Summary of recent advances
The immune response to virus infection is initiated when pathogen recognition receptors (PRRs) of the host cell recognize specific non-self motifs within viral products to trigger intracellular signaling events that induce innate immunity, the front line of defense against microbial infection. The replication program of all viruses includes a cytosolic phase of genome amplification and/or mRNA metabolism and viral protein expression. Cytosolic recognition of viral infection by specific PRRs takes advantage of the dependence of viruses on the cytosolic component of their replication programs. Such PRR-PAMP interactions lead to PRR-dependent non-self recognition and the downstream induction of type I interferons and proinflammatory cytokines. These factors serve to induce innate immune programs and drive the maturation of adaptive immunity and inflammation for the control of infection. Recent studies have focused on identifying the particular viral ligands recognized as non-self by cytosolic PRRs, and on defining the nature of the PRRs and their signaling pathways involved in immunity. The RIG-I-like receptors, RIG-I and MDA5, have been defined as essential PRRs for host detection of a variety of RNA viruses. Novel PRRs and their signaling pathways involved in detecting DNA viruses through non-self recognition of viral DNA are also being elucidated. Moreover, studies to identify the PRRs and signaling factors of the host cell that mediate inflammatory signaling through inflammasome activation following virus infection are currently underway and have already revealed specific NOD-like receptors (NLRs) as inflammatory triggers. This review summarizes recent progress and current areas of focus in pathogen recognition and immune triggering by cytosolic PRRs.
Introduction
The immune response to virus infection begins with the recognition of viral pathogen associated molecular patterns (PAMPs) as “non-self” signatures. This recognition occurs through host pattern recognition receptors (PRRs). Toll-like receptors (TLRs) are a class of PRRs that recognize viral motifs presented at the cell surface or within the endosomal compartment but are not known for mediating cytosolic pathogen recognition. On the other hand, cytosolic PRRs have been identified that play major roles in the recognition of viral nucleic acid. These include the RIG-I-like receptors (RLRs), novel DNA-binding factors, and the nucleotide-binding domain-leucine-rich repeat-containing molecules (NLRs).
Following recognition of viral RNA or DNA, the PRRs undergo conformation changes or specific modifications that drive their signaling-active state and their downstream induction of type I interferon (IFN) and proinflammatory cytokine expression by the infected cell. Type I IFN is subsequently secreted and binds the type I IFN receptor on the cell surface in an autocrine and paracrine manner to activate Jak/STAT signaling and lead to the production of hundreds of interferon stimulated genes (ISGs). ISGs function to directly inhibit viral infection, trigger apoptosis of infected cells, and they play an important role to modulate adaptive immunity [1–2]. Overall, PRR signaling and the initiation of innate antiviral defenses represent our first line immune response to virus infection.
In this review, we summarize recent understanding of cytosolic recognition of viral nucleic acids leading to immunity and inflammation to limit virus infection.
