Abstract
Although caspase-2 represents the most conserved caspase across species and was the second caspase identified, its precise function remains enigmatic. In several cell types we show that knockdown of caspase-2 specifically impaired DNA damage-induced p21 expression, whereas overexpression of a caspase-2 mutant increased p21 levels. Caspase-2 did not influence p21 mRNA transcription; moreover, various inhibitors targeting proteasomal or non-proteasomal proteases, including caspases, could not restore p21 protein levels following knockdown of caspase-2. As, however, silencing of caspase-2 impaired exogenous expression of p21 constructs containing 3′-UTR sequences, our results strongly indicate that caspase-2 regulates p21 expression at the translational level. Intriguingly, unlike depletion of caspase-2, which prevented p21 expression and thereby reverted the γ-IR-induced senescent phenotype of wild-type HCT116 colon carcinoma cells into apoptosis, knockdown of none of the caspase-2-interacting components RAIDD, RIP or DNA-PKcs was able to mimic these processes. Together, our data suggest that this novel role of caspase-2 as a translational regulator of p21 expression occurs not only independently of its enzymatic activity but also does not require known caspase-2-activating platforms.
Keywords: apoptosis, cell cycle, translational control, 3′-UTR, p53, RAIDD
Although caspases were initially identified as key apoptotic enzymes, they are now known to also exert essential functions in diverse cellular processes, including inflammation, differentiation and proliferation.1 Among them, caspase-2 is not only the most conserved caspase across species but intriguingly also the most enigmatic, as it combines features of both initiator and executioner caspases.2, 3 It contains a long caspase activation and recruitment domain (CARD), a characteristic trait of initiator caspases, whereas its cleavage specificity resembles that of effector caspases. Similar to initiator caspases such as caspase-8, -9 and -10, caspase-2 is activated by dimerization by an ‘induced-proximity' mechanism.4, 5 In contrast, as caspase-2 is a poor activator of effector caspases, it surely lacks an essential feature of initiator caspases. Thus, contradictory findings were reported that place caspase-2 either upstream or downstream of the mitochondria, or even fail to assign any specific function to this enzyme in apoptosis.6, 7
Casp2-knockout mice are viable and develop normally and show no obvious apoptosis defects;8 but exhibit signs of premature aging.9 However, certain Casp2-deficient cell types such as neurons or embryonic fibroblasts (MEFs) show a reduced or delayed apoptotic response toward certain stimuli,6, 7 indicating both redundant and non-redundant functions for caspase-2 in apoptosis. In addition, caspase-2 was reported to be involved in the regulation of DNA damage responses and cell-cycle progression, and may even function as a tumor suppressor.10, 11 However, the underlying mechanisms are completely unresolved mainly because of the fact that only few caspase-2-specific substrates have been identified until now.
Activation of caspase-2 is achieved by the formation of three functionally different multi-protein complexes. The first caspase-2-activating complex identified that is formed in response to genotoxic stressors was the RAIDD–PIDDosome, consisting of RAIDD (receptor-interacting protein (RIP)-associated ICH/CED-3-homologous protein with death domain), PIDD (p53-induced protein with death domain) and caspase-2.12 However, caspase-2 was also found to be processed and capable of inducing apoptosis in the absence of RAIDD or PIDD, suggesting the existence of additional caspase-2-activating platforms.13, 14 In addition, caspase-2 was proposed to form a cytosolic complex with RIP and TRAF-2 (TNF-receptor-associated factor-2) that does not induce apoptosis but strongly activates NF-κB and p38.15 Surprisingly, this was independent of the proteolytic activity of caspase-2, and instead required only the prodomain containing the CARD oligomerization motif.
Finally, a third caspase-2-activating complex formed in the nucleus was recently implicated in the maintenance of a G2/M DNA damage checkpoint and in DNA repair executed by the NHEJ pathway.10 Thereby, the DNA-dependent serine/threonine protein kinase (DNA-PKcs) and PIDD form the backbone of the so-called DNA-PKcs–PIDDosome that is constitutively present even in unstimulated cells. Following DNA damage, caspase-2 is recruited and activated in this complex by the DNA-PKcs-mediated phosphorylation of its Ser122 residue. Even though identification of the DNA-PKcs–PIDDosome resolved another enigma surrounding caspase-2, namely its constitutive nuclear localization, it remains unanswered how nuclear caspase-2 is integrated in the DNA damage-induced G2/M checkpoint and NHEJ pathways.
Although originally identified as a cyclin-dependent kinase (CDK) inhibitor mediating p53-induced cell-cycle arrest, p21 is now known to also participate in diverse biological processes, including transcription, DNA repair, differentiation, senescence and apoptosis.16, 17, 18 Thus, mechanisms are required that tightly control its expression and functional diversity. This is achieved, for instance, by multiple phosphorylations that result either in its stabilization or in an increased degradation by the proteasome.19 In addition, expression of p21 is also controlled at the transcriptional and posttranscriptional level,20, 21 giving the cell multiple opportunities to interfere with p21 function.
Here, we uncovered an unprecedented role of caspase-2 as an essential translational cofactor for DNA damage-induced p21 expression. Thereby, our findings add a new level of complexity to the enigma surrounding the role(s) of caspase-2 and establish a novel apoptosis-independent involvement of this protease in cellular DNA damage signaling pathways.
Results
Caspase-2 knockdown sensitizes wild-type HCT116 cells to γIR-induced apoptosis by reducing p21 protein levels
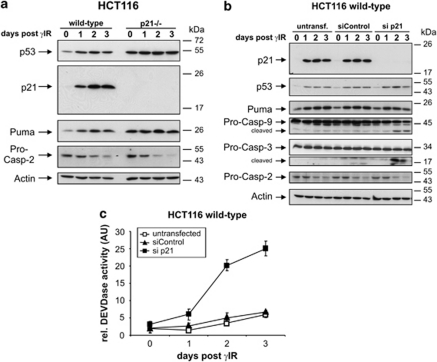
We recently reported that, besides induction of p53-dependent senescence, p21 is also required to simultaneously inhibit caspase activation and apoptosis downstream of mitochondria.22 This conclusion was drawn from our finding that only checkpoint-deficient HCT116 cells (p21−/− and p53−/−) succumb to apoptosis upon exposure to ionizing radiation (γ-irradiation (γIR)), whereas similarly treated wild-type cells are driven into senescence. To verify that the hypersensitivity of p21-deficient HCT116 cells is indeed due to the loss of p21 and not caused primarily by elevated levels of p53 and Puma (Figure 1a),22 we silenced p21 expression in wild-type cells by RNA interference. Knockdown of p21, but not a control short interfering RNA (siRNA), reverted the senescent phenotype of irradiated wild-type HCT116 cells and sensitized them to apoptosis, as evidenced by the processing and activation of caspase-9 and -3 (Figures 1b and c).
