Abstract
RNAi technology has brought a new category of treatments for various diseases including genetic diseases, viral diseases, and cancer. Despite the great versatility of RNAi that can down regulate almost any protein in the cells, the delicate and precise machinery used for silencing is the same. The major challenge indeed for RNAi-based therapy is the delivery system. In this review, we start with the uniqueness and mechanism of RNAi machinery and the utility of RNAi in therapeutics. Then we discuss the challenges in systemic siRNA delivery by dividing them into two categories--kinetic and physical barriers. At the end, we discuss different strategies to overcome these barriers, especially focusing on the step of endosome escape. Toxicity issues and current successful examples for lipid-based delivery are also included in the review.
Keywords: lipid, liposome, nanoparticles, siRNA, systemic delivery, endosome escape
2. Introduction
2.1 Discovery and uniqueness of RNAi
Post-transcriptional gene silencing by RNA interference, popularly known as RNAi, is a phenomenon that is exploited by cells to conduct gene regulation. This was first observed in plants by Napoli et al. [1]. The mechanism of gene suppression was finally established by Fire et al. [2] with the demonstration that when short stretches of 23mer to 25mer nucleotide sequence of double stranded RNA molecules were injected in C .elegans, complete degradation occurred of mRNA that was complementary to one of the strands of the injected double stranded RNA. This type of gene silencing, though initially shown to be a transient phenomenon, was later shown to be of a more stable type when short hairpin RNA containing the complementary feature of the double stranded RNA could be expressed in organisms as varied as C. elegans, Drosophila and plants [3–5]. The initially used double stranded RNA is now called siRNA and the later synthesized single stranded short hairpin RNA are called shRNA.
The uniqueness and impact of this discovery can be gauged from the fact that the initial discovery was done in the 1990s, the mechanism established by Fire et al. [2] in 1998, “breakthrough of the year” declared by the Science magazine in 2002 and the Nobel Prize for Medicine or Physiology awarded to its discoverers, Andrew Fire and Craig Mello, in 2006. What distinguishes this mode of gene silencing from the already established anti-sense mode of gene silencing is that though in both the methods the sequence complementarity feature on the mRNA is utilized for target recognition, the RNAi mechanism has a catalytic component incorporated into it, in the sense that a single siRNA/ shRNA molecule can silence or destroy thousands of copies of mRNA molecules. This machinery is extremely efficient as compared to the anti-sense technology, where a single anti-sense nucleic acid molecule can bind to and thereby silence only a single mRNA molecule.
2.2 Mechanism of action
In mammalian cell, siRNA is generated by the cleavage of larger double-stranded RNA (dsRNA) precursors by the RNAse III endonuclease Dicer [6]. Dicer is complexed with the TAR-RNA binding protein (TRBP) and hands off the siRNA to the RNA-induced silencing complex (RISC). RISC contains the protein that carries out the silencing activity by cleaving the target mRNA molecule between bases 10 and 11 relative to the 5’ end of the antisense siRNA strand. In the core of RISC lies the Argonaute (Ago) family members and in humans, Ago-2 carries out the catalytic cleavage activity [7, 8]. It has been observed that although the siRNA transferred to RISC are double stranded, Ago-2 cleaves and releases the “passenger” strand, leading to an activated form of RISC that contains a single-stranded “guide” RNA molecule. It is this guide RNA molecule that confers the specificity to the RISC and helps it in target recognition by intermolecular base pairing [9]. The selectivity of strand loading into RISC is governed by differential thermodynamic stabilities of the end of the siRNA [10, 11]. The less thermodynamically stable end is chosen for unwinding of the 5’ end of the guide strand and this binds to Ago-2. Ago-2 is composed of three domains: PAZ, MID, and PIWI domains. The function of PAZ and MID domains are docking and anchoring the RNA. PIWI domain is the one with slicer activity [12]. Messenger RNA molecule that displays perfect or near-perfect complementarity to the guide RNA are recognized and cleaved by Ago-2. Partial complementarity between a siRNA and target mRNA may in some cases repress translation or destabilize the transcripts if the binding mimics microRNA (miRNA) interactions with target sites. miRNA is actually the endogenous substrates for the RNAi machinery. It is initially expressed as long primary transcript (pre-miRNA) which is processed within the nucleus into 60–70bp hairpin by the microprocessor complex that consists of Drosha-DGCR8 [13, 14]. Finally, the loop is removed by further processing in the cytoplasm by the RNAse III Dicer and only one of the two strands is loaded into RISC in the cytoplasm. The mature miRNA can share only partial complementarity with sequences in the 3’UTR of target mRNA. The primary mechanism of action of miRNA is translation repression, although this can be accompanied by message degradation [15].
3. Utility of RNAi in therapeutics
It is possible to exploit this native gene silencing pathway for regulating gene(s) of choice. There are a number of methods for doing so. If a siRNA effector molecule with one strand having a complementarity match with a target nucleotide sequence is delivered into a cell, it can activate RISC directly and can result in specific silencing of the targeted mRNA. It is the reason why the procedure has become the method of choice for silencing specific gene expression in mammalian cells. Control of disease-associated genes can therefore make RNAi an attractive choice for future therapeutics. It can be envisaged that every human disease is caused by activity from one or a few genes and some of these genes should be amenable for RNAi intervention. The list of such disease includes cancer, cardiovascular and autoimmune disorders, dominant genetic disorders and viral infections. Moreover, the fact that miRNA can function both as tumor repressor and oncogene, endogenous miRNA can also become therapeutic target [16]. Although RNAi therapy is of great potentials, the technology still has one major limitation--it can only achieve “lost of function” therapy. If the disease is caused by loss function of genes, the strategy should be introducing gene expression by virus or plasmid. Thus, the traditional gene therapy approach should be complementary to the RNAi approach.
In addition to the activities in utilizing the promise of RNAi technology in creating gene knockouts by stable transfection of short hairpin RNA (shRNA) [17, 18] or cre-lox mediated knockdown of genes in transgenics [19], there has been an increasing amount of efforts in harnessing this powerful technology for therapeutic purposes. But in order to become a therapeutic modality, certain issues of drug development have to be addressed. First of all, the drug should be manufactured at reasonable cost and be suitable for administration in a safe and convenient manner. Second, it must be stable and accumulate to an effective level in the target cell. Third, it must be restricted in its effect on or within the target cell. Finally, it must not pose any toxicity to other cells. Keeping these fundamental issues in mind, some of the areas where siRNA can be used as a potent therapeutic molecule are as follows:
3.1 Genetic Diseases
There is a big promise that siRNA can be successfully used to treat a large number of genetic diseases. Preliminary studies have revealed that single nucleotide polymorphism in mutant allele transcripts can be used as effective targets for RNAi [20, 21]. One of the challenging targets for siRNA based therapeutics is the disease causing polyglutamine proteins encoded by the CAG repeat containing transcripts. Such repeats are found in several neurological diseases. But the problem is that the repeats are also found in many normal transcripts as well. As a result, they are difficult to be selectively targeted by siRNA. Alternatively, single nucleotide polymorphisms (SNP) are very often found in mutant allele transcripts and represent potential selective targets. The challenge is therefore to find a siRNA/SNP combination that is highly selective. This has been accomplished by systematic analyses of siRNA in which the polymorphic nucleotide is complementary to the mid region of the siRNA. In some cases, the siRNA directs selective degradation of only the mutant transcripts, leaving the wild type transcripts intact despite having only a single mismatch with the wild type sequence [20, 21]. An additional application of siRNA targeting a SNP has been reported in the studies of amyotrophic lateral sclerosis (ALS) [22]. This disease is caused by mutations in the Cu, Zn superoxide dismutase (SOD1) gene. Recently, siRNA has been systematically tested for their ability to discriminate wild type from mutant alleles for the SOD1 and the huntingtin (HTT) gene [23]. These studies have supported the notion that single nucleotide polymorphisms may indeed suffice to make mutant specific siRNA if mismatches are placed rationally within their sequence. Particular purine-purine mismatches at positions 10 and 16 relative to the 5’ end of the guide strand can provide selectivity. Since the wild type SOD1 performs important functions it is important to selectively eliminate the expression of only the mutant allelic transcript. Based on the above studies, it has been possible to achieve selective degradation of a mutant allele encoding SOD1, thereby providing a potential therapeutic application for the treatment of ALS. Furthermore, since siRNA and viral vectors encapsulating siRNA can be technically delivered to affected regions of the brain [24], the promise of clinical use of RNAi for treatment of degenerative, neurological diseases is expected to become a reality soon.
3.2 Viral Diseases
Initial application of the RNAi technology had been directed in the treatment of hepatitis B [25]. A significant knockdown (99%) of the hepatitis B virus (HBV) core antigens could be achieved in liver hepatocytes by using shRNA directed against them.