Recognition of RNA viruses by RLRs
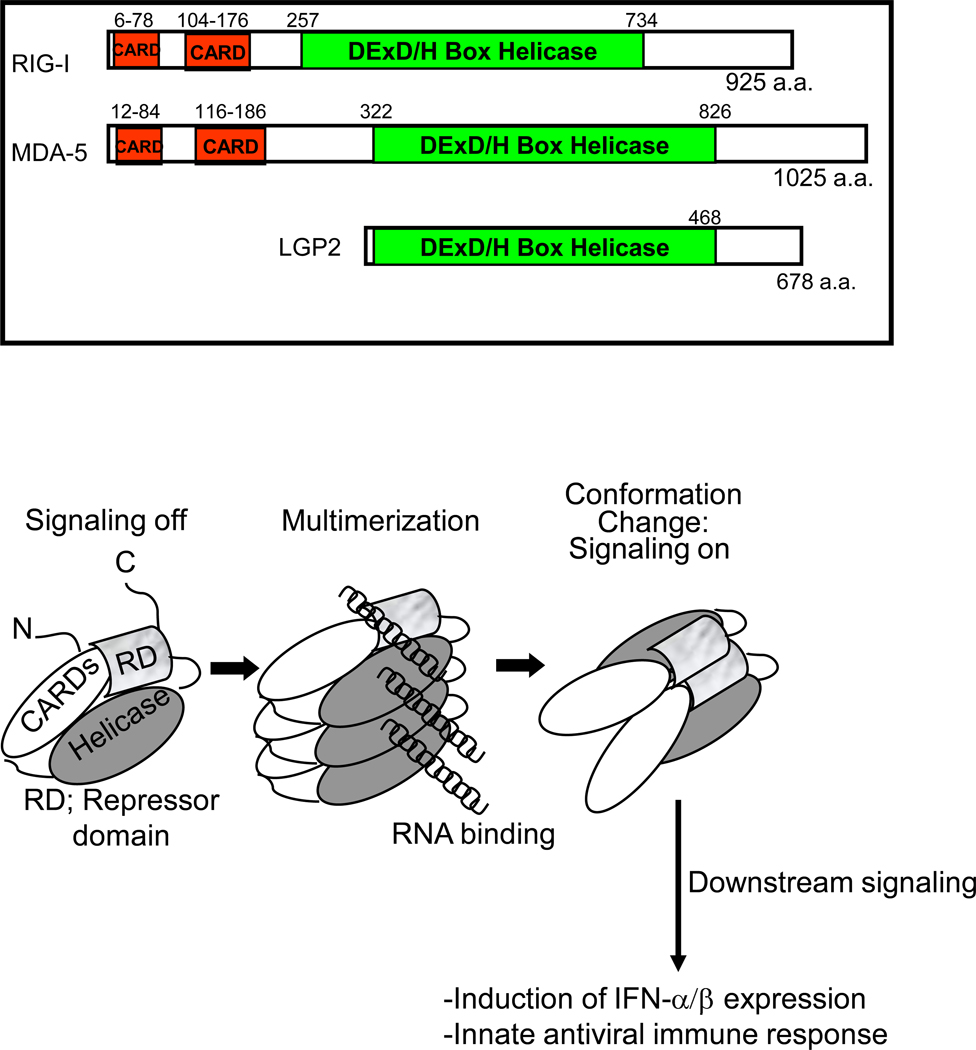
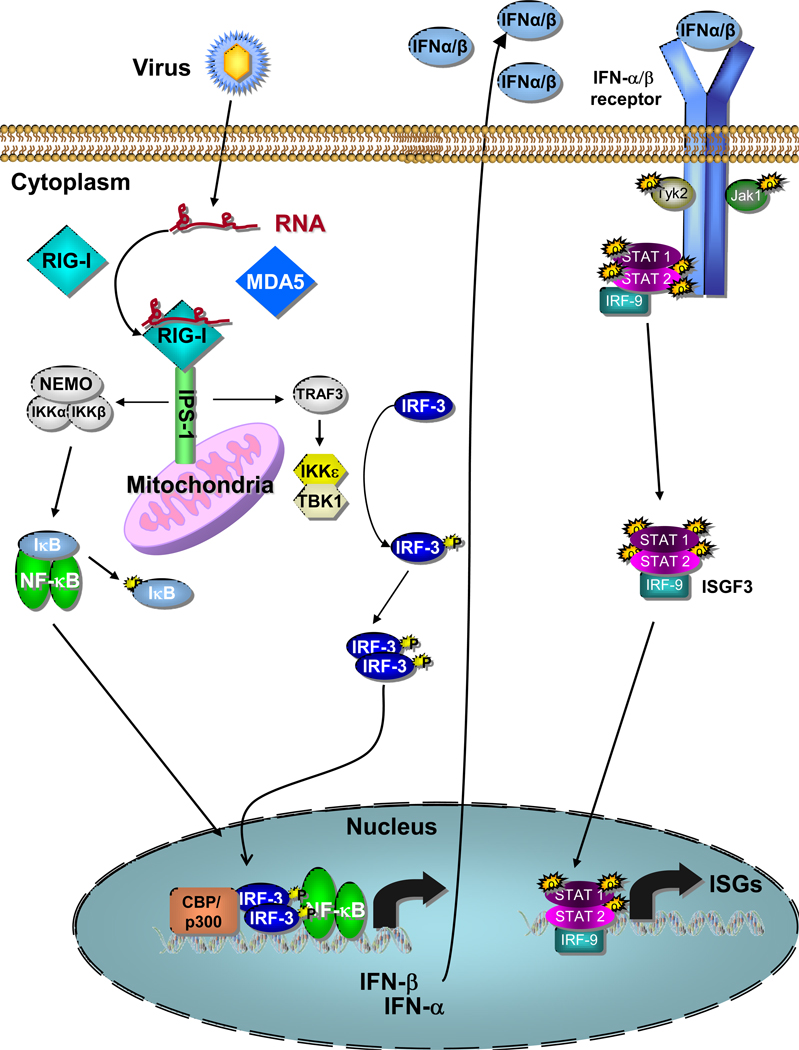
The RLR family consists of three members: retinoic acid inducible gene I (RIG-I), melanoma differentiation associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) [1]. RIG-I and MDA5 contain 2 N-terminal caspase activation and recruitment domains (CARDs) which are essential for their signaling activity. All three molecules have an internal DExD/H-box RNA helicase domain with ATPase activity. This ATPase activity, which is activated by ligand binding, does not appear to be required for RNA binding though it is necessary for signaling [3–4]. Finally, the C-terminus of RIG-I and LGP2 have been shown to act as a repressor domain (RD) that holds the molecule in an inactive conformation until RNA binding triggers an ATP-dependent conformational change that releases the CARD domains for signaling. In terms of RIG-I, this signaling initiation program has been defined through biochemical, genetic, and virologic studies (Figure 1) [5]. After binding non-self ligand, RIG-I and MDA5 interact via their CARD domains with the signaling-adaptor molecule, IPS-1 (also known as MAVS, VISA and Cardif), which recruits a signaling complex to activate transcription factors, including IRF3 and NF-κB. These events lead to the induced expression of IFN-β, IRF-3-target genes, and NF-κB target genes to drive antiviral and inflammatory responses against infection. (Figure 2) [6–9].
Figure 1.
