Abstract
The beta-lactams are the most important class of antibiotics in clinical use. Their lethal targets are the transpeptidase domains of penicillin binding proteins (PBPs), which catalyze the crosslinking of bacterial peptidoglycan (PG) during cell wall synthesis. The transpeptidation reaction occurs in two steps, the first being formation of a covalent enzyme intermediate and the second involving attack of an amine on this intermediate. Here we use defined PG substrates to dissect the individual steps catalyzed by a purified E. coli transpeptidase. We demonstrate that this transpeptidase accepts a set of structurally diverse D-amino acid substrates and incorporates them into PG fragments. These results provide new information on donor and acceptor requirements as well as a mechanistic basis for previous observations that non-canonical D-amino acids can be introduced into the bacterial cell wall.
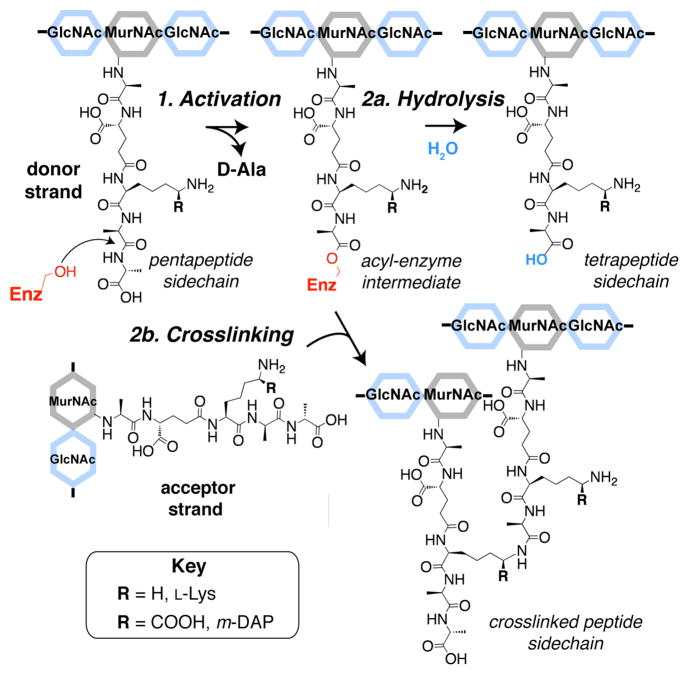
Peptidoglycan (PG) is an essential polymer that surrounds bacterial cells. It is composed of linear glycan chains coupled through peptide crosslinks. The glycan strands are synthesized by peptidoglycan glyco-syltransferases (PGTs). The peptide crosslinks are formed by enzymes called transpeptidases (TPs),1 which are the lethal targets of the most widely used class of antibiotics: the beta-lactams.2 The interactions of beta-lactams with TPs are far better characterized than transpeptidation reactions involving natural substrates.3 This is mainly due to difficulties in obtaining suitable substrates to study the TPs. TPs catalyze a two-step reaction that begins with the activation (Step 1, Figure 1) of a donor peptide through formation of an acyl-enzyme intermediate with concomitant release of a terminal D-Ala residue. This activated donor can either be hydrolyzed by water (Step 2a)4 or react with an amino group on an acceptor peptide attached to an adjacent glycan strand. While the amino acid residue containing the reactive amine varies depending on the bacterial species,5 the overall mechanism is conserved.
Figure 1.
Stepwise mechanism of peptidoglycan (PG) transpeptidases (TPs). TPs activate PG donor strands (Step 1) resulting in release of terminal D-Ala and formation of an acyl-enzyme intermediate. Either hydrolysis of this activated donor (Step 2a) or peptide crosslinking to an acceptor strand (Step 2b) can then occur. For most Gram-negative bacteria, including E. coli, R=COOH (meso-diaminopimelic acid, m-DAP), while R = H (L-Lys) for most Gram-positive bacteria.5 (MurNAc: N-acetylmuramic acid; GlcNAc: N-acetylglucosamine)
Here we report a method to dissect the individual steps of transpeptidation using substrates that act only as either donors or acceptors. By examining donor peptides following post-reaction cleavage from the glycan strand, we have established that the donor must be a polymeric fragment of PG. In contrast, the acceptor can be a single amino acid as long as the amine nucleophile is found in a D-stereocenter like that in the native substrate. Since single amino acids serve as acceptors, it is possible to efficiently incorporate a variety of non-canonical D-amino acids into peptidoglycan via transpeptidation. In addition to providing new tools to monitor the transpeptidation reaction, these results emphasize the range of enzymatic activities of TPs and suggest a role for these proteins in biological signaling through modifications of the cell wall.
