The compliance and dimensionality of the ECM regulate distinct changes in microtubule growth speed and growth persistence.
Abstract
During angiogenesis, cytoskeletal dynamics that mediate endothelial cell branching morphogenesis during vascular guidance are thought to be regulated by physical attributes of the extracellular matrix (ECM) in a process termed mechanosensing. Here, we tested the involvement of microtubules in linking mechanosensing to endothelial cell branching morphogenesis. We used a recently developed microtubule plus end–tracking program to show that specific parameters of microtubule assembly dynamics, growth speed and growth persistence, are globally and regionally modified by, and contribute to, ECM mechanosensing. We demonstrated that engagement of compliant two-dimensional or three-dimensional ECMs induces local differences in microtubule growth speed that require myosin II contractility. Finally, we found that microtubule growth persistence is modulated by myosin II–mediated compliance mechanosensing when cells are cultured on two-dimensional ECMs, whereas three-dimensional ECM engagement makes microtubule growth persistence insensitive to changes in ECM compliance. Thus, compliance and dimensionality ECM mechanosensing pathways independently regulate specific and distinct microtubule dynamics parameters in endothelial cells to guide branching morphogenesis in physically complex ECMs.
Introduction
Cell branching morphogenesis is critical to establishing a functional vascular system. During angiogenesis, vascular endothelial “tip cells” lead a migrating chain of endothelial cells (ECs). Tip ECs extend cell branches in response to directional cues that guide their migration through the ECM to establish the vascular network (Gerhardt et al., 2003, 2004; Gerhardt and Betsholtz, 2005). Similar morphogenetic processes occur during the establishment of the nervous system. In neurons, cell branches (neurites) also extend from the cell body to form axons and dendrites, both tipped by growth cones that are guided by extracellular cues toward target cells to establish a functional neuronal network (Dickson, 2002; Kalil and Dent, 2005). It is well established that during neurite initiation, cell branching is mediated by the coordinated remodeling of the actomyosin and microtubule (MT) cytoskeletons (Dehmelt et al., 2003; Dent and Gertler, 2003; Dehmelt and Halpain, 2004; Rösner et al., 2007), but the mechanisms underlying EC branching morphogenesis are less well understood. Myosin II contractility in the cortical actin cytoskeleton is a negative regulator of neurite initiation and elongation, as inhibition of myosin II or its upstream activators promotes these processes, whereas myosin II overexpression inhibits them (Kollins et al., 2009). Similarly, in ECs, myosin II contractility negatively regulates branch initiation, as indicated by the formation of branches at sites of local myosin II depletion in the cortex (Fischer et al., 2009). In neurons, MTs and their dynamic instability are required for neurite initiation and extension, elaboration of the growth cone, and axonal branching (Dent and Kalil, 2001; Dehmelt et al., 2003). In contrast to the wealth of information on the role of MTs in neuronal elaboration, the role of MTs in EC branching morphogenesis is not well defined.
Although soluble and diffusible guidance cues have long been known to regulate the cytoskeleton in cell morphogenesis and migration during angiogenesis, regulation by the physical/mechanical attributes of the ECM, a process termed “ECM mechanosensing,” is now emerging as an important mechanism (Ingber, 2002; Ghosh et al., 2008; Mammoto et al., 2009). Two specific physical properties of the ECM, compliance (stiffness/softness) and topology, can influence cell signaling and the organization of the cytoskeleton to drive changes in cell morphology. In “compliance mechanosensing,” cells sense ECM stiffness through cell–ECM focal adhesions, and respond by modulating myosin II activity such that cell contractile forces match the resistive compliance of the ECM (Pelham and Wang, 1997; Olson, 2004; Discher et al., 2005; Saez et al., 2005). Accordingly, compliant (soft) ECMs promote down-regulation of myosin II activity and extension of cell branches in both ECs and neurons, whereas stiff ECMs enhance myosin II activity to limit cell branching (Flanagan et al., 2002; Fischer et al., 2009).
Cells were known to exhibit differential morphological responses to 2D versus 3D ECM engagement (Beningo et al., 2004; Even-Ram and Yamada, 2005). This topology-dependent phenomenon is referred to as “ECM dimensionality mechanosensing.” When cells engage a planar 2D ECM in tissue culture, this defines their ventral surface and leaves their dorsal surface unengaged. Here, many cells including ECs display a spread, flattened morphology with actin stress fibers and peripheral lamellipodia. In contrast, ECs embedded in a 3D ECM (Fischer et al., 2009) or fibroblasts plated on planar 2D ECMs with local dorsal ECM engagement (Beningo et al., 2004) display a spindle-shaped morphology, with long cell extensions tipped by tiny lamellipodia (Cukierman et al., 2002; Doyle et al., 2009). Thus, compliance and dimensionality mechanosensing induce major changes in cell morphology that are likely mediated by specific regulation of the cytoskeletal systems.
We showed recently that 3D ECM engagement by ECs synergizes with ECM compliance to enhance cell branching, which suggests that compliance and dimensionality mechanosensing may mediate cell branching through distinct pathways (Fischer et al., 2009). One possibility is that compliance and dimensionality mechanosensing in ECs may affect cell branching morphogenesis differentially through myosin II– or MT-dependent pathways. Indeed, evidence in other cell types suggests that MTs, like myosin II, may be regulated by ECM mechanosensing, and may also mediate the morphological responses to mechanosensing (Kaverina et al., 2002; Rhee et al., 2007). For example, MTs grow toward sites of local ECM stiffening and retract in response to locally applied contraction inhibitors (Kaverina et al., 2002). In compliant 3D ECMs, MTs are required for branching morphology of fibroblasts, whereas in stiff 3D ECMs, MTs are not required for branching but are needed for cell polarization (Rhee and Grinnell, 2007). However, whether MT assembly dynamics are regulated by independent compliance or topology mechanosensing pathways to mediate EC branching morphogenesis is not known.
