chemiluminescence (ECL) reagent and the band intensities were measured with Kodak Image Station 4000R pro. The P-value was obtained using Student's t-test. These data are representative of at least two independent experiments
Ectopic expression of ULK1 and rapamycin treatment has additive effect on autophagy
We have shown that ULK1 is an important transcriptional target of p53 for autophagy induction. To test whether ectopic expression of ULK1 alone leads to autophagy, we generated ULK1-inducible expression stable lines in U2OS cells (Figure 4d). As shown in Figure 4c, ectopic expression of ULK1 alone did not result in significant increase of autophagy as tested by LLPD assay (lanes 3 and 7).
Recent studies have shown that mTOR functions directly upstream of endogenous ULK1/ATG13 complex to inhibit their autophagy-initiating activity.17, 18, 19 It is plausible that mTOR blocks the activation of exogenously expressed ULK1. We tested this hypothesis by adding rapamycin to the cells in undergoing ULK1 induction. Rapamycin treatment alone resulted in about 50% increase of LLPD (Figure 4c, lanes 2 and 6) compared with non-treatment control (Figure 4c, lanes 1 and 5), whereas ectopic expression of ULK1 plus rapamycin treatment caused 75% increase of LLPD (Figure 4c, lanes 4 and 8). Similarly, ULK1 ectopic expression alone did not have any impact on LC3 processing (Figure 4e, compare lane 3 with lane 1), whereas addition of rapamycin plus ectopically expressed ULK1 increased the ratio of LC3-II/LC3-I conversion from 2 to 3.4 (Figure 4e, compare lane 4 with lane 1). Decrease in LC3-I is not due to the translational inhibition effect of rapamycin, as treatment with rapamycin alone did not have such a dramatic effect (Figure 4e, lane 2). Taken together, these data suggest that treatment of rapamycin can remove the inhibition of mTOR on ULK1, which results in an additive effect on autophagy with ectopically expressed ULK1.
ATG13 knockdown promotes cell survival after DNA damage
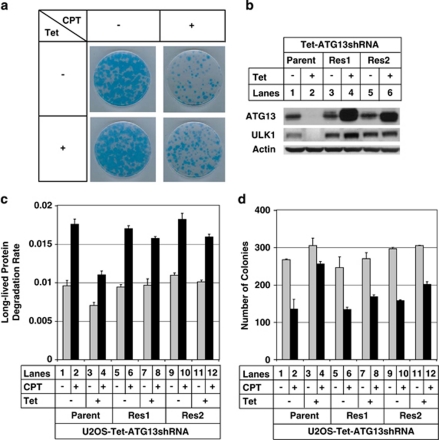
Long-term cell survival assay was performed in ATG13 knockdown stable cell line with CPT treatment, as ATG13 is required for the activation of both ULK1 and ULK2, and ATG13 knockdown resulted in destabilization of endogenous ULK1 (Figure 5b and Supplementary Figure 3). As shown in Figures 5a and d, ATG13 knockdown resulted in more surviving colonies after CPT treatment, suggesting that autophagy facilitates cell death induced by DNA damage. Exogenously expressed ATG13 restored endogenous ULK1 level (Figure 5b, lanes 4 and 6) and also rescued the LLPD rate induced by CPT treatment (Figure 5c, compare lane 4 with lane 8/12). For the long-term cell survival assay, there were fewer survival colonies in the rescue lines after CPT treatment (Figure 5d, lanes 8 and 12) compared with the knockdown line (Figure 5d, lane 4). ATG13 knockdown also promoted cell survival in short-term treatment (after 3 days of CPT treatment, Supplementary Figure 9), although the effect is not as strong as in long-term survival assay.
Figure 5.