Figure 1.
Loss of p21 radiosensitizes HCT116 wild-type cells toward γIR-induced apoptosis. (a) Western blot analyses for the status of the indicated proteins in HCT116 wild-type and p21−/− cells that were γ-irradiated and cultured for the indicated days. (b) HCT116 wild-type cells were either left untransfected or transfected with control and p21 siRNAs. Following γ-irradiation, cells were cultured for the indicated days before the extracts were subjected to western blot analyses. For panels a and b the blots shown represent results from two and three independent experiments, respectively. (c) Determination of DEVDase activities in HCT116 wild-type cells transfected and treated as described in panel b and analyzed after the indicated days. The arbitrary units (AU) shown are the mean of three independent experiments±(S.D.)
More interestingly, following γIR exposure, procaspase-2 levels decreased in a time-dependent manner in both HCT116 lines, regardless of whether they succumbed to apoptosis (Figures 1a and b), implying processing of caspase-2 (see section Discussion for further details). As, in contrast, procaspase-9 and -3 are processed and activated exclusively in apoptotic checkpoint-deficient cells, and because γIR induces the activation of mitochondria even in HCT116 wild-type cells that become senescent upon this treatment,22 our data imply an important upstream role for caspase-2 in DNA damage signaling.
Therefore, we compared the fate of irradiated HCT116 wild-type cells in the presence and absence of caspase-2. Surprisingly, siRNA-mediated silencing of caspase-2 reverted the senescent phenotype of irradiated wild-type cells and resulted in processing and activation of caspase-3, as well as in the death-associated release of lactate dehydrogenase (LDH) (Figures 2a–c). This phenomenon could not be explained by an elevated expression of p53 or Puma, because their γIR-induced protein levels did not differ significantly in the absence or presence of caspase-2 (Figure 2a). Instead, wild-type cells sensitized to γIR-induced apoptosis by knockdown of caspase-2 showed a remarkable decrease in p21 protein expression (Figure 2a). This was also observed when individual siRNAs were used that comprise the caspase-2 SMARTpool siRNA, ruling out off-target effects (Supplementary Figure 1). On the other hand, p21 levels increased following overexpression of caspase-2; however, this was observed only in unstressed cells and when a catalytically inactive caspase-2 mutant was used (Figure 2d). This probably reflects the facts that irradiation per se induces maximum p21 expression in HCT116 wild-type cells, which cannot be further enhanced by exogenous caspase-2, and that cells transfected with the wild-type enzyme undergo massive apoptosis (not shown), explaining not only its weak detection but also the minor effect on p21 expression. Together, these results are consistent with our previous observation that p21 protects HCT116 wild-type cells from γIR-induced apoptosis,22 and additionally suggest that caspase-2 modulates DNA damage responses also independently of its catalytic activity by positively regulating p21 expression.
Figure 2.
Modulation of p21 expression by caspase-2. (a) Western blot analyses for the status of the indicated proteins in HCT116 wild-type cells that were either left untransfected or transfected with caspase-2 siRNA before they were exposed to γIR and cultured for the indicated days. (b) Determination of DEVDase activities in HCT116 wild-type cells that were either left untransfected or transfected with control or caspase-2 siRNAs before they were γ-irradiated and cultured for the indicated days. Refer to Figures 3a and 7c for additional independent results. (c) Cell death assessment of HCT116 wild-type cells that were either left untransfected or transfected with control or caspase-2 siRNAs before γ-irradiation. The release of LDH into their supernatants was determined 5 days after γIR. The arbitrary units (AU) shown in panels b and c are the mean of two independent experiments±S.D. (d) Western blot analyses showing the influence of overexpression of a Flag-tagged caspase-2 wild-type protein and a catalytically inactive caspase-2 mutant on p21 expression in control and γ-irradiated wild-type cells. Blots shown in panels a and d are representative results out of two and three independent experiments, respectively
Caspase-2 is commonly required for DNA damage-induced p21 expression
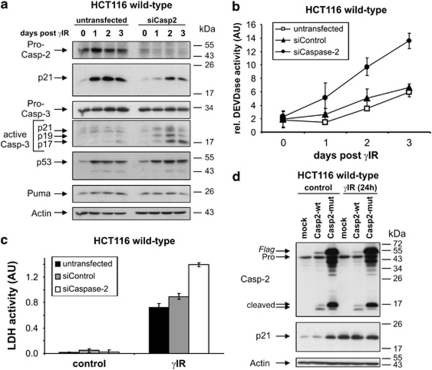
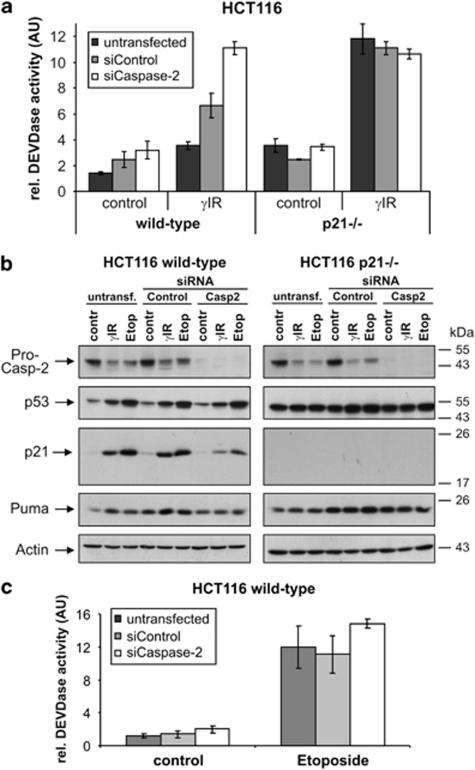
To analyze this observation in more detail, we compared the effects of caspase-2 knockdown on the apoptotic responses of wild-type and p21-deficient HCT116 cells. Although a comparable caspase-2 knockdown was achieved in both cell lines (Figure 3b), this measure only increased the amount of γIR-induced caspase-3-like DEVDase activity in wild-type cells, whereas it did not further radio-sensitize p21−/− cells (Figure 3a). This shows that suppression of p21 is an integral part of the mechanism by which caspase-2 deficiency increases the radio-sensitivity of HCT116 wild-type cells.
Figure 3.
