CD4+ T helper lymphocytes that express interleukin-17 (Th17 cells) have critical roles in mouse models of autoimmunity, and there is mounting evidence that they also influence inflammatory processes in humans. Genome-wide association studies in humans have linked genes involved in Th17 cell differentiation and function with susceptibility to Crohn's disease, rheumatoid arthritis, and psoriasis1-3. Thus, the pathway towards differentiation of Th17 cells and, perhaps, of related innate lymphoid cells with similar effector functions4, 5, is an attractive target for therapeutic applications. Mouse and human Th17 cells are distinguished by expression of the retinoic acid receptor-related orphan nuclear receptor RORγt, which is required for induction of IL-17 transcription and for the manifestation of Th17-dependent autoimmune disease in mice6. By performing a chemical screen with an insect cell-based reporter system, we identified the cardiac glycoside digoxin as a specific inhibitor of RORγt transcriptional activity. Digoxin inhibited murine Th17 cell differentiation without affecting differentiation of other T cell lineages and was effective in delaying the onset and reducing the severity of autoimmune disease in mice. At high concentrations, digoxin is toxic for human cells, but non-toxic synthetic derivatives, 20,22-dihydrodigoxin-21,23-diol (Dig(dhd)) and digoxin-21-salicylidene (Dig(sal)), specifically inhibited induction of IL-17 in human CD4+ T cells. Using these small molecule compounds, we demonstrate that RORγt is important for the maintenance of IL-17 expression in mouse and human effector T cells. These data suggest that derivatives of digoxin can be used as chemical probes for development of RORγt-targeted therapeutic agents that attenuate inflammatory lymphocyte function and autoimmune disease.
To identify small molecules that specifically inhibit transcriptional activity of RORγ and RORγt isoforms, we prepared Drosophila S2 cells stably expressing fusions of the GAL4 DNA binding domain (DBD) and the ligand binding domains (LBDs) of murine RORγ, RORα (mouse homolog of RORγ), and DHR3 (Drosophila orthologue for ROR family proteins), as well as the activation domain of the general transcriptional activator VP16. Induction of RORγ and the other fusion proteins led to robust expression of a firefly luciferase reporter (Supplementary Fig. 1a). Next, we investigated whether RORγ activity in the Drosophila system is dependent on a functional LBD and is ligand-dependent. A single amino acid change in the putative ligand binding pocket7 of RORγ completely abrogated its function as a transcriptional activator despite comparable level of protein expression both in S2 cells and in transgenic fly models (Supplementary Fig. 1b and c). In addition, Drosophila cells grown in serum-free media completely lacked RORγ activity, unless serum or cholesterol metabolites were supplemented into the cell culture (Supplementary Fig. 1d), suggesting that yet-to-be-identified ligands are required for RORγ reporter activity. These data justify utilization of the heterologous system to identify small molecules that modulate RORγ activity.
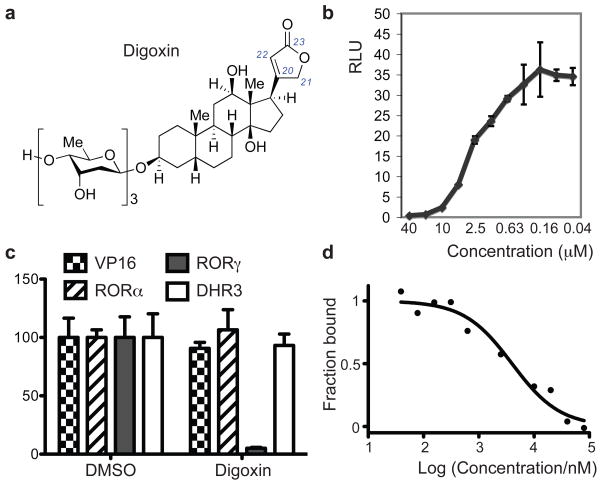
We next performed a chemical screen with 4,812 compounds and identified digoxin as a specific inhibitor for RORγ transcriptional activity (Fig. 1a). Digoxin inhibited RORγ (Fig. 1b and Supplementary Fig. 2a) with an IC50 (half-maximal inhibitory concentration) value of 1.98 μM. Inhibition of RORγ activity by digoxin was specific, as there was no effect on the transcriptional activity of VP16 or of the related nuclear hormone receptors RORα and DHR3 (Fig. 1c). Digoxin did not inhibit the activity of other nuclear hormone receptors, including C. elegans Daf12, human androgen receptor, and LXRα (Supplementary Fig. 2b and c). Digitoxin and β-acetyldigoxin also selectively inhibited RORγ (Supplementary Fig. 2d and e) with similar IC50 values. Next, we examined if digoxin targets RORγ directly. 