To better understand the mechanism of tip EC guidance during angiogenesis, we sought to determine if MT dynamics were regulated by ECM compliance and dimensionality, and in turn, to explore the role of regionally regulated MT dynamics in mechanosensing-mediated modulation of EC branching morphogenesis. To accomplish this, we relied on a recently developed computational image analysis method that tracks the position of fluorescently tagged MT plus end–tracking proteins to derive independently regulated parameters of MT dynamic instability, including growth rate and growth persistence (1/catastrophe frequency). Our systematic measurements show that in both 2D and 3D ECMs, compliance mechanosensing regulates the MT growth rate through a myosin II–dependent pathway. In contrast, compliance mechanosensing in 2D ECMs regulates MT growth persistence, whereas in 3D ECMs, MT growth persistence is insensitive to compliance. These results indicate that distinct pathways regulate specific parameters of MT dynamic instability via compliance and topology mechanosensing to guide branching morphogenesis of ECs.
Results
MT dynamics inhibit frequent cell branching and promote rapid directional migration
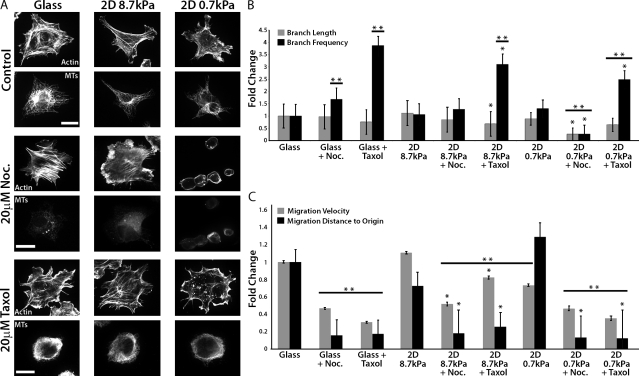
We previously demonstrated that EC branching is enhanced by softer (more compliant) ECMs (Fischer et al., 2009). To determine if MT dynamics mediate compliance-induced EC branching morphogenesis and migration, we investigated the response of human umbilical vein ECs (HUVECs) to MT-perturbing drugs and 2D ECMs of different stiffness. HUVECs were cultured on collagen-coated glass or on either 8.7 kPa (stiff) or 0.7 kPa (soft) collagen-coupled polyacrylamide (PA) substrates (Fig. 1 A). We analyzed branch frequency, defined as the number of pseudopodial protrusions >10 µm in length per unit cell area, as well as branch length. We first examined the role of MTs in EC branching by incubating HUVECs with 20 µM nocodazole to inhibit MT assembly and promote disassembly. When cultured on glass or a stiff ECM, this treatment increased branch frequency but had no effect on branch length. Both branch length and branch frequency were reduced in nocodazole-treated cells on soft ECMs (0.7 kPa), likely because the cells spread poorly (Fig. 1, A and B). To determine the role of MT disassembly in cell branching, we investigated the effects of stabilizing MTs with 20 µM taxol. On both glass and compliant ECMs, taxol treatment reduced branch length and significantly increased branch frequency (P < 0.05; Fig. 1 B). Because both nocodazole and taxol promoted frequent branching in well-spread cells on stiff or soft ECMs, this suggests that MT assembly/disassembly dynamics negatively regulate cell branching, independent of ECM compliance.
Figure 1.
Perturbation of MT growth or shortening promotes HUVEC branching and inhibits rapid directional migration. (A) Immunolocalization of MTs and fluorescent phalloidin staining of actin in HUVECs cultured on collagen-coated glass, or stiff (8.7 kPa) or soft (0.7 kPa) compliant ECMs. Samples were treated with DMSO (control), 20 µM nocodazole (Noc.), or 20 µM taxol for 90 min. Bars, 20 µm. (B) Analysis of the effects of the treatments in A on cell branch frequency and length. (C) Analysis of the effects of the treatments in A on cell migration velocity and distance to origin. *, P < 0.05 comparing compliance versus compliance + drug. **, P < 0.05 compared with glass (one-way ANOVA). Error bars indicate standard deviation.
We next sought to determine the role of MT assembly dynamics in ECM compliance-mediated modulation of cell migration (Fig. 1 C). Unlike other studies (Lo et al., 2000; Fischer et al., 2009; Petrie et al., 2009), we found that increasing ECM compliance did not significantly influence HUVEC migration velocity or directional persistence (defined as distance from origin), an effect that is likely caused by the relatively high variability in directional movement and instantaneous velocities of HUVECs compared with other cell types. Nevertheless, treatment of cells on either stiff or soft ECMs with either nocodazole or taxol significantly reduced both migration velocity and directional persistence (Fig. 1 C and Videos 1–3). Together, these data suggest that MT assembly/disassembly dynamics restrict excessive cell branching to promote fast, directional cell migration independent of ECM compliance.