DNA damage-induced p21 expression requires caspase-2. (a) Determination of DEVDase activities in HCT116 wild-type and p21−/− cells that were either left untransfected or transfected with the indicated siRNAs before they were γ-irradiated and cultured for 2 days. (b) Western blot analyses for the status of the indicated proteins in HCT116 wild-type and p21−/− cells that were either left untransfected or transfected with control or caspase-2 siRNAs before they were exposed to γIR or etoposide. Cells were harvested 36 h (etoposide) or 48 h (γIR) after treatment. The blots shown represent results from two independent experiments. (c) Determination of DEVDase activities in HCT116 wild-type cells that were either left untransfected or transfected with control or caspase-2 siRNAs before they were exposed to etoposide. The arbitrary units (AU) shown in panels a and c are the mean of three independent experiments±(S.D.)
To determine whether caspase-2-dependent p21 expression is confined to γIR-induced signaling, we used the topoisomerase-II inhibitor etoposide that also induces processing of procaspase-2 (Figure 3b). Consistently, the absence of caspase-2 in etoposide-treated wild-type cells was also accompanied by a marked reduction of p21, whereas expression of p53 and Puma remained unaffected (Figure 3b). However, only a marginal increase in DEVDase activity was observed when compared with etoposide-treated cells that were either untransfected or transfected with a control siRNA (Figure 3c). The failure of p21 to affect etoposide-induced apoptosis is probably explained by our observation that wild-type and p21-deficient HCT116 cells show similar apoptosis sensitivities toward various chemotherapeutic drugs, including etoposide (not shown), suggesting that p21 is not sufficient to protect cells under those conditions.
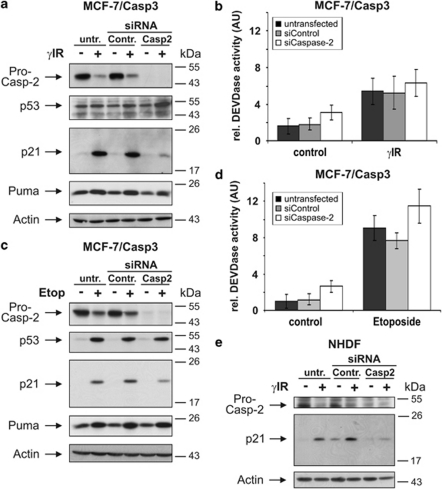
Next, we analyzed whether caspase-2 is also linked to p21 expression in other cell types. Therefore, MCF-7/Casp3 cells that, similar to HCT116 wild-type cells, also undergo γIR-induced senescence23 were transfected with control or caspase-2 siRNAs. Also in these cells, procaspase-2 was readily processed following γIR exposure (Figure 4a) despite their apoptosis resistance toward this treatment.23 More importantly, they too showed an almost complete loss of γIR-induced p21 expression following caspase-2 knockdown, whereas expression of p53 and Puma remained unchanged (Figure 4a). Similar results were obtained with etoposide-treated MCF-7/Casp-3 cells (Figure 4c) or when we exposed caspase-2-depleted primary normal human dermal fibroblasts (NHDFs) to γIR (Figure 4e). In contrast to irradiated wild-type HCT116 cells, however, knockdown of caspase-2 could not alter their apoptosis susceptibilities (Figures 4b,d; data not shown), suggesting that other mechanisms besides p21 determine these stress responses. Nevertheless, our results show that expression of p21 commonly requires caspase-2.
Figure 4.
Caspase-2 is commonly required for p21 expression. (a, c and e) Western blot analyses for the status of the indicated proteins in MCF-7/Casp3 cells and NHDFs that were either left untransfected or transfected with control or caspase-2 siRNAs before γ-IR (a and e) or etoposide treatment (c). Cells were harvested 2 (e) to 3 (a) days after γIR and 24 h after etoposide treatment. The blots shown represent results from three independent experiments. (b and d) Determination of DEVDase activities in MCF-7/Casp3 cells that were transfected and treated as described in panels a and c. The arbitrary units (AU) shown are the mean of three independent experiments±(S.D.)
Caspase-2 is essential for DNA damage-induced checkpoint control
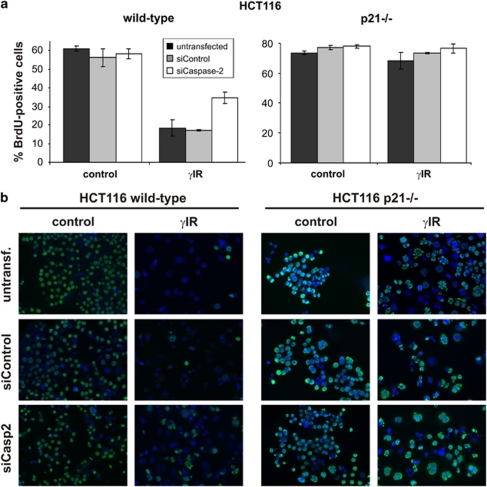
Next, we analyzed the proliferative capability of HCT116 cells in the presence and absence of caspase-2. Wild-type and p21-deficient HCT116 cells were transfected with control or caspase-2 siRNA, and incubated for 4 h with BrdU 2 days after their exposure to γIR in order to label cells actively synthesizing DNA. As expected, regardless of the presence or absence of caspase-2, p21-deficient cells were unable to undergo γIR-induced cell-cycle arrest, as similar numbers of BrdU-positive cells were detected in irradiated and non-irradiated samples (Figure 5a). In contrast, untransfected or control siRNA-transfected wild-type cells showed significantly lower numbers of BrdU-stained cells after γIR (Figure 5a), indicating that the observed cell-cycle arrest of wild-type cells depends mainly on p21. Consistently, diminishing p21 levels by caspase-2 siRNA increased the number of irradiated wild-type cells capable of incorporating 5-bromo-2′-deoxyuridine (BrdU) (Figure 5a).
Figure 5.
Caspase-2-deficient HCT116 wild-type cells show an increased proliferation rate following γ-IR. (a) Determination of the percentage of BrdU-positive HCT116 wild-type and p21−/− cells that were transfected with control or caspase-2 siRNAs before γ-IR. BrdU and DAPI staining procedures were performed 2 days after γIR. The data shown are the mean of two independent experiments±S.D. (b) Representative micrographs of HCT116 wild-type and p21−/− cells that were transfected and treated as described in panel a. Note that an increased number of irradiated wild-type cells show an aberrant nuclear structure only following knockdown of caspase-2. This phenomenon can also be observed in irradiated p21-deficient cells, however, regardless of whether they were transfected with the caspase-2 siRNA
Remarkably, similar to irradiated p21-deficient cells, irradiated wild-type cells transfected with the caspase-2 siRNA harbor extremely aberrant shaped nuclei that are characteristic of cells lacking an important cell-cycle checkpoint and hence try to enter mitosis despite an extensive damage to their DNA (Figure 5b). As such, an abnormal nuclear structure was not observed in irradiated wild-type cells that were left untransfected or transfected with control siRNA (Figure 5b); these data show that caspase-2 is crucial for an orderly executed stress response pathway upon DNA damage.