ATG13 knockdown enhances cell survival after camptothecin (CPT) treatment. (a) U2OS cells harboring tetracycline (Tet)-responsive ATG13shRNA were treated with or without 1 μg/ml of Tet for 5 days. The cells were then treated with or without 1 μM CPT for 1 day and then were washed with fresh media and allowed to grow for 3 more weeks in the presence or absence of 1 μg/ml of Tet. The cells were then stained with methylene blue as described in Materials and Methods. (b) Two ATG13 rescue cell lines (Res1 and Res2) and the parental ATG13-inducible knockdown cell line (Parent) were treated with or without Tet for 2 weeks. The cells were collected and lysed to perform western blot. The blots were probed with the antibodies against ATG13, unc-51-like kinase 1 (ULK1) and actin. (c) The same cell lines were treated as in (b) and were plated onto six-well plates to do long-lived protein degradation (LLPD) assay after being treated with or without 0.5 μM CPT for 2 days. (d) The experiments were repeated as in (a), in which parental ATG13 knockdown cell line and two rescue cell lines were used, except that the cells were left to grow for 2 more weeks after CPT treatment before counting colonies' number. Each condition was duplicated and the survival colonies were quantified. The data are presented as mean±standard deviation (S.D.) and these data are representative of at least two independent experiments
Discussion
Here we identified that both ULK1 and ULK2, the two autophagy-initiating kinases, are bona fide transcriptional targets of tumor suppressor p53. In response to DNA damage, p53 elevates the transcription level of ULK1/ULK2 and initiates sustained autophagy. The autophagy induced in this scenario leads to increased non-apoptotic cell death.
Relationship between autophagy and cell death is complicated.11, 12, 23, 24 It was reported that antiapoptotic proteins like Bcl-2 and Bcl-XL12, 25 can inhibit autophagy and some proapoptotic protein like BNIP326, 27 can induce autophagic cell death. On the contrary, autophagy inhibition was also reported to promote cancer cell death after chemotherapy (reviewed by Chen and Debnath28). These seemingly opposite observations could be due to the fact that autophagy exhibits distinct functions under different settings. For example, during the initiation of tumorigenesis, cancer cells rapidly outgrow their blood supply, and portions of cells encounter ischemia condition. Autophagy helps cancer cells cope with ischemia stress, making them survive long enough for angiogenesis and further tumor growth. In this scenario, autophagy facilitates tumorigenesis. On the other hand, autophagy defects have also been related to cancer formation. For instance, beclin 1 haplo-insufficiency mouse results in liver cancer.29, 30 ATG4c knockout mice are also prone to cancer formation.31 These could be owing to the fact that autophagy defects result in chronic necrosis, and necrosis in turn triggers inflammatory responses and angiogenesis, finally leading to tumor growth (a situation analogous to wound-healing response).32 In addition, lack of autophagy could result in the accumulation of damaged mitochondria and high-level reactive oxygen species generation and exposure, a stress condition that favor cancer cell growth, as they mainly rely on glycolysis for energy generation. Therefore, autophagy may participate in either inhibition or acceleration of tumor cell death, but under different circumstances: it may serve as a tumor suppressor mechanism before cancers form, whereas it is kidnapped and transformed to maintain tumor survival before angiogenesis. After tumor cells acquire sufficient blood supply, autophagy may again be inhibited for the sake of more rapid tumor growth.
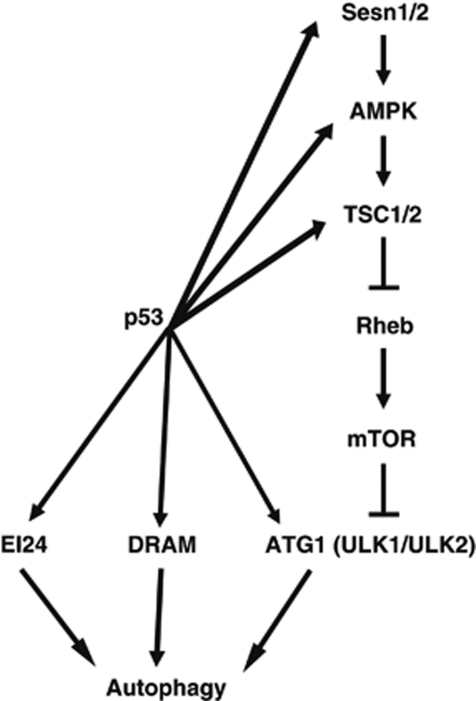
The role of p53 in regulating autophagy is complicated. Nucleus-localized p53 has been reported to induce autophagy through upregulating AMPK-β1/2, PTEN, TSC1/2, Sesn1/2 or DRAM, whereas cytosolic p53 is reported to inhibit autophagy.33 In our system, either p53 induced by DNA damage or ectopic expression of p53 resulted in the upregulation of ULK1/ULK2 and autophagy, which can be attenuated by pretreatment with 3-MA, an autophagy inhibitor. These data indicate that p53 can directly regulate autophagy via increasing the expression level of important components of autophagy machinery. As shown schematically in Figure 6, p53 is able to regulate autophagy through many layers at various steps. By upregulating ULK1/2, p53 facilitates the generation of more ULK1 (ULK2) kinase complexes with ATG13, FIP200 and other autophagy-initiating components, and primes cells for autophagy induction. P53 also upregulates AMPK and Sesn1/2, which inhibits the activity of mTOR kinase, resulting in the removal of upstream inhibition by mTOR. The de-inhibition of mTOR further facilitates the autophagy flux, causing sustained high level of autophagy.
