Abstract
Translation and mRNA stability are enhanced by the presence of a poly(A) tail. In vivo, the tail interacts with a conserved polypeptide, poly(A) binding protein (Pab1p). To examine Pab1p function in vivo, we have tethered Pab1p to the 3′ UTR of reporter mRNAs by fusing it to MS2 coat protein and placing MS2 binding sites in the 3′ UTR of the reporter. This strategy allows us to uncouple Pab1p function from its RNA binding activity. We show that mRNAs that lack a poly(A) tail in vivo are stabilized by Pab1p, and that the portions of Pab1p required for stabilization are genetically distinct from those required for poly(A) binding. In addition, stabilization by Pab1p requires ongoing translation of the mRNA. We conclude that the primary, or sole, function of poly(A) with respect to mRNA stability is simply to bring Pab1p to the mRNA, and that mRNA stabilization is an intrinsic property of Pab1p. The approach we describe may be useful in identifying and assaying 3′ UTR regulatory proteins, as it uncouples analysis of function from RNA binding.
Keywords: Poly(A), poly(A) binding protein, mRNA stability, translation, 3′ UTRs
The cellular functions of proteins that act on nucleic acids require that their activities be brought to specific sequences in RNA or DNA. In some cases, as with many RNA and DNA polymerases, one protein with enzymatic activity is directed to the correct location through its interaction with another polypeptide. Alternatively, as with many regulatory factors, a single polypeptide can possess distinct nucleic acid-binding and functional domains. Separation of these two domains from one another facilitates their detailed analysis, liberating each from the constraints of the other.
A poly(A) tail is a nearly universal feature of mRNAs in eukaryotes, and exists as an RNA-protein complex with poly(A)-binding protein (Pab1p) in the cytoplasm. This highly conserved protein consists of four tandem RNA recognition motif domains (RRM 1–4) and a carboxy-terminal region without recognizable functional motifs. RRMs 1 and 2 are required for high affinity binding to poly(A) in vitro (Nietfeld et al. 1990; Burd et al. 1991; Kühn and Pieler 1996; Deardorff and Sachs 1997). The presence of a poly(A) tail typically stabilizes mRNAs and can enhance their translation both in vivo and in vitro (for review, see Sachs et al. 1997; Wickens et al. 1997). It is unclear whether, in the cell, these biological functions reside solely in the protein, or instead require the poly(A)/Pab1p complex (see below).
A role for Pab1p in mRNA stability has been shown in the budding yeast Saccharomyces cerevesiae. The yeast PAB1 gene is required for viability (Sachs et al. 1986). The lethality of a pab1 deletion can be suppressed by mutations in various genes associated with mRNA turnover, such as xrn1 and dcp1, indicating that Pab1p likely has an essential function in mRNA decay (Hatfield et al. 1996). Degradation of most yeast mRNAs is initiated by shortening of the 3′ poly(A) tail, followed by Dcp1p cleavage of the 5′7mGpppN cap (decapping), which permits 5′ to 3′ exonucleolytic digestion by the XRN1 gene product (Decker and Parker 1993; Muhlrad et al. 1994; Beelman et al. 1996). Decapping occurs only after the poly(A) tail has been shortened to 10–12 adenosine residues (Muhlrad et al. 1994), which corresponds provocatively with the minimal Pab1p-binding site of 12 adenosines. These data suggest that eviction of the last Pab1p molecule triggers turnover. Consistent with this view, decapping becomes independent of poly(A) in mutant strains that lack Pab1p, such that mRNAs with long poly(A) tails are decapped (Caponigro and Parker 1995). The effects of pab1 mutations on cellular mRNA levels may be complex, with contributions from turnover and/or nuclear processes, such as mRNA 3′-end formation (Caponigro and Parker 1995; Amrani et al. 1997; Keller and Minvielle-Sebastia 1997; Minvielle-Sebastia et al. 1997; J. Morrisey and A. Sachs, pers. comm.).
The molecular link between poly(A) and the cap in yeast may be a tripartite protein bridge between the two ends of the mRNA. Pab1p, when bound to the poly(A) tail, interacts in vitro with two yeast proteins, TIF4631 and TIF4632, homologs of the translation initiation factor, eIF-4G (Tarun and Sachs 1996). eIF-4G in turn interacts with eIF-4E, a cap-binding protein and an initiation factor itself. The association of Pab1p and eIF-4G is consistent with earlier work showing biochemical interactions between poly(A) and complexes containing eIF-4F in vitro (Gallie and Tanguay 1994). The same tripartite bridge may explain both poly(A)’s stimulation of translation in vitro and in vivo and its ability to protect the cap.
The interaction between Pab1p and eIF-4G, shown in vitro, requires the presence of poly(A), implying that the poly(A)/Pab1p complex likely is essential. Similarly, deadenylation in crude yeast extracts is dependent on the presence of Pab1p, implying that the yeast deadenylase recognizes the poly(A)/Pab1p complex rather than naked poly(A) (Lowell et al. 1992). In this paper, we examine in vivo whether Pab1p alone is sufficient to stabilize an mRNA to which it is bound. To uncouple Pab1p’s function from its ability bind poly(A), we tether Pab1p via MS2 coat protein to a binding site placed in the 3′-untranslated region (UTR) of a reporter mRNA. This strategy allows us to monitor Pab1p function using a nonessential reporter, and to analyze the effect of mutations in Pab1p that do not support viability. We show that Pab1p alone can stabilize mRNAs, and that this process requires translation. These results imply that the role of the poly(A) tail in mRNA stability is passive, serving only to recruit Pab1p. The experimental strategy may be useful in identifying and dissecting proteins that regulate mRNA function, particularly those that act through the 3′ UTR.