Caspase-2 modulates p21 expression at the translational level
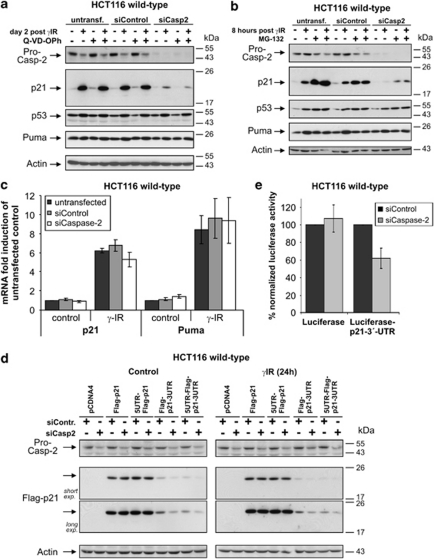
Several mechanisms could account for the loss of p21 following knockdown of caspase-2, including proteasomal degradation, cleavage by proteases, including caspases, or transcriptional repression of the p21 gene.16, 19, 24 However, irradiating caspase-2-depleted wild-type cells in the presence of the pan-caspase inhibitor Q-VD-OPh could not prevent the decrease of p21 (Figure 6a), showing that this event is not caused by caspase cleavage. In addition, Q-VD-OPh failed to inhibit procaspase-2 processing in irradiated control cells, and was completely ineffective in reducing γIR-induced p21 levels in untransfected wild-type cells or in cells transfected with control siRNA (Figure 6a). Similar results were obtained with z-VDVAD-fmk (not shown), another caspase inhibitor preferentially targeting caspase-2. On the basis of the effectiveness of Q-VD-OPh and z-VDVAD-fmk to completely suppress caspase-3 processing and activity (Supplementary Figure 2; data not shown), and our finding that a catalytically inactive caspase-2 mutant substantially increased p21 expression (Figure 2d), these data strongly suggest that caspase-2 modulates p21 expression independently of its enzymatic activity.
Figure 6.
Caspase-2 modulates p21 expression at the translational level. (a and b) HCT116 wild-type cells were either left untransfected or transfected with control or caspase-2 siRNAs before they were γ-irradiated in the presence or absence of the pan-caspase inhibitor Q-VD-OPh (a) or after a 1-h pre-incubation with the proteasomal inhibitor MG-132 (b). Cells were harvested 2 days (a) or 8 h (b) after γIR, and their cellular extracts were analyzed by western blotting for the expression of the indicated proteins. The blots shown represent results from two (a) and three (b) independent experiments. (c) Real-time PCR for the determination of the expression levels of the p21 and Puma mRNAs in HCT116 wild-type cells that were either left untransfected or transfected with control or caspase-2 siRNAs before γ-IR. Total RNA was isolated 4 h after γIR and analyzed with transcript-specific probes from Applied Biosystems. The data shown are the mean of three independent experiments±S.D. (d) Western blot analyses showing the influence of a caspase-2 knockdown on the expression of the indicated Flag-tagged p21 constructs containing either 5′- or 3′-UTRs, or both, in unstressed and irradiated HCT116 wild-type cells. As control, a plasmid encoding only the Flag-p21 open reading frame was used. Representative blots out of three independent experiments are shown. (e) Luciferase reporter assay showing the influence of a caspase-2 knockdown on the expression of luciferase cDNA constructs with or without the p21-3′-UTR. In each sample, the obtained luciferase activity was normalized to the amount of luciferase mRNA. The percentage of luciferase activities obtained from caspase-2 siRNA-treated cells as compared with that from control siRNA-treated cells is shown. The values are derived from three independent experiments (±S.D.)
We also used the proteasomal inhibitor MG-132, which, because of its high cytotoxicity in HCT116 cells, was only applied for a maximum of 8 h after γIR, a time point clearly sufficient to impair p21 expression in the absence of caspase-2 (Figure 6b). Although MG-132 caused, as expected, a rapid accumulation of p53 and p21 even in the absence of γIR, it was unable to restore the p21 levels of non-irradiated and irradiated caspase-2-knockdown cells to those observed in control siRNA-transfected or untransfected cells (Figure 6b). Similar results were obtained using other inhibitors targeting the proteasome (calpain inhibitor-I), calpains and cathepsins (calpain inhibitor-II), or interfering with lysosomal acidification such as ammonium chloride (not shown). Thus, neither caspases nor any other proteases investigated are involved in the reduction of p21 following caspase-2 knockdown. Furthermore, real-time PCR analyses for p21 and Puma mRNA showed no significant caspase-2-dependent differences in either their control or γIR-induced expression levels (Figure 6c). Therefore, our data not only argue against a direct off-target effect of the caspase-2 siRNAs on the p21 mRNA but additionally imply that caspase-2 controls p21 expression at the translational level.
To evaluate this possibility, wild-type cells were transfected with Flag-tagged p21 cDNA constructs containing either 5′- or 3′-UTR sequences, or both, or with a construct that only consists of the p21 coding region. Remarkably, although the two Flag-p21 constructs harboring 3′-UTR sequences were less well expressed than the p21 constructs that contained no untranslated regions (UTRs) or only the 5′-UTR, knockdown of capase-2 further compromised their expression in stressed and unstressed cells (Figure 6d). In contrast, expression of p21 constructs that lack both UTRs or only include the 5′-UTR was not affected in either condition and remained unaltered even in the absence of caspase-2. Obviously, when compared with the effect of a caspase-2 knockdown on expression of the endogenous p21 (Figure 2a), expression of Flag-p21-3′-UTR was less affected by the depletion of caspase-2 (Figure 6d). This is probably because of an important technical difference in the setup of these two experiments. Whereas endogenous p21 was only induced by γIR at a time point at which caspase-2 was already depleted from the cells (48 h after siRNA transfection), the Flag-p21-UTR constructs were simultaneously co-transfected with the caspase-2 siRNA, as two individual successive transfections proved to be detrimental to the cells. As siRNA-mediated caspase-2 knockdown requires up to 48 h, this co-transfection procedure denotes that translation of the exogenous Flag-p21 constructs will be initially supported by caspase-2. Thus, expression of Flag-p21 will only be impaired at later time points at which caspase-2 is sufficiently downregulated. Nevertheless, as depletion of caspase-2 also resulted in a severe reduction of luciferase activity when the p21-3′-UTR was cloned downstream from the luciferase cDNA (Figure 6e),25 our data imply that caspase-2 controls p21 expression at the translational level involving 3′-UTR sequences.