Increased ECM compliance promotes fast-growing, dynamically unstable MTs
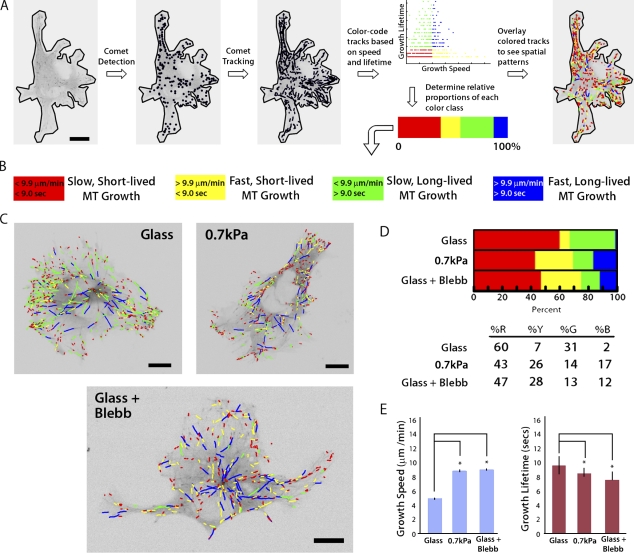
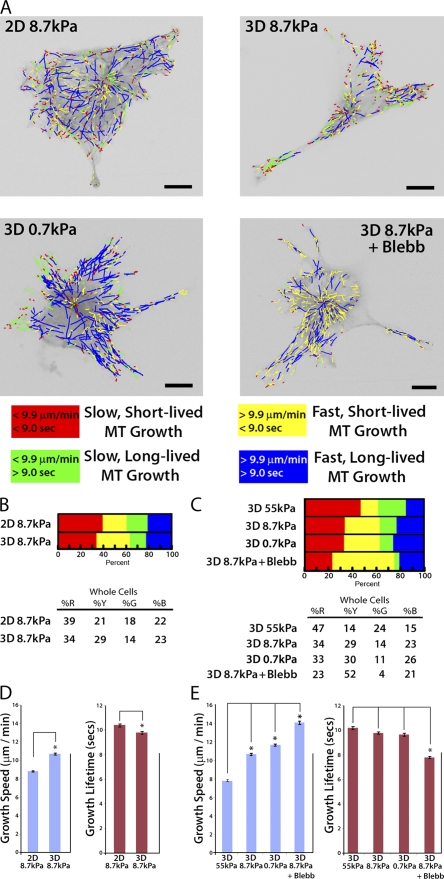
Because both MT assembly/disassembly dynamics and ECM compliance (Fischer et al., 2009) influence cell branching, we reasoned that ECM compliance mechanosensing could regulate MT assembly dynamics to modulate cell branching. To test the effects of substrate compliance on MT dynamics, we performed high-resolution live-cell imaging of fluorescently labeled EB3, which tracks with the growing plus ends of MTs. We used our recently developed software package plusTipTracker (Matov et al., 2010), which enables high-throughput measurement of MT growth speed and growth persistence (1/catastrophe frequency), in time-lapse image series of fluorescent MT plus end–binding proteins (Fig. 2 A). Data output from plusTipTracker includes image overlays of MT plus end motion tracks, with the magnitudes of MT growth speed and growth lifetime color-coded to allow qualitative visualization of regional differences in these values throughout the cell (Fig. 2, A and C). In addition, MT dynamics data from individual whole cells or user-defined subcellular regions can be pooled to allow quantitative measurements of variations in MT track density (Fig. S1 A), as well as statistical analysis of changes in MT dynamics behavior under different experimental conditions or in different subcellular regions that may not be readily apparent by eye (Video 4 and Fig. S1, B and C). Total MT track numbers were correlated with the mean cell area for individual experimental conditions (Fig. S2), and thresholds for classifying EB3 tracks as “slow” versus “fast” or “short-lived” versus “long-lived” were based on the mean value for each parameter from the entire population of cells analyzed over all experimental conditions (Fig. 2 B and Table S1).
Figure 2.
Down-regulation of myosin II by compliance mechanosensing promotes fast, short-lived MT growth excursions. (A) Workflow used in the plusTipTracker software package for detecting fluorescent EB3 comets, tracking their movement, and classifying MT growth dynamics based on growth speed and growth lifetime. (B) Color scheme for the four subpopulations of MT growth tracks derived by plusTipTracker software and depicted in C and D. (C) Color-coded MT growth track subpopulation overlays from 2 min time-lapse movies of GFP-EB3 (frame rate = 2 s) on representative cells for comparison between a cell plated on glass with (Glass + Blebb) or without (Glass) 20 µM blebbistatin treatment, or on a more compliant ECM (0.7 kPa). Bars, 10 µm. (D) Percentage of the population of MTs whose dynamics were categorized in the four subpopulations described in B in cells under the conditions described in C. (E) Comparison of percentages of mean MT growth speeds and growth excursion lifetimes in cells under the conditions described in C. *, P < 0.001. Error bars indicate standard error of the mean.
To test the hypothesis that ECM compliance mechanosensing regulates MT assembly dynamics, we analyzed GFP-EB3 dynamics in HUVECs cultured on substrates of varying compliance. To maximize the sensitivity of these experiments, we compared MT dynamics in cells cultured on collagen-coated glass with cells cultured on soft (0.7 kPa) collagen-coated PA substrates (Table I and Table S1). In cells adhered to glass, MT growth excursions were primarily slow (Fig. 2 D, red + green), and the majority of slow excursions were also short-lived (red), whereas on more compliant ECMs, a greater proportion of MTs had fast growth excursions (Fig. 2 D, yellow + blue). Qualitative examination of image overlays of color-coded MT plus end motion tracks for individual cells suggested that fast, long-lived growth may be concentrated in the cell center (Fig. 2 C, blue tracks), and that fast, short-lived growth may be concentrated in the cell periphery (Fig. 2 C, yellow tracks). These results were confirmed via analysis of MT dynamics in specific subcellular regions (Fig. 3 and Fig. S1). Together, the effects of increased compliance in ECs cultured on 2D ECMs resulted in a significantly greater mean MT growth speed and a significantly lower mean MT growth lifetime (P < 0.001; Fig. 2 E and Table I). Thus, ECM compliance mechanosensing reduces MT growth persistence (i.e., promotes dynamic instability) and promotes fast MT assembly.