To study the activation step of transpeptidation, we needed uncrosslinked peptidoglycan polymer that would serve as a donor but not an acceptor in the reaction (Step 1, Figure 1). We have shown that such polymers can be obtained by PGT-mediated polymerization of synthetic Lipid II (1).7 Most Gram-negative bacteria, including E. coli, make peptidoglycan with crosslinks to meso-diaminopimelic acid (m-DAP) as shown in Figure 1 (R = COOH).5 However, lysine-containing (R = H) precursor (1), which is found in most Gram-positive bacteria, is simpler to obtain. In order to study donor activation, we needed to ensure that crosslinking does not occur. Therefore, PG precursor Lipid II (1)7a, 8 was acylated on the reactive epsilon-amine of the lysine side chain to produce Lipid II analog 2 (Figure 2a).7e Radiolabeled acetic anhydride was used for the acylation to enable detection of future peptide modifications. Uncrosslinked PG (3, PG) was then produced by polymerizing [14C]Ac-Lipid II using the E. coli PBP1A PGT domain.9 Following heat inactivation of the PGT, 3 was incubated with or without E. coli PBP1A* (Figure 2b), which contains an inactive PGT domain and an active TP domain (See SI, Figures S1–2).3d The reaction was quenched after 20 min and a previously characterized amidase was added to cleave the peptides from the MurNAc residues of the glycan polymers (Figure 2a, dashed red line).6 As shown in Figure 2c, reactions containing PBP1A* produce two products upon amidase cleavage: the unmodified pentapeptide and a tetrapeptide that lacks the terminal D-Ala (lane 4) as confirmed by mass spectrometry analysis (Figure S3). The tetrapeptide is not observed when penicillin G, a general PBP inhibitor, is included in the reaction (lane 6) nor is it observed when the TP is absent from the reaction (lane 2). Furthermore, it was not observed if only Ac-Lipid II (2) was incubated with PBP1A* (Figure S4), which is not capable of producing glycan polymers. We conclude that the formation of tetrapeptide results from TP-dependent activation of pentapeptide followed by hydrolysis, a reaction expected to occur in the absence of acceptor substrate (Step 2a, Figure 1). Similar results were obtained for other TPs from both Gram-negative and Gram-positive organisms (Figure S5). These results show that polymerized N-acyl Lipid II (3) serves as a donor substrate for TPs from various organisms.
Figure 2.
TPs activate and hydrolyze PG donor strands. (a) Chemical structure of Lipid II analogs and nascent (uncrosslinked) peptidoglycan (PG) where n ~ 30. Amidases cleave peptides from MurNAc residues (dashed red line) of PG.6 (b) Schematic depicting detection method for modifications to the PG peptide. PG activated by PBP1A* results in release of terminal D-Ala followed by hydrolysis of the acyl-enzyme intermediate, which is indicated by a mixture of tetrapeptides and unreacted pentapeptides upon amidase cleavage. (c) SDS-PAGE analysis showing that E. coli PBP1A* incubated with PG followed by amidase treatment produces pentapeptides and tetrapeptides (lane 4), while PBP1A* inhibited by penicillin G (penG) produces only pentapeptides (lane 6) as seen in the reaction lacking PBP1A* (lane 2). (PBP1A* contains an inactivating mutation, E86Q, in the PGT domain, see SI.)
The ability of activated PG donors to undergo transpeptidation was investigated by adding amino acids as potential acceptors to the PBP reaction (Figure 3a, top). For the initial studies, amino acids were chosen for their similarity to natural peptide acceptors, which contain reactive amines on m-DAP or lysine residues (Figure 1).5 E. coli PBP1A, which contains an active PGT and an active TP domain, was incubated with [14C]Ac-Lipid II (2) and either L-Lys, D-Lys or m-DAP. The reactions were treated with amidase and analyzed by SDS-PAGE. m-DAP and D-Lys were found to be substrates, but L-Lys was not (Figure S6a–b). m-DAP contains two stereocenters, one L and one D, and in peptidoglycan the L-stereocenter is incorporated into the peptide backbone while the crosslinking side chain contains the D-stereocenter. Since D-Lys, but not L-Lys reacts, we concluded that reaction with D-Lys occurs on the alpha-amino group rather than the epsilon sidechain. These results suggest that the enzyme recognizes alpha-amino acids but discriminates against the L configuration.10 Consistent with this conclusion, both Gly and D-Ala were found to be good substrates but L-Ala did not react (Figure S6c).