p53 determines the role of caspase-2 in DNA damage responses
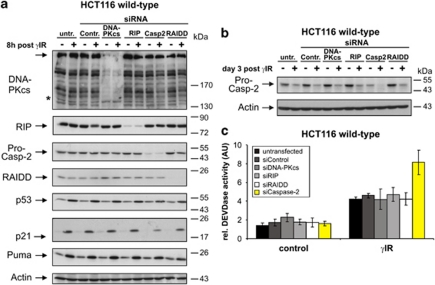
We finally asked whether regulation of p21 requires caspase-2 to associate with any of the three known caspase-2 complexes. Although a sufficient knockdown was achieved for DNA-PKcs, RIP and RAIDD (Figure 7a), none of these prevented the processing of procaspase-2 (Figure 7b) or resulted in a decrease in p21, as it was observed following caspase-2 knockdown (Figure 7a). Consistently, only irradiated wild-type cells transfected with caspase-2 siRNA showed an increase in DEVDase activity (Figure 7c), indicating that none of the known complexes are involved in this novel function of caspase-2.
Figure 7.
Caspase-2 modulates p21 expression independently of known caspase-2-activating complexes. (a) Western blot analyses for the status of the indicated proteins in HCT116 wild-type cells that were either left untransfected or transfected with the indicated siRNAs before γ-IR. Cells were analyzed 8 h after γIR. The blots shown represent results from three independent experiments. The asterisk denotes a band of unknown origin that was still detectable following DNA-PKcs knockdown serving as an additional loading control. (b) To show caspase-2 processing in these cells, an event not evident 8 h after γIR (see panel a), HCT116 wild-type cells were treated as described in panel a, but were analyzed for caspase-2 processing 3 days after γIR. Similar results were obtained 2 days after γIR (data not shown). (c) Determination of DEVDase activities in HCT116 wild-type cells that were treated as described in panel a. Cells were analyzed 3 days after γIR. The arbitrary units (AU) shown are the mean of three independent experiments±(S.D.)
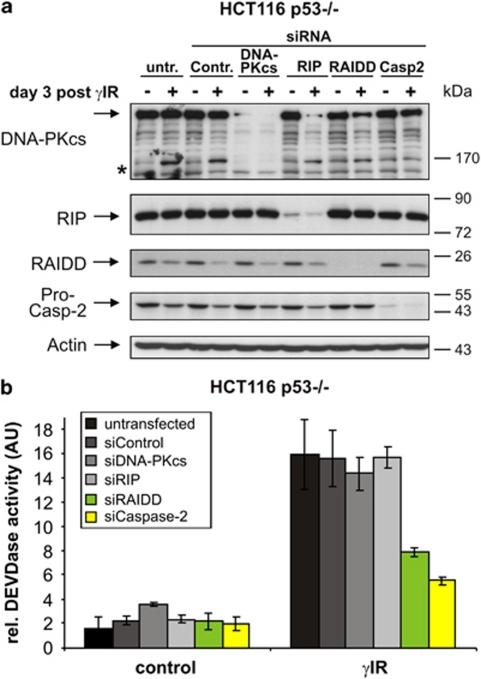
Intriguingly, a completely different picture emerged when we performed similar experiments with cells lacking p53. In sharp contrast to the responses of irradiated wild-type and p21-deficient HCT116 cells, caspase-2 knockdown rendered p53-deficient cells resistant toward γIR-induced apoptosis and inhibited DEVDase activity (Figure 8b). Moreover, siRNA-mediated inhibition of RAIDD expression, but not that of RIP or DNA-PKcs (Figure 8a), resulted in a similar reduction of γIR-induced DEVDase activity in these cells, as observed following caspase-2 knockdown (Figure 8b). Together with the observation that only RAIDD deficiency, but not that of RIP or DNA-PKcs, prevented the processing of procaspase-2 in p53-deficient cells (Figure 8a), these data indicate that, in the absence of p53, caspase-2 signals apoptosis most likely through formation of the RAIDD-PIDDosome.
Figure 8.
RAIDD and caspase-2 are required for γ-IR-induced apoptosis in p53-deficient cells. (a) Western blot analyses for the status of the indicated proteins in HCT116 p53−/− cells that were either left untransfected or transfected with the indicated siRNAs before they were γ-irradiated. Cells were analyzed 3 days after γIR. The blots shown represent results from three independent experiments. The asterisk denotes a band of unknown origin (see also figure legend Figure 7a). (b) Determination of DEVDase activities in HCT116 p53−/− cells that were treated as described in panel a. Cells were analyzed 3 days after γIR. The arbitrary units (AU) shown are the mean of three independent experiments±(S.D.)
Discussion
We describe an unprecedented role of caspase-2 as an essential cofactor for DNA damage-induced expression of the CDK inhibitor p21. This function might allow caspase-2 to participate in diverse signaling pathways modulated by p21, including cell-cycle progression, gene transcription, DNA repair, differentiation and apoptosis.16, 17, 18 Consistently, we observed that the diminished p21 protein pool due to the knockdown of caspase-2 resulted in an increased proliferation rate of irradiated HCT116 wild-type cells, a phenomenon recently reported also with caspase-2-deficient MEFs.11
Our data also suggest that it is the p21-modulating capability, rather than its apoptosis-inducing potential, that constitutes the primary function of caspase-2. This assumption is based on the observation that knockdown of caspase-2 was not sufficient to protect several p53-expressing cells from DNA damage-induced apoptosis. Even the radio-sensitizing effect observed in caspase-2-depleted wild-type HCT116 cells is probably not caused by a direct influence of caspase-2 on apoptosis signaling, as it rather appears to be a consequence of p21 downregulation. As this sensitization occurs only in cells susceptible to the antiapoptotic function of p21, such as irradiated HCT116 wild-type cells,22 our data are consistent with a previous report showing that p21 is not the only determinant in stress-induced p53 responses.26 Surprisingly, and in contrast to other reports,14, 27 the proapoptotic function of caspase-2 was revealed in our study only upon loss of p53. Thus, our data suggest that caspase-2 is additionally part of an apoptotic backup system that signals apoptosis only in the absence of a functional p53 pathway. Although cell type and stimulus-dependent differences clearly contribute to this controversial issue,14 our view is further supported by the observation that knockdown of RAIDD, but not that of RIP or DNA-PKcs, successfully abrogated DNA damage-induced processing and signaling of caspase-2; however, only in p53-deficient HCT116 cells. On the other hand, DNA damage-induced caspase-2 processing and modulation of p21 expression was not affected in wild-type cells by knockdown of any of these components, suggesting the existence of additional caspase-2-activating platforms.