Figure 6.
Model: p53 regulates autophagy in multiple nodes
ULK1 and ULK2 are differently regulated by p53. Ectopic expression of p53 is sufficient to elevate expression of ULK1, but not ULK2. Some extra transcription factors may be necessary to upregulate ULK2 in addition to p53. For example, p53 has two homologues, p6334, 35 and p73,36, 37 which can bind to the same p53-responsive elements in response to DNA damage.38 It is plausible that ULK kinases are also under the regulation of p63/p73 during DNA damage response. Moreover, ULK2 upregulation after DNA damage was only observed in human osteosarcoma U2OS cells, but not in various human colon cancer cell lines, suggesting that there may exist tissue-specific regulations for ULK1/ULK2.
DNA damage reagents trigger autophagy in a way different from rapamycin (inhibiting mTOR) and nutrient starvation. The primary toxicity of CPT occurs during the S phase,39 so its toxic effect is cell cycle dependent and mainly occurs after cells enter the S phase. After 2 days of sublethal CPT treatment, majority of the cells were damaged and we observed significant increase of autophagy. This may explain the deferred effect on autophagy by CPT treatment. As to rapamycin or nutrient starvation, these conditions are independent of cell cycle; therefore, the maximum autophagy effect can be observed in relatively shorter time. Prolonged treatment with rapamycin did not lead to sustained autophagy induction. The autophagy rate actually decreased to nearly basal level after 2 days of rapamycin treatment (only 10% above basal level). The mTOR activity was still suppressed with prolonged rapamycin treatment (data not shown). One explanation for these rapamycin-induced phenotypes could be that cells are depleted of autophagy-executing proteins owing to the translational inhibition effects of rapamycin.
It has been reported that 56% of glioblastoma patients exhibited more than 50% reduction in the expression levels of both ULK1 and ULK2 (according to the data deposited in The Cancer Genome Atlas). Downregulation of both ULK1 and ULK2 will surely result in the attenuation of autophagy, making tumors grow much more uncontrolled. Accordingly, our experiments show that knocking down ATG13 promoted cell survival after DNA damage, which suggests that sustained increase of autophagy leads to cell death. In this scenario, untamed autophagy induced by DNA-damaging agents may disrupt cells' homeostasis, contributing to cell death. Taken together, treatment of sublethal doses of DNA-damaging agents (Supplementary Figure 9) should provide us a good system to study autophagy-mediated non-apoptotic cell death.
Materials and Methods
Reagents
Opti-MEM I, Lipofectamine 2000 reagent and blasticidin were from Invitrogen (Carlsbad, CA, USA). Blasticidin was dissolved in H2O at 10 mg/ml. Tet-system-approved FBS and pPur were from Clontech (Mountain View, CA, USA). Puromycin and G418 were from Calbiochem (Gibbstown, NJ, USA). Puromycin was dissolved in H2O at 10 mg/ml and G418 was dissolved in PBS at 500 mg/ml. Tet was from Sigma (St Louis, MO, USA) and dissolved in 50% ethanol at 10 mg/ml. Cloning cylinder was from Bellco Glass Inc (Vineland, NJ, USA). ULK1 and p53 antibodies were from Santa Cruz (Santa Cruz, CA, USA). Autophagy antibody kits and p21 antibody were from Cell Signaling (Beverly, MA, USA). Actin antibody was from Sigma. Cell Titer-Glo assay reagents and Dual-Glo luciferase assay system were from Promega (Madison, WI, USA). Rapamycin was from Calbiochem. Wt and mt p53 plasmid, HCT 116 and HCT 116/p53KO were gifts from Dr. Bert Vogelstein.