Table I.
MT growth dynamics in 2D ECMs
| ECM condition | n | Mean speed | Mean lifetime |
| µm/min | s | ||
| Whole cell | |||
| Glass | 4,623 | 5.02 ± 0.254 | 9.65 ± 0.596 |
| 2D 0.7 kPa | 3,874 | 8.90 ± 0.282 | 8.71 ± 0.366 |
| Glass + Blebb | 3,235 | 9.10 ± 0.323 | 7.66 ± 0.354 |
| Branches | |||
| Glass | 18 | 5.10 ± 1.08 | 9.56 ± 1.17 |
| 2D 0.7 kPa | 173 | 8.52 ± 0.344 | 11.6 ± 0.488 |
| Glass + Blebb | 251 | 8.62 ± 0.299 | 9.82 ± 0.394 |
| Periphery | |||
| Glass | 2,285 | 4.93 ± 0.237 | 13.3 ± 0.747 |
| 2D 0.7 kPa | 1,888 | 9.55 ± 0.425 | 11.8 ± 0.667 |
| Glass + Blebb | 1,029 | 9.39 ± 0.535 | 10.5 ± 0.744 |
MT growth speeds and MT growth lifetimes in HUVECs cultured on 2D ECMs calculated for the entire cell area (whole cell) and subcellular regions (cell branches [Branches] vs. peripheral cell body [Periphery]). ECM condition refers to collagen-coated glass with (Glass + Blebb) or without (Glass) treatment with 20 µM blebbistatin or collagen coupled to PA of 0.7 kPa shear modulus (0.7 kPa). Mean values reported are ± standard error of the mean. n, number of MT growth tracks.
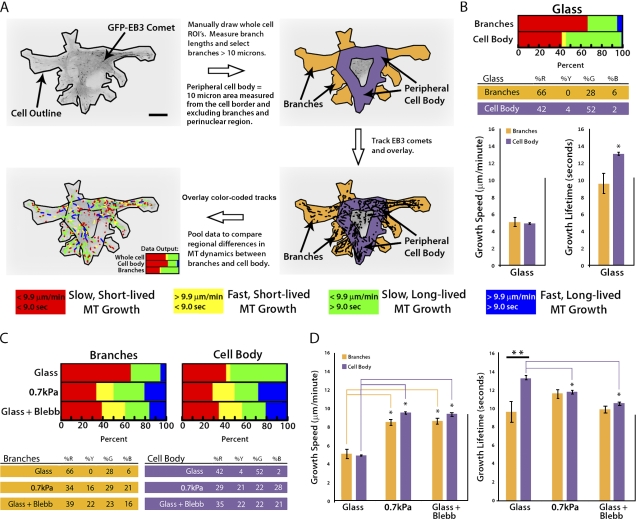
Figure 3.
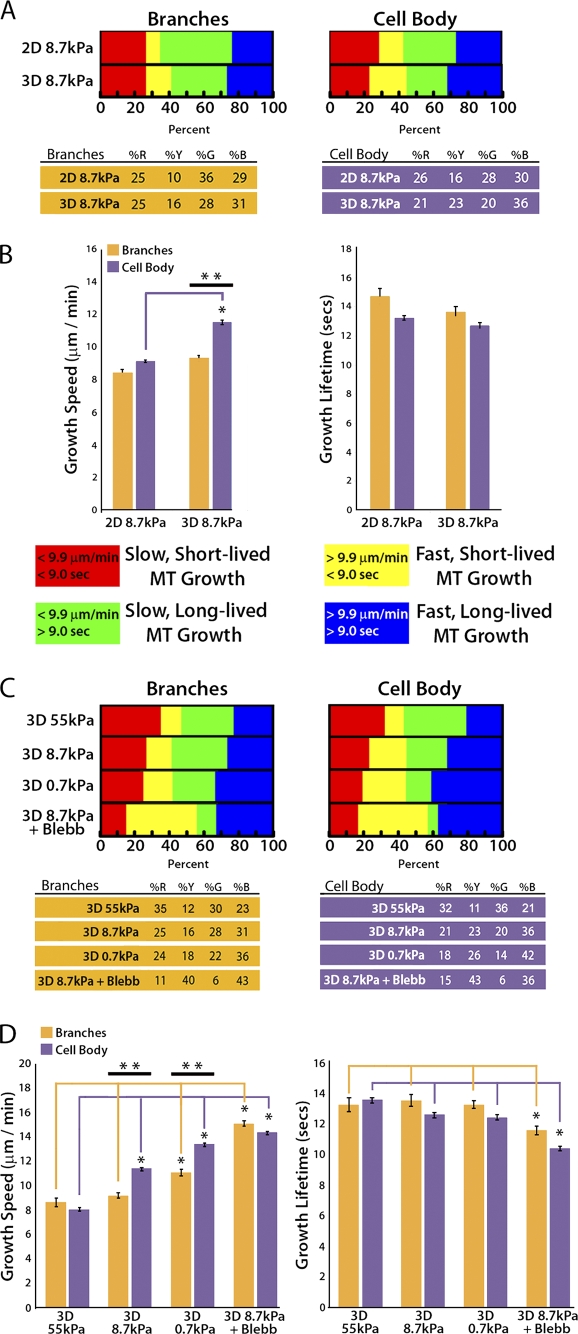
Down-regulation of myosin II by compliance mechanosensing promotes fast MT growth globally and short-lived growth excursions in the peripheral cell body. (A) Workflow for categorizing GFP-EB3 comet tracks of MT growth into specific subcellular regions including cell branches and the peripheral cell body. A color code key for the classification of MT growth excursion subpopulations is shown below. Bar, 10 µm. (B) Comparison of percentages (top) of the population of MTs whose growth dynamics were categorized in the four subpopulations described in A, and MT growth speeds and growth excursion lifetimes (bottom) in branch and peripheral cell body regions of cells plated on glass coverslips. (C) Comparison of percentages of the population of MTs whose growth dynamics were categorized in the four subpopulations described in A in branch and peripheral cell body regions. Cells were plated on glass with or without the addition of 20 µM blebbistatin (Glass + Blebb) or on compliant ECMs (0.7 kPa). Percentage values are shown below. (D) Comparison of MT growth speeds (left) and growth excursion lifetimes (right) in branch and peripheral cell body regions of cells plated on glass with or without the addition of 20 µM blebbistatin (Glass + Blebb) or on compliant ECMs (0.7 kPa). **, branches versus cell body; *, between group comparison, P < 0.001. Error bars indicate standard error of the mean.