Results
Experimental strategy
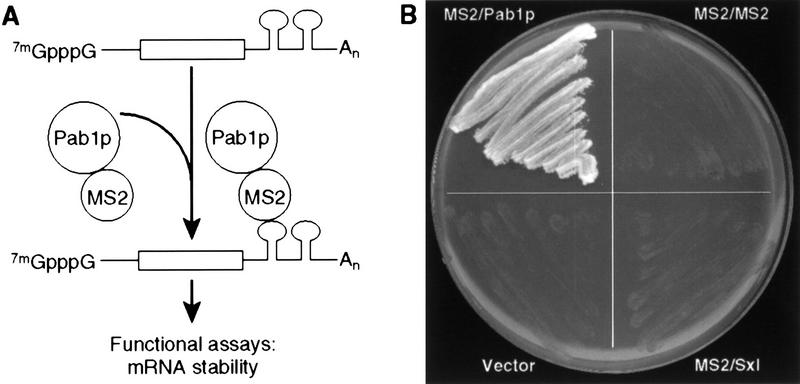
Pab1p is essential for viability in yeast (Sachs et al. 1986). To assess its function independent of effects on viability, we exploited the strategy diagrammed in Figure 1A. A chimeric protein consisting of MS2 bacteriophage coat protein linked to yeast Pab1p (MS2/Pab1p) was expressed in yeast; the same strain also carried a second plasmid expressing a reporter mRNA with MS2 recognition sites in its 3′ UTR. Two recognition sites were inserted, as binding of MS2 coat protein to adjacent sites is cooperative (Witherell et al. 1991). This approach separated analysis of the function of Pab1p from its ability to bind poly(A) or support cell growth.
Figure 1.
Experimental strategy. (A) The assay. To uncouple analysis of Pab1p function from the presence of a poly(A) tail, Pab1p was tethered to the 3′ UTR of reporter mRNAs by fusing it to MS2 coat protein and placing MS2-binding sites in the 3′ UTR of the reporter. For simplicity, a single-bound protein molecule is depicted; however, because MS2 coat protein binds as a dimer, at least two MS2/Pab1p molecules are likely bound per site. (B) MS2/Pab1p complements the essential functions of Pab1p in vivo. MS2/Pab1p alone complements a pab1 deletion mutation following 5-FOA selection against a plasmid bearing both the URA3 and PAB1 genes.
To use this strategy, it first was necessary to determine whether the MS2/Pab1p fusion protein could provide Pab1p’s normal essential function(s) in vivo. To this end, we tested whether a gene encoding the MS2/Pab1p fusion protein could complement a pab1 deletion (Sachs et al. 1987). A plasmid carrying the wild-type PAB1 gene and a URA3 marker was introduced into yeast harboring a pab1 deletion. This strain was then transformed with a second plasmid expressing the MS2/Pab1p fusion or various control plasmids. Transformants were then analyzed on medium containing 5-fluoro-orotic acid (5-FOA), which selects for loss of the URA3 plasmid carrying wild-type PAB1. Cells expressing the MS2/Pab1p fusion protein grew on 5-FOA (Fig. 1B). In contrast, cells expressing fusion proteins in which MS2 coat protein was fused to the Drosophila Sex-lethal protein (MS2/Sxl) or to another copy of MS2 coat protein (MS2/MS2) did not support viability, nor did the parental expression vector (Fig. 1B). Cells expressing the MS2/Pab1p fusion exhibited a generation time in minimal medium of 2.4 hr, which is comparable to the wild-type rate of 2.0 hr. These observations show that the MS2/Pab1p fusion protein can provide Pab1p’s essential function(s) in vivo.
mRNA stability is conferred by MS2/Pab1p tethered to the 3′ UTR
To examine the effects of Pab1p on mRNA decay, we inserted MS2 coat protein recognition sites into the 3′ UTR of MFA2 mRNA. MFA2 mRNA is among the least stable mRNAs in yeast, with a half-life of ∼4 min (Herrick et al. 1990). To measure mRNA turnover rates in vivo, we used a transcriptional pulse-chase protocol (Decker and Parker 1993). Transcription of the MFA2/MS2 reporter mRNA was placed under control of the GAL1 promoter. Cells were grown in raffinose, then incubated briefly in galactose to induce transcription. After 10 min, transcription was repressed by addition of glucose and by thermal inactivation of a temperature-sensitive allele of RNA polymerase II (Nonet et al. 1987). The decay of the newly synthesized mRNA was then monitored by Northern blotting.
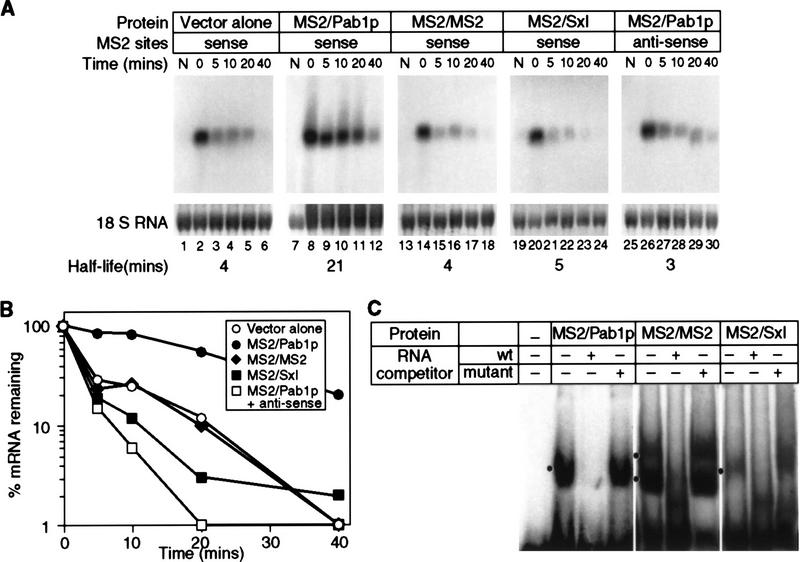
The MS2/Pab1p fusion protein specifically stabilized the reporter mRNA carrying MS2 sites (Fig. 2A,2B). mRNA half-life increased from 4 to 21 min in the presence of MS2/Pab1p protein (Fig. 2A, lanes 1–12, and B). This stabilization specifically required the Pab1p portion of the fusion protein: Its replacement with either MS2 coat protein (Fig. 2A, lanes 13–18; MS2/MS2) or the Drosophila Sex lethal (Sxl) protein (Fig. 2A, lanes 19–24; MS2/Sxl) eliminated the stabilization. Both Sxl protein and Pab1p contain multiple RRMs, strongly suggesting that the Pab1p moiety is specifically required, and that RRMs with other specificities do not suffice. mRNA stabilization is also specific with respect to RNA sequence: An mRNA in which the MS2-binding sites are in the antisense orientation is not stabilized by MS2/Pab1p (Fig. 2A, lanes 25–30). Similarly, MS2/Pab1p did not effect the stability of an MFA2 reporter lacking MS2 sites (data not shown). This result shows that the effect of MS2/Pab1p operates only in cis, and that expression of MS2/Pab1p does not stabilize all mRNAs.
