Abstract
The bHLH–PAS transcription factor SIM1 is expressed during the development of the hypothalamic–pituitary axis in three hypothalamic nuclei: the paraventricular nucleus (PVN), the anterior periventricular nucleus (aPV), and the supraoptic nucleus (SON). To investigate Sim1 function in the hypothalamus, we produced mice carrying a null allele of Sim1 by gene targeting. Homozygous mutant mice die shortly after birth. Histological analysis shows that the PVN and the SON of these mice are hypocellular. At least five distinct types of secretory neurons, identified by the expression of oxytocin, vasopressin, thyrotropin-releasing hormone, corticotropin-releasing hormone, and somatostatin, are absent in the mutant PVN, aPV, and SON. Moreover, we show that SIM1 controls the development of these secretory neurons at the final stages of their differentiation. A subset of these neuronal lineages in the PVN/SON are also missing in mice bearing a mutation in the POU transcription factor BRN2. We provide evidence that, during development of the Sim1 mutant hypothalamus, the prospective PVN/SON region fails to express Brn2. Our results strongly indicate that SIM1 functions upstream to maintain Brn2 expression, which in turn directs the terminal differentiation of specific neuroendocrine lineages within the PVN/SON.
Keywords: SIM, hypothalamus, paraventricular nucleus, supraoptic nucleus, hormones
The hypothalamus integrates a variety of signals from the brain and the periphery to modulate secretion by the pituitary of peptidic hormones involved in maintaining homeostasis. A large body of work, starting with the discovery of oxytocin and vasopressin (Du Vigneaud 1955), has led to the identification of numerous neuropeptides secreted by hypothalamic and pituitary cells. The availability of these markers has allowed a detailed characterization of the different cell types found in the hypothalamus and the pituitary and the delineation of their physiological role. This fundamental work has made the hypothalamic–pituitary axis an advantageous system to study the mechanisms involved in generating cell diversity in the central nervous system (CNS).
The neuroendocrine system consists of two sets of hypothalamic neurons: the magnocellular and the parvocellular neurons (for review, see Swanson and Sawchenko 1983; Swanson 1986; Sawchenko et al. 1992). The magnocellular neurosecretory system projects to the posterior pituitary where it releases vasopressin (AVP) and oxytocin (OT) directly into the general circulation. Vasopressin participates in the control of blood volume, osmolality, and pressure, whereas OT promotes parturition and lactation. The magnocellular neurons are located in two nuclei of the anterior hypothalamus, the paraventricular (PVN) and the supraoptic (SON) nuclei. Within the PVN and the SON, AVP and OT are produced by mutually exclusive sets of neurons. The sum of AVP- and OT-producing cells corresponds to the total number of magnocellular neurons, indicating that AVP and OT define the two cell types of this neurosecretory system.
The parvocellular neurosecretory system projects to the medial eminence where it releases hormones that are carried to the anterior pituitary by the portal vasculature. These hypophysiotropic hormones play an important role in modulating the secretion of anterior pituitary hormones, which include adrenocorticotropin (ACTH), thyrotropin (TSH), growth hormone, and the gonadotropins. Five peptidic hormones produced by the parvocellular neurons have been fully characterized, and the specific locations of the neurons that secrete them have been identified. Both corticotropin-releasing hormone (CRH) and thyrotropin-releasing hormone (TRH) are produced by neurons in the PVN, whereas growth hormone-releasing hormone (GHRH) is synthesized by neurons of the arcuate nucleus and the adjacent ventromedial nucleus, and gonadotropin-releasing hormone (GnRH) is synthesized by neurons located in the preoptic region. Somatostatin (SS) is mainly synthesized by parvocellular neurons in the anterior periventricular nucleus (aPV), which is ventrally contiguous with the PVN, although some SS is also synthesized in the PVN. Within the PVN and aPV, CRH, TRH, and SS each defines a distinct type of parvocellular neuron.
The PVN, which contains both magnocellular and parvocellular neurons, and the SON, which is mainly composed of magnocellular neurons, originate from a small patch of neuroepithelium located at the level of the ventral diencephalic sulcus. Cells that form the PVN remain near the ventricular zone, whereas those that form the SON migrate laterally to reach the surface of the hypothalamus (Altman and Bayer 1978). Considerable insight into the development of these nuclei was gained recently when it was shown that CRH, AVP- and OT-producing cells of the SON and PVN fail to develop in mice bearing a mutation in the POU domain transcription factor BRAIN-2 (BRN2) (Nakai et al. 1995; Schonemann et al. 1995). Mutant analysis and the fact that BRN2 binds and activates the CRH promotor suggest that it controls the development of these lineages at the terminal stage of their differentiation (Li et al. 1993; Schonemann et al. 1995).
A large family of transcription factors, characterized by the presence of a basic helix–loop–helix (bHLH) and PAS (for per, ARNT, and sim, the first genes shown to contain this motif) domains, has emerged during the past few years. Most bHLH–PAS proteins share the same primary structure (for review, see Hankison 1995; Schmidt and Bradfield 1996; Crews 1998). The basic domain, usually found at the amino terminus, contributes to sequence-specific DNA binding. Immediately carboxy-terminal to the basic domain, the HLH and the PAS domains together form a dimerization interface. Sequences carboxy-terminal to the bHLH–PAS domain appear to participate in activation or repression of the target genes.
Several vertebrate members of the bHLH–PAS family have attracted considerable attention during the past few years, as they have been shown to control physiological processes in response to the environment. These include CLOCK (King et al. 1997), which participates in the control of circadian rhythms, hypoxia-inducible factor (HIF) (Wang et al. 1995), and endothelial PAS domain protein (EPAS) (Ema et al. 1997; Tian et al. 1997), which regulate response to hypoxia, and the aryl hydrocarbon receptor (AHR) (Burbach et al. 1992), which mediates the effects of dioxin. Although AHR is the only member of this family for which a ligand has been identified, it has been postulated that other, if not all, bHLH–PAS proteins are also ligand-modulated transcription factors (Michaud and Fan 1997). Notably, members of a subgroup of bHLH– PAS proteins act as coactivators of nuclear receptors (for review, see Glass et al. 1997).
Drosophila Single-minded, one of the founding members of the bHLH–PAS protein family, is a key regulator of the development of CNS midline cells (Crews et al. 1988; Thomas et al. 1988; Nambu et al. 1990, 1991). Sim1 and Sim2, two murine homologs of this gene, have also been documented to display regionalized expression within the developing CNS (Ema et al. 1996; Fan et al. 1996; Moffett et al. 1996; Yamaki et al. 1996). However, their function in the CNS has not yet been determined. Here, we show that Sim1 RNA and protein are expressed in the PVN, SON, and aPV, from the early stages of their development to the neonatal period. Using a gene targeting approach, we have established that Sim1 is essential for the development of both the magnocellular and parvocellular neurosecretory lineages of the PVN, SON, and aPV. Moreover, we provide evidence that SIM1 controls the final stages of differentiation of a subset of these neurons by acting upstream to maintain Brn2 expression.
Results
Sim1 is expressed in the PVN, aPV, and SON
The PVN lines the third ventricle at the most dorsal aspect of the anterior hypothalamus as illustrated in Figure 1a. Originating from the prospective PVN region, the SON neurons migrate to the lateral surface of the hypothalamus (Fig. 1a). In newborn mice, both Sim1 transcript and protein are strongly expressed in the PVN (Fig. 1b,c) and the SON (Fig. 1d,e). The magnocellular and parvocellular neurons are segregated into distinct populations occupying specific regions within the PVN. Sim1 is expressed in the entire PVN, therefore including regions that contain either magnocellular or parvocellular neurons. Sim1 expression was also detected in the ventrally contiguous aPV (see below). No expression was detected in the preoptic area or in the arcuate nucleus that synthesize GnRH and GHRH, respectively.
Figure 1.