Hepatitis C virus (HCV) infects an estimated 3% of the world’s population and is a major cause of chronic liver disease, leading to the development of liver cirrhosis and hepatocellular carcinoma. The HCV genome is a positive-strand RNA molecule with a single open reading frame encoding a polyprotein that is processed post-translationally to produce at least ten proteins. The efficacy of siRNA mediated inhibition of virus replication has been studied in a number of replicon systems [26–28]. One of the encouraging results has been the one where siRNA has been targeted against the internal ribosomal entry site (IRES) and against non-structural protein NS3 and NS5b [27]. Similarly, anti-HCV siRNA has been shown to cure Huh-7.5 cells bearing persistently replicating HCV replicons [26]. In another study, both synthetic and Pol III promoter expressed anti-HCV siRNAs have been shown to direct efficient cleavage of HCV sequences in a HCV-luciferase fusion construct in vivo in mouse hepatocytes [29]. In a different in vivo study, siRNA has been used to treat fulminant hepatitis induced by an agonistic Fas-specific antibody in mice. Delivery of anti-Fas siRNA resulted in a survival rate of 82% in the treated mice after 10 days of observation, whereas the un-treated control mice died within 3 days [30].
HIV is another infectious agent that can be targeted by RNAi. This may be because of the fact that the life cycle of HIV is well understood together with its pattern of gene expression. Synthetic and expressed siRNAs have been used to target a number of early and late HIV-encoded RNAs and these include the TAR element [31], tat [32–34], rev [32, 34], gag [35, 36], env [36], vif [31], nef [31] and reverse transcriptase [34]. Cellular cofactors, such as NF-kb [34], the HIV receptor CD4 [35] and co-receptors CXCR4 and CCR5 [37] have also been successfully down-regulated by RNAi resulting in an inhibition of HIV replication. Moreover, inhibition of HIV replication has been achieved in numerous human cell lines and primary cells including T lymphocytes and hematopoietic stem cell derived macrophages [32, 34, 37–40].
Despite the success of in vitro RNAi-mediated inhibition of HIV-1, for future clinical applications, targeting the virus directly represents a substantial challenge since the high viral mutation rate will certainly lead to escape of mutants [41]. RNAi-mediated down-regulation of cellular co-factors required for HIV infection is an attractive alternative or complementary approach. One such target that holds particular promise is the macrophage-tropic chemokine (C-C motif) receptor 5 (CCR5) [42] that HIV uses as the most important co-receptor during the infection. CCR5 is non-essential for normal immune function. Individuals homozygous for a 32 bp deletion in this gene show resistance to HIV infection and individuals who are heterozygous for this deletion have delayed progression to AIDS [43, 44]. Based on this, Pol III expressed anti-CCR5 shRNA have been used to knocking off the expression of the gene in human lymphocytes [45]. Down-regulation of CCR5 resulted in a somewhat modest, but nevertheless significant 3 to 7 fold reduction in viral infectivity relative to controls. Despite this down-regulation, the anti-CCR5 shRNA treated cells were still susceptible to infection by T-tropic CXCR4 utilizing virus. This emphasizes the need to combine several shRNA against co-receptors and the virus in order to achieve a potent inhibition of HIV-1. Clinical trials have been undertaken where T lymphocytes from HIV infected individuals were utilized and transduced ex vivo with a lentiviral vector that encoded an anti-HIV antisense RNA. The transduced cells were subsequently expanded and reinfused into patients [46, 47]. This type of therapeutic approach is certainly applicable to vectors harboring genes that encode siRNA. A different approach is to transduce isolated hematopoietic progenitor or stem cells with the vector. This approach has the advantage that all the hematopoietic cells capable of being infected by the virus are derived from a multi-potent stem cell that can be transduced with an appropriate viral vector. Hematopoietic stem cells can be mobilized from the patients and transduced ex vivo prior to reinfusion.
3.3 Cancer
The use of RNAi for cancer therapeutics has the potentiality of revolutionizing the treatment of this devastating disease. The challenges for cancer are not dissimilar to those faced by other diseases. In most cases, mouse xenograft models have been used in studying the effect of the RNAi treatments [48]. In one case, shRNA targeting Hec1 has been delivered to show efficacy for treating adenocarcinoma-induced tumor growth [49]. Generally, RNAi mediated cancer therapies include: 1. Inhibit angiogenesis via inhibiting VEGF-signaling [50–55]; 2. Inhibit tumor survival and induce apoptosis [56] via inhibiting Wnt pathway [57, 58], NF-κB pathway [59, 60], EGFR [61], Her-2/ neu [62], telomerase [63, 64], MDM2 and p53 [65, 66], or Bcl-2 [67]; 3. Enhance radio- or chemo-sensitivity by inhibit MDR1 [63, 64]. Combination siRNA therapy that inhibits multiple pathways shows great anti-tumor effect and could be a novel approach for anti-cancer therapy [68, 69].
4. Delivery of RNAi
4.1 Challenges in delivery
For a successful siRNA delivery, the final destination is the cytoplasm of the target cells. Although this is one barrier (the nucleus membrane) less than delivering plasmid, it still remains a great challenge. The first concern is the stability of siRNA itself. Naked siRNA is prone to degradation and has a half life less than five minutes in plasma [70]. Their relatively small size also leads to rapid clearance by kidney filtration after systemic injection. Together with the negatively charged nature of siRNA that prevent it from having association with cell membrane, it’s unlikely to simply use siRNA itself as a drug formulation. When formulate into a delivery system, the stability of the delivery formulation is another issue and it also needs to escape from the non-specific uptake by the reticuloendothelial system (RES), especially the Kupffer cells in the liver and the macrophages in the spleen. Even if the siRNA stays in the blood circulation long enough, the siRNA formulation has to extravasate and gain access to the target tissue if the target is not blood cells or blood vessel cells. All the above is considered as the “kinetic barriers” which prevent the siRNA formulation from having “the access to the target cells”. The other category of barriers, i.e., the “physical barrier”, is the hurdles that impede “the access to the cytoplasm”. First, the siRNA needs to get across the cell membrane. The most commonly used delivery strategy for macromolecules including siRNA is to take advantage of the receptor-mediated endocytosis. Second, endocytosed siRNA will need to escape the endosome or caveosome to reach the cellular cytoplasm where the RISC locates.
In the case of expressing shRNA in the target cells by delivering a plasmid, plasmid vectors [71–75] or viral vectors [76] are commonly used. Plasmid vectors have been successfully used in mammalian cells in vitro by simple transfection. Plasmid vectors need to cross the nuclear membrane for transcription. This is not a serious problem for cells actively growing in culture, because the nuclear membrane disappears during mitosis. For primary cells that are usually not actively dividing, delivering siRNA is more favorable than the plasmid. Viral vectors that can transduce non-dividing cells [77, 78] are usually used for introducing shRNA in primary cells. Lentiviral and other retroviral vectors have been used extensively for stable expression of shRNA due to their ability to insert into the host genome. Adeno associated virus is non-pathogenic and can also achieve long term gene expression. Adenoviruses is known for its robust expression and herpes simplex virus is useful for its neuron-tropism [79]. However, immunogenicity and other safety issues are still great concerns of using viral vectors in human. On the other hand, molecular encapsulation of siRNA by non-viral vectors such as liposomes and other nanoparticles are preferred approach because of their low immunogenicity and high biocompatibility.
4.2 Strategies in lipid-based systemic siRNA delivery
There are many different types of non-viral delivery strategies. They include physical methods, such as hydrodynamic injection [80–82], particle bombardment [83] and electroporation [84–86], and chemical methods. Polymer, including proteins and peptides, mediated delivery of siRNA has been attempted by many investigators [87–90]. But this review will concentrate on the lipid-based vectors. Furthermore, only systematic deliveries will be discussed; local or topical deliveries will not be included. The following discussion is designed to address the two major barriers mentioned above, mainly using solid tumor as examples.
4.2.1 Kinetic barriers
Since safety and efficient delivery of siRNA is a necessary pre-condition for the delivery of a potential siRNA based therapy, the stability of the siRNA molecule becomes a major lingering issue [91]. In this regards, the in vivo stability of the siRNA molecule has been achieved by chemically modifying the basic RNA structure, and several methodologies are currently available for changing the backbone of the RNA molecule. It can not only lead to enhanced in vivo stability but also reduce in the dose of siRNA that is typically required for achieving a desired therapeutic effect. The strategies involve substituting the 2’-OH group on the ribose ring with –F, -O-Me, or simple –H groups to improve the stability of siRNA [92]. It has been observed that this type of selective substitution of the 2’-OH group does not reduce the efficiency of the RNAi effect [93]. Yet another modification is a phosphorothioate modification of the RNA backbone [70]. It has also been shown that linking a cholesterol molecule to the 3’ end of the sense strand of a siRNA molecule can suppress the siRNA degradation in a cell culture system [29, 80, 94]. This type of chemical modification has also been used widely on aptamer research. In fact, the most simple siRNA delivery design is the one that simply links siRNA with a RNA aptamer targeting prostate specific membrane antigen (PSMA) [95]. But the in vivo anti-cancer effect was only done with intra-tumor injection. Until now, no successful systemic delivery using the aptamer-siRNA chimera has been reported. It’s also worthy to note that in one study, simultaneous application of siRNA and aptamers against the transcription factor NF-κB could produce 90% knockdown, whereas either modality alone could only produce 60% inhibition [96]. Since aptamers bind and inhibit signaling protein in a different mechanism from that of RNAi, it provides another strategy for combination therapy with RNAi.