Upper: RLR structure diagram showing positions of functional domains. Lower: 3-step model of RIG-I activation from a monomeric resting form (left) to RNA-ligand bound, dimeric form (center) and the final active form (right). The positions of the CARDs, helicase domain, and repressor domain (RD) are indicated. Ligand binding by RIG-I facilitates a conformation change that releases it from autorepression by the RD, thus driving downstream signaling of innate antiviral immunity. Model adapted from reference 5.
Figure 2.
The RLR signaling pathway showing RIG-I bound to ligand RNA and signaling downstream to IRF-3 and NF-kB to induce IFN-α/β production from a virus-infected cell. IFN-α/β is then shown signaling through the IFN-α/β receptor and the Jak-STAT pathway to drive ISG expression and an innate immune response. Details are described in the text.
A large research effort has focused on understanding the ligands that are recognized by each of the RLRs. Despite the structural similarity between RIG-I and MDA5, they were found to be responsible for IFN-induction by distinct sets of viruses. While RIG-I recognizes a number of both positive and negative stranded viruses (including Hepatitis C virus, respiratory syncytial virus and related paramyxoviruses, vesicular stomatitis virus and influenza A virus), MDA5 is responsible for recognition of picornaviruses and is the primary sensor of the dsRNA mimetic poly(I:C) [10–11]. Interestingly, both sensors appear to respond to reoviruses (a segmented dsRNA virus), and West Nile virus and Dengue virus (positive-stranded RNA viruses) (Table 1) [11–12]. Much of the virus specificity between the two PRRs has been found to be due to the particular RNA structures or nucleotide composition recognized by each.
Table 1.