Effects of ECM compliance on MTs are myosin II dependent
It is well established that cell engagement of compliant ECMs down-regulates myosin II contractility (Discher et al., 2005; Fischer et al., 2009). Therefore, we sought to determine if ECM compliance-induced effects on MT dynamics were caused by compliance mechanosensing-mediated down-regulation of myosin II activity. In HUVECs cultured on glass and treated with 20 µM blebbistatin to inhibit myosin II ATPase activity (Fig. 2 D), MTs displayed an increase in the proportion of both fast, short-lived (yellow) and fast, long-lived (blue) growth excursions, and a reduction in the proportions of both slow, short-lived (red) and slow, long-lived (green) growth, resulting in an overall trend that was similar to the effects of compliant ECMs on MT dynamics (Fig. 2 D, compare 0.7 kPa and Glass + Blebb). Blebbistatin treatment and ECM compliance also similarly increased mean MT growth speed and decreased mean growth lifetime compared with cells on glass (P < 0.001; Fig. 2 E and Table I). Together, our results suggest that down-regulation of myosin II by ECM compliance mechanosensing promotes fast-growing, dynamically unstable MTs.
Regional modulation of myosin II by compliance mechanosensing promotes fast MT growth globally, but differentially regulates MT growth persistence in cell branches and the cell body
Qualitative examination of the spatial distribution of MT growth tracks (Fig. 2 C, colored tracks) suggested that cells may respond to increased ECM compliance by differentially modulating MT assembly dynamics in subcellular regions (Wittmann and Waterman-Storer, 2005). To test this hypothesis, we compared measurements of MT dynamics in cell branches (Fig. 3 A, orange regions) to measurements of MT dynamics in the peripheral cell body (Fig. 3 A, purple region). Because MTs growing from the centrosome to the peripheral cell body are known to have dynamics different from those in membrane-proximal regions (Komarova et al., 2002; Wittmann and Waterman-Storer, 2005), we excluded data from the cell center to focus our comparison on local regulation of membrane-proximal cell regions. We first analyzed regional MT regulation in cells on ECM-coated glass where branches are few and short. This revealed that the majority of MT growth excursions within branches were slow and short-lived, whereas in the peripheral cell body, MT growth excursions were similarly slow, but with significantly longer growth lifetimes (*, P < 0.001; Fig. 3 B). Thus, on glass, MT dynamics are regionally regulated such that more persistent growth occurs in the peripheral cell body, whereas in cell branches, MTs tend to be dynamically unstable.
Comparison of regional MT dynamics in cells on compliant (0.7 kPa) ECMs to those in cells on glass revealed an increase in the proportion of fast MT growth excursions, both short- and long-lived (yellow and blue), in both cell branches (31% increase) and in the cell body (43% increase; Fig. 3 C). This resulted in increased mean growth speeds in both branches and the cell body compared with the same regions of the cells on glass (*, P < 0.001; Fig. 3 D, left; and Table I). In contrast, ECM compliance tended to increase MT growth lifetime in branches and significantly decrease growth lifetime in the cell body compared with the same regions of cells on glass. As a result, growth lifetimes that were regionally distinct on glass substrates (**, P < 0.001; Fig. 3 D, right) became similar in branches and the cell body on compliant substrates (Fig. 3 D, right; and Table I). These results suggest that compliance mechanosensing promotes fast MT assembly globally, and reduces growth lifetimes specifically in the cell body.
To determine if compliance-induced regional effects on MT assembly dynamics were caused by down-regulation of myosin II activity (Fischer et al., 2009), we compared regional MT dynamics in blebbistatin-treated cells plated on glass to those in cells on 0.7 kPa ECMs. This revealed that ECM compliance and myosin II inhibition produced similar effects on MT dynamics in both the cell body and cell branches (P = 0.022; Fig. 3 D), enhancing growth speed globally and specifically reducing growth lifetime in the cell body compared with the same regions of cells on glass. Together, these data suggest that down-regulation of myosin II by ECM compliance mechanosensing promotes fast MT growth globally throughout the cell, but homogenizes regional differences in MT growth lifetimes by locally increasing MT dynamic instability in the cell body and locally increasing persistent MT growth in cell branches.
MT dynamic instability regulates cell branching and migration similarly in 2D and 3D
Our previous studies in primary mouse ECs showed that the combined effects of increased substrate compliance and 3D ECM engagement synergistically promote cell branching, which suggests that compliance and 3D ECM engagement promote branching via distinct pathways (Fischer et al., 2009). We hypothesized that 3D-specific effects on cell branching may be mediated through the MT cytoskeleton. To control the compliance of the in vitro 3D environment, we covalently coupled 3D collagen matrices to PA gels of defined shear modulus and affixed these to microscope coverslips to make collagen/PA/glass “sandwich cultures” (Fischer et al., 2009). ECs at the PA–collagen interface engage collagen on their ventral surface, which is covalently coupled to PA of a defined stiffness, but also engage dorsal collagen in the 3D matrix. Although the cells experience anisotropic stiffness (the collagen matrices are softer than the underlying PA), their behavior is dominated by the maximum stiffness encountered (Lo et al., 2000; Guo et al., 2006), which is that of the collagen coupled to the PA.