Cationic lipid formulations such as Lipofectamine 2000 [97, 98] or cardiolipin analogs [99, 100] have been widely used for in vitro plasmid DNA or siRNA delivery. By charge complementarities, negatively charged nucleic acids and positively charged lipids spontaneously form nanoparticles (called lipoplex) of 50 nm to 200 nm in diameter [101-103]. With optimal N/P (nitrogen to phosphate, or +/−) ratio (usually around 2 or 3), the complex has an overall positively charged surface and thus favors their interaction with cell membrane. Lipid-mediated siRNA transfection is at least 1000 fold more efficient than that of naked siRNA [98].
Though the information on the bio-distribution of the complex of siRNA with cationic liposomes is rather scarce [104–106], it depends on the colloidal properties of the complex as well as their interaction with blood components. Surface charge of the complex plays an important role in determining the final destination of the complex. Complexes with a strong anionic charge are usually absorbed by scavenger cells, resulting in a rapid elimination of the injected dose from the blood. A strong positive charge can also create problems. For example, following intravenous administration in mice, 60% of the dose of plasmid DNA delivered as a complex with cationic liposome accumulated in the liver. However, the level of transgene expression per microgram of DNA taken up in the tissue was a 1000-fold lower in the liver than in the lung. The decreased transgene expression in the liver may be due to the rapid degradation of DNA following phagocytosis of the lipoplex by the Kupffer cells [107]. This phenomenon falls into the same category of delivery problem for other colloidal carriers, i.e. strong uptake by the reticuloendothelial system (RES), which include the Kupffer cells in the liver [108]. The reason why unprotected lipoplex is prone to RES uptake is because opsonins such as IgM, IgG, fibronectins, or complement C3 attaching to the bare surface of lipoplex can attract phagocytic cells. One way to deal with this problem is to use stealth liposome strategy with surface grafted carbohydrate or polyethylene glycol (PEG) for protection and shielding the surface charge [109]. PEGylation of liposomes can increase the circulation half life of the i.v. injected liposomes to 6–10h in mice and to about 40h in human [110].
In a recent study, lipid-mediated delivery of a siRNA against apolipoprotein B (apoB) has been used to target apoB mRNA in the liver. Administration of the apoB-specific siRNA without a formulation or chemical conjugation showed no in vivo silencing activity even at doses higher than 50 mg/kg [92]. However, more than 80% silencing of apoB mRNA and apoB-100 protein could be achieved with a single 1 mg/kg dose of lipid-formulated siRNA in a non-human primate [104]. The lipid formulation in this case contained the lipids 3-N-[(ω-methoxypoly(ethylene glycol)2000) carbamoyl]-1,2-dimyristoyloxy-propylamine (PEG-C-DMA), 1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane, (DLinDMA), 1,2-distearoyl-sn-glycerol-3-phosphocholine (DSPC) and cholesterol, in a 2:40:10:48 molar ratio. A major fraction of the siRNA delivered intravenously using this vector accumulated in the liver, followed by spleen and small intestine. How much of the liver uptake was due to the hepatocytes and how much due to the Kupffer cells was not known.
For delivering siRNA to solid tumor cells, it is important to review the work of Matsumura and Maeda [111]. They discovered that macromolecules and colloidal nanoparticles could accumulate in the solid tumor due to the leaky neovasculature of the tumor [112–115]. Accumulation of the Stealth liposomes [61] and other PEGylated colloidal systems [116, 117] in the tumor due to the so called Enhanced Permeability and Retention (EPR) effect is also contributed by the fact that tumor lacks the lymphatic drainage like the normal tissues [111, 118, 119]. It must be remembered that the degree of leakiness of the neovasculature varies among different tumors [120]. Although the EPR effect has been demonstrated in human [121–124], not all human tumors are equally leaky. For tumors with a not-so-leaky neovasculature, nanoparticles with small size, e.g. less than 30 nm in diameter would be desirable. Unfortunately, most of the self-assembled nanoparticles, lipid-based or polymer-based, are larger than this size. Formulations with a small particle size may penetrate the liver fenestrae and promote the uptake by the hepatocytes [125]. To provide improved capacity and protection to siRNA, advanced core/shell lipid–based nanoparticles (lipopolyplex) have been designed. The core consists of DNA (with or without siRNA) condensed by a polycation by polyelectrolyte coacervation. The first such example was the LPD (liposome-polycation-DNA) nanoparticles designed in our lab in the mid 90s [126]. To deliver siRNA, we have used FDA approved protamine, a cationic polypeptide, to condense siRNA with the help of the high molecular weight calf thymus DNA. The self-assembled LPD nanoparticles were further modified by post-inserting a PEG-phospholipid conjugate for surface protection [127]. Taking advantage of the EPR effect, the PEGylated LPD nanoparticles can accumulate a high dose (60–80% injected dose per g of tissue) in the H460 lung cancer xenograft model [61]. Such very high tumor accumulation was not ligand dependent, because particles with or without a targeting ligand, anisamide in this case [128], the tumor uptake of the encapsulated siRNA was about the same [61]. However, due to the presence of the sigma receptor on the H460 tumor cells, nanoparticles with anisamide were internalized by the tumor cells. Nanoparticles without a targeting ligand stayed outside the cells and accumulated in the interstitial fluid space in the tumor. siRNA delivered intracellularly by the anisamide targeted LPD nanoparticles almost completely silenced the target gene after three daily injections [61]. Although the result is already promising, confocal microscopy of tumor sections revealed that most of the fluorescently labeled siRNA were still retained in the endosomes in the tumor cells and were thus not bio-available [61]. Gene silencing could be accomplished by injecting a lower dose if the formulation can efficiently release the encapsulated siRNA from the endosomes to the cytoplasm. This subject is discussed in greater detail in the following section. In a similar approach, a plasmid DNA encoding a short hairpin RNA (shRNA) condensed by poly-L-lysine (PLL) into a Multifunctional Envelope type Nano Device (MEND) has been developed by the Harashima group [129]. This system produced 96% inhibition of the marker luciferase gene expression in the target cells.
It is important to mention that the LPD formulation contained 10 mol % surface grafted PEG-phospholipid conjugate [61]. It is known that lipid bilayer can maximally tolerate only about 5–6 mol % of PEGylation. This is due to the detergent-like activity of the PEG-phospholipid conjugate which lyses liposomes at high concentrations. The LPD formulation could be modified with a high amount of the conjugate because the lipid bilayer on the surface of the LPD is stabilized by charge-charge interaction with the negatively charged core. The high degree of PEGylation is probably the reason why the formulation is not taken up by the liver and spleen to a significant degree [61]. The injected dose could then extravasate into the tumor via the leaky neovasculature, i.e. the EPR effect.
4.2.2 Physical barriers
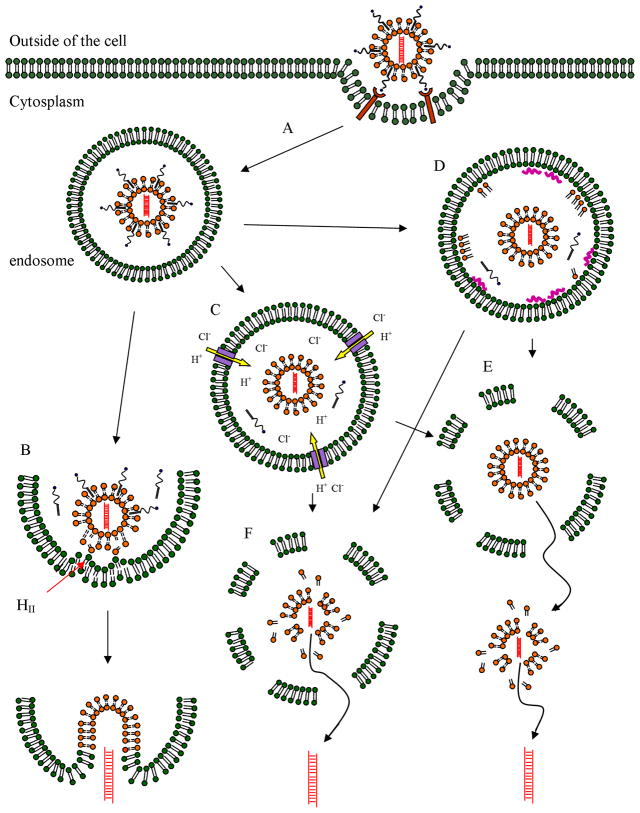
After a siRNA formulation gains access to the target cells, the next step is to cross the cell membrane and enter into their action site—cytoplasm. Different ligands such as cell penetrating peptides, proteins, antibodies (including Fab, scFv, etc.), aptamers, and small molecular weight ligands have been used for binding and triggering endocytosis. But only being endocytosed is not sufficient. In fact, the most challenging part in siRNA delivery field now is how to get the siRNA out of the endosomes intactly. This requires both endosome escape and sufficient de-assembling of the formulation. Fig. 1 shows the overall endosome escape mechanisms for lipid mediated siRNA delivery. The details will be discussed below.
Fig. 1. Endosome escape in lipoplex mediated siRNA delivery.
A. Lipoplex containing siRNA (shown as orange lipid bilayer and red siRNA) with PEG and targeting ligand on the tip (shown as blue circle) are taken up by target cell via receptor mediated endocytosis.