25-Hydroxycholesterol has been shown to bind to the RORγ LBD8, and conjugation of fluorescein to this surrogate ligand did not affect its ability to bind to the human RORγ LBD (with a Kd of 109 nM). Addition of digoxin led to a dose-dependent decrease in fluorescence polarization values, demonstrating that digoxin can displace the sterol ligand with an IC50 of 4.1 μM (Fig. 1d). In addition, circular dichroism (CD) analysis showed that digoxin increased the thermal stability of the RORγ-LBD, indicating that it interacts directly with RORγ (Supplementary Fig. 3a)9. Digoxigenin, the aglycone of digoxin, did not inhibit RORγt activity in Drosophila cells and did not bind to the RORγt LBD in the CD and competition assays (data not shown and Supplementary Fig. 3b). We further investigated whether digoxin binds inside the ligand binding pocket of RORγ. We performed random mutagenesis on the LBD and screened 200 clones to identify those that were resistant to digoxin-mediated inhibition. Two clones with this property were identified and shared mutation of amino acid 290 (L290P/A494T and L290F/C318S). RORγ harboring mutations at all three residues (RORγ/γt(triple)) exhibited much less sensitivity to digoxin, in spite of transcriptional activity similar to that of the wild-type molecule (Supplementary Fig. 3c and d). Two of the mutations mapped to the ligand binding pocket (L290 and C318) and one to helix 11 (A494)8, consistent with digoxin binding inside the pocket.
Figure 1). Digoxin binds to RORγ and inhibits its transcriptional activity.
a, Chemical structure of digoxin. b, Digoxin demonstrates dose-dependent inhibition of RORγ transcriptional activity in the Drosophila S2 cell luciferase reporter system. Ratio of firefly to Renilla luciferase activity is shown as relative luciferase unit (RLU) on the y-axis. c, Digoxin (10 μM) selectively inhibits RORγ dependent transcriptional activity without affecting that of RORα, DHR3, or VP16. Percentages of relative luciferase units compared to DMSO-treated reporter cells are shown on the y-axis. Error bars indicate standard deviation. d, In vitro competition assay. Recombinant human RORγ LBD was loaded with fluorescently-labeled 25-hydroxycholesterol in the presence of the indicated concentrations of digoxin, and fluorescence polarization was measured.
When naïve mouse CD4+ T cells were cultured under Th17 polarizing conditions (IL-6 and TGF-β), treatment with digoxin led to markedly reduced expression of IL-17a protein (Fig. 2a). Transcriptional up-regulation of genes encoding IL-23 receptor (IL-23R), IL-17a, IL-17f, or IL-22, was also strongly inhibited (Supplementary Fig. 4a and b). Expression of RORγt-independent Th17 signature genes, such as IL-21, cMaf, RORα, Batf, and IRF4, was not affected by dixogin (Supplementary Fig. 4c and d). Reduction of Th17 cell differentiation following treatment of wild-type cells with digoxin was similar to that observed upon targeted inactivation of Rorc(t) (Fig. 2a). IL-23– induced Th17 cell differentiation10 was also inhibited in the presence of digoxin (Supplementary Fig. 4e). Importantly, digoxin had no effect on differentiation of naïve CD4+ T cells into other lineages, including Th1, Th2, and regulatory T cells (Supplementary Fig. 4f and g). Other cardiac glycosides with structures related to digoxin, including proscillaridin A, deslanoside, erysimoside, oleandrin, ouabain, ouabagenin, digitoxigenin, digoxigenin, and lanatoside C, had no significant effect on RORγ transcriptional activity or Th17 cell differentiation (Supplementary Fig. 5a and b).