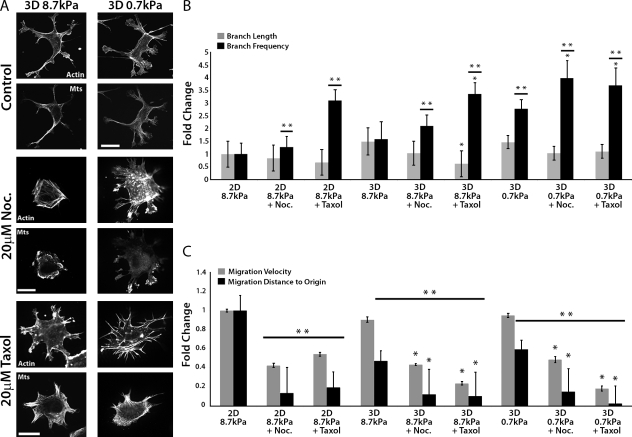
To determine if MT assembly/disassembly dynamics are important for the effects of compliance and 3D ECM engagement on cell branching morphogenesis and migration, we analyzed HUVEC branching in 3D sandwich cultures of variable compliance (Fig. 4 A). We first confirmed that, like primary mouse ECs (Fischer et al., 2009), engagement of compliant (0.7 kPa) 3D ECMs in sandwich cultures strongly enhanced HUVEC branching frequency compared with stiffer (8.7 kPa) 2D ECMs and 3D sandwich cultures (P < 0.05; Fig. 4 B). Pharmacological perturbations revealed that, similar to results in 2D ECMs (Fig. 1, B and C), treatment of cells in 3D sandwich cultures of different stiffnesses with either taxol or nocodazole increased cell branching frequency but had little effect on branch length (Fig. 4 B), and reduced both migration velocity and directional persistence (Fig. 4 C and Videos 5 and 6). Together these data suggest that MT assembly/disassembly dynamics suppress excessive EC branching in both 2D and 3D ECMs, and promote fast, directional migration, independent of compliance and dimensionality.
Figure 4.
Perturbation of MT growth or shortening affects cell branching and migration similarly in 2D and 3D ECMs. (A) Immunolocalization of MTs and fluorescent phalloidin staining of actin in HUVECs cultured in 8.7 kPa or 0.7 kPa compliant 3D ECMs and treated for 90 min with DMSO vehicle (control), 20 µM nocodazole (Noc.), or 20 µM taxol. Bars, 20 µm. (B) Analysis of the effects of the treatments in A on cell branch frequency and length compared with similar drug treatments of cells plated on 2D 8.7 kPa ECMs. (C) Analysis of the effects of the treatments in A on cell migration velocity and distance to origin compared with similar drug treatments of cells cultured on 2D 8.7 kPa ECMs. *, P < 0.05 comparing compliance versus compliance + drug. **, P < 0.05 compared with 2D 8.7 kPa ECMs (one-way ANOVA). Error bars indicate standard deviation.
Compliance mechanosensing promotes fast MT growth in both 2D and 3D ECMs
To test the hypothesis that 3D ECM engagement modulates MT dynamics, we compared MT dynamics in cells on 2D ECMs with cells in 3D sandwich cultures of the same PA stiffness (8.7 kPa; Fig. 5 A). Compared with 2D ECMs, in 3D sandwich cultures a greater proportion of MTs had fast, short-lived growth excursions (Fig. 5 B and Fig. S3), which appeared concentrated near the cell periphery (3D 8.7 kPa; Fig. 5 A). This resulted in a significantly greater mean MT growth speed and a significantly lower mean MT growth lifetime in cells in 3D compared with 2D ECMs (P < 0.001; Fig. 5 D and Table II). These data suggest that compared with 2D ECMs of the same stiffness, 3D ECM engagement promotes fast and dynamically unstable MT growth.
Figure 5.
Compliance mechanosensing promotes fast MT assembly in both 2D and 3D, but 3D ECM engagement makes MT growth persistence insensitive to compliance. (A) Color-coded MT growth track subpopulation overlays from 2-min time-lapse movies of GFP-EB3 (frame rate = 2s) on representative cells cultured on 8.7 kPa or 0.7 kPa 2D ECMs, for comparison with 8.7 kPa or 0.7 kPa 3D ECMs or with cells cultured in 8.7 kPa 3D ECMs and treated with 20 µM blebbistatin (3D 8.7 kPa + Blebb). A color code key for MT growth dynamics classifications is shown below. Bars, 10 µm. (B and C) Comparison of percentages of the population of MTs whose growth dynamics were categorized in the four subpopulations described in A in cells under the conditions described in A or on 55 kPa 3D ECMs. Percentage values shown below. (D and E) Comparison of mean MT growth speeds and growth excursion lifetimes in cells under the conditions described in A or on 55 kPa 3D ECMs. *, P < 0.001. Error bars indicate standard error of the mean.
Table II.