B. The cationic lipid of the lipoplex forms ion pairs with the anionic endosomal lipid (PEG molecules may leave the lipoplex spontaneously or under appropriate design) and can further form the inverted hexagonal phase (HII). This leads to the fusion of the lipoplex with endosomal membrane and release the siRNA into cytoplasm.
C. Lipoplex containing molecules having buffer capacity in endosomal pH range can trigger proton sponge effect that causes the influx of Cl− and swelling of the endosome.
D. Free highly positive charged molecules (shown with orange colored cationic lipid and purple colored PEI or oligo-arginine) can interact with anionic endosomal membrane and destabilize it by excluding water.
E. Intact lipoplex may escape from the ruptured endosome and de-assemble in the cytoplasm and release siRNA if the particle is not too large for the “holes” of the ruptured endosome.
F. Lipoplex may also de-assemble inside the endosome and directly release siRNA out of the ruptured endosome.
5. Endosome escape
5.1 Ion-pair formation (fusogenic property of liposomes & inverted hexagonal phase)
The mechanism by which cationic lipoplex triggers endosome release has been proposed by Xu & Szoka [130]. After endocytosis, cationic lipids form ion pairs with the anionic lipids in the endosome membrane and thus destabilize the endosomal membrane by excluding the surface bound water (Fig.2) [130]. Electrostatic interaction between cationic lipid and anionic lipid could further promote the formation of the inverted hexagonal (HII) phase as purposed by Cullis et al [131]. Lipids with small and/or less hydrophilic head group and bulky acyl or alkyl chains favor HII phase formation. Thus, cationic lipid with a C18:2 alkyl chains showed higher delivery efficiency than the one with C18:1 chains [131]. DOTAP, containing two C18:1 acyl chains, is a commonly used cationic lipid for transfection. DSTAP, a close analog of DOTAP but with two C18:0 chains, is not. HII phase is an intermediate structure when two lipid bilayers fuse with each other [132, 133]. In the process of fusion, both bilayers are destabilized. Thus, the ion-pair formation between the cationic lipids in the lipoplex and the anionic lipids in the endosome membrane not only destabilizes the endosome membrane, but also promote the de-assembly of the lipoplex. Ion-pair formation requires close contact of the lipids with opposite charges. PEGylation of liposomes or LPD may significantly reduce the interactions between the cationic lipids and the endosomal lipids due to steric hindrance. Since PEGylation of the vector is often required to overcome the kinetic barriers as discussed above, this is a dilemma in the delivery of nucleic acid therapeutics. Tunable stealth liposomes [134], cleavable PEG-lipid linker [135], or acid labile PEG molecule design could be the answers to this problem.
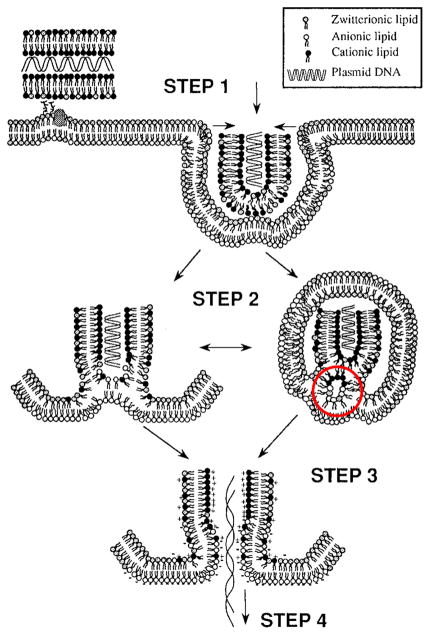
Fig. 2.
Proposed mechanism of endosome escape by lipoplex (figure taken from reference 128 with permission). When lipoplex is endocytosed (step 1), the cationic lipids can interact with the anionic membrane lipids in the early endosome by ion-pair formation (step 2). Thus, it destabilizes the endosome membrane (step 3) with the formation of the inverted hexagonal phase (HII), indicated by the red circle, as proposed by Cullis et al [131]. At the end, it causes the release of cargos (showed as plasmid DNA) to the cytoplasm (step 4).
Besides cationic lipids, other protein transduction domain such as the famous HIV-1 Tat, Drosophila Antennapedia transcription factor, herpes simplex virus type-1 VP22 transcription factor, or even just simple oligo-arginine (R8, R9) peptides also show the ion-pair effect. It is interesting to note that many protein transduction domains contain arginine, and not lysine. For example, oligo-arginine, but not oligo-lysine, shows a membrane penetration activity [136]. Sakai & Matile [137] showed that the presence of multiple guanidinium cations is important to their ion-pair formation in endosome that leads to efficient endosome escape. The reason why oligo-lysine, which also has one positive charge on each amino acid residue, does not work as well as oligo-arginine is a very interesting question. Sakai & Matile proposed that this is because a guanidinium cation contains a delocalized positive charge that can interaction with an anionic group which also contains a delocalized negative charge. It is interesting to note that naturally occurring anionic groups in biological molecules, including phosphate, carboxyl and sulfate, all contain the delocalized negative charge. Fig. 3 demonstrates the ion-pair formation between the delocalized positive charge of arginine and the delocalized negative charge of the phosphate group. Please note that in additional to the charge-charge interaction, H-bondings are also involved in the ion-pair. Note that the natural DNA condensing molecule in the sperm, i.e., protamine, is a polypeptide containing many arginines.
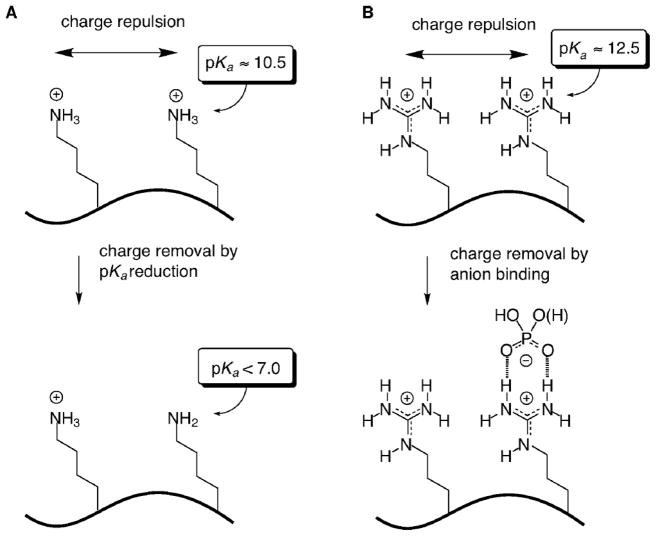
Fig. 3.
Difference between oligo-arginine and oligo-lysine in ion-pairing with negatively charged group. Oligo-arginine has delocalized positively charged guanidinium (B) groups that can interact strongly with delocalized anion phosphate groups. Note that H-bondings are also involved in the interaction. On the other hand, the positive charged ammonium groups (A) of oligo-lysine are not delocalized. Figure is reproduced from reference [137] with permission.
5.2 Proton sponge effect
Unlike lipoplex that relies on the fusogenic property of the lipid bilayer to mediate endosomal escape, polymeric carriers such as polyethyleneimine (PEI) [138–141] are supposed to use the so-called “proton sponge” effect to enhance endosomal release of the endocytosed polyplex [142–145]. According to this mechanism, the unprotonated 1°–, 2°- and 3°-amines in a polymer will show different pKa values due to crowding of the amines and will confer a buffering effect over a wide range of pH. The buffering may protect the siRNA from degradation in the endosomal compartment during the maturation of the early endosomes to late endosomes and their subsequent fusion with the lysosomes. The buffering property allows the polycation polyethyleneimine (PEI) to escape from the endosome. This is because at the typically lower pH of the endosomes, the buffering capacity of PEI causes an influx of protons, chloride ions and water into the endosomes, which eventually leads to swelling and burst the endosome due to increased osmotic pressure. PEI is not the only polymer has this buffering property, polymers containing crowded histidines (imidazoles) or morpholinos also show the same effect [146]. In fact, the problem of PEI is its high cell toxicity (especially the high MW PEI [141, 143, 147, 148]), although the purified PEI polyplex show reduced toxicity and could be useful in vivo [149].
Although Verkman et al. did a beautiful piece of work showing chloride accumulation in the endosomes containing the internalized polyplex and chloride release after osmotic swelling [145], they did not provide direct evidence showing that the polyplex itself was released to the cytoplasm. In fact, the protonation of the 2°- and 3°-amines in PEI may lead to even stronger association with the bound DNA or siRNA and make it more difficult to de-assemble. Boeckle et al. showed that the presence of free unbound PEI 4h after transfect with purified polyplex can increase gene expression, probably by helping polyplex release form the endosomes [150]. Thus, free PEI may form ion pair with negatively charged endosome lipids and destabilize endosome membrane by excluding the surface water just like cationic lipids as discussed above. If so, we predict that free oligo-arginine would perform a better job than free PEI for reasons mentioned above. It is likely that proton sponge effect itself might not be sufficient for endosome escape of the endocytosed DNA or siRNA. Ion-pair formation may play a more important role in this case.