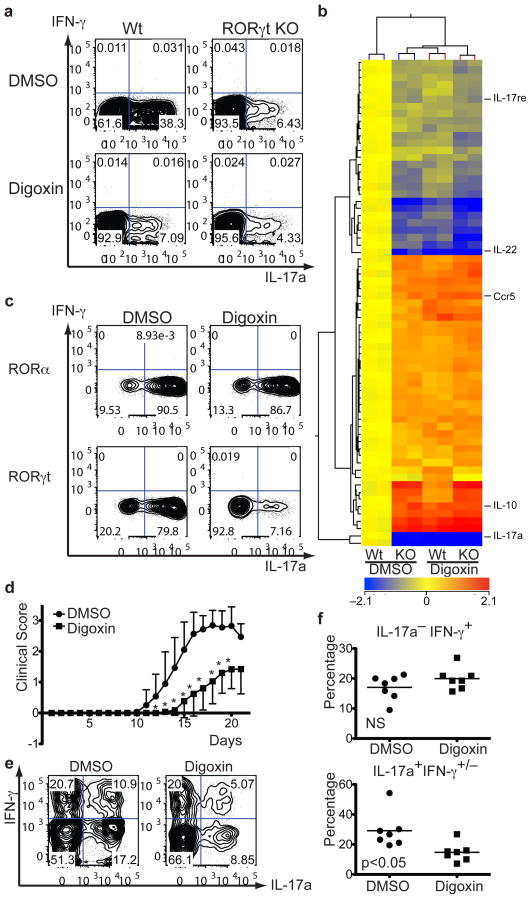
Figure 2). Digoxin inhibits mouse Th17 cell differentiation and ameliorates Th17-mediated autoimmune disease.
a, Flow cytometry of intracellular staining for IL-17a and IFN-γ in sorted naïve T cell populations (from RORγtfl/fl mice following transduction with control-IRES-GFP (WT) or CRE-IRES-GFP (KO) retrovirus) activated and expanded in the presence of mouse Th17 polarizing cytokines. DMSO or 10 μM digoxin was added at 6 hours after viral transduction on day 1 and GFP expressing cells were gated for analysis on day 5. b, Two-dimensional hierarchical clustering of the 67 genes (including redundant probe sets and genes of unknown function) identified to be significantly affected by 2-way ANOVA analysis (DMSO versus digoxin treament, p<0.05). Each row corresponds to a gene and each column corresponds to an experimental sample. c, FACS-sorted naïve T cells were transduced with retroviral vectors encoding murine RORα-IRES-GFP or RORγt-IRES-GFP on Day 1 (16 h after TCR stimulation) and GFP expressing cells were gated for anyalsis on Day 5. DMSO or 10 μM digoxin was added 6–8 h after transduction. d, EAE disease course in B6 wild-type mice that were IP injected with either DMSO or digoxin (40 μg/mouse) every day starting from day 2 after disease induction with MOG(35-55)/CFA. Shown is averaged curve shape from seven experiments (10 or 20 mice were used per trial). * indicates statistical significance (p<0.05). Error bars represent standard deviation. e, f, Th1 and Th17 cells in spinal cord of EAE mice treated with DMSO or digoxin. Lymphocytes were isolated on day 21 after disease induction. The cells were stimulated for 4h with PMA/Ionomycin and stained for surface markers and intracellular cytokines. Representative FACS plots (gated on CD45+CD11b−CD4+ cells) from mice from each group are shown (top). T cells isolated from spinal cords of DMSO (n=7) or digoxin treated mice (n=7) were stained intracellularly for IFN-γ or IL-17a. Statistical analysis was by a two-tailed unpaired Student's t test; NS, not significant and p=0.014 (bottom).