Generation of RNAi stable cell lines
The Tet repressor stable cell line was first generated and cultured in DMEM containing 5 μg/ml blasticidin. The 19 bp of functional siRNA sequences were adapted into pSuperior vector (from Oligoengine, Seattle, WA, USA) according to the manufacturer's protocol. The cassettes of H1 promoter together with the shRNA sequence were cut out and cloned into a modified pBSK with BamHI and BglII sites flanking both sides. Four copies of the cassettes were generated using the compatibility of BamHI and BglII sites as described in ref. 40 and were then cut out and cloned into pPur vector using BamHI site. The resulting four copies of shRNA cassette-pPur plasmids were linearized with NdeI and transfected into U2OS-TetR to select stable colonies in Tet-free medium with 2 μg/ml puromycin. Usually 24 colonies were enough to screen for the stable expression cell lines. The 19-bp shRNA sequences were as follows: ATG13siRNA1, AGACCATCTTTGTCCGAAA; ATG13siRNA2, GAAGAATGTCCGCGAGTTT; and ULK1siRNA, GGAGAAAACTTGTAGGTGT.
Long-lived protein degradation assay
The protocol was modified from Gronostajski et al.'s method.14 On day 0, the cells were seeded in six-well plates at 40 000 cells per well in DMEM containing 65 μM leucine and 1 μCi/ml 3H-labeled leucine. Each condition has three independent wells. On day 3, the cells were washed three times with normal DMEM and replaced with 2 mM cold leucine in DMEM. On day 4, the cells were washed again with normal DMEM and replaced with required medium. After 2 h, 1 ml of medium was taken out from each sample and added with 112 μl of 100% TCA. The samples were then vortexed and centrifuged at 14 000 r.p.m. for 5 min. A measure of 0.4 ml aliquots of the supernatant was measured for the content of tritium with a scintillation counter and the number was converted to the total amounts in the supernatant and was labeled as A. The cells were washed with PBS once and 10% TCA was then added and incubated at room temperature for 5 min. Next, the cells were washed with 10% TCA twice and 2 ml of 0.2 N NaOH (37°C) was added and incubated at 37°C for 10 min. A measure of 0.4 ml of the lysed sample was used to measure tritium content using a scintillation counter and the number was converted to total amounts in the pellet and was labeled as B. The ratio of A/(A+B) was the rate of long-lived protein degradation rate at 2 h. Every experiment was repeated at least twice.
siRNA transfection
On day 0, one million U2OS cells were seeded onto each 6-cm tissue culture dish. siRNA (600 pmol) was used to transfect the cells with 9 μl of Lipofectamine 2000 reagent (Invitrogen) at the same time of plating cells. On day 1, the cells were transfected with the same amount of siRNA again for 4 h. The cells were then trypsinized and half of them were used to do long-lived protein degradation assay. The other half were left to grow for 3 more days to test knockdown efficiency.
Quantitative PCR
Total RNA was extracted from five million U2OS cells, using Trizol reagent from Invitrogen, according to the manufacturer's instruction. The first strand of cDNA was generated from 2 μg total RNA using the SuperScript III Reverse Transcriptase kit from Invitrogen according to the protocol. The forward and reverse primers for ULK1 were as follows: TCGAGTTCTCCCGCAAGG and CGTCTGAGACTTGGCGAGGT. The probe for ULK1 was CACCGCGAGAAGCACGATTTGGA. The forward and reverse primers for ULK2 were as follows: TGGGTCCTCCCAACTATCTACAAGT CGAGATGTTGTGTGGCACCAA. The probe for ULK2 was: TCTGCCAGTACTAGTAGCAAG. The quantitative PCR was carried out according to the manufacturer's protocol.
Luciferase reporter assay
Oligonucleotides containing four copies of either wt or mt ULK1-p53BS were synthesized by Integrated DNA Technology (IDT) DNA technology and cloned into pGL3-promoter between KpnI and XhoI sites. The sequence of the wt ULK1-p53BS (four copies) was: AAACATGTCCTTCCTCGGTGGGCCTAG TAAAAACATGTCCTTCCTCGGTGGGCCTAGTAAAAA CATGTCCTTCCTCGGTGGGCCTAGTAAAAACATGTCCTTCCTCGGTGGGCCTAGTAA; the sequence of the mt ULK1-p53BS (four copies) was: AAAaATaTCCTTCCTCGGTGGGCaTAa TAAAAAaATaTCCTTCCTCGGTGGGCaTAaT AAAAAaATaTCCTTCCTCGGTGGGCaTAaTAAAAAaATaTCCTTCCTCGGTGGGCaTAaTAA (lower case alphabets represent mutations different from the wt ULK1-p53BS). Transfection of U2OS cells were performed in 48-well plate using 0.3 μg of luciferase reporter plasmid, 0.1 μg of pRL-CMV and 0.2 μg of pcDNA3.1 encoding either wt p53 or mt p53R175H. The Renilla luciferase containing pRL-CMV plasmid was used as a control for transfection efficiency. Firefly luciferase reporter assay and Renilla luciferase reporter assays were performed 24 h after transfection using the Dual-Glo luciferase assay system from Promega. Each condition was performed three times independently with the error bars representing the standard deviation.