5.3 De-assembly
The siRNA complex must de-assemble sufficiently in the endosome or in the cytoplasm so that the released siRNA could access the RISC complex for gene silencing. The requirement is the same in any other lipid- or polymer-based DNA/siRNA condensing formulation. If the complex or condensed core is still bind tightly in the endosome, the size of the complex may be too large for the membrane holes that generated during the endosome rupture and will not escape. Strategies like enzyme active linker, acid labile cross-linkers [151], pH-sensitive detergent [152], thermal sensitive liposomes [153], or reductive environment sensitive disulfide cross-linker [154–156] have been explored to improve efficient self de-assembly of the nanoparticles. Clever design with efficient de-assembly could also involve the release of free membrane destabilizing molecules like cationic lipid, PEI or cationic peptides (for example, R8) and thus helps endosome destabilization. Ideally, sufficient de-assembly and endosome escape should occur at the same time. If successful, the required dose for siRNA based therapy could be reduced dramatically.
6. Toxicity
6.1 siRNA-mediated immune stimulation
Toxicity is a major concern when a formulation is designed for human therapeutic purpose. siRNA duplex (generally 19–21 base pairs) was originally thought to be immunologically inert. It is sufficiently short to bypass the cell’s intrinsic anti-viral response that mediated by the recognition of PKR (dsRNA-binding protein kinase) [157], the RNA helicase RIG-I (retinoic acid-inducible gene-I) [158], and MDA-5 (melanoma differentiation-associated protein-5) [159] in the cytoplasm. But recently, it has been shown that siRNA is capable of induce the production of type I interferon (IFN-α and IFN-β) via activating of PKR [160–162] or recognition by toll-like receptor 7, 8 and 9 (TLR 7/8/9) [163–165] especially with the aid of delivery systems such as lipoplex [166]. This can further lead to the production of other inflammatory cytokines such as interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), and other pro-inflammatory chemokines [167] as well. Judge and MacLachlan et al. showed that the recognition of siRNA by innate immune system is sequence specific [168]. Some investigators misinterpreted their siRNA therapeutics results because they used a GFP siRNA as a control which showed unusually low immuno-stimulatory activity. Kleinman et al. also showed that non-targeted control siRNA could suppress angiogenesis as good as siRNAs targeting vascular endothelial growth factor-A (VEGFA) or its receptor VEGFR1 via cell-surface TLR3 [169]. Non-specific angiogenesis inhibition and immune stimulation are the toxicities which can confuse the data interpretation. It may be advantageous that siRNA itself can induce immune response as a synergistic effect in cancer treatment [170] or viral infection therapy. But it can be problematic for many other therapeutic purposes. Strategies such as selective incorporation of 2’-O-methyl uridine or guanosine nucleosides into one strand of siRNA duplex to overcome the immune response stimulated by siRNA have been reported [171] and reviewed by Judge and MacLachlan [167].
6.2 Carrier-mediated side effect (focused on lipid-based systems)
Although lipid-based drug delivery systems are far less toxic than the viral systems, they are not without overt toxicity. First, opsonins can attach to the charged surface of lipoplex as previously mentioned and thus activate the complement pathway. Studies showed that protecting the charged surface with PEG can prevent this side effect [172]. Second, in many lipoplex delivery systems, pulmonary inflammation and increased TNF-α, IL-6, IL-12, and interferon γ (INF-gamma;) have been observed [173–182]. This is mainly attributed to the unmethylated CpG motifs in the plasmid DNA. To minimize the cytokine response, strategies such as using methylated DNA [183] or non-bacterial amplification fragments [184] and incorporating anti-inflammatory molecule into liposomes have been reported [178, 185, 186]. For the LPD formulation [61], calf-thymus DNA was used as a carrier for siRNA. Calf-thymus DNA was selected because it contains fewer unmethylated CpG motifs than the bacterial plasmid DNA and was thus less immuno-stimulatory. For human use it is important to replace the DNA with other more acceptable carriers. Hyaluronic acid (HA) is a highly negatively charged natural polymer. The new Liposome-protamine-HA (LPH) nanoparticles significantly reduced the serum cytokine levels and still showed similar characteristics and siRNA delivery capabilities comparable to the previous LPD formulation [187]. Furthermore, recent data from this lab also showed that a particular cationic lipid, 1,2-dioleoyl-3-trimethylammonium propane (DOTAP) not only played the role in transfection but also stimulated dendritic cells and thus functioned as an adjuvant for vaccines [188, 189]. DOTAP at low concentration stimulated pERK which is an anti-apoptotic protein. When DOTAP was a part of the LPD formulation for siRNA, it protected the tumor cells from death (Chen et al. unpublished). The undesirable side effect of the formulation was successfully eliminated by replacing DOTAP with another cationic lipid.
7. Examples of lipid-based siRNA delivery
Although there is still much to improve, lipid-based delivery system has been successfully used in siRNA therapy. For example, Zimmermann et al. [104] developed a stable nucleic acid lipid particle (SNALP) for delivering siRNA against apoB to the liver of monkeys. Significant apoB protein, low-density lipoprotein, and cholesterol level reduction can be achieved and lasted for up to 11 days. Halder et al. used their neutral liposomal formulation to deliver anti-FAK siRNA to three different human ovarian tumor bearing nude mice. Four days inhibition of FAK in tumor tissue and up to 72% mean tumor weight reduction was observed [190]. Recently, they reported that anti IL-8 siRNA delivery could reduce the micro-vessel density and the mean tumor weight by 32% and 52% in HeyA8 and SKOV3ip1 mouse models [191]. Since their lipid formulation was neutral, it is unclear how endosome escape could take place. Morrissey et al. showed that siRNA against HBV encapsulated by the polyethylene glycol (PEG) modified liposomes could reduce HBV titer in the mouse liver [192]. Li et al. [61, 69, 193] developed a modified LPD (liposome-polycation-DNA) formulation which was PEGylated and targeted with anisamide. The formulation was very effective in delivering siRNA to sigma receptor expressing tumor cells. Specifically, MDM2, c-myc, and VEGF siRNA combination was delivered to the pulmonary metastasis model of melanoma. Significant tumor growth and inhibition was observed (Fig.4).
Fig. 4.
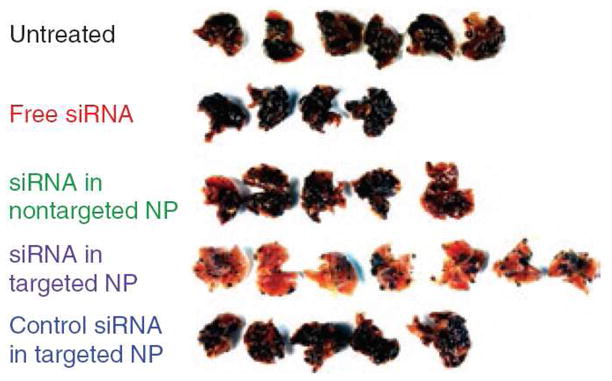
LPD nanoparticles (NP) deliver siRNA for metastatic tumor growth inhibition. Mice were i.v. injected with lung-homing B16F10 melanoma cells. After 10 days, mice were intravenously injected twice with siRNA (0.45 mg/kg, MDM2/c-myc/VEGF = 1:1:1, weight ratio). Six days later, the mice were sacrificed for examination of tumor growth. The lungs of individual mice were photographed. Only the mice injected with the targeted NP containing therapeutic siRNA showed significant reduction in tumor load. Figure is reproduced from reference [69] with permission.