Figure 2.
mRNA stability is conferred by MS2/Pab1p tethered to the 3′ UTR. (A) The decay of mRNAs in the presence of MS2/Pab1p, analyzed by transcriptional pulse-chase experiments and Northern blotting. The RNA and protein present in each strain is indicated above each group of six lanes, as are the orientation of MS2 sites, and the time (in min) following transcriptional repression. (Vector alone) Strains not expressing a fusion protein, but still transformed with the parental protein vector; (N) not induced sample taken prior to galactose induction. Half-lives are presented at the bottom of each panel. (B) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, shown below each lane in A. (C) RNA-binding activities of fusion proteins in yeast extracts. Gel retardation analyses were performed with crude extracts of yeast strains containing the proteins indicated at top of each group of lanes. The labeled RNA contains two MS2 coat protein recognition sites. The unlabeled competitor RNAs either contain high (wt) or low (mutant) affinity MS2 recognition sites. Mutant sites bind in vitro with an 100-fold reduction in affinity(Witherell et al. 1991). Black dot is at left of each group of three lanes indicates positions of specific complexes.
To ensure that each fusion protein was expressed and capable of RNA binding, we performed Western blotting and gel retardation experiments. Cross-reacting species of the expected size and comparable concentrations were observed in Western blotting experiments by use of an anti-MS2 coat protein monoclonal antibody to probe yeast extracts (data not shown). Moreover, in gel retardation experiments (Fig. 2C), each fusion protein present in yeast extracts formed complexes with RNAs carrying the MS2 recognition element; these complexes were sequence specific, as they were eliminated by the inclusion of excess unlabeled RNA containing the MS2 recognition elements, but not by an excess of RNA containing elements with point mutations that reduce binding affinity (Fig. 2C; Lowary and Uhlenbeck 1987).
We conclude that MS2/Pab1p bound to a 3′ UTR stabilizes an otherwise unstable mRNA. Stabilization specifically requires the Pab1p portion of the molecule. For simplicity, we refer to the MS2/Pab1p molecule bound to the 3′ UTR as tethered Pab1p.
Tethered Pab1p stabilizes partially and fully deadenylated mRNAs
Deadenylation rates of many mRNAs are directly related to their rates of decay in vivo (Chen and Shyu 1995; for review, see Caponigro and Parker 1996). Thus, tethered Pab1p could stabilize an mRNA by effecting the rate either of deadenylation or of turnover subsequent to deadenylation. To distinguish between these possibilities, we determined poly(A) lengths as a function of time in a transcriptional pulse-chase experiment. To do so, RNAs were separated in polyacrylamide gels, then analyzed by Northern blotting (Fig. 3). As expected, the MFA2 mRNA bearing MS2 recognition sites was stabilized dramatically (Fig. 3A,B). After 6 min, little reporter mRNA remains in cells lacking MS2/Pab1p; those mRNAs that are detected possess short poly(A) tails (Fig. 3B). At the same time point, the reporter mRNA is abundant in cells containing MS2/Pab1p (Fig. 3A), and the population of stabilized mRNAs includes mRNAs that have undergone partial or complete deadenylation. To confirm that mRNAs that had undergone deadenylation could be stabilized by MS2/Pab1p, we analyzed poly(A) tail lengths after a protracted chase of 60 min (Fig. 3C). mRNAs present after 60 min in the presence of MS2/Pab1p possess little or no poly(A), as they comigrate with mRNAs whose poly(A) tails have been removed with oligo(dT) and RNase H (Fig. 3C). These results suggest that tethered Pab1p stabilizes an mRNA that has had its tail largely, or completely, removed.
Figure 3.

Tethered Pab1p stabilizes deadenylated mRNAs but does not slow poly(A) removal . A transcriptional pulse-chase experiment was performed with strains carrying MS2/Pab1p or the comparable vector. RNAs were separated through a polyacrylamide gel to resolve different lengths of poly(A) tails. Time (in min) following transcriptional repression are given above each lane. Poly(A) tail lengths of the mRNAs are shown at left; the position of the fully deadenylated RNA (A0) was determined by RNaseH/oligo (dT) cleavage of the full-length reporter. (A) RNA prepared from strain expressing MS2/Pab1p. (B) RNA prepared from strain containing the vector alone (no MS2/Pab1p). (C) Poly(A) length of mRNAs that accumulate after 60 min in the presence of MS2/Pab1p. RNA was isolated either 0 or 60 min after repression from strains with or without MS2/Pab1p, as indicated above each lane. An aliquot of RNA isolated immediately after repression (zero time) was treated with oligo(dT) and RNase H to remove poly(A) tails, and thereby provides a standard for the deadenylated reporter mRNA. Note that after 60 min, the mRNA that accumulates in the presence of MS2/Pab1p comigrates with the deadenylated mRNA standard. No mRNA is detectable in the strain lacking MS2/Pab1p after this long chase interval.
Tethered Pab1p stabilizes mRNAs that do not receive a tail
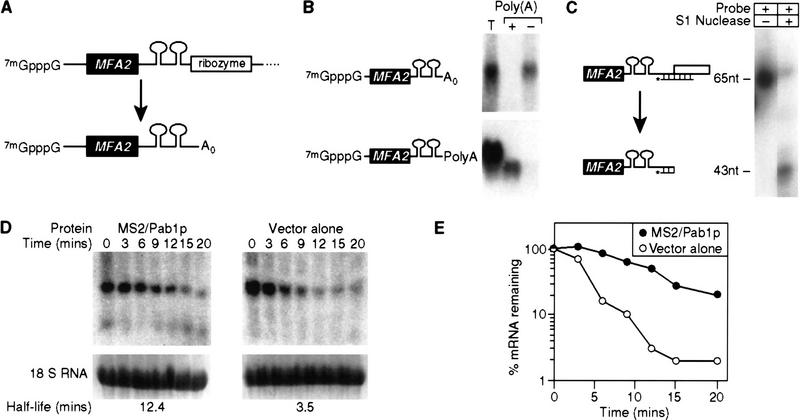
To test directly whether tethered Pab1p can stabilize an mRNA lacking a poly(A) tail, we created a reporter mRNA that did not receive poly(A) in vivo. To do so, a self-cleaving ribozyme sequence was inserted into the pre-mRNA (Fig. 4A). The ribozyme was derived from hepatitis δ virus (HDV) ribozyme, and has been shown previously to function in yeast (Quadt et al. 1995).
Figure 4.