Long-term cell survival assay
On day 0, 500 U2OSATG13shRNA cells were seeded in 100 mm plates in the absence or presence of 1 μg/ml Tet. On day 5, the cells were treated with 1 μg/ml of CPT. On day 6, the cells were washed and replaced with fresh DMEM with or without Tet. The cells were replaced with fresh media every 3 days thereafter. After 2–3 weeks, cells were washed with PBS once and stained with 0.1% methylene blue for 10 min. The cells were then washed with pure water several times, air dried and imaged. The colonies were counted in three independent plates for each condition and the error bars represent the standard deviation.
Electrophoretic mobility shift assay
The binding of recombinant p53 to the putative p53 binding sites of ULK1 and ULK2 were analyzed by electrophoretic mobility shift assay according to the manufacturer's protocol using the LightShift Chemiluminescent EMSA kit (Catalogue no. 20148) from Pierce. Oligonucleotides containing two duplicated p53 binding sites were synthesized by IDT with biotin at 5′ end. The wt of putative p53 binding site for ULK1 was: AAACATGTCCTTCCTCGGTGGGCCTAGTAA; and the mt p53 binding site for ULK1 was: AAAaATaTCCTTCCTCGGTGGGCaTAaTAA; the wt of putative p53 binding site for ULK2 was: AAGCATGTTTAGACACACATGCAT and the mt p53 binding site for ULK2 was: AAGaATaTTTAGACACAaATaCAT.
Cell Titer-Glo assay
To knock down ATG13, the Tet-ATG13shRNA-stable knockdown cells were pretreated with 1 μg/ml Tet for 5 days and the control cells were not pretreated. On day 0, 100 μl of cells from each treatment were plated into 96-well plates at 5000 cells per well in triplicate. On day 1, different concentrations of CPT were added and the cells were collected after 1 day, 2 days or 3 days of treatment. To do the CellTiter-Glo assay, the plates were taken out to cool to room temperature and 25 μl of CellTiter-Glo reagents were then added to each well. The plates were shaken vigorously at room temperature for 10 min and incubated for another 10 min at rest. The plates were then taken to measure luminescence.
Acknowledgments
We thank Dr. Bert Vogelstein for kindly providing the p53 constructs, HCT116 and HCT116/p53KO cells. We acknowledge Drs. Qinghua Liu and Chunyang Liang for providing Tet-LashRNA stable cell lines. We thank Mr. Gregory Kunkle for critically reading the manuscript. The work was also supported by a grant from the Welch Foundation (I-1214).
Glossary
- 3-MA
3-methyladenine
- mTOR
mammalian target of rapamycin
- CPT
camptothecin
- Eto
etoposide
- LLPD
long-lived protein degradation
- p53BS
p53 binding site
- Rapa
rapamycin
- Tet
tetracycline
- ULK1
unc-51-like kinase 1
- ULK2
unc-51-like kinase 2
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by V De Laurenzi
Supplementary Material
References
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, et al. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics. 1998;51:76–85. doi: 10.1006/geno.1998.5340. [DOI] [PubMed] [Google Scholar]
- Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, et al. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun. 1998;246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, et al. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Gronostajski RM, Pardee AB. Protein degradation in 3T3 cells and tumorigenic transformed 3T3 cells. J Cell Physiol. 1984;119:127–132. doi: 10.1002/jcp.1041190120. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Daido S, Kanzawa T, Yamamoto A, Takeuchi H, Kondo Y, Kondo S. Pivotal role of the cell death factor BNIP3 in ceramide-induced autophagic cell death in malignant glioma cells. Cancer Res. 2004;64:4286–4293. doi: 10.1158/0008-5472.CAN-03-3084. [DOI] [PubMed] [Google Scholar]
- Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–991. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- Chen N, Debnath J. Autophagy and tumorigenesis. FEBS Lett. 2010;584:1427–1435. doi: 10.1016/j.febslet.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci. 2000;113 (Part 10:1661–1670. doi: 10.1242/jcs.113.10.1661. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.