Figure 3.
PG fragments synthesized by E. coli PBP1A are labeled with excess D-Ala via transpeptidation. (a) Top: Chemical reaction depicting transpeptidation of an acyl-enzyme intermediate (donor) and D-amino acid (acceptor) to produce a pentapeptide side chain on PG. Bottom: Schematic of assay to analyze TP product using D-Ala as an acceptor. PG is treated with hydrolases (PBP5 or amidase) followed by paper chromatography to separate unreacted PG (low) from cleaved D-Ala or peptide (high). (c) Assay results: PBP1A (400 nM) was incubated with unlabeled Lipid II (1, 40 μM) and [14C]-D-Ala (1 mM) to produce [14C]-PG. The [14C]-label is removed by treatment with PBP5 or amidase (4 μM each ), but not PBP5 inactivated by penG (5 kU/ml ). Background was not subtracted. Error bars indicate the standard deviation of triplicate experiments.
We further examined incorporation of D-Ala into activated PG polymers to verify the position of labeling. Lipid II (1) was incubated with [14C]-D-Ala and PBP1A, and the resulting radiolabeled PG polymer ([14C]-PG) was treated with amidase. The reaction was analyzed by paper chromatography to separate PG polymers from low molecular weight cleavage products (Figure 3a, bottom and SI). 6, 7a–b, 11 Amidase treatment resulted in release of radiolabel from the polymer (Figure 3b).6 Labeled PG was also incubated with PBP5 in the presence and absence of penicillin G. PBP5 is a known carboxypeptidase that hydrolyzes the terminal D-Ala residue from the pentapeptide sidechain of PG (Figure S7).12 Paper chromatography analysis showed that PBP5 causes release of radiolabel from [14C]-PG only in the absence of penicillin G, which inhibits its activity. These data confirm that PBP1A introduces [14C]-D-Ala into the fifth position of the peptide sidechain of PG. The ability to efficiently make labeled PG via transpeptidation should enable further mechanistic studies of the transpeptidase reaction.
The ability of transpeptidases to accept both D-Ala and D-Lys suggested that other amino acids would be tolerated. Notably, there have been several reports describing the incorporation of non-canonical D-amino acids into existing peptidoglycan of both Gram-negative and Gram-positive bacteria.13 The incorporation of these D-amino acids affects bacterial cell shape,13f as well as biofilm formation,13g and is proposed to increase sensitivity of cells to beta-lactams and serve as a means of biological regulation.13h The mechanism(s) for introduction of D-amino acids into cell wall has not been established. Since our results indicate that D-amino acids can be introduced into PG via transpeptidation, we evaluated the ability of PBP1A to incorporate several non-canonical amino acids, including phenylalanine, tyrosine, and tryptophan, into PG. SDS-PAGE analysis of reactions containing these amino acids revealed new bands of higher molecular weight than the substrate pentapeptide in all lanes containing D- (lanes 4, 6, 8), but not L- (lane 3, 5, 7) isomers (Figure 4a). Liquid chromatography-mass spectrometry (LC-MS) analysis confirmed that D-Phe, D-Tyr and D-Trp replace one of the alanine residues in these PG pentapeptides (Figure 4b, Table S1).14 MS/MS fragmentation of the D-Tyr peptide (4) confirmed that the added amino acid was in the fifth position (Figure 4c). Hence, even D-amino acids with large aromatic substituents function as acceptors. We found that D-Met, D-Thr, D-Asp, D-Gln, and D-Pro, but not the corresponding L-isomers, also function as substrates for transpeptidation (Figures S6d-e, S8; Table S1). Hence, all classes of amino acids can be incorporated into nascent (uncrosslinked) PG polymer by transpeptidation.
Figure 4.