In this context, the most important questions remaining are how does caspase-2 affect the expression of p21 and does this process require a processed and active caspase-2 enzyme? A variety of mechanisms known to regulate p21 expression at the protein and DNA/RNA level have been elucidated in recent years.16, 19, 20 However, post-translational regulation could be excluded here, as various inhibitors targeting proteasomal or other proteases, including caspases, could not restore p21 levels following caspase-2 knockdown. In addition, combined with our finding that overexpression of a catalytically inactive caspase-2 mutant was sufficient to increase p21 expression in unstressed wild-type cells, these data strongly suggest that the enzymatic activity of caspase-2 is dispensable for this event. A similar conclusion was reached with regard to its processing, as impaired p21 expression as a consequence of caspase-2 depletion was already observed 8 h after γIR, a time point at which decrease of pro-caspase-2 was not even evident in irradiated control cells. However, with regard to this, it is presently unclear whether the observed γIR-induced decrease of pro-caspase-2 indeed reflects its processing, as caspase-inhibitory peptides neither compromised this process nor the concomitant generation of mature caspase-2 fragments (Supplementary Figure 2). Using two different caspase-2 antibodies, the mature large caspase-2 subunit was only generated in irradiated p53- and p21-deficient HCT116 cells, whereas the appearance of a fragment probably resembling the processed prodomain was also detected in wild-type cells. As this fragment might have also arisen from an alternative splicing event,28 it remains possible that the decrease of caspase-2 in wild-type cells does not reflect its processing, but rather a p53-mediated suppression, as reported recently.29 However, real-time PCR analyses showed comparable amounts of caspase-2 mRNA transcripts in unstressed and irradiated wild-type and p53-deficient HCT116 cells (not shown), clearly arguing against this possibility.
Whereas therefore a direct intervention of caspase-2 with p53-mediated p21 transcription seemed reasonable to assume, irradiated caspase-2-depleted cells exhibited only a slight decrease in p21 mRNA transcripts. Instead, as lack of caspase-2 affects exogenous expression of p21 constructs only if they contain 3′-UTR sequences, we favor a model in which caspase-2 controls p21 expression at the translational level. As caspase-2 does not harbor an obvious RNA-binding domain, we would like to suggest an indirect mechanism by which caspase-2 affects p21 mRNA translation, perhaps by association with RNA-binding proteins or modulation of related pathways. Thereby, we can surely exclude participation of certain RNA-binding proteins such as members of the Hu family that were shown to interact and stabilize p21 mRNA,30 or CUGBP1 and calreticulin, which compete with each other for binding to the p21-5′-UTR.31 However, our results leave open the possibility that caspase-2 modulates the function of a different class of RNA-binding proteins known to stimulate or inhibit p21 mRNA translation without interfering with their stabilities, such as musashi or the heterogeneous nuclear ribonucleoprotein-K (hnRNP-K).32, 33
Another possible mechanism by which caspase-2 may modulate p21 translation involves repression of certain microRNAs (miRs)34 known to prevent p21 mRNA translation without interfering with its stability. Although there might exist an overwhelming number of at least 100 miRs that theoretically could target the p21 mRNA (predicted from microRNA.org), we have analyzed the expression levels of miR-17, miR-20a and miR-106b because these were not only shown to efficiently block p21 mRNA translation25, 35, 36 but were also found to be repressed by p53.37 However, expression of these miRs did not change 16 h following exposure of HCT116 wild-type cells to γIR, and also remained unaltered in the presence or absence of caspase-2 (not shown). Although these observations clearly argue against an involvement of these miRs, the possibility remains that other miRs contribute to the caspase-2-dependent expression of p21.
In summary, we have uncovered an all-new function for caspase-2 as an essential cofactor for DNA damage-induced p21 expression that neither requires its activation nor its recruitment into known caspase-2-containing complexes. It will now be challenging to identify the components involved and to elucidate the mechanism of how exactly caspase-2 regulates p21 translation.
Materials and Methods
Cell lines, reagents and antibodies
HCT116 wild-type colon carcinoma cells and their checkpoint-deficient variants (p53−/− and p21−/−) were maintained in McCoy's 5A medium (PromoCell, Heidelberg, Germany), whereas MCF-7/Casp3 breast carcinoma cells were cultured in RPMI 1640 (PAA Laboratories, Linz, Austria) in the presence of 400 μg/ml neomycin. The primary NHDFs were obtained from PromoCell and were cultured in low-glucose DMEM supplemented with 1% MEM nonessential amino acid solution and 0.1 mM β-mercapto-ethanol. Only passages below P20 were used. All media were supplemented with 10% heat-inactivated fetal calf serum, 10 mM glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin (PAA Laboratories). The pan-caspase-inhibitor peptide Q-VD-OPh (Q-Val-Asp-CH2-O-Ph) and the caspase-2-inhibitor peptide z-VDVAD-fmk (z-Val-Asp-Val-Ala-Asp-fluoromethyl-ketone) were obtained from MP Biomedicals (Irvine, CA, USA). The fluorometric caspase-3 substrate DEVD-AMC (N-acetyl-Asp-Glu-Val-Asp-aminomethylcoumarin) was from Biomol (Hamburg, Germany). The polyclonal antibodies against caspase-3 and caspase-9 were from R&D Systems (Wiesbaden, Germany) and from Cell Signaling Technology (Danvers, MA, USA), respectively. The rat caspase-2 mAb was from Alexis Biochemicals (Lausen, Switzerland). The p53 monoclonal Ab-6 antibody and the polyclonal antibody for Puma were from Calbiochem (Bad Soden, Germany), whereas the mAbs recognizing p21 and RIP were from BD Biosciences (Heidelberg, Germany). The mAb to RAIDD was from MBL International (Woburn, MA, USA). The polyclonal antibody recognizing the catalytic subunit of DNA-PK was obtained from Santa Cruz (Heidelberg, Germany). The actin mAb, the nuclear stain 4′,6-diamidino-2-phenylindole (DAPI), the topoisomerase-II inhibitor etoposide, the two calpain inhibitors I (ALL-N) and II (ALL-M) and the protease inhibitors PMSF, aprotinin, leupeptin and pepstatin were from Sigma-Aldrich (Deisenhofen, Germany). From Enzo Life Sciences (Biozol; Eching, Germany) we obtained the proteasome inhibitor MG-132, and the peroxidase-labeled secondary antibodies were from Promega GmbH (Mannheim, Germany).