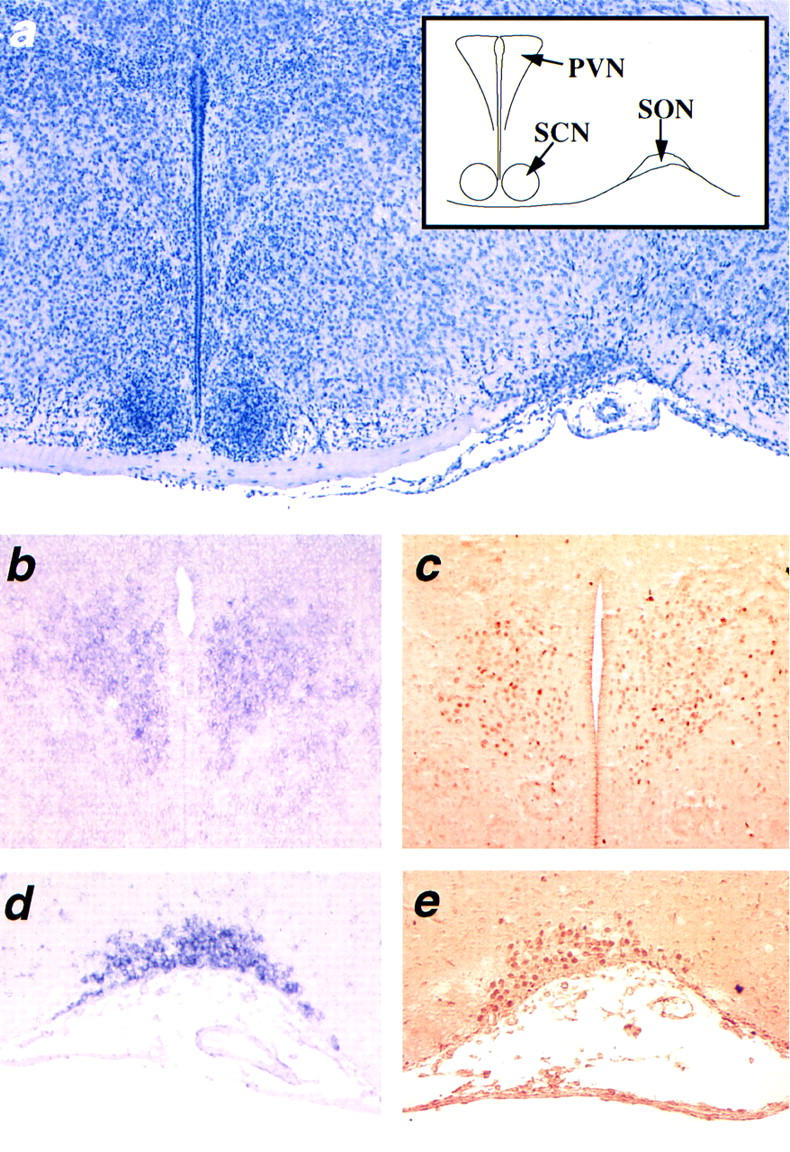
Sim1 expression in the PVN and SON. Coronal sections of wild-type newborn brains through the anterior hypothalamus. (a) Section stained with hematoxylin and scheme showing the location of the paraventricular nucleus (PVN), the suprachiasmatic nucleus (SCN), and the supraoptic nuleus (SON). (b,c) Expression of Sim1 transcript (b) and protein (c) in the PVN. (d,e) Expression of Sim1 transcript (d) and protein (e) in the SON. The Sim1 transcript was detected by in situ hybridization using a digoxygenin–UTP-labeled probe, whereas the SIM1 protein was detected by immunocytochemistry using a polyclonal SIM1 antiserum.
Sim1 mutant mice die shortly after birth
To gain insight into the function of Sim1 in the neuroendocrine hypothalamus, we generated mice homozygous for a null allele at the Sim1 locus, Sim1−, by gene targeting in embryonic stem (ES) cells. In the targeting vector we constructed (Fig. 2a), 750 bp of the 5′ region, the translation initiation codon, and the sequence coding for the basic domain of SIM1 were replaced by a Pgk–neo cassette (see Materials and Methods). Homologous recombinant ES cell clones, in which the altered Sim1 sequences replaced one copy of the wild-type gene, were identified by Southern analysis as described in Materials and Methods. These clones were obtained at a frequency of 7% (14 positives out of 200 clones assayed). After blastocyst injection and mating of the resulting chimeras to C57BL/6 mice, a line of mice heterozygous for the Sim1− allele was established. A 5′ and a 3′ external probe were used to confirm that these mice indeed carry the homologously recombined Sim1 mutant allele (Fig. 2b).
Figure 2.
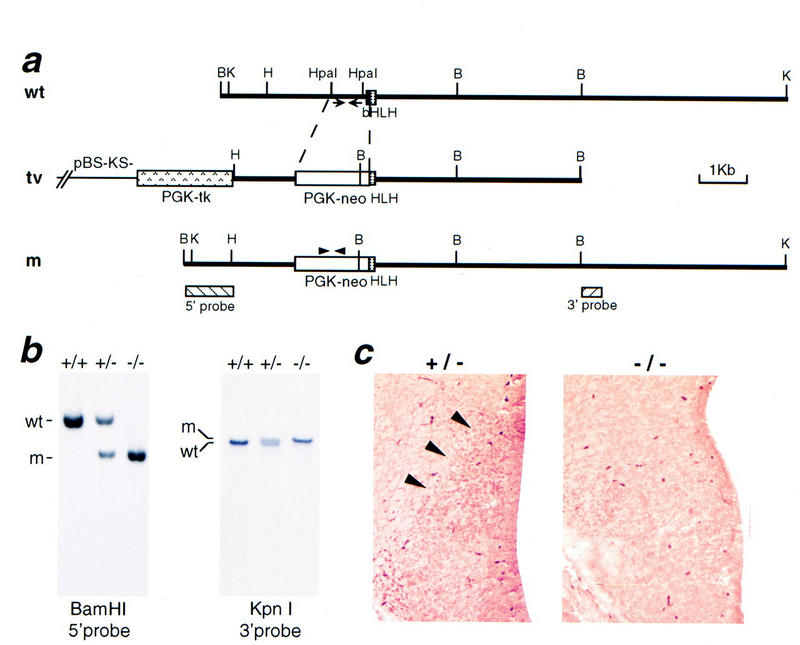
Targeted disruption of the mouse Sim1 locus. (a) Schematic representation of the Sim1 locus (wt), of the targeting vector (tv), and of the Sim1 mutant allele (m). Homologous recombination replaces 750 bp of the 5′ region, the initiation codon, and the sequence corresponding to the basic domain of Sim1 by a Pgk–neo cassette. The 5′ and 3′ external probes used in Southern blot analysis are represented by boxes at the bottom of the panel. The position of the primers used to amplify a Sim1 fragment present only in the wild-type allele or a neo fragment present only in the mutant allele are represented by arrows and arrowheads, respectively. Restriction enzymes: (B) BamHI, (H) HindIII, (K) KpnI. (b) Southern blot analysis of genomic DNA from Sim1+/+, Sim1+/−, Sim1−/− mice. The 5′ probe detects a wild-type (wt) 5.2-kb BamHI fragment and a mutant (m) 3.4-kb BamHI fragment. The 3′ probe detects a wild-type (wt) 11.2-kb KpnI fragment and a mutant (m) 12.6-kb KpnI fragment. (c) Coronal sections through the prospective PVN/SON of heterozygous control and mutant E13.5 embryos labeled with an antiserum recognizing SIM1. Production of SIM1 (arrowheads) is not detectable in the mutant.
Sim1− heterozygotes show no obvious physical or behavioral abnormalities and are fertile. Heterozygous siblings were bred to obtain homozygous mice. Up to birth, an expected Mendelian inheritance was observed [+/+, 44 (22%); +/−, 110 (55%); −/−, 44 (22%)], indicating that the mutation does not result in embryonic lethality. After birth, however, homozygous mice begin to die and none survive for more than 24 hr.
Disrupting Sim1 did not appear to significantly affect its transcription or the stability of its message, as Sim1 transcripts are expressed abundantly in various tissues of Sim1 mutant embryos (see below), albeit at a slightly lower level than in control embryos. To confirm that the targeted Sim1 allele did not produce any protein, we tested whether SIM1 protein is present in the prospective PVN/SON of embryonic day 13.5 (E13.5) Sim1 mutant embryos, where Sim1 transcripts are expressed. As shown in Figure 2c, no obvious SIM1 protein was detected in this tissue using an antiserum raised against a peptide corrresponding to a region of the carboxyl terminus of SIM1 (see Materials and Methods), indicating that the targeting resulted in a null allele.
Loss of Sim1 function affects development of the SON, PVN, and aPV
Histological sections of Sim1−/− and Sim1+/− littermate newborn brains revealed that both the PVN and SON are strikingly hypocellular in mutant mice, whereas surrounding structures such as the suprachiasmatic nucleus and the zona incerta have a normal appearance (Fig. 3). No significant changes were observed in the mutant aPV, possibly because this nucleus is morphologically more difficult to distinguish than the PVN and SON. Interestingly, although the Sim1 transcript is produced in Sim1−/− embryos, it is no longer detectable in the PVN, SON, or aPV of homozygous newborn mice (data not shown). This observation along with the histological analysis suggest that, in mutant mice, the neurons that normally express Sim1 in the PVN, SON, and aPV either changed fate or died.
Figure 3.