E. coli PBP1A incorporates D-, but not L-, non-canonical amino acids into the termini of PG peptides via transpeptidation. (a) SDS-PAGE analysis of PBP1A (400 nM) reactions with aromatic amino acids (1 mM) and labeled Ac-Lipid II (2, 4 μM). Amidase treatment of PG products reveals a new peptide band in the D-amino acid reactions (lanes 4, 6, and 8) but not the corresponding L-isomer reactions (lanes 3, 5, and 7) or with added penG (1 kU/ml, lane 1). (b) Overlaid LC-MS extraction traces of amidase-treated PG produced by incubation of D-Phe, D-Tyr, or D-Trp (each 1 mM) with Lipid II (1, 40 μM) and PBP1A (400 nM). Peaks represent mass values of pentapeptides containing aromatic amino acids in place of terminal D-Ala ([M+H]+ calculated, observed m/z values for each, respectively: 565.2980, 565.2984; 581.2930, 581.2933; 604.3089, 604.3085). (c) MS/MS fragmentation of the D-Tyr reaction trace at t = 18.7 min confirms the sequence of the D-Tyr pentapeptide (4) indicated by the chemical structure (inset with calculated m/z values).
We have reported the development of methods to probe the transpeptidation step of peptidoglycan synthesis. We can observe substrate activation and hydrolysis by post-reaction detection of tetrapeptide product. Using this method we have established that activation requires polymeric substrate since Lipid II does not react. Although E. coli PBP1A normally utilizes substrates containing m-DAP in the peptide side chain, activation does not depend on the presence of this amino acid since polymer containing N-acyl lysine (3) can be used as a donor. By monitoring the formation of new pentapeptides, we have also shown that single amino acids undergo transpeptidation; however, only amines in alpha-amino acids react and there is a stringent selection against the L-configuration. Since transpeptidation using amino acid acceptors is the reverse of activation and D-Ala is released during activation,15 the preference for the D-configuration is unsurprising, but the tolerance for a wide range of side chains was unexpected. This finding provides a possible mechanism to explain the observation that non-canonical D-amino acids are incorporated in vivo into the peptidoglycan of bacteria;13 there is good agreement between promiscuity seen for D-amino acid incorporation in vivo to what we have observed in vitro.13b–e,16 The susceptibility of nascent PG containing non-canonical D-amino acids to subsequent hydrolysis or crosslinking by PG biosynthetic enzymes is unknown, but can now be studied in vitro since our results have provided a facile method to obtain the required substrates. The method also allows for incorporation of radiolabeled D-Ala into substrates for future PBP rate studies.17
Our results explain the observation that L-Lys-D-Ala-D-Ala analogs, commonly used substrates to study PBP-catalyzed PG hydrolysis,10a,12b do not undergo transpeptidation with E. coli PBPs.3b,18 This tripeptide cannot serve as a TP donor since polymeric PG substrates are required. Furthermore, it cannot serve as a TP acceptor because the lysine side-chain amine does not contain an alpha-D-amino acid.19 In closing, we note that the ability to reconstitute TP activity using defined substrates and purified proteins will enable detailed studies into the cross-linking step of the transpeptidation reaction. Understanding the chemistry of these enzymes will provide insight into the various roles that TPs play in building new cell wall, editing existing cell wall, and perhaps transmitting cellular signals through these chemical modifications.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (R01 GM076710; R01 GM066174). All LC-MS data were acquired on an Agilent 6520 Q-TOF spectrometer supported by the Taplin Funds for Discovery Program (P.I.: S.W.). We thank the Bernhardt Lab (Harvard Medical School) for overexpression plasmids (see SI).