MT growth dynamics in 2D versus 3D ECMs
| ECM Condition | n | Mean speed | Mean lifetime |
| µm/min | s | ||
| Whole cell | |||
| 2D 8.7kPa | 5,417 | 8.82 ± 0.222 | 10.4 ± 0.394 |
| 3D 8.7kPa | 4,604 | 10.7 ± 0.409 | 9.79 ± 0.476 |
| 3D 0.7kPa | 4,644 | 11.7 ± 0.463 | 9.67 ± 0.460 |
| 3D 8.7kPa + Blebb | 6,234 | 14.0 ± 0.480 | 7.80 ± 0.387 |
| Branches | |||
| 2D 8.7kPa | 301 | 8.42 ± 0.200 | 14.7 ± 0.551 |
| 3D 8.7kPa | 644 | 9.30 ± 0.207 | 13.7 ± 0.353 |
| 3D 0.7kPa | 956 | 11.2 ± 0.220 | 13.4 ± 0.270 |
| 3D 8.7kPa + Blebb | 709 | 15.2 ± 0.233 | 11.7 ± 0.267 |
| Periphery | |||
| 2D 8.7kPa | 2,661 | 9.14 ± 0.296 | 13.2 ± 0.629 |
| 3D 8.7kPa | 2,178 | 11.5 ± 0.523 | 12.7 ± 0.673 |
| 3D 0.7kPa | 1,969 | 13.5 ± 0.613 | 12.5 ± 0.678 |
| 3D 8.7kPa + Blebb | 2,234 | 14.5 ± 0.519 | 10.5 ± 0.506 |
MT growth speeds and MT growth lifetimes in HUVECs cultured on 2D ECMs or in 3D collagen-PA-glass sandwich cultures calculated for the entire cell area (whole cell) and subcellular regions (cell branches [Branches] vs. peripheral cell body [Periphery]). ECM condition refers to collagen coupled to PA of 8.7 kPa shear modulus (2D 8.7 kPa) or a 3D collagen gel coupled to PA of a defined shear modulus with (3D 8.7 kPa + Blebb) or without treatment with 20 µM blebbistatin (3D 8.7 kPa or 3D 0.7 kPa). Mean values reported are ± standard error of the mean. n, number of MT growth tracks.
To determine if compliance mechanosensing modulates MT dynamics in cells in 3D ECM, we compared MT dynamics in cells in 3D sandwich cultures of three different stiffnesses (55 kPa, 8.7 kPa, and 0.7 kPa). For 3D studies, we used very stiff (55 kPa) PA substrates instead of glass to evaluate the effects of 3D ECM engagement because it is necessary to conjugate the collagen sandwich gel to a PA substratum. Examining the proportions of MTs in different dynamics classes revealed that, similar to effects in 2D (Fig. 2 D), increasing compliance in 3D increased the proportion of MTs exhibiting fast growth (Fig. 5 B), which resulted in a significant increase (P < 0.001) in mean MT growth speed compared with MTs in cells in stiffer 3D ECMs (Fig. 5 D). Given that changes in compliance in 3D sandwich cultures alter myosin II activity (Fischer et al., 2009), it seemed likely that myosin II activity may contribute to the increase in MT growth speed observed in compliant 3D ECMs. To confirm this, we compared MT dynamics in cells in compliant 3D sandwich cultures with and without blebbistatin. This revealed that, as in 2D (Fig. 2 D), myosin II inhibition in 3D ECMs increased the proportion of fast, short-lived MT growth excursions and reduced the proportion of both long-lived and short-lived slow MT growth excursions (Fig. 5 C and Fig. S3). This resulted in a significantly greater mean MT growth speed (Fig. 5 E and Table II). These results suggest that down-regulation of myosin II by compliance mechanosensing promotes fast MT growth in both 2D and 3D ECMs.
3D ECM engagement uncouples compliance mechanosensing from myosin II–mediated regulation of MT growth persistence
Because we observed in 2D cultures that compliance significantly reduced MT growth lifetimes (Fig. 2 E and Table S1, compare glass vs. 0.7 kPa), we also compared MT growth lifetimes in 3D sandwich cultures of three different stiffnesses. Interestingly, unlike in 2D, when comparing cells in compliant 3D ECMs to cells in stiffer 3D ECMs, the proportion of MTs exhibiting short-lived or long-lived MT growth excursions were similar (Fig. 5 C and Fig. S3), and the mean MT growth lifetimes were not significantly different (Fig. 5 E and Table II). Thus, 3D ECM engagement makes MT growth persistence insensitive to compliance. However, despite the insensitivity of MT growth persistence to compliance in 3D sandwich cultures, we found that in 3D ECMs, growth persistence was still sensitive to direct inhibition of myosin II via blebbistatin. As shown in Fig. 5 C, myosin II inhibition in 3D promoted an increase in the proportion of fast, short-lived MT growth excursions, and reduced the proportion of both long-lived and short-lived slow MTs, similar to the effects of blebbistatin in 2D cultures (Fig. 2 D and Fig. S3, compare glass vs. glass + Blebb). Also similar to cells in 2D, blebbistatin treatment of cells in 3D ECMs produced these effects throughout the cell and caused mean MT growth lifetime to decrease (P < 0.001; Fig. 5 E and Table II). Thus, global inhibition of myosin II activity promotes global increases in MT growth speed and dynamic instability in both 2D ECMs and 3D sandwich cultures. These results suggest that 3D ECM engagement uncouples compliance mechanosensing from myosin II–mediated regulation of MT growth persistence. Together, our results suggest that ECM compliance and dimensionality mechanosensing regulate distinct, specific parameters of MT dynamic instability; MT growth speed (i.e., assembly rate) is regulated by ECM compliance independent of dimensionality, but MT growth persistence (i.e., catastrophe frequency) is differentially regulated by ECM compliance and dimensionality.