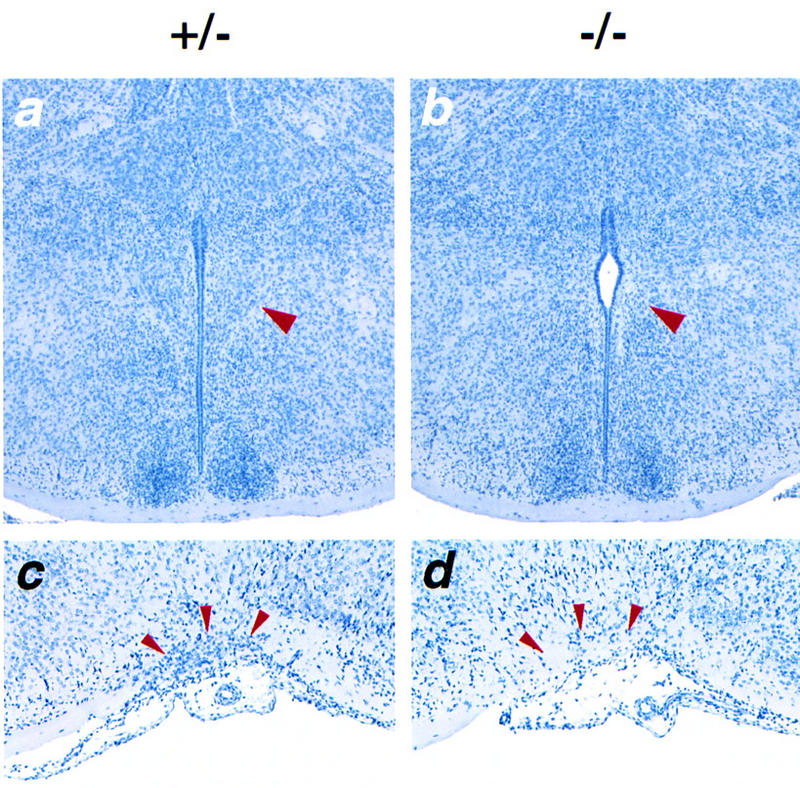
The morphology of the PVN and SON is altered in Sim1 mutant mice. Coronal sections through the anterior hypothalamus of Sim1+/− (a,c) and Sim1−/− (b,d) newborn mice, stained with hematoxylin. (a,b) Sections through the PVN (red arrowhead). The PVN has the shape of a triangle lining the third ventricle. In the mutant hypothalamus, no such structure is recognizable. The region normally occupied by the PVN appears hypocellular. (c,d) Sections through the SON (red arrowheads). The SON of Sim1−/− mice is strikingly hypocellular compared to that of Sim1+/− mice.
To characterize further the defects in the PVN and SON of Sim1−/− mice at the molecular level, we examined the magnocellular neurons by studying the production of AVP (Figs. 4a, and b and 5a, and b) and OT (Figs. 4c,d and 5c,d) and the parvocellular neurons by studying the expression of Crh (Fig. 4e,f) and Trh (Fig. 4g,h). Remarkably, the expression of none of these neuropeptides could be detected in mutant PVN or SON. SS-producing neurons of the aPV were also absent in the Sim1 mutant brain (Fig. 4i,j). We conclude that Sim1 disruption affects development of multiple neurosecretory lineages centered in the SON, PVN, and aPV. These defects are specific in that expression of the markers studied here could be detected in structures surrounding the PVN and SON (e.g., CRH in the lateral hypothalamus and AVP in the suprachiasmatic nucleus).
Figure 4.
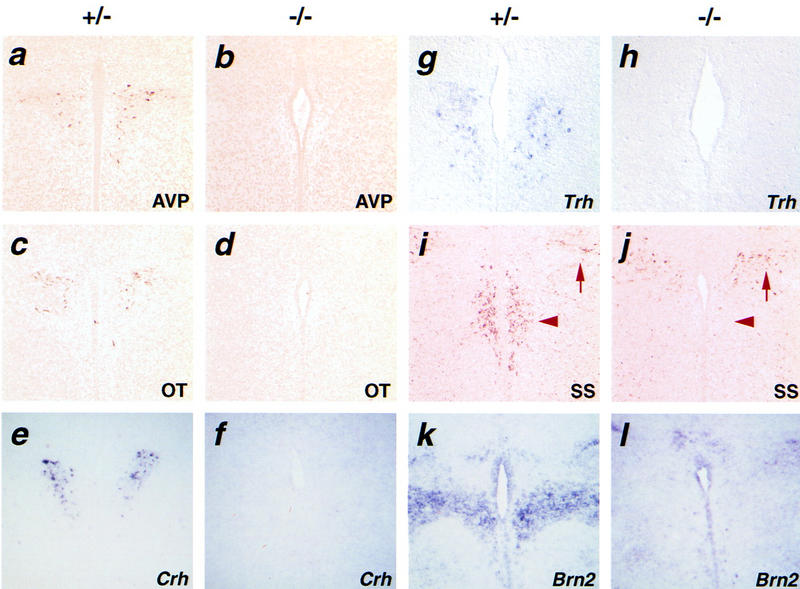
Molecular markers of the PVN are not detected in Sim1 mutant mice. Coronal sections through the PVN of Sim1+/− (a,c,e,g,i,k) or Sim1−/− (b,d,f,h,j,l) newborn mice. AVP (a,b), OT (c,d), Crh (e,f), Trh (g,h), SS (i,j), and Brn2 (k,l) are readily detected in sections of mutant heterozygotes but not mutant homozygotes. (i,j) Arrowheads indicate the location of the aPV and arrows show the production of SS in the zona incerta. AVP, OT, and SS were detected by immunocytochemistry, whereas Crh, Trh, and Brn2 were detected by in situ hybridization using digoxigenin–UTP-labeled probes.
The magnocellular neurons of the PVN and the SON send axonal projections to the posterior lobe of the pituitary. It has been proposed previously that these axons produce trophic factors for the pituicytes, the astroglial cells of the posterior lobe (Schonemann et al. 1995). To verify whether the absence of magnocellular neurons in Sim1 mutant mice affects the pituicytes, histological analysis was performed. Examination of sagittal sections of pituitaries showed obvious abnormalities in the posterior lobe of Sim1 homozygous newborns (Fig. 6a,b). Notably, the mutant pituicytes are packed more densely compared to those of the controls, presumably due to the lack of magnocellular axons originating from the PVN/SON. This is consistent with the fact that the mediolateral length of these mutant posterior pituitaries was, on average, significantly decreased when compared to that of control littermates (172 ± 41 μm vs. 289 ± 60 μm, P < 0.05; Fig. 6c). To address whether the pituicytes are affected by the Sim1 mutant allele, we counted the cells in the posterior lobe. We found that the average number of cells in mid-sagittal sections of the mutant posterior lobe was decreased (178 ± 12 vs. 346 ± 31, P < 0.005; Fig. 6d), indicating that the absence of magnocellular neurons in Sim1 mutant mice led to either a decreased proliferation or survival of pituicytes. In contrast to the posterior lobe, the anterior and intermediate lobes appear to develop properly in the mutant; even in the absence of TRH- and CRH-producing neurons in the Sim1 mutant PVN, production of ACTH (Fig. 6e,f) and TSH (not shown) was comparable to that of control pituitaries.
Figure 6.