Treatment of cells and measurement of cell death
Cells were exposed to γIR (routinely 20 Gy at 200 kV) using a Gulmay RS 225 X-ray system from IsodoseControl (Bochum, Germany) or were treated with etoposide (50 μM) for the indicated time periods. Cell death was assessed either microscopically or by determination of LDH activity in the supernatants of 105 cells according to the protocol of the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). The values obtained are given in arbitrary units (AU).
Preparation of cell extracts and western blotting
Total cell extracts were prepared in lysis buffer (50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1% NP-40) containing protease inhibitors as described.22 Protein concentrations were determined by the Bio-Rad protein assay, followed by separation of the extracts in SDS-polyacrylamide gels and electroblotting onto polyvinylidene difluoride membranes (Amersham Biosciences, Braunschweig, Germany). Following antibody incubation, the proteins were visualized by enhanced chemiluminescent staining using ECL reagents (Amersham Biosciences).
Fluorometric determination of caspase-3-like-activity (DEVDase assay)
For the detection of caspase-3-like activities, 50 μg of cell extracts was incubated for 3 to 4 h with 50 μM of the caspase-3 substrate DEVD-AMC in 200 μl buffer containing 50 mM HEPES (pH 7.4), 100 mM NaCl, 10% sucrose, 0.1% CHAPS and 10 μM DTT. Release of fluorogenic AMC was measured at an excitation wavelength of 346 nm and an emission wavelength of 442 nm using an Infinite M200 microplate reader (Tecan, Langenfeld, Germany). The detected fluorometric signal that directly correlates to the caspase activity in the cell extracts is expressed in arbitrary units (AU).
Plasmids and generation of p21-UTR constructs
The plasmids encoding Flag-tagged wild-type caspase-2 or an inactive mutant (C320G) (pEF-3x-Flag-Casp2-WT and –MUT) were gifts from A Villunger (Innsbruck, Austria). The plasmids pCEP-Waf138 and pMT5-Flag-21-WT39 were purchased from Addgene (Cambridge, MA, USA) and served as templates for cloning of the different p21-UTR constructs, all of which contain an N-terminal Flag epitope (see below). The open reading frame of Flag-p21 was amplified from the pMT5-Flag-p21-WT plasmid using primers harboring BamHI and EheI restriction sites at the 5′ end and SacII and EcoRI sites at the 3′ end. Following digestion with BamHI and EcoRI, the amplified sequence (∼650 bp) was cloned into the pcDNA4-Myc-His-A vector (Invitrogen, Carlsbad, CA, USA), yielding the pcDNA4-Flag-p21 plasmid. The 5′-UTR of p21 (∼80 bp) was amplified from the pCEP-WAF1 plasmid using primers with a HindIII-site at the 5′ end and an EheI site at the 3′ end. After digestion with HindIII and EheI, this amplificate was cloned into the pcDNA4-Flag-p21 plasmid, yielding the pcDNA4-5-UTR-Flag-p21 plasmid. Similarly, the 3′-UTR of p21 (∼1500 bp) was amplified by PCR from the pCEP-WAF1 plasmid using primers containing a SacII site at the 5′ end and a NotI site at the 3′ end, and subsequently cloned into pcDNA4-Flag-p21 following digestion with these two enzymes, thereby generating the pcDNA4-Flag-p21-3-UTR plasmid. To finally produce a plasmid expressing the full-length p21 mRNA, the 3′-UTR was cut out of the pcDNA4-Flag-p21-3-UTR plasmid with SacII and NotI and cloned into the pcDNA4-5-UTR-Flag-p21 plasmid. The generated plasmid was termed pcDNA4-5-UTR-Flag-p21-3-UTR.
Transfection of siRNAs and plasmids
ON-TARGETplus SMARTpool siRNAs were purchased from Dharmacon RNA technologies (Lafayette, CO, USA) and knockdown was performed according to the instructions of the manufacturer. Twenty-four to forty-eight hours after transfection, cells were harvested and divided equally to receive either no treatment or exposure to γIR or etoposide. At the indicated time points following DNA damage, cells were harvested and directly analyzed by western blotting and by the fluorometric caspase substrate assay for a successful knockdown of the target protein and DEVDase activity, respectively. For the simultaneous transfection of plasmids and siRNAs, the Dharmafect DUO transfection reagent was used according to the manufacturer's protocol. Briefly, HCT116 wild-type cells seeded in six-well plates were transfected with 2 μg of plasmid and 200 nmol siRNA using 4 μl Dharmafect DUO in 2 ml antibiotic-free medium. One day after transfection the cells were trypsinized, divided equally into two wells and directly γ-irradiated or left untreated as control. One day after irradiation the cells were harvested and analyzed by western blotting.
Real-time PCR
Total RNA was isolated using the RNeasy kit (Qiagen, Hilden, Germany) according to the protocol of the manufacturer. Reverse transcription was performed using the High Capacity cDNA kit from Applied Biosystems (Darmstadt, Germany). Taqman gene expression probes for human p21, Puma and actin mRNA (Applied Biosystems) were used to analyze their relative expression levels using the 7300 Real-Time PCR system (Applied Biosystems). The actin mRNA served as an endogenous normalization control for every sample. The fold induction of the analyzed RNAs was calculated by the 2^(Δ(ΔCt) method, thereby normalizing all samples to the level of the analyzed RNA in untransfected control HCT116 wild-type cells.
Luciferase reporter assay
Cells were transfected with a vector containing luciferase cDNA with or without the p21-3′-UTR,25 together with either caspase-2 or control siRNA. Two days after transfection, cells were harvested and divided into two aliquots. Luciferase activities were determined using the Dual-luciferase reporter assay (Promega) and a Centro LB 960 luminometer (Berthold Technologie, Bad Wildbad, Germany). For normalization of the obtained activities, real-time PCR was performed with a specific firefly luciferase mRNA probe (Custom Taqman Assay; Applied Biosystems).
BrdU labeling
Cells actively synthesizing DNA were labeled with the BrdU Labeling and Detection Kit I from Roche Molecular Biochemicals according to the manufacturer's protocol. Briefly, following siRNA transfection, cells were seeded on 15-mm coverslips and irradiated. Two days after γIR, cells were labeled for 4 h with BrdU before they were fixed, incubated with a monoclonal antibody recognizing BrdU, followed by incubation with a fluorescence-coupled antibody directed against the primary antibody. Total DNA was stained with DAPI before the coverslips were mounted on glass slides. Subsequently, 6–10 representative pictures were taken from every sample using a × 20 objective of the Zeiss Axio Observer A1 and the corresponding AxioVision Software (Carl Zeiss MicroImaging GmbH, Göttingen, Germany). The number of cells containing green (BrdU-positive) and blue (DAPI) nuclei was counted from all representative pictures taken. Using this method, 500–1200 blue cells were analyzed for every condition in each single experiment.