Specificity of RIG-I like receptors for virus recognition. Summarized from reference 11.
| Virus | Genome RNA | Host cytosolic PRR |
|---|---|---|
| Vesicular stomatitis virus | Non-segmented negative-sense, single strand | RIG-I |
| Respiratory syncytial virus | Non-segmented negative-sense, single strand | RIG-I |
| Influenza A virus | 8 RNA segments, negative-sense, single strand | RIG-I |
| Ebola virus | Non-segmented negative-sense, single strand | RIG-I |
| Reovirus | 10 double-stranded segments | RIG-I and MDA5 |
| Hepatitis C virus | Non-segmented positive-sense, single strand | RIG-I |
| Dengue virus | Non-segmented positive-sense, single strand | RIG-I and MDA5 |
| West Nile virus | Non-segmented positive-sense, single strand | RIG-I and MDA5 |
| Polio virus | Non-segmented positive-sense, single strand | MDA5 |
RNA recognition by RIG-I requires the presence of a free 5’-triphosphate structure [13]. This requirement allows for differential recognition of non-self/viral RNA versus self RNAs, as host RNA is either capped or posttranslationally modified to remove the 5’-triphosphate. The exact nature of the RNA recognized by RIG-I is still rather controversial. RIG-I was first reported as binding dsRNA [14]. Although it has been reported that ssRNA bearing a 5’-triphosphate is recognized by RIG-I [15], a recent report suggests that the RNA requires some double-strandedness and that previous results were due to unintended hairpins produced by T7 in vitro transcription [16]. However, this assertion requires validation from other groups in addition to strict biochemical proof. RIG-I also recognizes particular sequences within the RNA. Uridine and adenosine-rich regions of RNA are preferentially utilized as RIG-I substrates, and are found within the PAMP structure of Hepatitis C virus and a number of other viruses recognized by RIG-I [17–18]. RIG-I is also able to recognize cleavage products produced by the host endonuclease RNase L during virus infection, which potentially serves to amplify RLR signaling responses [19].
The ligand of MDA5 is not yet well-defined. Picornavirus RNA is specifically recognized by MDA5, as the VPg molecule attached to the 5’ end of the viral RNAs blocks the 5’-triphosphate required for RIG-I recognition [10,15]. It is now thought that MDA5 preferentially binds long dsRNAs (greater than 1kb in length) while RIG-I is responsible for binding shorter dsRNA fragments and specific homopolymeric nucleotide sequences [20]. Further studies will be required to more accurately define the motifs recognized by MDA5 for activation of signaling.
Among the RLRs the role of LGP2 and the nature of its ligand recognition properties are comparably less well understood. The lack of a CARD domain led to the belief that LGP2 functions as a negative regulator of RLR signaling, and this was backed up by initial in vitro experiments [21]. However, intital studies of LGP2 knockout mice suggested that this molecule can also serve as a positive regulator of RLR signaling depending on the virus and likely the specific cell type involved in the infection [22]. More detailed analyses are needed to identify the exact role of LGP2 in cytosolic PAMP recognition and RLR signaling programs.
PRR recognition of cytoplasmic DNA
Recent studies have revealed specific factors expressed in mammalian cells that participate in the non-self recognition of B-form DNA, including viral DNA. While TLR9 has been known as a PRR that recognizes unmethylated/microbial DNA in the endosome, B-form DNA has been shown to induce type I interferon when introduced into the cytoplasm of host cells. These DNAs include DNA from viruses, bacteria, apoptotic host cells and synthetic B-form DNAs (particularly poly(dA:dT) and interferon stimulatory DNA (ISD)) [23–24]. The first reported cytoplasmic DNA receptor was identified as DNA-dependent activator of IFN-regulatory factors (DAI, also known as ZBP-1) [25]. DAI is an interferon-inducible gene whose product is able to bind B-form DNA and trigger signaling to type I IFN through downstream induction of NF-κB and IRF3 transcriptional activities. While DAI appears to be responsible for DNA-dependent IFN induction in particular cell types, other cell types and DAI-knockout mice exhibit normal responses to DNA vaccines, indicating the presence of other as yet unidentified DNA sensor(s) responsible for immune induction in response to microbial DNA [26]. It should be noted that recent studies now show that synthetic poly(dA:dT) or DNA encoded by Epstein-Barr virus (EBV), Herpes Simplex virus 1 (HSV-1), adenovirus and Legionella pneumophila are actually recognized indirectly through RIG-I [27–28]. In this case, the cellular RNA polymerase III was shown to transcribe AT-rich dsDNA of the microbe into dsRNA containing a 5’-triphosphate which subsequently activates RIG-I signaling through IPS-1. Pathogen recognition will occur through RIG-I recognition of homopolymeric U and A motifs in the 5’-ppp RNA products. However, DNA sensing occurs in many cell types in the absence of IPS-1 signaling, and the physiological relevance of RNA polymerase III-dependent recognition remains to be elucidated.