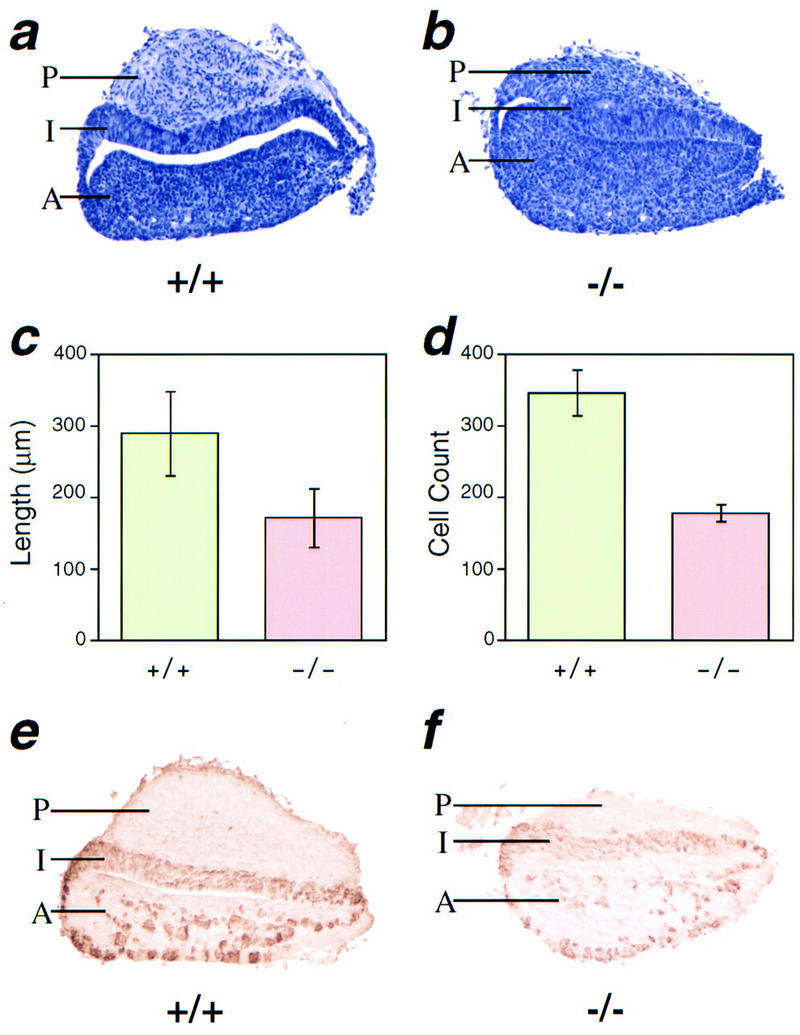
Posterior pituitary lobe is reduced in size in Sim1 mutant mice. Pituitaries collected from Sim1+/+ and Sim1−/− animals were sagittally sectioned for analysis. Staining with hematoxylin revealed that the anterior (A) and intermediate (I) lobes of mutant pituitaries appear normal, but that their posterior (P) lobe is reduced in size (b) when compared to that of wild-type littermates (a). To quantify the difference between the mutants and controls, the mediolateral length of the posterior lobe was measured (c) and cells were counted (d) (see Materials and Methods). These morphometric studies confirmed that the posterior lobe of mutant pituitaries is hypoplasic. Antibodies to ACTH detected normal expression of this antigen in the anterior and intermediate pituitary in mutants (f), when compared to wild-type littermates (e).
Neurons of the prospective PVN/SON do not terminally differentiate in Sim1 mutant embryos
In the mouse, the neurons of the PVN and SON are born between E10.5 and E12.5 (Karim and Sloper 1980; Okamura et al. 1983). Terminal differentiation indicated by the production of hormones starts at E13.5 for CRH-producing neurons, at E15.5 for AVP-producing neurons, and at later stages for the neurons producing the other markers studied here (Hyodo et al. 1991; Seasholtz et al. 1991). Importantly, Crh expression and AVP production were not detectable at E13.5 and E15.5, respectively, in the prospective PVN of Sim1 mutant embryos (data not shown), indicating that PVN/SON neurons do not proceed to terminal differentiation in the mutant.
In the developing anterior hypothalamus, Sim1 expression begins at E10.5, a stage coinciding with the birth of the first PVN/SON neurons, and is restricted to the mantle layer, indicating that neuronal precursors of the PVN/SON begin to express Sim1 only after they leave the ventricular zone and stop proliferating (Fan et al. 1996). We took advantage of the persistence of Sim1 transcripts in mutant mice to determine the fate of the presumptive PVN/SON region during development. Whole-mount in situ hybridization was performed on bisected brains, collected from E12.5 to E15.5, at 24-hr intervals. In these bisected brains, the Sim1 domain has the shape of a stripe that extends from the ventral diencephalic sulcus to the optic recess (Fig. 7a,c,g,k). The prospective PVN/SON occupies the most dorsal region of this stripe. Up to E14.5, a slight but stable decrease in the level of Sim1 expression in this domain was noted in mutant embryos (Fig. 7c,d,g,h). At E14.5, a dramatic down-regulation of Sim1 expression was observed in the most dorsal portion of this domain in half of the mutant embryos studied (2 of 4) (Fig. 7k,l). At E15.5, all of the mutant embryos examined showed this pattern (4 of 4; data not shown). We conclude that in mutant mice, Sim1-expressing cells are born initially in the prospective PVN, but either change fate or die around E15.5 in the absence of terminal differentiation.
Figure 7.
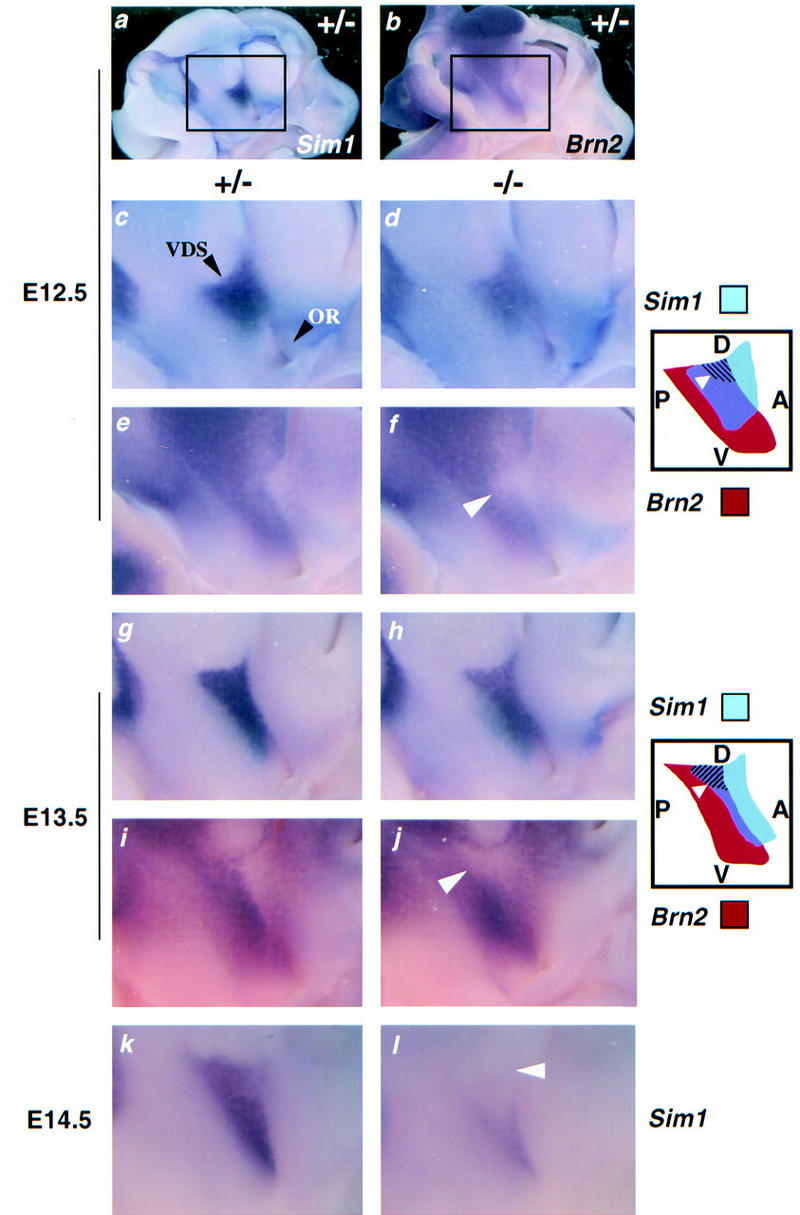
Expression of Sim1 and Brn2 overlaps during development of the hypothalamus. Whole-mount in situ hybridization performed on bisected heads from Sim1+/− (a–c,e,g,i,k) and Sim1−/− (d,f,h,j,l) embryos at the stages indicated at the left. The region of the anterior hypothalamus in which Sim1 (a) and Brn2 (b) are expressed is indicated by a box (a,b), and is magnified (c–l). The ventral diencephalic sulcus (VDS) and the optic recess (OR) are indicated by black arrowheads in c. The diagrams illustrate the spatial relationship of the expression domains of Sim1 (blue) and Brn2 (red) and the regions in which they overlap (purple). The dorsal (D)–ventral (V) and the anterior (A)–posterior (P) axes are indicated in the diagrams. (c,f) At E12.5, the level of Sim1 expression (c,d) is slightly lower in the mutant embryo. Brn2 expression (e,f) is significantly decreased in the mutant in a small patch (arrowhead in f and hatched region in the diagram) localized in the dorsal portion of the region in which the Sim1 and Brn2 expression domains overlap. (g–j) At E13.5, the level of Sim1 expression is slightly lower in the mutant embryo (g,h). Brn2 expression is significantly decreased in the mutant in a small patch (arrowhead in j and hatched region in the diagram) that occupies the entire dorsal portion of the region in which the Sim1 and Brn2 expression domains overlap. (k,l) At E14.5, the level of mutant Sim1 expression is decreased dramatically in the dorsal aspect of its anterior hypothalamic domain (arrowhead).