Acknowledgments
We are grateful to B Vogelstein and A Villunger for the HCT116 cell lines and the caspase-2 plasmids, and to C Disselhoff for excellent technical assistance. The luciferase cDNA constructs with or without the p21-3′-UTR were gifts from X He. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 728) and the Forschungskommission of the Heinrich-Heine-University of Düsseldorf.
Glossary
- CARD
caspase activation and recruitment domain
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- γIR
γ-irradiation
- MEFs
mouse embryonic fibroblasts
- NHDF
normal human dermal fibroblasts
- NHEJ
non-homologous end joining
- PIDD
p53-induced protein with death domain
- RAIDD
RIP-associated ICH/CED-3-homologous protein with death domain
- RIP
receptor-interacting protein
- siRNA
short interfering RNA
- TRAF2
TNF-receptor-associated factor-2
- UTR
untranslated region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by W El-Deiry
Supplementary Material
References
- Schwerk C, Schulze-Osthoff K. Non-apoptotic functions of caspases in cellular proliferation and differentiation. Biochem Pharmacol. 2003;66:1453–1458. doi: 10.1016/s0006-2952(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Kitevska T, Spencer DM, Hawkins CJ. Caspase-2: controversial killer or checkpoint controller. Apoptosis. 2009;14:829–848. doi: 10.1007/s10495-009-0365-3. [DOI] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen's view. Cell Death Differ. 2009;16:195–207. doi: 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga BC, Read SH, Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–1241. doi: 10.1038/sj.cdd.4401492. [DOI] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SW, Dillon CP, et al. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell. 2009;35:830–840. doi: 10.1016/j.molcel.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L. The role of caspase-2 in stress-induced apoptosis. J Cell Mol Med. 2010;14:1212–1224. doi: 10.1111/j.1582-4934.2010.01037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more. Nat Rev Cancer. 2009;9:897–903. doi: 10.1038/nrc2745. [DOI] [PubMed] [Google Scholar]
- O'Reilly LA, Ekert P, Harvey N, Marsden V, Cullen L, Vaux DL, et al. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 2002;9:832–841. doi: 10.1038/sj.cdd.4401033. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Padalecki SS, Chaudhuri AR, De Waal E, Goins BA, Grubbs B, et al. Caspase-2 deficiency enhances aging-related traits in mice. Mech Ageing Dev. 2007;128:213–221. doi: 10.1016/j.mad.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Vivian CJ, Lee KJ, Ge C, Morotomi-Yano K, Manzl C, et al. DNA-PKcs–PIDDosome: a nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance. Cell. 2009;136:508–520. doi: 10.1016/j.cell.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, et al. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu H, Olsson M, Orrenius S, Zhivotovsky B. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene. 2006;25:5683–5692. doi: 10.1038/sj.onc.1209569. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, D'Hondt K, Vande Walle L, van Gurp M, Denecker G, Demeulemeester J, et al. A novel caspase-2 complex containing TRAF2 and RIP1. J Biol Chem. 2005;280:6923–6932. doi: 10.1074/jbc.M411180200. [DOI] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- Jänicke RU, Sohn D, Essmann F, Schulze-Osthoff K. The multiple battles fought by antiapoptotic p21. Cell Cycle. 2007;6:407–413. doi: 10.4161/cc.6.4.3855. [DOI] [PubMed] [Google Scholar]
- Child ES, Mann DJ. The intricacies of p21 phosphorylation: protein/protein interactions, subcellular localization and stability. Cell Cycle. 2006;5:1313–1319. doi: 10.4161/cc.5.12.2863. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chen X. Posttranscriptional regulation of p53 and its targets by RNA-binding proteins. Curr Mol Med. 2008;8:845–849. doi: 10.2174/156652408786733748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn D, Essmann F, Schulze-Osthoff K, Jänicke RU. p21 blocks irradiation-induced apoptosis downstream of mitochondria by inhibition of cyclin-dependent kinase-mediated caspase-9 activation. Cancer Res. 2006;66:11254–11262. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- Essmann F, Engels IH, Totzke G, Schulze-Osthoff K, Jänicke RU. Apoptosis resistance of MCF-7 breast carcinoma cells to ionizing radiation is independent of p53 and cell cycle control but caused by the lack of caspase-3 and a caffeine-inhibitable event. Cancer Res. 2004;64:7065–7072. doi: 10.1158/0008-5472.CAN-04-1082. [DOI] [PubMed] [Google Scholar]
- Gervais JL, Seth P, Zhang H. Cleavage of CDK inhibitor p21(Cip1/Waf1) by caspases is an early event during DNA damage-induced apoptosis. J Biol Chem. 1998;273:19207–19212. doi: 10.1074/jbc.273.30.19207. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- Polyak K, Waldman T, He TC, Kinzler KW, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis. Proc Natl Acad Sci USA. 2008;105:1937–1942. doi: 10.1073/pnas.0711800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droin N, Beauchemin M, Solary E, Bertrand R. Identification of a caspase-2 isoform that behaves as an endogenous inhibitor of the caspase cascade. Cancer Res. 2000;60:7039–7047. [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C. Caspase 2 is both required for p53-mediated apoptosis and downregulated by p53 in a p21-dependent manner. Cell Cycle. 2008;7:1133–1138. doi: 10.4161/cc.7.9.5805. [DOI] [PubMed] [Google Scholar]
- Joseph B, Orlian M, Furneaux H. p21(waf1) mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J Biol Chem. 1998;273:20511–20516. doi: 10.1074/jbc.273.32.20511. [DOI] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, et al. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J Biol Chem. 2005;280:12690–12699. doi: 10.1074/jbc.M411119200. [DOI] [PubMed] [Google Scholar]
- Beitzinger M, Meister G. MicroRNAs: from decay to decoy. Cell. 2010;140:612–614. doi: 10.1016/j.cell.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgdorff V, Lleonart ME, Bishop CL, Fessart D, Bergin AH, Overhoff MG, et al. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1) Oncogene. 2010;29:2262–2271. doi: 10.1038/onc.2009.497. [DOI] [PubMed] [Google Scholar]
- Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, et al. Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J. 2009;28:2719–2732. doi: 10.1038/emboj.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Spohn B, Lee MH, Hung MC. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat Cell Biol. 2001;3:245–252. doi: 10.1038/35060032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.