Cytosolic PRRs and virus-induced inflammasome activation
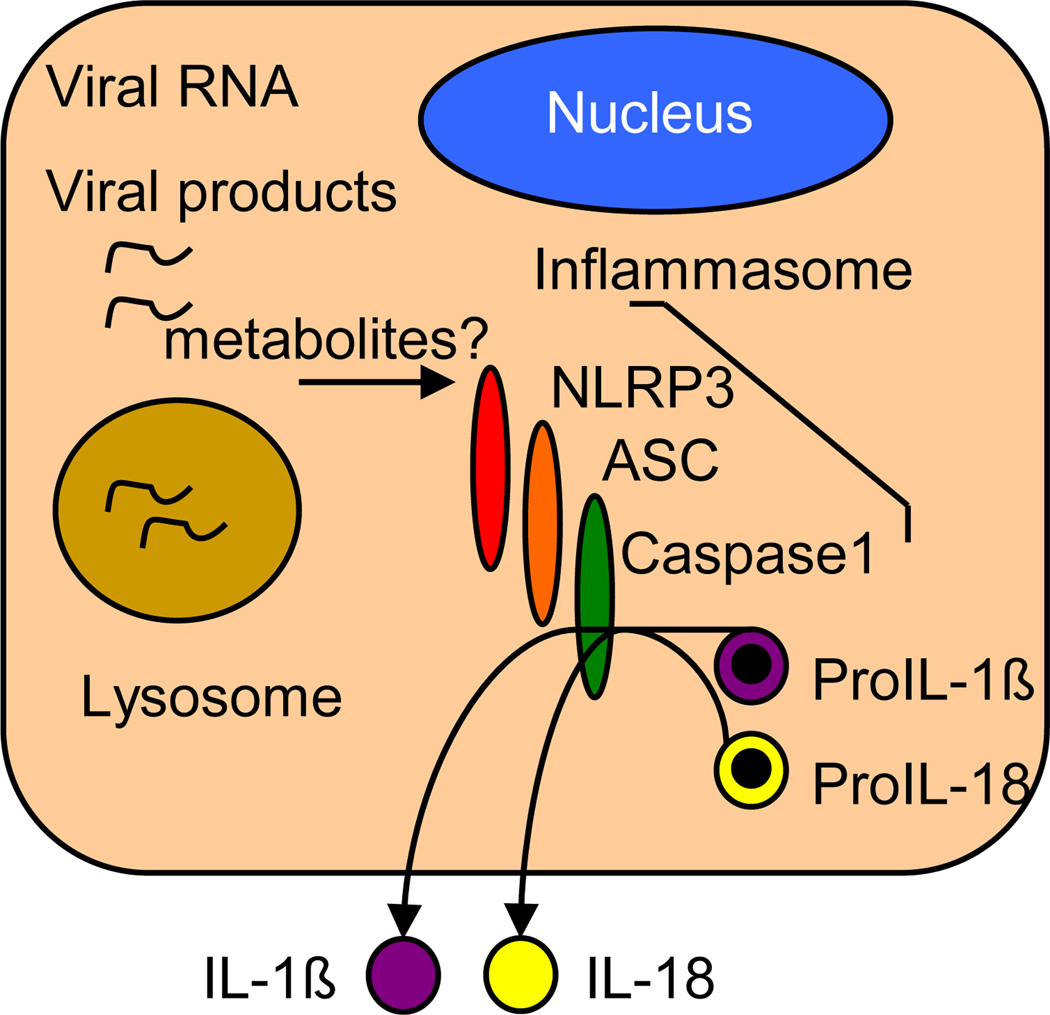
In addition to triggering the expression of type I IFN and IFN-induced innate immune defenses from infected cells, viruses are also able to induce activation of proinflammatory cytokines including IL-1β and IL-18 in a variety of cell types. Activation of IL-1β and IL-18 occurs through triggering of the inflammasome, a complex composed of specific NLRs that oligomerize upon stimulation (often requiring the presence of CARD-containing adaptor molecules) and recruit caspase 1 through CARD-CARD interactions. This interaction induces the self-cleavage and maturation of caspase 1, which then cleaves pro-IL-1β and pro-IL-18 to release the mature forms of these proinflammatory cytokines for secretion, thus driving the inflammatory response that serves to recruit immune cells to the site of infection [29]. The PRR role of NLRs and their functions in inflammasome signaling have been primarily studied in the context of bacterial infection through their recognition of microbial PAMP components such as peptidoglycan and flagellin [30]. However, recent work has highlighted a role for NLR inflammatory pathways in controlling DNA or RNA virus infection and immunity.