To investigate if RORγt is the major target of digoxin or if another dominant cellular target exists, we performed gene expression profiling with total RNA samples isolated from DMSO- or digoxin-treated wild-type or RORγt-deficient cells cultured in Th17 conditions. Treatment with digoxin resulted in changes in gene expression that were very similar to those observed in RORγt-deficient cells: 2-way ANOVA analysis of differential gene expression revealed 67 genes that were significantly affected by the compound (DMSO vs. digoxin) irrespective of genotype (p<0.05) (Fig. 2b) and 323 that were affected by the genotype (WT vs. KO) irrespective of compound treatment (p<0.05) (Supplementary Fig. 6a). 94% of genes affected by digoxin treatment were similarly affected by RORγt deficiency. Importantly, no genes were significantly affected by the combination of gene inactivation and digoxin treatment. These results indicate that the effects of digoxin are predominantly mediated through RORγt. Induction of RORγt mRNA and protein expression was not affected by digoxin (Supplementary Fig. 4a and d). To rule out the possibility that digoxin blocks steps downstream of RORγt activity during Th17 cell differentiation (e.g. IL-17a production), we examined its effect upon ectopic expression of RORγt or RORα in naive CD4+ T cells. Both nuclear receptors were previously shown to be sufficient to induce IL-17a expression11, presumably by binding to the same cis-acting elements12. Digoxin suppressed RORγ- and RORγt-, but not RORα-mediated induction of IL-17a (Fig. 2c and Supplementary Fig. 6b), confirming that it acts selectively on RORγt in mouse T cells. However, digoxin (10 μM) failed to inhibit RORγt(triple) mutant- mediated IL-17a production (Supplementary Fig. 3e). Digitoxin and β-acetyldigoxin also selectively inhibited RORγt-dependent Th17 cell differentiation (Supplementary Fig. 6c). The aryl hydrocarbon receptor (AHR) is another ligand-dependent transcription factor that augments Th17 responses13. Its activity was unaffected by digoxin, as addition of the AHR ligand FICZ increased RORα-dependent IL-17a expression even in the presence of digoxin (Supplementary Fig. 6d). RORγt is predominantly found in the nucleus of Th17 cells14. Digoxin treatment did not inhibit its nuclear localization in Drosophila cells or in in vitro differentiated Th17 cells (Supplementary Fig. 7a and b). These data raise the question of how digoxin suppresses RORγt transcriptional activity. Chromatin immunoprecipitation-sequencing (ChIP-Seq) analysis with an anti-RORγt antibody (Supplementary Fig. 7c) was used for genome-wide identification of its transcriptional target sites in Th17 cells (M. Ciofani and D.R.L., unpublished results). We evaluated the effect of digoxin on binding of RORγt to sites in two relevant loci, Il17a/f and Il23r. RORγt binding to these sites was substantially reduced upon treatment with digoxin (Supplementary Fig. 7d), demonstrating one mode of its activity. In vitro, digoxin not only reduced the binding of RORγt onto its target, but also displaced SRC3-1b co-activator peptides (IC50 of 1.8 μM) from the RORγt LBD and facilitated its interaction with co-repressor NCOR2 peptides (IC50 of 3.9 μM) (Supplementary Fig. 7e, 8a, and b).
We next examined if digoxin can exert an anti-inflammatory effect in mice. We induced experimental autoimmune encephalomyelitis (EAE), a Th17-mediated autoimmune inflammatory disease, in C57BL/6 wild-type mice15, 16 in conjunction with intra peritoneal (IP) injections of digoxin or carrier each day from day 2. Digoxin treatment not only delayed onset, but also reduced severity of EAE progression (Fig. 2d). Also, the total number of mononuclear cells infiltrating the spinal cord was markedly reduced in mice treated with digoxin (Supplementary Fig. 9). Importantly, the percentage of IL-17-producing T cells infiltrating the spinal cord in digoxin-treated mice was reduced by more than 50%, as compared to DMSO-treated mice, whereas that of IFN-γ-producing Th1 cells was approximately the same (Fig. 2e and f). Administration of digoxigenin had no effect on progression of EAE (data not shown), indicating that the cardiac glycoside activity17 has no role in the observed amelioration of disease.