Tethered Pab1p stabilizes mRNAs that do not receive a tail. (A) Strategy. The HDV self-cleaving ribozyme was inserted into the 3′ UTR of MFA2/MS2 RNA, yielding an mRNA without a poly(A) tail (A0) in vivo. (B) Oligo(dT) cellulose chromatography. RNAs were extracted from cells carrying the ribozyme-containing construct depicted in A, or the control plasmid used in previous figures. RNAs were fractionated by oligo(dT) cellulose chromatography, then analyzed by Northern blotting. The ribozyme-containing RNA is not retained by the column, whereas the control RNA is retained. (T) Total RNA prior to fractionation; (+) retained by the column; (−) not retained by the column. The mRNA runs slightly faster in the oligo(dT) retained (+) sample than in the total (T) RNA, because less RNA is loaded per lane. (C) S1 nuclease analysis. The 3′ end of mRNA prepared from a strain carrying the ribozyme-containing RNA was determined by S1 nuclease mapping. The position of undigested probe (65 nucleotides) and of the protected species (43 nucleotides) are shown. The prominent 3′ terminus lies at the expected position for ribozyme cleavage. (D) Decay of ribozyme-cleaved RNA by a transcriptional pulse-chase experiment and Northern blotting. The turnover rate of ribozyme-cleaved reporter mRNAs was determined in strains with (left) or without (right) MS2/Pab1p. Time (in min) following transcriptional repression are given directly at top; half-lives are at bottom. (E) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, at bottom of each lane in D. (•) MS2/Pab1p; (○) vector alone.
A series of control experiments showed that the ribozyme functioned as expected in vivo. The ribozyme generated mRNAs of the predicted length in Northern blots Fig. 4B); further, as expected, these mRNAs lacked poly(A), as they were not retained by oligo(dT) cellulose (Fig. 4B, top), whereas mRNAs generated by normal mRNA 3′ processing were retained (Fig. 4B, bottom). In addition, nuclease S1 mapping experiments showed that the 3′ termini generated by ribozyme cleavage were within 3 nucleotides of the sites predicted (Fig. 4C). To ensure that cleavage occurred in vivo and not during extraction of the RNA, care was taken to prevent ribozyme cleavage during RNA preparation. These precautions included extracting and storing RNA at low pH and in the presence of EDTA (Donahue and Fedor 1997). To determine directly the extent of ribozyme cleavage during preparation, a radiolabeled in vitro transcript containing the HDV ribozyme was added to pelleted yeast cells prior to RNA extraction; RNA was then prepared following the usual protocol. The labeled RNA remained uncleaved, as shown previously (Donahue and Fedor 1997; data not shown). We deduce that the ribozyme cleaved in vivo to form nonpolyadenylated 3′ termini.
The nonpolyadenylated MFA2/MS2 mRNA produced by the ribozyme is unstable, exhibiting a half-life of ∼3 min (Fig. 4D,E). The presence of the MS2/Pab1p fusion protein stabilized the mRNA approximately fourfold. We conclude that mRNA stabilization by tethered Pab1p does not require the presence of a poly(A) tail.
Tethered Pab1p does not prevent decay through mRNA surveillance
The presence of a premature nonsense codon triggers rapid mRNA decay without deadenylation, through a process termed mRNA surveillance (Pulak and Anderson 1993). To determine whether tethered Pab1p could prevent degradation through this poly(A)-independent pathway, we examined the stability of a PGK1 transcript carrying a nonsense mutation and bearing MS2-binding sites in its 3′ UTR. The presence of the nonsense mutation greatly accelerates degradation of this normally stable transcript, reducing its half-life from 35 min to 4 min (Muhlrad and Parker 1994). Transcriptional pulse-chase analysis showed that the presence of tethered Pab1p did not significantly stabilize the mutant mRNA (Fig. 5A,B). These results strongly suggest that tethered Pab1p stabilizes only mRNAs whose decay is deadenylation dependent.
Figure 5.

Tethered Pab1p does not prevent decay through mRNA surveillance. (A) Decay of nonsense mutant-containing PGK1 mRNA in a transcriptional pulse-chase experiment and Northern blotting. Stability of a PGK1/MS2 transcript harboring a nonsense mutation at position 103 was assayed in cells expressing MS2/Pab1p (left) or vector alone (right). Time (in min) following transcriptional repression are given at top of each lane; half-lives are at bottom. (B) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, shown at bottom of each lane in A. (•) MS2/Pab1p; (○) vector alone.
Translation is not impaired by tethered Pab1p
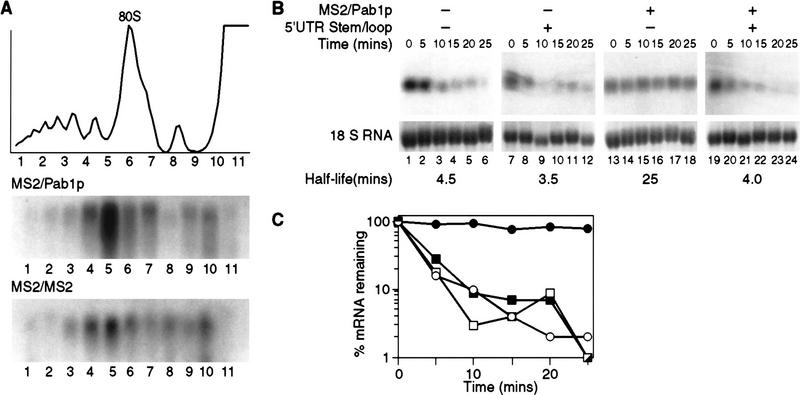
Because perturbation of translational activity can significantly affect stability (Muhlrad et al. 1995; Jacobson and Peltz 1996), we determined whether translation of MFA2/MS2 mRNA was affected by the binding of MS2/Pab1p. To do so, we fractionated polyribosomes by sucrose gradient centrifugation from cells expressing the normal MFA2 reporter, which receives a poly(A) tail. The polysomal distribution of this RNA was unaffected by tethered Pab1p (Fig. 6A). This result suggests that mRNAs bound to MS2/Pab1p are functional, and are not sequestered into a stable but inactive pool. This implies that effects of MS2/Pab1p are not the result of changes in the mRNA’s translation status.
Figure 6.