SIM1 and BRN2 function along the same genetic pathway
The phenotype observed at birth in brains of Sim1 mutant mice is strikingly similar to that reported for Brn2 mutants. Moreover, in Sim1 mutant mice, Brn2 expression in the newborn PVN and SON is undetectable, whereas its expression in the surrounding tissues, including the ventricular layer lining the anterior hypothalamus, is unaffected (Fig. 4k, and l and 5e, and f). Sim1 is expressed essentially in all cells of the PVN, whereas Brn2 is only expressed in a subset of these cells (see Fig. 1 and below). In the SON, Sim1 and Brn2 are expressed by all magnocellular neurons (Schonemann et al. 1995; Malik et al. 1996; and below). It follows that Sim1 acts in all Brn2-expressing cells of the PVN/SON. Therefore, the loss of Brn2-expressing cells in Sim1 mutant PVN/SON is most likely the result of a cell autonomous effect.
The lack of Brn2 expression in the PVN/SON of Sim1 mutant neonates may be accounted for by its hypocellularity and does not necessarily suggest the existence of a genetic interaction between the two genes. To test whether Sim1 and Brn2 interact along the same genetic pathway or in parallel pathways to specify the hypothalamus, we compared their expression during development of Sim1+/− and Sim1−/− embryos by performing whole-mount in situ hybridization on bisected brains collected at E11.5, E12.5, and E13.5. At these stages, Sim1 and Brn2 are each expressed in the presumptive anterior hypothalamus, in a stripe that extends from the ventral diencephalic sulcus to the optic recess (Fig. 7a–c,g,i). These stripes do not overlap completely, as the Sim1 domain extends more anteriorly and the Brn2 domain extends more posteriorly. At E11.5, the Brn2 expression pattern was not affected by the Sim1−/− allele (not shown). In E12.5 and E13.5 Sim1 mutant embryos, however, Brn2 becomes undetectable in a patch immediately ventral to the sulcus (Fig. 7f,j), whereas Sim1 expression remains unchanged (Fig. 7d–h). Although at E12.5 this patch does not occupy the entire dorsal portion of the domain where Sim1 and Brn2 expression overlaps, at E13.5, Brn2 is down-regulated in the whole dorsal aspect of this overlapping domain. The down-regulation of Brn2 in a region of the prospective PVN/SON that continues to produce Sim1 transcripts indicates that SIM1 is necessary for the maintenance of Brn2 expression.
During development of the anterior hypothalamus, the dorsal aspect of the Sim1-expression domain can be divided into a posterior Brn2-positive region and an anterior Brn2-negative region. Interestingly, when the expression of Sim1 and Brn2 was compared on adjacent sections of the newborn PVN, a similar correlation was also observed (Fig. 8). In the most anterior part of the PVN, Sim1 expression is detected but Brn2 expression is not (Fig. 8a,b). More posteriorly, Brn2 expression is present, representing a subset of the Sim1 expression domain (Fig. 8c–f). In the most posterior part of the PVN, Sim1 and Brn2 expression domains are similar, occupying the entire PVN (Fig. 8g,h). The observation raises the possibility that the dorsal Sim1 expression domain contains all the neurons of the prospective PVN from its earliest stage of development. Sim1 would then control the development of the entire PVN. In Sim1 mutants, the loss of Sim1 expression in the whole dorsal aspect of the Sim1 domain at E15.5 (see Fig. 7l) and the absence of expression of all the PVN markers examined here are consistent with this possibility. Because Brn2 expression coincides with the posterior portion of the Sim1 domain, Brn2 may only control the development of the posterior portion of the PVN (see Discussion).
Figure 8.
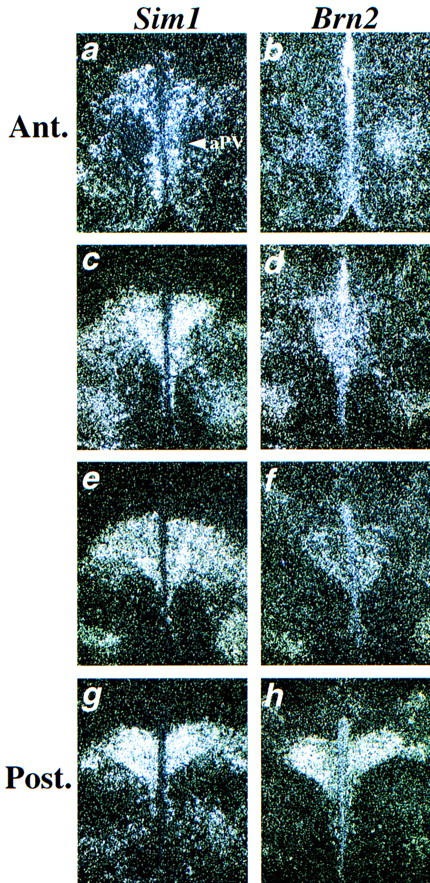
Comparison of Sim1 and Brn2 expression in the PVN. Adjacent sections through the PVN were hybridized either with the Sim1 (a,c,e,g) or the Brn2 (b,d,f,h) probe. Each set of adjacent sections was collected at an interval of 30 μm starting anteriorly. In the most anterior (Ant.) part of the PVN (a,b), Sim1 expression is detected in the PVN and in the aPV (indicated by an arrowhead) but Brn2 expression is not. More posteriorly, the Brn2 expression domain appears to represent a subset of the Sim1 expression domain (c–f). At the posterior (Post.) end of the PVN, Sim1 and Brn2 expression patterns are similar (g,h).
Discussion
Our study shows that Sim1 is essential for the development of specific neuronal lineages of the PVN, aPV, and SON. The entire magnocellular neurosecretory system, which secretes AVP and OT, and three major types of parvocellular neurosecretory cells, identified by the synthesis of TRH, CRH, and SS, fail to develop (Fig. 9). Disruption of the PVN, aPV, and SON is most likely the cause of death of Sim1 mutant mice. Although mutations in CRH, TRH, AVP, or OT do not compromise survival (Schmale and Richter 1984; Muglia et al. 1995; Young et al. 1995; Nishimori et al. 1996; Yamada et al. 1997), the simultaneous loss of multiple cell types within the PVN and SON would likely have a major physiological impact.
Figure 9.
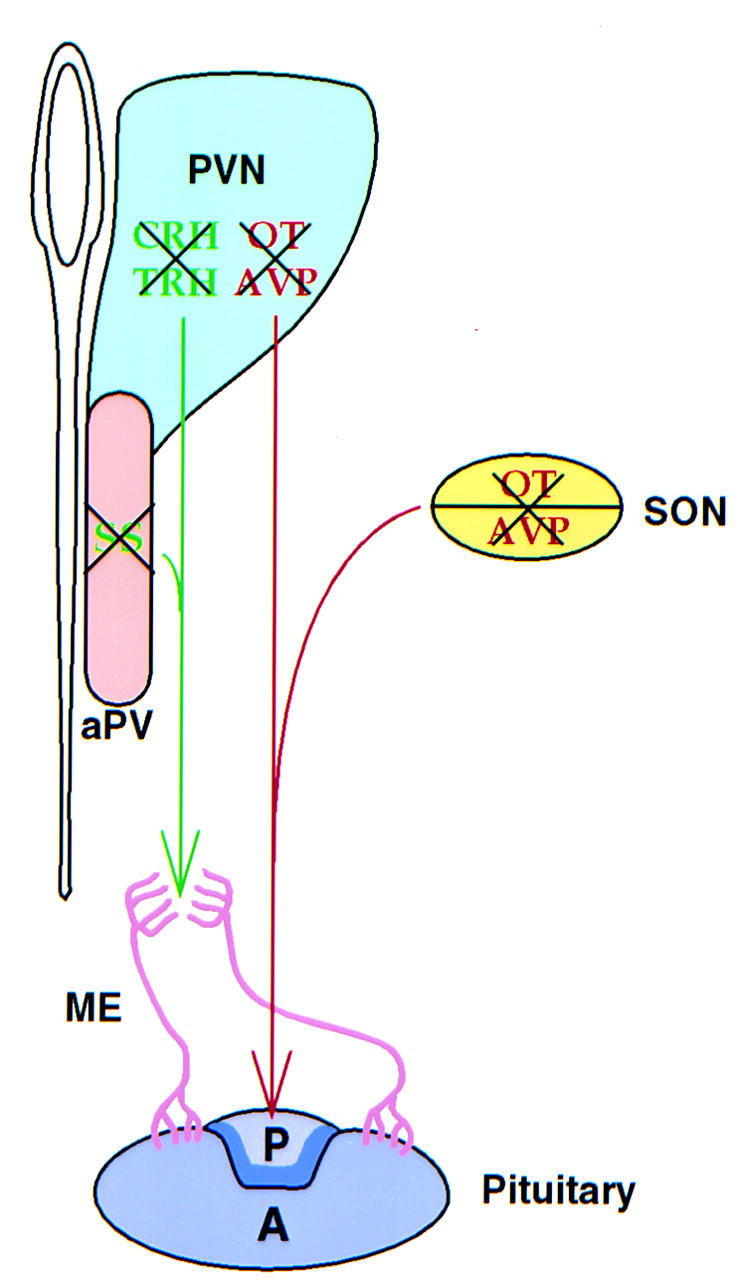
Summary of the consequences of the loss of Sim1 function on the hypothalamic–pituitary axis . Loss of Sim1 function affects the development of both the magnocellular and parvocellular lineages. The magnocellular neurons are located in the paraventricular nucleus (PVN) and the supraoptic nucleus (SON) and project their axons (red) to the posterior pituitary where they secrete AVP and OT. The parvocellular neurons of the PVN and of the anterior periventricular nucleus (aPV) project their axons (green) to the medial eminence (ME) where they release TRH, CRH, and SS. These hormones are transported by the ME portal vasculature (pink) to the anterior pituitary; there they modulate the secretion of peptidic hormones. Expression of the peptides studied here, AVP, OT, CRH, TRH, and SS, defines at least five distinct cell types. Morphological and marker analysis indicates that these lineages are absent in Sim1 mutant mice (crossed-through by black X). (Adapted from Schonemann et al. 1995.)