Digoxin, an inhibitor of the Na+/K+-ATPase, has long been used for treatment of congestive heart failure17, 18, but is toxic for human cells at concentrations (>300 nM)19 well below those required for RORγt inhibition. Expression of the catalytic α1 subunit of murine Na+/K+-ATPase, which binds digoxin poorly, rendered human cells much less sensitive to digoxin-mediated cytotoxicity20, 21. Thus, we ectopically expressed in human cord blood CD4+ T cells the αl subunit of murine Na+/K+-ATPase in the presence of cardiac glycosides. Lanatoside C (Supplementary Fig. 5a), which has inhibitory activity on Na+/K+-ATPase similar to digoxin17, but does not inhibit RORγt activity (data not shown), had no effect on IL-17a expression. However, digoxin suppressed IL-17a production (Supplementary Fig. 10a). Next, human T cells expressing the murine Na+/K+-ATPase were further transduced with lentivirus encoding human RORαd, β, or γt, all of which are sufficient to induce IL-17 expression22. Digoxin inhibited only RORγt-mediated induction of IL-17a (Supplementary Fig. 10b), demonstrating its direct and selective activity on human RORγt.
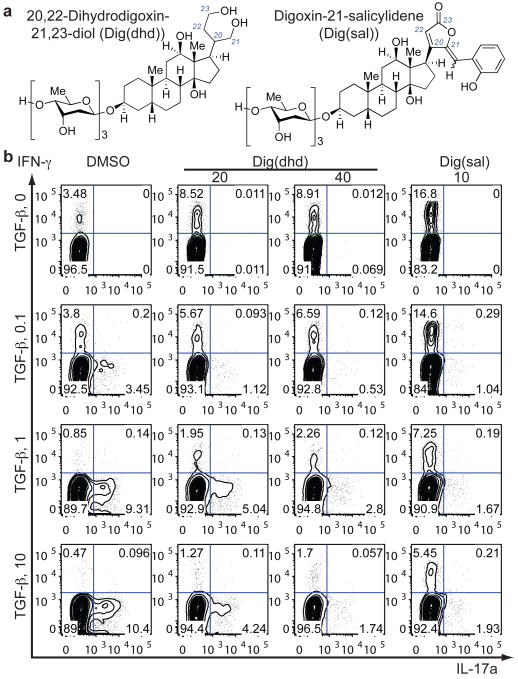
Cardiac glycosides of the cardenolide class have three common structural motifs, namely a central steroidal core fused with a butenolide and various sugars23, 24. The glycans are dispensable, as digoxigenin still inhibits Na+/K+-ATPase17. 20,22-Dihydrodigoxin (Supplementary Fig. 2d), which was derived upon butenolide reduction of digoxin by the intestinal commensal Eubacterium lentum25, has weak cardiac glycoside activity with much lower binding affinity than that of digoxin for Na+/K+-ATPase17, 26, yet it inhibited RORγ activity in the S2 reporter system (data not shown). Since 20,22-dihydrodigoxin was still cytotoxic for human cells at 2.5 μM, digoxin was further modified by complete reduction of the butenolide to generate Dig(dhd) (Fig. 3a). Dig(dhd) lacked cytotoxic activity on human cells at concentrations up to 40 μM, but it still possessed RORγ inhibitory activity and displaced the sterol ligand from the RORγ LBD (IC50 of 12 μM) (Supplementary Fig. 11a). Additional derivatization of digoxin was achieved by aldol condensation of the butenolide with salicylaldehyde to produce Dig(sal) (Fig. 3a), which, similarly to digoxin, bound directly to RORγ in the CD assay (Supplementary Fig. 3a). These compounds selectively inhibited both mouse and human RORγt activities without affecting those of mouse RORα and human LXRβ (Supplementary Fig. 11b, c, and d). Moreover, Dig(sal) treatment reduced severity of EAE progression (Supplementary Fig. 11e). When tested on human CD4+ T cells transduced with viruses encoding RORαd or RORγt, Dig(dhd) or Dig(sal) treatment selectively suppressed RORγt-mediated IL-17a induction (Supplementary Fig. 11f). Intriguingly, addition of either compound blocked Th17 cell differentiation22 (Fig. 3b) and induced reciprocal increases of IFN-γ or FoxP3 expression in T cells (Fig. 3b and Supplementary Fig. 11g), suggesting that functional RORγt or its downstream events may normally suppress development into other T cell lineages. Expression of another human Th17 cell-associated surface marker, CCR6, was also reduced in Dig(dhd) treated cells (Supplementary Fig. 11h).