Translation is required in cis. (A) Polysome distribution of the MFA2/MS2 mRNA when bound by tethered Pab1p. The translational status of the MFA2/MS2 mRNA was determined by isolating polysomes from strains expressing either MS2/Pab1p or MS2/MS2. Sample optical trace is at top. Numbers at bottom indicate collected fractions. (B) Decay of MFA2/MS2 mRNA bearing a stem–loop in its 5′ UTR, analyzed by transcriptional pulse chase. The turnover rate of the MFA2/MS2 mRNA with or without the stem–loop in its 5′ UTR was determined in strains expressing (lanes 13–18 and lanes 19–24) or lacking (lanes 1–6 and lanes 7–12) MS2/Pab1p. Time (in min) following transcriptional repression are given at top of each lane; half-lives are presented at bottom. (C) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA, shown at bottom of each lane in B. (█) Vector alone, no stem–loop; (□) vector alone, stem–loop; (•) MS2/Pab1p, no stem–loop; (○) MS2/Pab1p, stem–loop.
Translation is required in cis
mRNA decay and translation are linked in yeast (for review, see Jacobson and Peltz 1996), and Pab1p may be essential in both processes (Sachs and Davis 1989; Hatfield et al. 1996). To determine whether mRNA stabilization by tethered Pab1p requires translation of the reporter, we prepared an MFA2 transcript containing a 11-nucleotide stem–loop structure stabilized by a UUCG tetraloop in its 5′ UTR, as well as tandem MS2-binding sites in its 3′ UTR. Previous work has shown that the presence of this secondary structure in the 5′ UTR reduces translation of MFA2 mRNA to <1% that of normal (Beelman and Parker 1994).
The presence of the 5′ UTR stem–loop structure had no significant effect on mRNA stability in the absence of MS2/Pab1p (Fig. 6B, lanes 1–6 vs. 7–12, and C). In contrast, however, the presence of the 5′ stem–loop entirely abrogated stabilization by tethered Pab1p, reducing the half-life from 25 to 4.5 min (Fig. 6B, lanes 13–18 vs. 19–24, and C). This implies that some aspect of translation is required for stabilization by Pab1p.
mRNA stabilization and poly(A) binding by Pab1p are genetically separable
To determine independently whether mRNA stabilization by Pab1p requires poly(A) binding, we removed segments of Pab1p from the MS2/Pab1p fusion protein, and then analyzed whether the mutant proteins stabilized the MFA2/MS2 reporter mRNA. Previous work has shown that RNA recognition motifs (RRMs) 1 and 2 of Pab1p bind specifically to the poly(A) tail, whereas RRMs 3 and 4 bind to poly(G) and poly(U) (Nietfeld et al. 1990; Burd et al. 1991; Kühn and Pieler 1996; Deardorff and Sachs 1997); RRM 2 also mediates interaction with eIF-4G in vitro (Kessler and Sachs 1998).
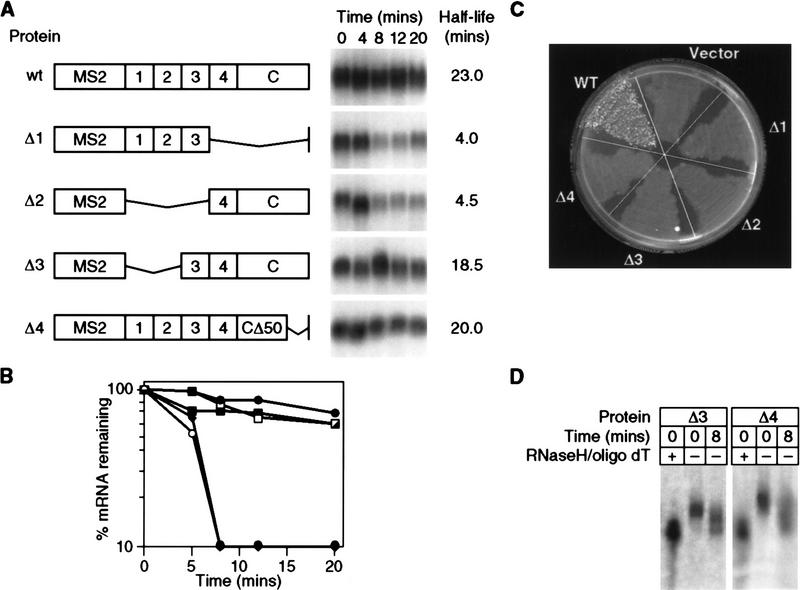
Plasmids encoding mutant forms of Pab1p (as fusions with the MS2 intramolecular dimer) were introduced into cells carrying a chromosomal, wild-type PAB1 gene; wild-type Pab1p was present to ensure cell viability (see below). Deletion of RRM 4 plus the carboxy-terminal domain (Δ1), or of RRMs 1–3 (Δ2), abrogated stabilization of the MFA2 reporter bearing MS2 sites (Fig. 7A,B). In contrast, removal of RRMs 1 and 2 had little effect on stabilization; a fusion protein lacking these domains stabilized the reporter mRNA to the same extent as did wild-type MS2/Pab1p (Δ3 vs. wt; Fig. 7A,B). Similarly, the final 50 amino acids of the carboxy-terminal domain were not required for stabilization (Δ; Fig. 7A,B). In certain genetic backgrounds, lack of Pab1p delays deadenylation for 30 min or more (Caponigro and Parker 1995). A comparable delay is not observed in strains containing Δ3 and Δ4: Even after 8 min, such strains contain partially and fully deadenylated reporter mRNAs (Fig. 7D).
Figure 7.
mRNA stabilization and poly(A) binding by Pab1p are genetically separable. (A) Decay of MFA2/MS2 mRNA in strains carrying MS2/Pab1p or deletion derivations, analyzed by the transcriptional pulse-chase protocol. The structure of each deletion form of MS2/Pab1p is depicted at left. Each RRM domain is designated by a number; (C) carboxy-terminal portion of Pab1p, (CΔ50) lacks the last 50 amino acids of Pab1p. Time (in min) following transcriptional repression are given at top of each lane. Half-lives are presented at right. (B) Quantitation of results. Amounts of mRNA were normalized to the level of 18S rRNA. (•) Wild type; (○) Δ1; (♦) Δ2; (█) Δ3; (□) Δ4. (C) Only full-length MS2/Pab1p complements the essential functions of Pab1p in vivo. The ability of plasmids carrying each form of MS2/Pab1p depicted in A to support cell growth in a strain lacking a functional PAB1 gene was determined by a plasmid shuffle assay. The assay was performed as described in Materials and Methods, and in the text (Fig. 1B). Only the wild-type construct complements a pab1 deletion following 5-FOA selection against a plasmid bearing both the URA3 and PAB1 genes. (D) Poly(A) length of mRNAs that accumulate after 8 min in the presence of Δ3 or Δ4. RNA was isolated either 0 or 8 min after repression, from strains with Δ3 or Δ4 as indicated above each lane. An aliquot of the RNA isolated immediately after repression (zero time) was treated with oligo(T) and RNase H to remove poly(A) tails, and thereby provide a standard for the deadenylated reporter mRNA.