Loss of magnocellular neurons resulted in a decreased number of pituicytes, the astroglial cells of the posterior lobe of the pituitary. In contrast, the loss of the parvocellular lineages studied here had no apparent impact on the development of the anterior pituitary. Although growth hormone-producing cells require the hypothalamic factor GHRH for development (Godfrey et al. 1993; Lin et al. 1993; Li et al. 1996), ACTH- and TSH-producing cells appear to develop normally in the absence of their regulator CRH (Muglia et al. 1995; Schonemann et al. 1995; Smith et al. 1998; Timpl et al. 1998) and TRH (Yamada et al. 1997), respectively. The fact that the production of ACTH and TSH is not perturbed in the Sim1 mutant pituitaries supports this conclusion.
Sim1 controls the later stages of PVN/SON development
Neurons that will constitute the PVN and SON are born between E10.5 and E12.5 (Karim and Sloper 1980; Okamura et al. 1983). Terminal differentiation indicated by the onset of production of the secreted neuropeptides occurs during the following days. In Sim1 mutant embryos, Sim1-expressing cells of the prospective PVN/SON are generated and appear to survive up to at least E14.5/15.5. However, they stop expressing Brn2 at E12.5 and they never terminally differentiate. Therefore, it seems that Sim1 controls the later stages of differentiation of the PVN/SON neurons, after the neuronal precursors have been produced. In the absence of terminal differentiation, Sim1-expressing cells either die or change fate. The correct regional expression of the Sim1 mutant transcript up to the stage of terminal differentiation and the later striking hypocellularity of the mutant PVN and SON suggest that these cells do indeed die.
SIM1 does not homodimerize but it does heterodimerize with aryl hydrocarbon receptor nuclear translocator (ARNT) and ARNT2, bHLH–PAS proteins that are considered to be the partner of many members of this family (Swanson et al. 1995; Ema et al. 1996; Probst et al. 1997). However, in contrast to Drosophila sim and to all other known bHLH–PAS proteins, which act as transcriptional activators, SIM1 and SIM2 behave as transcriptional repressors in cell culture/transfection assays (Ema et al. 1996; Moffett et al. 1997; Probst et al. 1997). This repressor activity has been mapped to a region located in the carboxy-terminal half of the protein. Consistent with their divergent activity, the carboxyl termini of fly and murine SIMs are not conserved. It is unclear whether SIM1 also functions as a repressor in vivo. The phenotype observed in Sim1 mutant mice is consistent with either SIM1 acting as a transactivator of genes involved in specifying the PVN/SON lineages or acting as a repressor of a gene that itself represses the PVN/SON differentiation program.
SIM1 is expressed at birth in the PVN and the SON, long after these nuclei have differentiated. This observation suggests that, in addition to specifying neuronal lineages, SIM1 could play a second role in the PVN and SON. For instance, SIM1 could also be involved directly in maintaining the expression of the secreted neuropeptides examined here. Alternatively, SIM1 could have a more dynamic function by controlling the transcription of various genes in response to specific physiological stimuli, possibly mediated by the production of a ligand. A precedent for this hypothesis is provided by the aryl hydrocarbon receptor, a bHLH–PAS protein whose activity is controlled by the ligand dioxin (Burbach et al. 1992). In addition to the PVN/SON, Sim1 has also been documented to be expressed in the developing somites and kidneys (Fan et al. 1996). However, no discernible phenotype has been observed in these tissues (or their derivatives) in mutant animals.
Sim1 and Brn2 interact genetically in the hypothalamus
Drosophila Sim is a master regulator of the CNS midline (Nambu et al. 1990, 1991). Loss of sim function results in the complete absence of midline development. Drifter, a POU domain transcription factor that binds the same DNA sequence as does BRN2, has also been implicated in controlling the development of CNS midline cells in the fly (Anderson et al. 1995). Expression and phenotypic analysis have shown that Sim acts upstream of Drifter. Our data indicate that in mice, SIM1 likewise acts upstream of a POU domain transcription factor BRN2. Specifically, we have found that Brn2 is down-regulated in a region of the prospective PVN/SON that continues to express the Sim1 mutant transcript, indicating that SIM1 and BRN2 function along the same pathway. The fact that Brn2 expression in the prospective hypothalamus of Sim1 mutant embryos is not altered until E12.5 suggests that Sim1 is not involved in initiating but in maintaining Brn2 expression. Whether SIM1 controls BRN2 transcription directly or indirectly remains an open question.
Consistent with the conclusion that Sim1 functions upstream to maintain Brn2 expression, we show here that all the hypothalamic lineages that are reported to be affected by the loss of Brn2 function are also affected in Sim1-deficient mice; the loss of Brn2 function affects the development of AVP-, OT-, and CRH-producing cells (Nakai et al. 1995; Schonemann et al. 1995), and the same cell types are affected by the loss of Sim1 function. In contrast, TRH- and SS-producing cells are missing in Sim1 mutant but are present in Brn2 mutant PVN and aPV (Schonemann et al. 1995). This is consistent with the observation that Trh and BRN2 expression share minimal overlap in the PVN (Schonemann et al. 1995; Malik et al. 1996). Similarily, Brn2 is not expressed in the aPV, where SS is produced abundantly (Schonemann et al. 1995; and our data). Although Nakai et al. (1995) have reported that TRH- and SS-producing cells fail to develop in Brn2 mutant mice, this observation has not been reproduced (Schonemann et al. 1995).
The loss of the five cell types studied here in Sim1 mutant mice raises the possibility that most, if not all, of the neuronal lineages constituting the PVN, SON, and aPV originate from the dorsal aspect of the prospective anterior hypothalamic Sim1 domain. This domain can be divided into an anterior region only expressing Sim1 and a posterior region expressing both Sim1 and Brn2. It is tempting to speculate that the CRH, AVP, and OT lineages, which are affected in both Sim1 and Brn2 mutant mice, are derived from the posterior region, whereas the TRH and SS lineages, which are only affected in Sim1 mutant mice, are derived from the anterior region. This is consistent with the observation that in the newborn hypothalamus, Brn2 is not expressed in the aPV or in the anterior end of the PVN, where SS- and TRH-producing cells, respectively, are found.
In Brn2 mutant mice, precursors of the PVN and SON survive up to E15.5 but fail to express the secreted neuropeptides (Schonemann et al. 1995). BRN2 binds and activates the CRH promotor, supporting a role for BRN2 in controlling the terminal stage of differentiation (Li et al. 1993). The survival of the PVN/SON precursors up to E15.5 in both Sim1 and Brn2 mutant embryos and the down-regulation of Brn2 in Sim1 mutant embryos would suggest that the loss of Brn2 expression mediates the effect of the Sim1 mutant allele on the development of CRH, AVP, and OT neuroendocrine lineages. Whether Sim1 controls the differentiation of TRH- and SS-expressing cells directly or indirectly, through activation of another POU domain transcription factor, remains to be determined.
Materials and methods
Targeting vector construction and gene targeting in ES cells
A Sim1 genomic clone was isolated from a 129/Sv mouse genomic library (EMBL4). A 1-kb fragment containing 750 bp of the 5′ region, the initiation codon, and the sequence coding for the basic domain (the first 17 amino acids) was replaced by a Pgk–neo cassette (Tybulewicz et al. 1991). The final targeting vector contained 1.2 kb of Sim1 sequence 5′ and 5 kb of Sim1 sequence 3′ of the Pgk–neo cassette, and a Pgk–tk cassette was inserted at the 5′ end of the construct (Fig. 2a). In the predicted targeted allele, an alternative initiation methionine (amino acids 52) is found after the bHLH domain. If this methionine were used, the protein product would presumably be nonfunctional.