8. Summary
Lipid-based drug delivery system has been widely accepted in the clinic, especially in chemotherapy, for more than two decades. Now, lipid-based delivery system also works promisingly and safely in systemic siRNA delivery. Although the recent success is encouraging, there still remains much room for improvement. If efficient tissue targeting and endosome escape can be achieved at the same time, the effective dose required for RNAi-based therapy may reduce significantly. This is important for two reasons. RNA is still an expensive drug. Lower dose would mean lower cost of manufacture. At the same time, the off-target side effect of siRNA [194, 195] would also be reduced. The challenge of siRNA delivery will remain the focus of much research in drug delivery in the near future. Acknowledgment: The original work in this lab has been supported by NIH grant CA129825. References:
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Napoli C, Lemieux C, Jorgensen R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell. 1990;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Tavernarakis N, et al. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24(2):180–3. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 4.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18(8):896–8. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 5.Smith NA, et al. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407(6802):319–20. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, et al. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 9.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30(2):106–14. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 11.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 12.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12(3):99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Martinez J, et al. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–74. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 17.Paddison PJ, et al. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–58. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2(3):243–7. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 19.Kasim V, Miyagishi M, Taira K. Control of siRNA expression using the Cre-loxP recombination system. Nucleic Acids Res. 2004;32(7):e66. doi: 10.1093/nar/gnh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller VM, et al. Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles. Nucleic Acids Res. 2004;32(2):661–8. doi: 10.1093/nar/gkh208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller VM, et al. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci U S A. 2003;100(12):7195–200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding H, et al. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2(4):209–17. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz DS, et al. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2(9):e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson BL, Paulson HL. Molecular medicine for the brain: silencing of disease genes with RNA interference. Lancet Neurol. 2004;3(3):145–9. doi: 10.1016/S1474-4422(04)00678-7. [DOI] [PubMed] [Google Scholar]
- 25.McCaffrey AP, et al. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21(6):639–44. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- 26.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100(1):235–40. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson JA, et al. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci U S A. 2003;100(5):2783–8. doi: 10.1073/pnas.252758799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci U S A. 2003;100(4):2014–8. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418(6893):38–9. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 30.Song E, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9(3):347–51. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 31.Jacque JM, Triques K, Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418(6896):435–8. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee NS, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20(5):500–5. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 33.Coburn GA, Cullen BR. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J Virol. 2002;76(18):9225–31. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surabhi RM, Gaynor RB. RNA interference directed against viral and cellular targets inhibits human immunodeficiency Virus Type 1 replication. J Virol. 2002;76(24):12963–73. doi: 10.1128/JVI.76.24.12963-12973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novina CD, et al. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8(7):681–6. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 36.Park WS, et al. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 2002;30(22):4830–5. doi: 10.1093/nar/gkf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez MA, et al. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16(18):2385–90. doi: 10.1097/00002030-200212060-00002. [DOI] [PubMed] [Google Scholar]
- 38.Capodici J, Kariko K, Weissman D. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J Immunol. 2002;169(9):5196–201. doi: 10.4049/jimmunol.169.9.5196. [DOI] [PubMed] [Google Scholar]
- 39.Banerjea A, et al. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol Ther. 2003;8(1):62–71. doi: 10.1016/s1525-0016(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 40.Li MJ, et al. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther. 2003;8(2):196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 41.Boden D, et al. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77(21):11531–5. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caldwell DJ, Evans JD. Developing clinical role of a CCR5 co-receptor antagonist in HIV-1 infection. Expert Opin Pharmacother. 2008;9(18):3231–42. doi: 10.1517/14656560802576324. [DOI] [PubMed] [Google Scholar]
- 43.Eugen-Olsen J, et al. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS. 1997;11(3):305–10. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- 44.Samson M, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 45.Qin XF, et al. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100(1):183–8. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dropulic B. Lentivirus in the clinic. Mol Ther. 2001;4(6):511–2. doi: 10.1006/mthe.2001.0501. [DOI] [PubMed] [Google Scholar]
- 47.Davis BM, Humeau L, Dropulic B. In vivo selection for human and murine hematopoietic cells transduced with a therapeutic MGMT lentiviral vector that inhibits HIV replication. Mol Ther. 2004;9(2):160–72. doi: 10.1016/j.ymthe.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 48.Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97(8):689–96. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurzov EN, Izquierdo M. RNA interference against Hec1 inhibits tumor growth in vivo. Gene Ther. 2006;13(1):1–7. doi: 10.1038/sj.gt.3302595. [DOI] [PubMed] [Google Scholar]
- 50.Hadj-Slimane R, et al. Short interfering RNA (siRNA), a novel therapeutic tool acting on angiogenesis. Biochimie. 2007;89(10):1234–44. doi: 10.1016/j.biochi.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today. 2006;11(1–2):67–73. doi: 10.1016/S1359-6446(05)03668-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Das B, et al. A hypoxia-driven vascular endothelial growth factor/Flt1 autocrine loop interacts with hypoxia-inducible factor-1alpha through mitogen-activated protein kinase/extracellular signal-regulated kinase 1/2 pathway in neuroblastoma. Cancer Res. 2005;65(16):7267–75. doi: 10.1158/0008-5472.CAN-04-4575. [DOI] [PubMed] [Google Scholar]
- 53.List AF, et al. Vascular endothelial growth factor receptor-1 and receptor-2 initiate a phosphatidylinositide 3-kinase-dependent clonogenic response in acute myeloid leukemia cells. Exp Hematol. 2004;32(6):526–35. doi: 10.1016/j.exphem.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Kou R, et al. Differential regulation of vascular endothelial growth factor receptors (VEGFR) revealed by RNA interference: interactions of VEGFR-1 and VEGFR-2 in endothelial cell signaling. Biochemistry. 2005;44(45):15064–73. doi: 10.1021/bi0509898. [DOI] [PubMed] [Google Scholar]
- 55.Takei Y, et al. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64(10):3365–70. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 56.Huang C, et al. Small interfering RNA therapy in cancer: mechanism, potential targets, and clinical applications. Expert Opin Ther Targets. 2008;12(5):637–45. doi: 10.1517/14728222.12.5.637. [DOI] [PubMed] [Google Scholar]
- 57.Wieczorek M, et al. Silencing of Wnt-1 by siRNA induces apoptosis of MCF-7 human breast cancer cells. Cancer Biol Ther. 2008;7(2):268–74. doi: 10.4161/cbt.7.2.5300. [DOI] [PubMed] [Google Scholar]
- 58.Rosell R, et al. Future directions in the second-line treatment of non-small cell lung cancer. Semin Oncol. 2006;33(1 Suppl 1):S45–51. doi: 10.1053/j.seminoncol.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Guo J, et al. Enhanced chemosensitivity to irinotecan by RNA interference-mediated down-regulation of the nuclear factor-kappaB p65 subunit. Clin Cancer Res. 2004;10(10):3333–41. doi: 10.1158/1078-0432.CCR-03-0366. [DOI] [PubMed] [Google Scholar]
- 60.Guo J, Fu YC, Becerra CR. Dissecting role of regulatory factors in NF-kappaB pathway with siRNA. Acta Pharmacol Sin. 2005;26(7):780–8. doi: 10.1111/j.1745-7254.2005.00140.x. [DOI] [PubMed] [Google Scholar]
- 61.Li SD, et al. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16(1):163–9. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshizawa T, et al. Folate-linked lipid-based nanoparticles for synthetic siRNA delivery in KB tumor xenografts. Eur J Pharm Biopharm. 2008;70(3):718–25. doi: 10.1016/j.ejpb.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 63.Wang R, et al. The therapeutic potential of survivin promoter-driven siRNA on suppressing tumor growth and enhancing radiosensitivity of human cervical carcinoma cells via downregulating hTERT gene expression. Cancer Biol Ther. 2007;6(8):1295–301. doi: 10.4161/cbt.6.8.4505. [DOI] [PubMed] [Google Scholar]
- 64.Shen Y, et al. hTERT-targeted RNA interference inhibits tumorigenicity and motility of HCT116 cells. Cancer Biol Ther. 2008;7(2):228–36. doi: 10.4161/cbt.7.2.5259. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, et al. Antisense anti-MDM2 oligonucleotides as a novel therapeutic approach to human breast cancer: in vitro and in vivo activities and mechanisms. Clin Cancer Res. 2001;7(11):3613–24. [PubMed] [Google Scholar]
- 66.Martinez LA, et al. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci U S A. 2002;99(23):14849–54. doi: 10.1073/pnas.222406899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hao JH, et al. Inhibition of the proliferation of human gastric cancer cells SGC-7901 in vitro and in vivo using Bcl-2 siRNA. Chin Med J (Engl) 2007;120(23):2105–11. [PubMed] [Google Scholar]
- 68.Chen SM, et al. Effect of blocking VEGF, hTERT and Bcl-xl by multiple shRNA expression vectors on the human laryngeal squamous carcinoma xenograft in nude mice. Cancer Biol Ther. 2008;7(5):734–9. doi: 10.4161/cbt.7.5.5722. [DOI] [PubMed] [Google Scholar]