To determine whether the various MS2/Pab1p mutants could provide Pab1p’s essential function(s), we repeated the plasmid shuffle assay described in Figure 1B. Of the five forms of MS2/Pab1p analyzed in Figure 7B, only the one carrying a wild-type Pab1p supported viability (Fig. 7C). In particular, Δ3, which lacks RRM domains 1 and 2 and is capable of stabilizing mRNAs to which it is tethered, does not support cell growth. This is consistent with the predicted inability of this protein to bind to poly(A), or with other essential functions of this region.
Discussion
The results presented here show that Pab1p can stabilize an mRNA in the absence of the poly(A) tail, and without the regions of the protein that bind specifically to poly(A). We conclude that one important function of poly(A) in mRNA stability is simply to tether Pab1p to the 3′ end of mRNA. Furthermore, the ability of Pab1p to stabilize an mRNA is blocked by the insertion of a stem-loop structure in the 5′ UTR, suggesting that Pab1p may act in part through the translational machinery to achieve its effects on stability.
Two lines of evidence establish that tethered Pab1p can stabilize an mRNA independent of the presence of a poly(A) tail. First, tethered Pab1p does not affect the deadenylation rate of mRNAs formed through the normal mRNA 3′-end formation pathway; rather, it stabilizes molecules that have undergone complete, or nearly complete, deadenylation. Second, tethered Pab1p stabilizes an mRNA that lacks a poly(A) tail, in which the 3′ terminus was formed by ribozyme cleavage. These two assays may reflect the same cellular function of Pab1p. On the other hand, the stabilization of initially polyadenylated and of ribozyme-cleaved mRNAs might occur through overlapping, but distinct, mechanisms. For example, Pab1p might be required for transport of the ribozyme-generated mRNA from the nucleus (Huang and Carmichael 1996). Escape from the nucleus might then permit stabilization.
Our results imply that the sole function of deadenylation is the simple elimination of Pab1p-binding sites: If they are effectively restored through the insertion of MS2 sites and an MS2/Pab1p chimera, then mRNAs without poly(A) become stable. Deadenylation rates vary among mRNAs, and are often controlled by sequences in the 3′ UTR (Muhlrad and Parker 1992; Herrick and Ross 1994; Chen et al. 1995). Such 3′ UTR elements have been suggested to provide alternative binding sites for Pab1p, shuttling it off the poly(A) tail, thereby allowing deadenylation and decay (Bernstein et al. 1989). Our data argue against such models, because Pab1p binding to the 3′ UTR stabilizes, rather than destabilizes, the mRNA.
The effects of tethered Pab1p on mRNA stability appear to require ongoing translation. Similarly, perturbation of mRNA translation stimulates rapid deadenylation and decapping of some transcripts in yeast (Muhlrad et al. 1995; Jacobson and Peltz 1996). In one model, a bridge linking Pab1p to eIF-4E through eIF-4G is critical both for translational stimulation and stabilization (Tarun and Sachs 1996). However, the in vitro association between Pab1p and eIF-4G is absolutely dependent on the poly(A) tail and requires RRM2 (Tarun and Sachs 1996; Kessler and Sachs 1998), both of which we show here are dispensable for mRNA stabilization by tethered Pab1p. Thus the Pab1p/eIF-4G linkage does not appear to be required for mRNA stabilization. Alternatively, other regions of Pab1p may recruit eIF-4G.
In principle, strong secondary structures inserted into the 5′ UTR could elicit an abnormal rapid decay phenotype that is dominant over MS2/Pab1p stabilization, complicating our interpretation of Figure 6, B and C. However, rapid turnover associated with stem–loop insertions appears to be caused by accelerated deadenylation followed by decapping (Muhlrad et al. 1995). mRNA stabilization by tethered Pab1p is independent of deadenylation (Figs. 3 and 4). Thus, if the only effect of the stem–loops were to accelerate deadenylation and decay, then they should not destabilize an mRNA carrying tethered Pab1p. The fact that they do, therefore, argues that stabilization by MS2/Pab1p requires some step in translation.
Although previous work suggested a single RRM from Pab1p was sufficient to restore viability of a PAB1 deletion (Sachs et al. 1987), more recent studies have suggested RRM domains 2 and 4 are vital (Deardorff and Sachs 1997). Our own data are consistent with these more recent findings. In addition, our results suggest that the carboxy-terminal 50 amino acids of Pab1p are required to support viability but are not required for mRNA stabilization. These results imply that those amino acids have an essential and distinct function in vivo. This function might be to facilitate ordered binding of Pab1p to poly(A), as implied by Kühn and Pieler (1996).
Viewed broadly, the tethered function approach may provide a general and facile assay for the function of 3′ UTR-binding proteins, independent of their ability to bind to RNA. Clearly, mRNA-binding proteins have been linked previously to foreign RNA binding specificities, with a variety of successful applications; we emphasize here the particular value of tethering to the 3′ UTR, and the opportunities it presents for the analysis of 3′ UTR function. The approach uncouples analysis of function from RNA binding; because multi-protein complexes appear to be common in 3′ UTR regulation, regulatory activity may not reside in the same protein that contacts the mRNA. Moreover, as illustrated by our studies of Pab1p, proteins placed at wrong locations can still function, and vital functions can be analyzed without the complications of inviability.
Materials and methods
Yeast strains
Yeast strains used in this study; yN218 (MATa, rpb1-1, ura3-52, his 3, leu2) (Nonet et al. 1987), yJD5ts (MATα, rpb1-1, ura3-52, his4-38, leu2-1, trp1-Δ) (gift of Dr. Allan Jackson, University of Massachusetts), and yAS320 (MATa, pab1::HIS3, pPAB1/URA3/CEN, his3, ura3, leu2, ade2, trp1) (gift of Dr. Alan Sachs, University of California at Berkeley).