Figure 3). 20,22-Dihydrodigoxin-21,23-diol and digoxin-25-salicylidene inhibit human Th17 cell differentiation.
a, Chemical structures of 20,22-dihydrodigoxin-21,23-diol and digoxin-21-salicylidene. b, Flow cytometry of the production of IL-17a and IFN-γ by human naïve cord blood T cells cultured for six days in the presence of IL-2, IL-23, and IL-1β, with various concentrations of TGF-β. DMSO, Dig(dhd), or Dig(sal) at indicated concentrations (μM) was added 16h after cytokine addition.
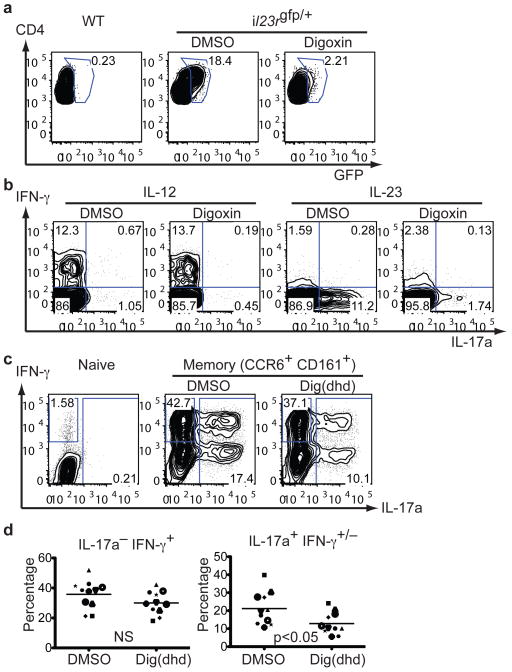
We next investigated if digoxin can inhibit IL-17 production from pre-differentiated Th17 cells. In vitro digoxin treatment of expanded mouse Th17 cells derived from immunized mice inhibited both IL-23R (Fig. 4a) and IL-17a expression without affecting IFN-γ expression (Fig. 4b). We also purified GFP-positive Th17 cells from MOG-immunized Il23rgfp/+ mice27 after 4-day in vitro culture with IL-23 and MOG peptide. More than 70% of the sorted GFP-positive cells expressed IL-17a (Supplementary Fig. 12a, day 0). GFP-positive cells were then treated with DMSO or digoxin for an additional 3 days. Digoxin treatment reduced IL-17a-expressing cells by more than 70% (Supplementary Fig. 12a, day 3), confirming that mouse Th17 cells generated in vivo and expanded in vitro require continuous RORγt activity to maintain their identity. To test if digoxin suppresses the activity of pre-existing Th17 cells in vivo, we transferred IFN-γ-deficient, MBP-specific Th17 cells into lymphopenic RAG-2-deficient mice and assessed EAE manifestation following daily administration of digoxin. Since the transferred cells lack IFN-γ, the EAE phenotype observed in these mice is entirely attributed to the function of Th17 cells. Digoxin treatment from day 2 delayed onset and reduced severity of Th17 cell transfer-mediated EAE, which further confirms a requirement for continuous RORγt activity in Th17 cells (Supplementary Fig. 12b). We then examined if RORγt activity is also important for sustained expression of IL-17a in human CD4+ T cells. Human memory Th17 cells were purified from peripheral blood samples and enriched by in vitro culture. Naïve CD4+ T cells cultured in the same cytokine conditions did not produce IL-17a (Fig. 4c, left plot). Dig(dhd) treatment led to 40–50% reduction of IL-17a-expressing cells with little effect on IFN-γ-expressing cells (Fig. 4c and d). These data demonstrate that human RORγt activity plays an important role in maintaining the human Th17 cell population.