Footnotes
Supporting Information. Experimental procedures, synthesis of substrates and compound analysis, protein construct descriptions, protein cloning and purification protocols, Q-TOF LC-MS and SDS-PAGE analysis of peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Scheffers DJ, Pinho MG. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. FEMS Microbiol Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]; (c) Vollmer W, Seligman SJ. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 2.(a) Tipper DJ, Strominger JL. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ghuysen JM, Frere JM, Leyh-Bouille M, Coyette J, Dusart J, Nguyen-Disteche M. Annu Rev Biochem. 1979;48:73–101. doi: 10.1146/annurev.bi.48.070179.000445. [DOI] [PubMed] [Google Scholar]; (c) Waxman DJ, Strominger JL. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]; (d) Spratt BG. J Gen Microbiol. 1983;129:1247–1260. doi: 10.1099/00221287-129-5-1247. [DOI] [PubMed] [Google Scholar]; (e) Wilke MS, Lovering AL, Strynadka NC. Curr Opin Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]; (f) Testero SA, Fisher JF, Mobashery S. β-Lactam Antibiotics. In Burger’s Medicinal Chemistry. In: Abraham DJ, Rotella DP, editors. Drug Discovery and Development. Vol. 7. Wiley & Sons; New York: 2010. pp. 259–404. [Google Scholar]
- 3.Natural substrates refer to PG (or PG fragments) rather than beta-lactams (see reference 2) or modified peptides (see reference 10). See the following for studies that utilize PG substrates: Waxman DJ, Yu W, Strominger JL. J Biol Chem. 1980;255:11577–11587.Schwartz B, Markwalder JA, Wang Y. J Am Chem Soc. 2001;123:11638–11643. doi: 10.1021/ja0166848.Bertsche U, Breukink E, Kast T, Vollmer W. J Biol Chem. 2005;280:38096–38101. doi: 10.1074/jbc.M508646200.Born P, Breukink E, Vollmer W. J Biol Chem. 2006;281:26985–26993. doi: 10.1074/jbc.M604083200.
- 4.The TP domains discussed in this work are fused to N-terminal PGT domains in bifunctional proteins. These TPs had not been shown to catalyze PG hydrolysis using natural substrates. For a review, see: Goffin C, Ghuysen JM. Microbiol Mol Biol Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002.
- 5.For many Gram-positive bacteria, the epsilon-amino group of L-Lys is acylated with other amino acids or peptides, which are involved in cross-links. See the following for a review on PG structure: Vollmer W, Blanot D, de Pedro MA. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x.
- 6.E. coli AmiA mature construct was used as the amidase in this work. See SI and the following for more details: Lupoli TJ, Taniguchi T, Wang TS, Perlstein DL, Walker S, Kahne DE. J Am Chem Soc. 2009;131:18230–18231. doi: 10.1021/ja908916z.
- 7.(a) Ye XY, Lo M-C, Brunner L, Walker D, Kahne D, Walker S. J Am Chem Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]; (b) Chen L, Walker D, Sun B, Hu Y, Walker S, Kahne D. Proc Natl Acad Sci USA. 2003;100:5658–5663. doi: 10.1073/pnas.0931492100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Barrett DS, Chen L, Litterman NK, Walker S. Biochemistry. 2004;43:12375–12381. doi: 10.1021/bi049142m. [DOI] [PubMed] [Google Scholar]; (d) Barrett D, Leimkuhler C, Chen L, Walker D, Kahne D, Walker S. J Bacteriol. 2005;187:2215–2217. doi: 10.1128/JB.187.6.2215-2217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Barrett D, Wang TS, Yuan Y, Zhang Y, Kahne D, Walker S. J Biol Chem. 2007;282:31964–31971. doi: 10.1074/jbc.M705440200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Proc Natl Acad Sci USA. 2007;104:5348–5353. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. J Am Chem Soc. 2007;129:12674–12675. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Wang TS, Manning SA, Walker S, Kahne D. J Am Chem Soc. 2008;130:14068–14069. doi: 10.1021/ja806016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Men H, Park P, Ge M, Walker S. J Am Chem Soc. 1998;120:2484–2485. [Google Scholar]; (b) Ha S, Chang E, Lo MC, Men H, Park P, Ge M, Walker S. J Am Chem Soc. 1999;121:8415–8426. [Google Scholar]; (c) Chen L, Men H, Ha S, Ye XY, Brunner L, Hu Y, Walker S. Biochemistry. 2002;41:6824–6833. doi: 10.1021/bi0256678. [DOI] [PubMed] [Google Scholar]
- 9.E. coli PBP1A PGT domain construct consists of residues M1-I291. See SI and the following review for sequence alignment: Vollmer W, Bertsche U. Biochim Biophys Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007.