Plasmid construction
RNA plasmids
RNA reporters were transcribed in vivo under the control of the GAL promoter on CEN plasmids. The reporter RNA in which the MS2 recognition elements are inserted into the natural MFA2 3′ UTR was created by inserting the BglII fragment of pLTAR17 (Bardwell and Wickens 1990) into to the BglII site of pRP688 (Muhlrad and Parker 1992), creating both sense (pMM2-1) and antisense (pMM2-1a) MFA2/MS2 RNA reporters. In the mRNAs produced from these plasmids, the MS2 elements are inserted upstream of the MFA2 instability elements, at a position known to have little effect on stability (Muhlrad and Parker 1992).
To create an RNA reporter that lacks poly(A) (Fig. 4), the natural MFA2 3′ UTR in the MFA2/MS2 reporter was deleted and replaced with the HDV ribozyme. This construct was generated by inserting the Acc65I–SmaI fragment of pMM2-1 into the SnaB1–StuI site of pB2RQ39 (Quadt et al. 1995) creating pMM2-6.
For studies of mRNA surveillance (Fig. 5), two MS2 sites were placed into the 3′ UTR of the PGK1 transcript containing a nonsense mutation at position 103. This was accomplished by cloning the BglII fragment of pMM2-1 into the ClaI site of pRP610 (Muhlrad and Parker 1994). This plasmid construct is called pPM2-2.
Translational effects on stability (Fig. 6) were tested by cloning MS2 sites into the 3′ UTR of the MFA2 transcript bearing a 5′ UTR stem-loop structure. This plasmid was constructed by digesting pMM2-1 with BamHI and inserting this fragment into the BamHI site of pRP456 (Beelman and Parker 1994; Beelman et al. 1994) showed that this stem–loop prevents translation of the MFA2 transcript. This plasmid construct is called pMM2-3
Fusion proteins
Each chimeric polypeptide was fused to the carboxyl terminus of the MS2 coat protein and expressed in vivo under control of the ADH1 promoter on high-copy plasmids carrying the 2μ plasmid origin of replication. Expression in yeast was achieved by cloning each fusion protein into pANA-1, a yeast expression vector based on Yeplac181 (Gietz and Sugino 1988). The MS2–Pab1p fusion was created by amplifying the gene for MS2 coat from pLexA–MS2 (SenGupta et al. 1996) by use of Pfu polymerase (Stratagene) and the primers 5′-caggtcatatgggtccgcgggcttctaactttactcagttcgtt-3′ and 5′-tgctactcgagggcgctagcgtagatgccggagtttgctgcgat-3′. The MS2 coat protein PCR product was then inserted into the NdeI–XhoI site of pET-15b (Novagen) generating pET–MS2. Pab1p was then fused in-frame to MS2 by moving the BamH1 fragment of pAS1p68 (Sachs et al. 1987) into the BamHI site of pET–MS2, generating pMS2–YPAB. Expression of MS2/Pab1p in yeast was achieved by cloning the XbaI–SpeI fragment of pMS2–YPAB into the NheI site of pANA-1, creating pA–MPII.
The MS2/MS2 coat protein dimer was created by PCR amplification of MS2 coat and fusing in-frame with a second copy of the MS2 gene in pLexA–MS2. The resulting MS2 intramolecular dimer was then transferred into pANA-1 containing a NcoI–XhoI polylinker. This plasmid is called pD–MS2.
The MS2–Sxl protein fusion (pA–Sxl) was created by amplifying Sxl from plasmid pGEX–TEV–Sxl (Valcárcel et al. 1993) by use of oligonucleotides 5′-atgtacggcgctagcaatccgggtagtaacaataataatggtgg-3′ and 5′-tcagataaagctagcagcatcgaaatagggatgcgagttttggaagcg- 3′ and inserting the resulting Sxl ORF into the NheI site of pA–MPII followed by XhoI–DraIII liberation of Pab1p. This plasmid is called pA–Sx1.
Pab1p deletions
Each Pab1p deletion was expressed under control of the ADH1 promoter on the high-copy, 2 μ plasmid, pANA-1. The Δ1 MS2/Pab1p deletion was created by subcloning the BamHI fragment of pAS1p68 into pGEM7z (Promega) creating pPAB7z. RRM4 and the carboxyl terminus were then deleted by digesting pPAB7z with BamHI–BalI. The deletion was then fused to MS2 by transferring the BamHI–AatII fragment into pD–MS2. The Δ2 MS2/Pab1p deletion was created by removing RRM1-3 by HindIII digestion of pPAB7z followed by transferring the BamHI–AatII fragment into pD-MS2. The Δ3 deletion was created by PCR amplification of RRM3-4 plus the carboxyl terminus with the primers 5′-ccagaaaggctagcgactctcaattggaagagac-3′ and 5′-gggaagtaggtgatgctagcgagcattaagcttgc-3′, followed by insertion into pD–MS2. The Δ4 deletion was also created by PCR amplification using the primers 5′-gcgccctcgaggatcccgctgaacaattgg-3′ and 5′-ttaccagctgcttctgctagcgaagtcttagcag-3′, followed by insertion into pD-MS2. These plasmids are called pD–PABΔ1, pD–PABΔ2, pD-PABΔ3, and pD–PABΔ4.
MS2/Pab1p complementation
Plasmid shuffle experiments were performed as described in Sachs et al. (1987). Yeast strain yAS320 containing a pab1 deletion and a plasmid expressing both PAB1 and URA3 gene, was transformed with pA–MPII, pANA-1, pD–MS2, or pA–SXL. Transformants were plated on selective medium and then grown overnight in liquid culture. Cells were then plated onto selective medium containing the drug 5-fluoro-orotic acid. Plates were incubated at 30°C for 3 days.
RNA preparation and analysis
Vectors containing the various reporter mRNAs were transformed into strains harboring a temperature-sensitive allele of RNA polymerase II (rpb1-1). Transcriptional pulse-chase analysis was performed as described previously (Herrick et al. 1990; Muhlrad and Parker 1992; Decker and Parker 1993), with the exception that cells in Figure 4B and C were grown in synthetic medium supplemented with 2% galactose and 2% sucrose.