Figure 4). RORγt activity is important for maintenance of mouse and human Th17 cells.
a, b, Flow cytometry of intracellular staining for IL-17a and IFN-γ by CD4+ T cells. Mononuclear cells were collected from draining lymph nodes of wild-type or IL-23R-GFP knock-in heterozygous mice 7 days after MOG(35-55)/CFA injection. Cells were cultured for four more days with MOG(35-55) peptide and exogenous IL-23 or IL-12, in the presence of DMSO or 10 μM digoxin. Without pre-immunization, addition of IL-23 and MOG(35-55) peptide to culture did not lead to de novo Th17 cell differentiation (data not shown). Digoxin treatment suppressed expansion of in vivo differentiated Th17 cells, assayed by IL-23R reporter GFP expression (a) or by IL-17a production (b). c, d, Human naïve (CD45RA+CD3+CD4+) or memory (CD45RO+CD45RA− CD3+CD4+CCR6+CD161+) cells were purified from healthy donor peripheral blood samples and were cultured in the presence of IL-1β, IL-23 and IL-2 for 6 days with or without 40 μM Dig(dhd). Intracellular staining for IFN-γ or IL-17a in memory CD4+ T cells from multiple donors (n=11) in the presence of IL-1β, IL-23, and IL-2, assessed on day 6. c, Representative FACS plots from one donor are shown. d, Each symbol indicates a separate donor. Statistical analysis was by a two-tailed unpaired Student's t test; IL-17a− IFN-γ+, not significant and IL-17a+IFN-γ+/−, p=0.02.
T cells and innate lymphocytes that produce IL-17a and IL-22 are recognized as having key roles in maintenance of barrier function at mucosal surfaces and also in the pathophysiology of autoimmune disease. All such cells, which include Th17 cells, other TCRαβ cells, TCRγδ cells, lymphoid tissue inducer cells, and NK-like cells (also referred to as NK22 cells) share in the property of requiring expression of RORγt for their differentiation. Abrogation of RORγt expression results in marked reduction or complete depletion of these cell types and in resistance to Th17-mediated autoimmune disease in mouse models4, 6, 28. Therefore, RORγt antagonists digoxin, Dig(dhd), and Dig(sal) may serve as good chemical templates for the development of potent therapeutic compounds to treat various diseases associated with inflammatory lymphocyte dysfunction.
The digitalis-like compounds were originally identified in plants. A body of evidence indicates the presence of endogenous digitalis-like compounds in mammals including humans18, 29. Identification of digoxin as a RORγt antagonist suggests that related molecules in mammals may modulate RORγ- and RORγt-mediated functions. However, it would be derivative compounds with better IC50 values that would have such roles. In light of recent findings of the roles of microbiota in generation of Th17 cells in the small intestine30, it is interesting that Eubacterium lentum, another commensal bacterium, has the capacity to metabolize digoxin into dihydrodigoxin. The possibility of the existence of endogenous digitalis-like compounds in host organisms and of their modification by microbes may present additional opportunities for modulating the function of RORγt and Th17 cell differentiation.
Methods Summary
Chemical screen
10,000 Drosophila stable S2 cells with genomic integration of the Cu2+-inducible G4DBD-mouse RORγ construct were transfected with 5 ng of pUAST-firefly luciferase and 7 ng of Pol III-Renilla luciferase and dispensed into white bottom-tissue culture 384-well plates (Corning). Two days later, small compounds (total 4,812 compounds from the ICCB chemical libraries, including Bioactives and Prestwick collections) were added and, after 6 hours, Cu2+ was added to the wells (700 μM). The following morning, Stop-glo luciferase substrates (Promega) were used to measure luciferase activity. Initial hits including digoxin were tested against three different control S2 reporter cell lines.
Cell culture
Mouse and human CD4+ T cell culture and viral transduction were performed as described previously6, 22, unless indicated otherwise in the text.
Identification of non-toxic digoxin derivatives
Various digoxin derivatives were synthesized and first tested for toxicity on human embryonic kidney 293T cells at various concentrations. Compounds exhibiting reduced toxicity compared to digoxin were further tested for their RORγ inhibitory activities with the insect cell reporter lines.
General
All DNA constructs were generated by PCR-based methodology and confirmed by sequencing. Retroviral production and transduction, EAE experiments, and gene chip analysis were performed as described previously6. IL-17a, IFN-γ, IL-4, Foxp3, and CCR6 protein expression was examined by intracellular or surface staining according to the manufacturer's protocol.