- 69.Li SD, Chono S, Huang L. Efficient oncogene silencing and metastasis inhibition via systemic delivery of siRNA. Mol Ther. 2008;16(5):942–6. doi: 10.1038/mt.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Layzer JM, et al. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10(5):766–71. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 72.Sui G, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci U S A. 2002;99(8):5515–20. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paul CP, et al. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20(5):505–8. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 74.Yu JY, et al. Simultaneous inhibition of GSK3alpha and GSK3beta using hairpin siRNA expression vectors. Mol Ther. 2003;7(2):228–36. doi: 10.1016/s1525-0016(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 75.Guo Y, et al. Effect of vector-expressed shRNAs on hTERT expression. World J Gastroenterol. 2005;11(19):2912–5. doi: 10.3748/wjg.v11.i19.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar LD, Clarke AR. Gene manipulation through the use of small interfering RNA (siRNA): from in vitro to in vivo applications. Adv Drug Deliv Rev. 2007;59(2–3):87–100. doi: 10.1016/j.addr.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 77.Hama S, et al. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol Ther. 2006;13(4):786–94. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 78.Hama S, et al. Quantitative and mechanism-based investigation of post-nuclear delivery events between adenovirus and lipoplex. Nucleic Acids Res. 2007;35(5):1533–43. doi: 10.1093/nar/gkl1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cattaneo R, et al. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat Rev Microbiol. 2008;6(7):529–40. doi: 10.1038/nrmicro1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 81.Stoll SM, et al. Epstein-Barr virus/human vector provides high-level, long-term expression of alpha1-antitrypsin in mice. Mol Ther. 2001;4(2):122–9. doi: 10.1006/mthe.2001.0429. [DOI] [PubMed] [Google Scholar]
- 82.Suda T, Suda K, Liu D. Computer-assisted hydrodynamic gene delivery. Mol Ther. 2008;16(6):1098–104. doi: 10.1038/mt.2008.66. [DOI] [PubMed] [Google Scholar]
- 83.Belyantseva IA. Helios Gene Gun-mediated transfection of the inner ear sensory epithelium. Methods Mol Biol. 2009;493:103–23. doi: 10.1007/978-1-59745-523-7_7. [DOI] [PubMed] [Google Scholar]
- 84.Liu F, Huang L. A syringe electrode device for simultaneous injection of DNA and electrotransfer. Mol Ther. 2002;5(3):323–8. doi: 10.1006/mthe.2002.0540. [DOI] [PubMed] [Google Scholar]
- 85.Lee RC, et al. Surfactant-induced sealing of electropermeabilized skeletal muscle membranes in vivo. Proc Natl Acad Sci U S A. 1992;89(10):4524–8. doi: 10.1073/pnas.89.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu F, Huang L. Electric gene transfer to the liver following systemic administration of plasmid DNA. Gene Ther. 2002;9(16):1116–9. doi: 10.1038/sj.gt.3301733. [DOI] [PubMed] [Google Scholar]
- 87.Merdan T, Kopecek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Deliv Rev. 2002;54(5):715–58. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 88.Patil Y, Panyam J. Polymeric nanoparticles for siRNA delivery and gene silencing. Int J Pharm. 2008 doi: 10.1016/j.ijpharm.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bartlett DW, Davis ME. Physicochemical and biological characterization of targeted, nucleic acid-containing nanoparticles. Bioconjug Chem. 2007;18(2):456–68. doi: 10.1021/bc0603539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar P, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134(4):577–86. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59(2–3):75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Soutschek J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–8. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 93.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9(9):1034–48. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10(10):1735–7. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 95.McNamara JO, 2nd, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24(8):1005–15. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 96.Chan R, et al. Co-expression of anti-NFkappaB RNA aptamers and siRNAs leads to maximal suppression of NFkappaB activity in mammalian cells. Nucleic Acids Res. 2006;34(5):e36. doi: 10.1093/nar/gnj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dalby B, et al. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33(2):95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 98.Santel A, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13(16):1222–34. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 99.Chien PY, et al. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Ther. 2005;12(3):321–8. doi: 10.1038/sj.cgt.7700793. [DOI] [PubMed] [Google Scholar]
- 100.Pal A, et al. Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer. Int J Oncol. 2005;26(4):1087–91. [PubMed] [Google Scholar]
- 101.Hayes ME, et al. Assembly of nucleic acid-lipid nanoparticles from aqueous-organic monophases. Biochim Biophys Acta. 2006;1758(4):429–42. doi: 10.1016/j.bbamem.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 102.Khoury M, et al. Efficient new cationic liposome formulation for systemic delivery of small interfering RNA silencing tumor necrosis factor alpha in experimental arthritis. Arthritis Rheum. 2006;54(6):1867–77. doi: 10.1002/art.21876. [DOI] [PubMed] [Google Scholar]
- 103.Zuhorn IS, et al. Phase behavior of cationic amphiphiles and their mixtures with helper lipid influences lipoplex shape, DNA translocation, and transfection efficiency. Biophys J. 2002;83(4):2096–108. doi: 10.1016/S0006-3495(02)73970-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–4. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 105.Griesenbach U, et al. Inefficient cationic lipid-mediated siRNA and antisense oligonucleotide transfer to airway epithelial cells in vivo. Respir Res. 2006;7:26. doi: 10.1186/1465-9921-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Geary RS, et al. Pharmacokinetic properties of 2'-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J Pharmacol Exp Ther. 2001;296(3):890–7. [PubMed] [Google Scholar]
- 107.Kabanov AV. Taking polycation gene delivery systems from in vitro to in vivo. Pharm Sci Technolo Today. 1999;2(9):365–372. doi: 10.1016/s1461-5347(99)00186-8. [DOI] [PubMed] [Google Scholar]
- 108.Popielarski SR, et al. A nanoparticle-based model delivery system to guide the rational design of gene delivery to the liver. 2. In vitro and in vivo uptake results. Bioconjug Chem. 2005;16(5):1071–80. doi: 10.1021/bc0501146. [DOI] [PubMed] [Google Scholar]
- 109.Maruyama K, Kennel SJ, Huang L. Lipid composition is important for highly efficient target binding and retention of immunoliposomes. Proc Natl Acad Sci U S A. 1990;87(15):5744–8. doi: 10.1073/pnas.87.15.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woodle MC. Surface-modified liposomes: assessment and characterization for increased stability and prolonged blood circulation. Chem Phys Lipids. 1993;64(1–3):249–62. doi: 10.1016/0009-3084(93)90069-f. [DOI] [PubMed] [Google Scholar]
- 111.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46(12 Pt 1):6387–92. [PubMed] [Google Scholar]
- 112.Maeda H. The enhanced permeability and retention (EPR) effect in tumor vasculature: the key role of tumor-selective macromolecular drug targeting. Adv Enzyme Regul. 2001;41:189–207. doi: 10.1016/s0065-2571(00)00013-3. [DOI] [PubMed] [Google Scholar]
- 113.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliv Rev. 2004;56(11):1649–59. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 114.Modi S, et al. Exploiting EPR in polymer drug conjugate delivery for tumor targeting. Curr Pharm Des. 2006;12(36):4785–96. doi: 10.2174/138161206779026272. [DOI] [PubMed] [Google Scholar]
- 115.Iyer AK, et al. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov Today. 2006;11(17–18):812–8. doi: 10.1016/j.drudis.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 116.Zhang G, et al. Influence of anchoring ligands and particle size on the colloidal stability and in vivo biodistribution of polyethylene glycol-coated gold nanoparticles in tumor-xenografted mice. Biomaterials. 2009 doi: 10.1016/j.biomaterials.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Verbaan FJ, et al. Steric stabilization of poly(2-(dimethylamino)ethyl methacrylate)-based polyplexes mediates prolonged circulation and tumor targeting in mice. J Gene Med. 2004;6(1):64–75. doi: 10.1002/jgm.475. [DOI] [PubMed] [Google Scholar]
- 118.Park JW. Liposome-based drug delivery in breast cancer treatment. Breast Cancer Res. 2002;4(3):95–9. doi: 10.1186/bcr432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Allen TM, Cullis PR. Drug delivery systems: entering the mainstream. Science. 2004;303(5665):1818–22. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 120.Hashizume H, et al. Openings between defective endothelial cells explain tumor vessel leakiness. Am J Pathol. 2000;156(4):1363–80. doi: 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Konno T, et al. Effect of arterial administration of high-molecular-weight anticancer agent SMANCS with lipid lymphographic agent on hepatoma: a preliminary report. Eur J Cancer Clin Oncol. 1983;19(8):1053–65. doi: 10.1016/0277-5379(83)90028-7. [DOI] [PubMed] [Google Scholar]
- 122.Konno T, et al. Selective targeting of anti-cancer drug and simultaneous image enhancement in solid tumors by arterially administered lipid contrast medium. Cancer. 1984;54(11):2367–74. doi: 10.1002/1097-0142(19841201)54:11<2367::aid-cncr2820541111>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 123.Maki S, Konno T, Maeda H. Image enhancement in computerized tomography for sensitive diagnosis of liver cancer and semiquantitation of tumor selective drug targeting with oily contrast medium. Cancer. 1985;56(4):751–7. doi: 10.1002/1097-0142(19850815)56:4<751::aid-cncr2820560409>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 124.Seymour LW, et al. A novel dosage approach for evaluation of SMANCS [poly-(styrene-co-maleyl-half-n-butylate) - neocarzinostatin] in the treatment of primary hepatocellular carcinoma. Int J Oncol. 1998;12(6):1217–23. doi: 10.3892/ijo.12.6.1217. [DOI] [PubMed] [Google Scholar]
- 125.Lievens J, et al. The size of sinusoidal fenestrae is a critical determinant of hepatocyte transduction after adenoviral gene transfer. Gene Ther. 2004;11(20):1523–31. doi: 10.1038/sj.gt.3302326. [DOI] [PubMed] [Google Scholar]
- 126.Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4(9):891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- 127.Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release. 2008;126(1):77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Banerjee R, et al. Anisamide-targeted stealth liposomes: a potent carrier for targeting doxorubicin to human prostate cancer cells. Int J Cancer. 2004;112(4):693–700. doi: 10.1002/ijc.20452. [DOI] [PubMed] [Google Scholar]
- 129.Nakamura Y, et al. Octaarginine-modified multifunctional envelope-type nano device for siRNA. J Control Release. 2007;119(3):360–7. doi: 10.1016/j.jconrel.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 130.Xu Y, Szoka FC., Jr Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35(18):5616–23. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 131.Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8(15):1188–96. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 132.Hafez IM, Cullis PR. Roles of lipid polymorphism in intracellular delivery. Adv Drug Deliv Rev. 2001;47(2–3):139–48. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]