RNA purification and Northern analysis were performed as described in Forrester et al. (1992), with the exception that the RNA from ribozyme-containing strains was prepared as described by Donahue and Fedor (1997) to ensure that self cleavage did not occur during RNA isolation. For Northern analysis, 8–10 μg of total RNA was loaded onto either a 1.5 % formaldehyde agarose gel (Figs. 2, 4–7) or a 4% polyacrylamide/urea gel (Fig. 3). All Northerns were probed with 32P-labeled RNA complementary to the MS2-recognition elements transcribed from vector pMS2-2 (SenGupta et al. 1996). Results were quantitated and standardized to 18S ribosomal RNA, by use of a PhosphorImager and ImageQuant Software (Molecular Dynamics).
Poly(A) status of the ribozyme-containing MFA2/MS2 mRNA was determined by oligo(dT) cellulose chromatography (Cleaver et al. 1996) and S1 nuclease mapping (Goldrick et al. 1996). The S1 nuclease probe was generated by 3′ end labeling an oligonucleotide complementary to junction between the MFA2 3′ and HDV ribozyme, by use of terminal deoxynucleotidyl transferase (Promega), and [α32P]cordycepin-5′ triphosphate.
Polysome analysis was performed as describe din Snyder (1989) with the following modifications: Cells were transformed with either the normal MFA2 reporter, or either MS2/Pab1p or MS2/MS2, and then grown in synthetic medium supplemented with 2% galactose and 2% sucrose. Cells were harvested at mid-log phase and polysomes were extracted.
Extracts for gel retardation were prepared as described in Snyder (1989) with the following exceptions. Cells were pelleted, washed, and lysed in 24 mm HEPES (pH 7.6), 150 mm potassium acetate, 1.5 mm MgCl2, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 2 μg/ml chymostatin, 10 μg/ml aprotinin, 17.5 μg/ml PMSF, and 5% (wt/vol) glycerol. RNA probe and competitor were generated with T7 RNA polymerase from plasmids MSA–CAT and MSCU–CAT (Stripecke and Hentze 1992) after digestion with Xba I as described (Gray et al. 1993). Extracts were pretreated on ice for 5 min with 1 mm acetic acid and 1 mm DTT. Binding reactions were performed on ice for 1 hr in a buffer containing 200 mm Tris (pH 8.5), 160 mm potassium chloride, 20 mm magnesium acetate, and 160 μg/ml BSA. Where indicated, unlabeled competitor RNAs were added prior to the addition of the 32P-labeled probes. Heparin (final concentration 0.5 mg/ml) was added and followed by a 10 min incubation. RNA/protein complex formation was analyzed by nondenaturing gel electrophoresis and autoradiography.
Acknowledgments
We thank members of the Wickens laboratory for helpful discussions and comments on the manuscript. We appreciate strains and plasmids provided by Roy Parker and his colleagues (University of Arizona), as well as their willingness to communicate results prior to publication and to discuss the problem and approach. We thank Dan Celander for providing antibodies to MS2 coat protein. We also thank Paul Alhquist, Matthias Hentze, Allan Jacobson, Alan Sachs, and Richard Young for plasmids and strains. The University of Wisconsin Biochemistry Media Laboratory was helpful in preparing figures. This work was supported by National Institutes of Health grant GM50942 to M.W. and a Wellcome International Prize Travelling Research Fellowship to N.K.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL wickens@biochem.wisc.edu; FAX (608) 265-2603.
References
- Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell V, Wickens M. Purification of RNA and RNA-protein complexes by an R17 coat protein affinity method. Nucleic Acids Res. 1990;18:6587–6594. doi: 10.1093/nar/18.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C, Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- Beelman C, Stevens A, Caponigro G, Lagrandeur T, Hatfield L, Fortner D, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Bernstein P, Peltz S, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C, Matunis E, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- ————— Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Shyu AB. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen C, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte macrophage colony-stimulating factor transcripts: Different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver O, Patterson K, Krieg P. Isolation of poly-A+ RNA. In: Krieg P, editor. A laboratory guide to RNA. New York, NY: Wiley-Liss; 1996. pp. 51–63. [Google Scholar]
- Deardorff J, Sachs A. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- Decker C, Parker R. A turnover pathway for both stable and unstable mRNA in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Donahue C, Fedor M. Kinetics of hairpin ribozyme cleavage in yeast. RNA. 1997;3:961–973. [PMC free article] [PubMed] [Google Scholar]
- Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: Possible coupling of mRNA processing and chromatin structure. Genes & Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- Gallie D, Tanguay R. Poly(A) binds to initiation factors and increases cap-dependent translation in vitro. J Biol Chem. 1994;269:17166–17173. [PubMed] [Google Scholar]
- Gietz R, Sugino A. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goldrick M, Kessler D, Winkler M. Analysis by nuclease protection. In: Krieg P, editor. A laboratory guide to RNA. New York, NY: Wiley-Liss; 1996. pp. 105–131. [Google Scholar]
- Gray NK, Quick S, Goossen B, Constable A, Hirling H, Kühn LC, Hentze MW. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase: A functional assay for translational repression and direct demonstration of the iron switch. Eur J Biochem. 1993;218:657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- Harada K, Martin S, Frankel A. Selection of RNA-binding peptides in vivo. Nature. 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- Hatfield L, Beelman C, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D, Ross J. The half-life of c-myc mRNA in growing and serum-stimulated cells: Influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol Cell Biol. 1994;14:2119–2138. doi: 10.1128/mcb.14.3.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevesiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Carmichael G. Role of polyadenylation in nucleoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz S. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′ end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- Kessler S, Sachs A. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Pieler T. Xenopus poly(A) binding protein: Functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- Lowary P, Uhlenbeck O. An RNA mutation that increases the affinity of an RNA-protein interaction. Nucleic Acids Res. 1987;15:10483–10493. doi: 10.1093/nar/15.24.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell J, Rudner D, Sachs A. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes & Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker P, Keller W. The major yeast poly(A) binding protein functions in pre-messenger RNA 3′-end formation. Proc Natl Acad Sci. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes & Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- ————— Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker C, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′–3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- ————— Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietfeld W, Mentzel H, Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes & Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A, Davis R. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A, Bond M, Kornberg R. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: Domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sachs A, Davis R, Kornberg R. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Sengupta D, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA–protein interactions in vivo. Proc Natl Acad Sci. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. Yeast SPA2 protein localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripecke R, Hentze M. Bacteriophage and spliceosomal proteins function as position-dependent cis/trans repressors of mRNA translation in vitro. Nucleic Acids Res. 1992;20:5555–5564. doi: 10.1093/nar/20.21.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S, Sachs A. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J, Singh R, Zamore P, Green M. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362:171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: Messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- Witherell G, Gott J, Uhlenbeck O. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acids Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]