Twenty micrograms of the construct was linearized at a NotI site located at its 3′ end and electroporated into passage 13 R1 ES cells, which were grown as previously described (Nagy et al. 1993). To obtain negative and positive selection for homologous recombinants, gancyclovir and G418 were added to the culture medium at a final concentration of 0.55 μg/ml and 150 μg/ml, respectively. Double-resistant clones were further analyzed by Southern blot using a probe containing Sim1 genomic sequences 5′ of those used in the targeting vector. This probe hybridizes to a 5.2-kb BamHI fragment of the wild-type Sim1 allele and to a 3.4-kb BamHI fragment of the Sim1 mutant allele (Fig. 2b).
Generation of germ-line-transmitting mice and genotyping
Homologous recombinant ES cell clones were microinjected into C57BL/6 blastocysts to produce chimeric mice. The resulting male chimeras were backcrossed to C57BL/6 females. Germ-line heterozygotes were identified by PCR and Southern blot analysis. In addition to the 5′ probe described above, a probe containing Sim1 genomic sequences 3′ of those cloned into the targeting vector was used to confirm that the Sim1 locus was targeted correctly (Fig. 2b). This probe detects a wild-type 11.2-kb KpnI fragment and a mutant 12.4-kb KpnI fragment. For PCR, two sets of primers were used. The first set was designed to detect the mutant allele, and amplifies a 189-bp fragment of the neo gene. The second set was designed to detect the wild-type Sim1 allele, and amplifies a 287-bp fragment that is deleted in the mutant allele. The sequences of these primers are as follows: neo, CTCGGCAGGAGCAAGGTGAGATG and GTCAAGACCGACCTGTCCGGTGC; Sim1, ATACCCAGGGCTGTGACGAG and AATGCTGGAAGGACTCCCGA. The reaction was carried out at 94°C for 30 sec, 61°C for 45 sec, and 72°C for 45 sec with 10% DMSO for 33 cycles, using Taq polymerase.
Immunocytochemisty, in situ hybridization, and histology
Polyclonal rabbit antibodies were raised against a peptide corresponding to residues 542–560 (ESGDRYRTEQYQNSPHEPS) of SIM1. The DNA sequence encoding this peptide region is not deleted in the targeted allele. For immunocytochemistry, coronal sections of newborn brains fixed in Carnoy’s fluid and embedded in paraffin were boiled in an antigen-unmasking solution (Vector). Immunocytochemistry was performed with the ABC immunoperoxidase system (Vector), using peptide affinity-purified antibodies diluted to 1:200.
Rabbit antisera against AVP, OT, and SS were purchased from Peninsula Laboratories and used at a dilution of 1:500. Immunocytochemistry was performed on paraffin sections of newborn brains fixed in Bouin’s fluid, using the TSA amplification procedure (New England Nuclear) followed by standard HRP staining. The ACTH antibody (Peninsula Lab) and the TSH antibody [a gift from Dr. A.F. Parlow, National Institutes of Health (NIH)] were diluted to 1:500, applied to paraffin sections of pituitaries fixed in Bouin’s fluid and detected with the ABC system.
In situ hybridization on frozen and paraffin sections, and on whole embryos was performed as previously described (Fan et al. 1996). The Sim1 probe used in this study corresponds to a 1.2-kb sequence 3′ to the disrupted region, thus allowing hybridization to both the wild-type and mutant transcripts. The CRH probe was graciously provided by A.F. Seasholtz (Seasholtz et al. 1991). The Brn2 and Trh probes were generated by RT–PCR, using mouse newborn hypothalamus RNA extracts to prepare the substrate for PCR. The sequence of the primers used to amplify Brn2 and Trh is as follows: Trh, GTGCCAACCAAGACAAGGAT and CAATCCTACCCTTTCTGAGG; Brn2, AAGCAATTCAAGCAGAGGCG and CTACTTCATTGCCTGGGTAC.
For histological analysis, newborn brains and pituitaries were fixed in Bouin’s fluid, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin.
Cell counts
Pituitaries of mutant newborn mice and control littermates were sectioned sagittally. Sections in which the posterior lobe was present were then located. By multiplying, for each case, the number of sections in which the posterior lobe was present by the thickness of each section (6 μm), we obtained a measure of the mediolateral length of the posterior lobe. In addition, the nuclei of the posterior lobe were counted on five adjacent sections representing the medial region of the posterior pituitary. Results were analyzed with the Student’s t test.
Figure 5.
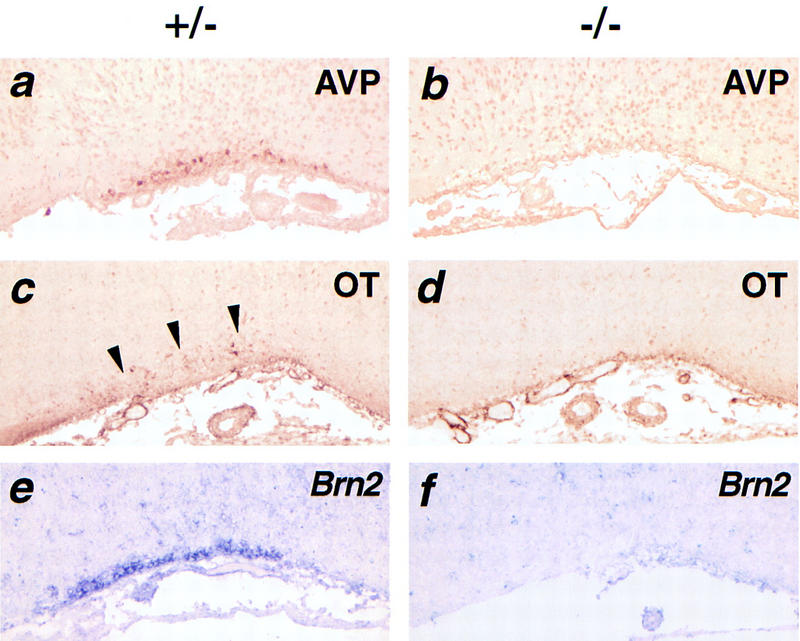
Molecular markers of the SON are not detected in Sim1 mutant mice. Coronal sections through the SON of Sim1+/− (a,c,e) or Sim1−/− (b,d,f) newborn mice. AVP (a,b) and Brn2 (e,f) are readily detected in sections of mutant heterozygotes but not mutant homozygotes. Although weak, OT (c,d) production is also detectable in mutant heterozygotes (arrowheads) but not in mutant homozygotes. AVP and OT were detected by immunocytochemistry, whereas Brn2 was detected by in situ hybridization using a digoxigenin–UTP-labeled probe.
Acknowledgments
We are especially grateful to Drs. Marc Tessier-Lavigne and Gail R. Martin for helping to initiate this work and to produce the Sim1 mutant mouse line, and for insightful comments on the manuscript. We also thank Dr. A.F. Seasholtz for the Crh probe, Dr. A.F. Parlow for the TSH antibody, Drs. D. Brown, D. Koshland, J. Rubenstein, Y. Zheng, and A. Spradling for critical reading of the manuscript. J.L.M. is a Clinician-Scientist fellow of the Medical Research Council of Canada. T.R. was supported by an NIH post-doctoral training grant (CA09043). This work is supported by the John Merck Fund, the NIH Shannon Award (R55 HD/OD35596-01), and by a NIH grant (RO1 HD35596-01) awarded to C.-M.F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL fan@mail1.ciwemb.edu; FAX (410) 243-6311.