Supplementary Material
Acknowledgments
We thank Caroline Shamu at ICCB-Longwood for screening small compound libraries. We also thank the New York Cord Blood Center for providing samples, the Developmental Studies Hybridoma Bank for β–tubulin antibody, the NYU Cancer Institute Genomics Facility for expert assistance with microarray experiments, V. Kuchroo and M. Oukka for the IL-23R GFP reporter mice, T. Iwaki, C. Thummel and R. Dasgupta for plasmids, M. Garabedian for R1881 and plasmids, D. Mangelsdorf for dafachronic acid and for plasmids, G. Diehl and M. Sellars for reading the manuscript, M. Menager and J. Johnson for sharing PBMC, and members of the Littman laboratory for their suggestions. Supported by the Jane Coffin Childs Memorial Funds (J.R.H.), the Howard Hughes Medical Institute (D.R.L.) and National Institutes of Health grants F32GM0860552 (M.R.K.), RO1GM058833 (D.Y.G.), RO1GM067659 (D.Y.G), 2RO1GM55217 (F.R.), and RO1AI080885 (D.R.L.).
Footnotes
Full methods are included within the Supplementary information.
Author Contributions J.R.H., J.J.L., H.E.X., D.Y.G., F.R., and D.R.L. designed the experiments. J.R.H. and D.R.L. wrote the manuscript with input from the co-authors. J.R.H. developed the screen and executed it with assistance from J.C. and A.C.. F.R.S. developed the serum-free system for S2 cell culture. M.C. performed the ChIP experiments, J.R.H., N.M., and S.V.K. performed the T cell culture experiments, and J.R.H. and M.W.L.L. did in vivo compound tests and the follow-up analyses. P.H. did in vitro competition and CD assays, R.M. performed ALPHA screen assays, and D.A.R. and M.R.K. synthesized and purified digoxin derivatives.
The microarray data sets are deposited in the Gene Expression Omnibus database under accession number GSE27241.
Reprints and permission information are available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Stehlin C, et al. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. Embo J. 2001;20:5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin L, et al. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 14.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 17.Paula S, Tabet MR, Ball WJ., Jr Interactions between cardiac glycosides and sodium/potassium-ATPase: three-dimensional structure-activity relationship models for ligand binding to the E2-Pi form of the enzyme versus activity inhibition. Biochemistry. 2005;44:498–510. doi: 10.1021/bi048680w. [DOI] [PubMed] [Google Scholar]
- 18.Nesher M, Shpolansky U, Rosen H, Lichtstein D. The digitalis-like steroid hormones: new mechanisms of action and biological significance. Life Sci. 2007;80:2093–2107. doi: 10.1016/j.lfs.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Johansson S, et al. Cytotoxicity of digitoxin and related cardiac glycosides in human tumor cells. Anticancer Drugs. 2001;12:475–483. doi: 10.1097/00001813-200106000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Price EM, Lingrel JB. Structure-function relationships in the Na,K-ATPase alpha subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry. 1988;27:8400–8408. doi: 10.1021/bi00422a016. [DOI] [PubMed] [Google Scholar]
- 21.Zavareh RB, et al. Inhibition of the sodium/potassium ATPase impairs N-glycan expression and function. Cancer Res. 2008;68:6688–6697. doi: 10.1158/0008-5472.CAN-07-6833. [DOI] [PubMed] [Google Scholar]
- 22.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7:926–935. doi: 10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 24.Mijatovic T, et al. Cardiotonic steroids on the road to anti-cancer therapy. Biochim Biophys Acta. 2007;1776:32–57. doi: 10.1016/j.bbcan.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Robertson LW, Chandrasekaran A, Reuning RH, Hui J, Rawal BD. Reduction of digoxin to 20R-dihydrodigoxin by cultures of Eubacterium lentum. Appl Environ Microbiol. 1986;51:1300–1303. doi: 10.1128/aem.51.6.1300-1303.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindenbaum J, Rund DG, Butler VP, Jr, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med. 1981;305:789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 27.Awasthi A, et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luci C, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 29.Bagrov AY, Shapiro JI. Endogenous digitalis: pathophysiologic roles and therapeutic applications. Nat Clin Pract Nephrol. 2008;4:378–392. doi: 10.1038/ncpneph0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.