NLRP3 (also known as NALP3 or cryopyrin) is a known mediator of bacterial and chemical triggers of inflammation, including ATP and uric acid [31]. NLRP3, with its adaptor molecule, apoptotic speck-like protein containing a CARD (ASC) and caspase 1, make up the NLRP3 inflammasome [31]. The NLRP3 inflammasome was found to be stimulated during acute infection by both adenovirus and vaccinia virus, leading to IL-1β secretion from the infected cells [32–33]. In the case of adenovirus, inflammasome activation was dependent on the presence of viral genomic dsDNA, lending support to the idea that NALP3 may directly or indirectly serve as a PRR for DNA-based pathogens to trigger inflammation [32].
RNA viruses also have the potential to activate inflammasomes, as influenza A virus was recently reported to trigger inflammasome signaling through NLRP3 [34–36]. NLRP3, ASC and caspase 1 were found to be essential for IL-1β activation in mouse infection of influenza A virus, and loss of these molecules led to increased mortality in a disease model. Two groups reported differing results on the requirement of inflammasome signaling for influenza virus-specific adaptive immune responses, though it was suggested that the disparity could be due to differential amounts of virus in inoculations [34–35]. There are likely additional NLRs that detect influenza A virus infection, as some cell types responded to the virus in an ASC- and caspase 1-dependent but NLRP3-independent manner [34].
The aforementioned studies indicate a role of NLRP3 inflammasomes in cytosolic detection of DNA and RNA viruses, leading to activation of proinflammatory cytokines. However, it is not yet known if NLRP3 directly detects nucleic acids or if these inflammasomes are activated indirectly following nucleic acid recognition by an unknown sensor that may associate with NLRP3. The broad range of NLRP3 activators suggests that it may respond to a less specific stimulus such as cellular stress or alterations of host metabolites downstream of pathogen infection.
As opposed to NLRP3, the absent in melanoma 2 (AIM2) protein, a HIN-200 protein family member, has been shown to directly bind and respond to cytosolic dsDNA [32,37–40]. Binding of DNA to AIM2 induces its oligomerization and recruitment of ASC through its pyrin domain, leading to caspase 1 activation as in the case of the NLRs. Inflammasome activation by AIM2 is responsible for IL-1β secretion following dsDNA transfection into the cytoplasm of host cells as well as by infection from various DNA viruses including adenovirus and vaccinia virus [32–33].
These studies identify the inflammasome as a mediator of proinflammatory responses to viral infection whose actions are triggered by cytosolic PRRs (Figure 3). Further studies in this field could potentially lead to novel therapeutics that target inflammasome processes for the effective and balanced induction of inflammation and the control of viral infection.
Figure 3.
PRR signaling of the inflammasome. Model shows signaling by NLRP3 during influenza virus infection. Host metabolite products and/or viral products such as nucleic acid or viral protein serve as specific NLR ligands to trigger NLRP3 signaling activation. Adapted from reference 31.