References
- Altman J, Bayer SA. Development of the diencephalon in the rat. II. Correlation of the embryonic development of the hypothalamus with the time of origin of its neurons. J Comp Neurol. 1978;182:973–994. doi: 10.1002/cne.901820512. [DOI] [PubMed] [Google Scholar]
- Anderson MG, Perkins GL, Chittick P, Shrigley RJ, Johnson WA. drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline cells. Genes & Dev. 1995;9:123–137. doi: 10.1101/gad.9.1.123. [DOI] [PubMed] [Google Scholar]
- Burbach KM, Poland A, Bradfield CA. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews ST. Control of cell lineage-specific development and transcription by bHLH–PAS proteins. Genes & Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- Crews ST, Thomas JB, Goodman CS. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- Du Vigneaud V. Hormones of posterior pituitary glands: Oxytocin and vasopressin. Harvey Lect Ser. 1955;L:1–26. [PubMed] [Google Scholar]
- Ema M, Morita M, Ikawa S, Tanhaka M, Gotoh O, Saijoh S, Fujii H, Hamada H, Fujii-Kuriyama Y. Two new members of murine sim gene family are transcriptional repressors and show different patterns during mouse embryogenesis. Mol Cell Biol. 1996;16:5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH–PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C-M, Kuwana E, Bulfone A, Fletcher CF, Copeland NG, Jenkins NA, Crews S, Martinez S, Puelles L, Rubenstein JLR, Tessier-Lavigne M. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol Cell Neurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rose D, Rosenfeld MG. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Godfrey P, Rahal JO, Beamer WG, Copeland NG, Jenkins NA, Mayo KE. GHRH receptor of little mice contains a missense mutation in the extracellular domain that disrupts receptor function. Nat Genet. 1993;4:227–232. doi: 10.1038/ng0793-227. [DOI] [PubMed] [Google Scholar]
- Hankinson O. The Aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Hyodo S, Yamada C, Takezawa T, Urano A. Expression of provasopressin gene during ontogeny in the hypothalamus of developing mice. Neuroscience. 1991;46:241–150. doi: 10.1016/0306-4522(92)90024-v. [DOI] [PubMed] [Google Scholar]
- Karim MA, Sloper JC. Histogenesis of the supraoptic and paraventricular neurosecretory cells of the mouse hypothalamus. J Anat. 1980;130:341–347. [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zeitler PS, Valerius MT, Small K, Potter SS. Gsh-1, an orphan Hox gene, is required for normal pituitary development. EMBO J. 1996;15:714–724. [PMC free article] [PubMed] [Google Scholar]
- Li P, He H, Gerrero MR, Mok M, Aggarwal A, Rosenfeld MG. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes & Dev. 1993;7:2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- Lin S-C, Lin CR, Gukovsky I, Lusis AJ, Sawchenko PE, Rosenfeld MG. Molecular basis of the little mouse phenotype and implications for cell type-specific growth. Nature. 1993;364:208–213. doi: 10.1038/364208a0. [DOI] [PubMed] [Google Scholar]
- Malik KF, Kim J, Hartman AL, Kim P, Young WS., III Binding preferences of the POU domain protein BRAIN-4: Implications for autoregulation. Mol Brain Res. 1996;38:209–221. doi: 10.1016/0169-328x(95)00308-f. [DOI] [PubMed] [Google Scholar]
- Michaud J, Fan C-M. Single-minded—Two genes, three chromosomes. Genome Res. 1997;7:569–571. doi: 10.1101/gr.7.6.569. [DOI] [PubMed] [Google Scholar]
- Moffett P, Dayo M, Reece M, McCormick MK, Pelletier J. Characterization of msim, a murine homolog of the Drosophila sim transcription factor. Genomics. 1996;35:144–155. doi: 10.1006/geno.1996.0333. [DOI] [PubMed] [Google Scholar]
- Moffett P, Reece M, Pelletier J. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol Cell Biol. 1997;17:4933–4947. doi: 10.1128/mcb.17.9.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia L, Jacobson L, Dikkes P, Majzoub JA. Corticotropin-releasing hormone deficiency reveals major fetal but not adult glucocorticoid need. Nature. 1995;373:427–432. doi: 10.1038/373427a0. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abranow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, Shiba K, Noda T. The POU domain transcription factor BRN2 is required for the determination of specific neuronal lineages in the hypothalamus of the mouse. Genes & Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Franks RG, Hu S, Crews ST. The single-minded gene of Drosophila is required for the expression of genes important for the development of CNS midline cells. Cell. 1990;63:63–75. doi: 10.1016/0092-8674(90)90288-p. [DOI] [PubMed] [Google Scholar]
- Nambu JR, Lewi JL, Wharton KA, Crews ST. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- Nishimori K, Young LJ, Guo Q, Wang Z, Insel TR, Matzuk MM. Oxytocin is required for nursing but is not essential for parturition or reproductive behavior. Proc Natl Acad Sci. 1996;93:11699–11704. doi: 10.1073/pnas.93.21.11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Fukui K, Koyama E, Tsutou HLO, Tsutoou T, Terubayashi H, Fujisawa H, Ibata Y. Time of vasopressin neuron origin in the mouse hypothalamus: Examination by combined technique of immunocytochemistry and [3H]thymidine autoradiography. Dev Brain Res. 1983;9:223–226. doi: 10.1016/0165-3806(83)90055-x. [DOI] [PubMed] [Google Scholar]
- Probst MR, Fan C-M, Tessier-Lavigne M, Hankinson O. Two murine homologs of the Drosophila Single-minded protein that interact with the mouse aryl hydrocarbon receptor nuclear translocator protein. J Biol Chem. 1997;272:4451–4457. doi: 10.1074/jbc.272.7.4451. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Imaki T, Vale W. Co-localization of neuroactive substances in the endocrine hypothalamus. In Functional anatomy of the neuroendocrine hypothalamus. Ciba Found Symp. 1992;168:16–42. doi: 10.1002/9780470514283.ch3. [DOI] [PubMed] [Google Scholar]
- Schimdt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Schmale H, Richter D. Single base deletion in the vasopressin gene is the cause of diabetes insipidus in Brattleboro rats. Nature. 1984;308:705–709. doi: 10.1038/308705a0. [DOI] [PubMed] [Google Scholar]
- Schonemann MD, Ryan AK, McEvilly RJ, O’Connell SM, Arias CA, Kalla KA, Li P, Sawchenko PE, Rosenfeld MG. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor BRN2. Genes & Dev. 1995;9:3122–3135. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- Seasholtz AF, Bourbonais FJ, Harnden CE, Camper SA. Nucleotide sequence and expression of the mouse corticotropin-releasing hormone gene. Mol Cell Neurosci. 1991;2:266–273. doi: 10.1016/1044-7431(91)90054-r. [DOI] [PubMed] [Google Scholar]
- Smith GW, Aubry J-M, Dellu F, Contarino A, Bilezikjian LM, Gold LH, Chen R, Marchuk Y, Hauser C, Bentley CA, Sawchenko PE, Koob GF, Vale W, Lee K-F. Corticotropin releasing factor receptor 1-deficient mice display decreased anxiety, impaired stress response, and aberrant neuroendocrine development. Neuron. 1998;20:1093–1102. doi: 10.1016/s0896-6273(00)80491-2. [DOI] [PubMed] [Google Scholar]
- Swanson HI, Chan WK, Bradfeld CA. DNA binding specificities and pairing rules of the Ah receptor, ARNT, and Sim proteins. J Biol Chem. 1995;280:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- Swanson LW. Organization of mammalian neuroendocrine system. In: Mountcastle VB, Bloom FE, Geiger SR, editors. Handbook of physiology, Section 1: The nervous system, Vol. IV, Intrinsic regulatory systems of brain. Bethesda, MD: American Physiological Society; 1986. pp. 317–363. [Google Scholar]
- Swanson LW, Sawchenko PE. Hypothalamic integration: Organization of the paraventricular and supraoptic nuclei. Annu Rev Neurosci. 1983;6:269–324. doi: 10.1146/annurev.ne.06.030183.001413. [DOI] [PubMed] [Google Scholar]
- Thomas JB, Crews ST, Goodman CS. Molecular genetics of the single-minded locus: A gene involved in the development of the Drosophila nervous system. Cell. 1988;52:133–141. doi: 10.1016/0092-8674(88)90537-5. [DOI] [PubMed] [Google Scholar]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes & Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- Timpl P, Spanagel R, Sillaber I, Kresse A, Reul JM, Stalla GK, Blanquet V, Steckler T, Holsboer F, Wurst W. Impaired stress response and reduced anxiety in mice lacking a functional corticotropin-releasing hormone receptor. Nat Genet. 1998;19:162–166. doi: 10.1038/520. [DOI] [PubMed] [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang B-H, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Saga Y, Shibusawa N, Hirato J, Murakami M, Iwasaki T, Hashimoto K, Satoh T, Wakabayashi K, Taketo MM, Mori M. Tertiary hypothyroidism and hyperglycemia in mice with targeted disruption of the thyrotropin-releasing hormone gene. Proc Natl Acad Sci. 1997;94:10862–10867. doi: 10.1073/pnas.94.20.10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki A, Noda S, Kudoh J, Shindoh N, Maeda H, Minoshima S, Kawasaki K, Shimizu Y, Shimizu N. The mammalian single-minded gene: Mouse cDNA structure and diencephalic expression indicate a candidate gene for Down syndrome. Genomics. 1996;35:136–143. doi: 10.1006/geno.1996.0332. [DOI] [PubMed] [Google Scholar]
- Young WS, III, Shepard E, Amico J, Hennighausen L, Wagner KU, LaMarca ME. Deficiency in mouse oxytocin prevents milk injection but not fertility or parturition. J Neuroendocrinol. 1995;8:847–856. doi: 10.1046/j.1365-2826.1996.05266.x. [DOI] [PubMed] [Google Scholar]