- 10.Preference of PBPs for D-amino acid acceptors is supported by previous work that used peptide or unnatural peptide substrates: Pollock JJ, Ghuysen JM, Linder R, Salton MR, Perkins HR, Nieto M, Leyh-Bouille M, Frere JM, Johnson K. Proc Natl Acad Sci US,A. 1972;69:662–666. doi: 10.1073/pnas.69.3.662.Adam M, Damblon C, Jamin M, Zorzi W, Dusart V, Galleni M, el Kharroubi A, Piras G, Spratt BG, Keck W, Coyette J, Ghuysen JM, Nguyen-Disteche M, Frere JM. Biochem J. 1991;279:601–604. doi: 10.1042/bj2790601.Kumar I, Pratt RF. Biochemistry. 2005;44:9971–9979. doi: 10.1021/bi050542z.
- 11.Anderson JS, Matsuhashi M, Haskin MA, Strominger JL. Proc Natl Acad Sci USA. 1965;53:881–889. doi: 10.1073/pnas.53.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholas RA, Strominger JL. J Biol Chem. 1988;263:2034–2040.van der Linden MP, de Haan L, Dideberg O, Keck W. Biochem J. 1994;303:357–362. doi: 10.1042/bj3030357.Hesek D, Suvorov M, Morio K, Lee M, Brown S, Vakulenko SB, Mobashery S. J Org Chem. 2004;69:778–784. doi: 10.1021/jo035397e.Zhang W, Shi Q, Meroueh SO, Vakulenko SB, Mobashery S. Biochemistry. 2007;46(35):10113–10121. doi: 10.1021/bi700777x.E. coli PBP5 soluble construct consisting of residues D30-N385 (see SI) was used based on the following report: Lee M, Hesek D, Suvorov M, Lee W, Vakulenko S, Mobashery S. J Am Chem Soc. 2003;125:16322–16326. doi: 10.1021/ja038445l.
- 13.(a) Lark C, Bradley D, Lark KG. Biochim Biophys Acta. 1963;78:278–288. doi: 10.1016/0006-3002(63)91638-x. [DOI] [PubMed] [Google Scholar]; (b) Tsuruoka T, Tamura A, Miyata A, Takei T, Iwamatsu K, Inouye S, Matsuhashi M. J Bacteriol. 1984;160:889–894. doi: 10.1128/jb.160.3.889-894.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Tsuruoka T, Tamura A, Miyata A, Takei T, Inouye S, Matsuhashi M. Eur J Biochem. 1985;151:209–216. doi: 10.1111/j.1432-1033.1985.tb09089.x. [DOI] [PubMed] [Google Scholar]; (d) Caparros M, Torrecuadrada JL, de Pedro MA. Res Microbiol. 1991;142:345–350. doi: 10.1016/0923-2508(91)90050-k. [DOI] [PubMed] [Google Scholar]; (e) Caparros M, Pisabarro AG, de Pedro MA. J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Cava F, Lam H, de Pedro MA, Waldor MK. Cell Mol Life Sci. 2011;68:817–831. doi: 10.1007/s00018-010-0571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uehara T, Parzych KR, Dinh T, Bernhardt TG. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Q, Meroueh SO, Fisher JF, Mobashery S. J Am Chem Soc. 2011;133:5274–5283. doi: 10.1021/ja1074739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The range of incorporation of amino acids in this study is broader than past reports. The location of the added amino acid in the peptide sidechain of PG can vary. Previous work using crude E. coli cell membranes suggests that added D-amino acids can replace the terminal peptide residue, see: Izaki K, Matsuhashi M, Strominger JL. Proc Natl Acad Sci USA. 1966;55:656–663. doi: 10.1073/pnas.55.3.656.
- 17.Preliminary experiments indicate that hydrolysis is significantly slower than transpeptidation using bifunctional PBP1A and Lipid II as substrate with or without D-amino acid acceptor. Access to radiolabeled substrates will allow further rate studies.
- 18.Jamin M, Damblon C, Millier S, Hakenbeck R, Frere JM. Biochem J. 1993;292:735–741. doi: 10.1042/bj2920735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.These findings about acceptor requirements suggested that it would not be necessary to use N-acyl-L-Lys-protected donors to prevent crosslinking, as was done in the experiments reported above. In fact, we have confirmed that L-Lys-containing PG polymers do not undergo crosslinking with E. coli PBP1A, although they are activated and will react with alpha-D-amino acid acceptors.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.