Viral evasion of host recognition
As the host has evolved factors and strategies for detecting and responding to viral infection, viruses have developed countermeasures to inhibit these processes. One of the best characterized of these evasion methods is the cleavage of IPS-1 by the Hepatitis C virus (HCV) NS3/4a protease [41]. This cleavage results in a blockade of RIG-I signaling of innate immunity, thereby indirectly attenuating PRR actions and modulating the immune response to infection to favor viral persistence.
RIG-I and MDA5 are also direct targets of viral inhibition. The influenza A virus NS1 protein has been shown to inhibit PRR signaling by RIG-I through direct interaction [42], while the paramyxovirus V protein binds and inhibits MDA5 to abrogate its PRR signaling actions [43]. Other viruses alter potential ligands of PRR recognition in order to avoid detection by PRRs. An interesting highlight of these actions is the viral-directed removal of 5’-triphosphates from the RNA nucleic acid of Hantaan, Crimean-Congo hemorrhagic fever and Borna disease viruses, thereby providing a mechanism to escape the RLRs [44].
IRF3 is another common target of viral inhibition of innate immune signaling due to its essential role as a downstream transcription factor of RLR and DNA sensor PRR signaling. Importantly, human herpesviruses and human immunodeficiency virus have both been shown to induce the specific degradation of IRF3 in infected cells to thus inhibit induction of antiviral genes downstream of the actions of their specific PRRs [45–48]. Other examples of the targeted degradation of downstream factors of cytosolic PRR signaling have been presented, revealing a theme in viral evasion [1]. Many pathogenic viruses employ additional mechanisms by which they block PRR actions. Therapeutic interventions that are designed to target the virus-host interface of such immune evasion strategies to restore the functions of PRRs and their signaling pathways will offer the potential to restore innate immune induction and inflammatory programs that initiate immunity for the control of virus infection [29,41].
Conclusion
Recent advances in cytoplasmic recognition of viral infection include the elucidation of specific PRRs involved in detecting viral nucleic acids as non-self moieties that trigger immunity to infection. Additionally, cytosolic PRRs that trigger the inflammasome have been identified. These new insights should pave the way for the development of improved vaccine adjuvants, vaccine strategies, and overall design of antiviral therapeutics to control virus infections. Such developments will be supported by continuing investigations aimed at specifically defining PRR/PAMP interactions and the nature of viral nucleic acids that confer PRR recognition. Finally, an increased understanding of the mechanisms by which pathogenic viruses regulate innate immune signaling should allow us to define key targets for future therapeutic strategies aimed at restoring PRR functions and amplifying PRR signaling actions to enhance immunity.
Acknowledgements
Work in the Gale laboratory is supported through NIH grants, the Burroughs Wellcome Fund, . CW is supported through NIH grant XXX.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 2.Lei Y, Moore CB, Liesman RM, O'Connor BP, Bergstralh DT, Chen ZJ, Pickles RJ, Ting JP. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4:e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Jr., Akira S, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 4.Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr., Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 7.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 10.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredericksen BL, Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80:2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 14.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 15.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e, Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5'-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by compositiondependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malathi K, Dong B, Gale M, Jr., Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acidinducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 22.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 23.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 24.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 26.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 27.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol. 2009;19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Shaw MH, Reimer T, Kim YG, Nunez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20:377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassel SL, Joly S, Sutterwala FS. The NLRP3 inflammasome: a sensor of immune danger signals. Semin Immunol. 2009;21:194–198. doi: 10.1016/j.smim.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 33.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, Petrilli V, Gomez CE, Perdiguero B, Tschopp J, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas PG, Dash P, Aldridge JR, Jr., Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 38.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 40.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mibayashi M, Martinez-Sobrido L, Loo YM, Cardenas WB, Gale M, Jr., Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci U S A. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, et al. Processing of genome 5' termini as a strategy of negativestrand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One. 2008;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melroe GT, DeLuca NA, Knipe DM. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol. 2007;81:3077–3086. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doehle BP, Hladik F, McNevin JP, McElrath MJ, Gale M., Jr Human immunodeficiency virus type 1 mediates global disruption of innate antiviral signaling and immune defenses within infected cells. J Virol. 2009;83:10395–10405. doi: 10.1128/JVI.00849-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]