- 133.Ewert KK, et al. Cationic lipid-DNA complexes for non-viral gene therapy: relating supramolecular structures to cellular pathways. Expert Opin Biol Ther. 2005;5(1):33–53. doi: 10.1517/14712598.5.1.33. [DOI] [PubMed] [Google Scholar]
- 134.Webb MS, et al. Comparison of different hydrophobic anchors conjugated to poly(ethylene glycol): effects on the pharmacokinetics of liposomal vincristine. Biochim Biophys Acta. 1998;1372(2):272–82. doi: 10.1016/s0005-2736(98)00077-7. [DOI] [PubMed] [Google Scholar]
- 135.Hatakeyama H, et al. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Ther. 2007;14(1):68–77. doi: 10.1038/sj.gt.3302843. [DOI] [PubMed] [Google Scholar]
- 136.Tunnemann G, et al. Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. J Pept Sci. 2008;14(4):469–76. doi: 10.1002/psc.968. [DOI] [PubMed] [Google Scholar]
- 137.Sakai N, Matile S. Anion-mediated transfer of polyarginine across liquid and bilayer membranes. J Am Chem Soc. 2003;125(47):14348–56. doi: 10.1021/ja037601l. [DOI] [PubMed] [Google Scholar]
- 138.Werth S, et al. A low molecular weight fraction of polyethylenimine (PEI) displays increased transfection efficiency of DNA and siRNA in fresh or lyophilized complexes. J Control Release. 2006;112(2):257–70. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 139.Urban-Klein B, et al. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12(5):461–6. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 140.Grayson AC, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23(8):1868–76. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 141.Thomas M, et al. Full deacylation of polyethylenimine dramatically boosts its gene delivery efficiency and specificity to mouse lung. Proc Natl Acad Sci U S A. 2005;102(16):5679–84. doi: 10.1073/pnas.0502067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92(16):7297–301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Thomas M, Klibanov AM. Enhancing polyethylenimine's delivery of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2002;99(23):14640–5. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Akinc A, et al. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7(5):657–63. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 145.Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J Biol Chem. 2003;278(45):44826–31. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 146.Martin ME, Rice KG. Peptide-guided gene delivery. AAPS J. 2007;9(1):E18–29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Thomas M, Klibanov AM. Conjugation to gold nanoparticles enhances polyethylenimine's transfer of plasmid DNA into mammalian cells. Proc Natl Acad Sci U S A. 2003;100(16):9138–43. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Thomas M, et al. Cross-linked small polyethylenimines: while still nontoxic, deliver DNA efficiently to mammalian cells in vitro and in vivo. Pharm Res. 2005;22(3):373–80. doi: 10.1007/s11095-004-1874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fahrmeir J, et al. Electrophoretic purification of tumor-targeted polyethylenimine-based polyplexes reduces toxic side effects in vivo. J Control Release. 2007;122(3):236–45. doi: 10.1016/j.jconrel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 150.Boeckle S, et al. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6(10):1102–11. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 151.Guo X, Szoka FC., Jr Chemical approaches to triggerable lipid vesicles for drug and gene delivery. Acc Chem Res. 2003;36(5):335–41. doi: 10.1021/ar9703241. [DOI] [PubMed] [Google Scholar]
- 152.Asokan A, Cho MJ. Exploitation of intracellular pH gradients in the cellular delivery of macromolecules. J Pharm Sci. 2002;91(4):903–13. doi: 10.1002/jps.10095. [DOI] [PubMed] [Google Scholar]
- 153.Needham D, et al. A new temperature-sensitive liposome for use with mild hyperthermia: characterization and testing in a human tumor xenograft model. Cancer Res. 2000;60(5):1197–201. [PubMed] [Google Scholar]
- 154.Saito G, Swanson JA, Lee KD. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv Drug Deliv Rev. 2003;55(2):199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 155.Austin CD, et al. Oxidizing potential of endosomes and lysosomes limits intracellular cleavage of disulfide-based antibody-drug conjugates. Proc Natl Acad Sci U S A. 2005;102(50):17987–92. doi: 10.1073/pnas.0509035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang J, et al. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proc Natl Acad Sci U S A. 2006;103(37):13872–7. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17(9):961–83. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 158.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 159.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 160.Sledz CA, et al. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–9. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 161.Bridge AJ, et al. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34(3):263–4. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 162.Kim DH, et al. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22(3):321–5. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 163.Hornung V, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–70. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 164.Heil F, et al. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur J Immunol. 2003;33(11):2987–97. doi: 10.1002/eji.200324238. [DOI] [PubMed] [Google Scholar]
- 165.Lee J, et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci U S A. 2003;100(11):6646–51. doi: 10.1073/pnas.0631696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ma Z, et al. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem Biophys Res Commun. 2005;330(3):755–9. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 167.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19(2):111–24. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 168.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–62. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 169.Kleinman ME, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452(7187):591–7. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Poeck H, et al. 5'-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14(11):1256–63. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 171.Judge AD, et al. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13(3):494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 172.Bradley AJ, et al. Inhibition of liposome-induced complement activation by incorporated poly(ethylene glycol)-lipids. Arch Biochem Biophys. 1998;357(2):185–94. doi: 10.1006/abbi.1998.0798. [DOI] [PubMed] [Google Scholar]
- 173.Scheule RK, et al. Basis of pulmonary toxicity associated with cationic lipid-mediated gene transfer to the mammalian lung. Hum Gene Ther. 1997;8(6):689–707. doi: 10.1089/hum.1997.8.6-689. [DOI] [PubMed] [Google Scholar]
- 174.Li S, et al. Effect of immune response on gene transfer to the lung via systemic administration of cationic lipidic vectors. Am J Physiol. 1999;276(5 Pt 1):L796–804. doi: 10.1152/ajplung.1999.276.5.L796. [DOI] [PubMed] [Google Scholar]
- 175.Whitmore M, Li S, Huang L. LPD lipopolyplex initiates a potent cytokine response and inhibits tumor growth. Gene Ther. 1999;6(11):1867–75. doi: 10.1038/sj.gt.3301026. [DOI] [PubMed] [Google Scholar]
- 176.Loisel S, et al. Contribution of plasmid DNA to hepatotoxicity after systemic administration of lipoplexes. Hum Gene Ther. 2001;12(6):685–96. doi: 10.1089/104303401300057405. [DOI] [PubMed] [Google Scholar]
- 177.Sakurai F, et al. Therapeutic effect of intravenous delivery of lipoplexes containing the interferon-beta gene and poly I: poly C in a murine lung metastasis model. Cancer Gene Ther. 2003;10(9):661–8. doi: 10.1038/sj.cgt.7700617. [DOI] [PubMed] [Google Scholar]
- 178.Liu F, Shollenberger LM, Huang L. Non-immunostimulatory nonviral vectors. FASEB J. 2004;18(14):1779–81. doi: 10.1096/fj.04-2187fje. [DOI] [PubMed] [Google Scholar]
- 179.Zhao H, et al. Contribution of Toll-like receptor 9 signaling to the acute inflammatory response to nonviral vectors. Mol Ther. 2004;9(2):241–8. doi: 10.1016/j.ymthe.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 180.Kuramoto T, et al. Use of lipoplex-induced nuclear factor-kappaB activation to enhance transgene expression by lipoplex in mouse lung. J Gene Med. 2006;8(1):53–62. doi: 10.1002/jgm.812. [DOI] [PubMed] [Google Scholar]
- 181.Sakurai H, et al. Comparison of gene expression efficiency and innate immune response induced by Ad vector and lipoplex. J Control Release. 2007;117(3):430–7. doi: 10.1016/j.jconrel.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 182.Sakurai H, et al. Innate immune response induced by gene delivery vectors. Int J Pharm. 2008;354(1–2):9–15. doi: 10.1016/j.ijpharm.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 183.Yew NS, et al. Reduced inflammatory response to plasmid DNA vectors by elimination and inhibition of immunostimulatory CpG motifs. Mol Ther. 2000;1(3):255–62. doi: 10.1006/mthe.2000.0036. [DOI] [PubMed] [Google Scholar]
- 184.Hofman CR, et al. Efficient in vivo gene transfer by PCR amplified fragment with reduced inflammatory activity. Gene Ther. 2001;8(1):71–4. doi: 10.1038/sj.gt.3301373. [DOI] [PubMed] [Google Scholar]
- 185.Liu F, et al. Novel nonviral vectors target cellular signaling pathways: regulated gene expression and reduced toxicity. J Pharmacol Exp Ther. 2007;321(2):777–83. doi: 10.1124/jpet.106.118117. [DOI] [PubMed] [Google Scholar]
- 186.Tan Y, Zhang JS, Huang L. Codelivery of NF-kappaB decoy-related oligodeoxynucleotide improves LPD-mediated systemic gene transfer. Mol Ther. 2002;6(6):804–12. doi: 10.1006/mthe.2002.0811. [DOI] [PubMed] [Google Scholar]
- 187.Chono S, et al. An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor. J Control Release. 2008;131(1):64–9. doi: 10.1016/j.jconrel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yan W, Chen W, Huang L. Mechanism of adjuvant activity of cationic liposome: phosphorylation of a MAP kinase, ERK and induction of chemokines. Mol Immunol. 2007;44(15):3672–81. doi: 10.1016/j.molimm.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 189.Chen W, Yan W, Huang L. A simple but effective cancer vaccine consisting of an antigen and a cationic lipid. Cancer Immunol Immunother. 2008;57(4):517–30. doi: 10.1007/s00262-007-0390-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Halder J, et al. Focal adhesion kinase targeting using in vivo short interfering RNA delivery in neutral liposomes for ovarian carcinoma therapy. Clin Cancer Res. 2006;12(16):4916–24. doi: 10.1158/1078-0432.CCR-06-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Merritt WM, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100(5):359–72. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morrissey DV, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–7. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 193.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3(5):579–88. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 194.Jackson AL, et al. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12(7):1179–87. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fedorov Y, et al. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12(7):1188–96. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]