MT growth speed, but not growth persistence, is regionally regulated by compliance mechanosensing in 3D ECMs
To determine if 3D ECM engagement modulates MT dynamics within distinct subcellular compartments, we compared regional MT dynamics between cells on 2D ECMs and 3D sandwich cultures of the same stiffness. First, we documented that like in 2D, in 3D, MTs in cell branches grew more slowly and persistently than MTs in the peripheral cell body (Fig. 6 A). Comparison of regional MT dynamics between cells on 2D ECMs or 3D sandwich cultures of the same stiffness (8.7 kPa) revealed that 3D ECM engagement promoted a significant increase in mean MT growth speed only in the cell body (P < 0.001), whereas growth speeds in branches and growth lifetimes in both cell regions were similar in cells in 2D ECMs and 3D sandwich cultures (Fig. 6 B and Table II). Thus, 3D ECM engagement promotes fast MT growth specifically in the cell body and not in cell branches.
Figure 6.
Compliance mechanosensing does not regionally regulate MT growth lifetimes in 3D ECMs. (A) Comparison of percentages of the population of MTs whose growth dynamics are categorized in four subpopulations (key and percentage values are shown below) in cell branch (left) and peripheral cell body (right) regions. (B) Comparison of mean MT growth speeds (left) and growth excursion lifetimes (right) of cells cultured on 2D or in 3D sandwich gels of the same stiffness (8.7 kPa). (C) Comparison of percentages of the population of MTs whose growth dynamics were categorized in the four subpopulations described in A in cell branch (left) and peripheral cell body (right) regions (percentage values are shown below). (D) A comparison of mean MT growth speeds (left) and growth excursion lifetimes (right) of cells cultured in 8.7 kPa 3D sandwich gels with (3D 8.7 kPa + Blebb) or without the addition of 20 µM blebbistatin (3D 8.7 kPa) or in more compliant 3D sandwich gels (3D 0.7 kPa). **, branches versus cell body; *, between group comparison, P < 0.001. Error bars indicate standard error of the mean.
We next sought to determine if compliance mechanosensing or myosin II activity regionally modulates MT assembly dynamics in cells in 3D ECMs. To first determine the effects of compliance mechanosensing, we compared regional MT dynamics in cells in 3D sandwich cultures of three different stiffnesses. This showed that increased compliance in 3D enhanced the proportion of fast-growing MTs in both cell branches and the peripheral cell body (Fig. 6 C). In contrast to 2D ECMs (Fig. 3), on the most compliant (0.7 kPa) 3D ECMs, mean growth speeds were significantly increased specifically within the peripheral cell body, but not in cell branches. This demonstrates a 3D ECM-specific effect on regional regulation of MT growth speed (**, P < 0.001; Fig. 6 D, left; and Table II). Furthermore, regional differences in MT growth speeds were only observed in 3D sandwich cultures of intermediate or greater compliance (8.7 kPa or 0.7 kPa; **, P < 0.001). In the stiffest (least compliant) 3D sandwich cultures (55 kPa), where myosin II activity is high, or when myosin II was inhibited directly with blebbistatin, no regional differences in MT growth speed were observed. This suggests that regional regulation of MT growth speed functions in a limited dynamic range of myosin II activity in 3D. Similar to analysis of MT dynamics in whole cells, compliance of 3D sandwich cultures had no effect on MT growth lifetimes in either cell body or branch regions (Fig. 6 D, right; and Table S1). Thus, compliance mechanosensing does not regionally regulate MT growth persistence in 3D ECM, but has region-specific effects on MT growth speed.
MT growth speed is directly correlated with branch frequency and inversely correlated with branch elongation
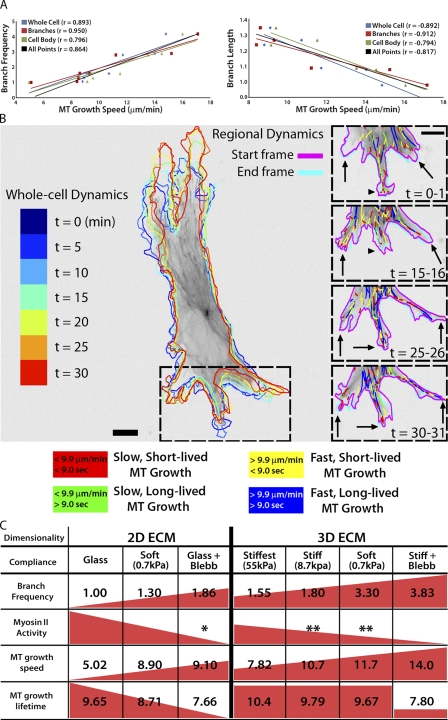
To determine if specific parameters of MT assembly/disassembly dynamics regulate cell branching morphogenesis, we examined the relationships between parameters of MT dynamics and cell branching across all experimental conditions. This revealed significant correlations between MT growth speed and branch frequency, with a direct correlation between MT growth speed and cell branching (ratio = 2.5:1; r = 0.893; P < 0.01), and an inverse correlation between MT growth speed and branch length (ratio = −19.5:1; r = −0.892; P < 0.01; Fig. 7 A). The correlation coefficients were even higher for data specifically from cell branches (Fig. 7 A, red trend lines). In contrast, we found no significant correlations between MT growth persistence and branch frequency or branch length (Fig. S4). These results show that fast MT growth correlates with frequent cell branching, whereas slow MT growth correlates with branch elongation.
Figure 7.
MT growth speed is directly correlated with branch frequency and inversely correlated with branch length. (A) Relationships between branch frequency (left; fold increase) or branch length (right; fold increase) and MT growth speed. r, Pearson correlation coefficient. (B, left) Outlines of a HUVEC expressing GFP-EB3 migrating in a 0.7 kPa 3D sandwich gel at 5 min intervals, color-coded by time as shown on the far left. (B, right) Color-coded MT growth track subpopulation overlays from 30-min time-lapse movies of GFP-EB3 (frame rate = 2 s) from the boxed region of selected time points during branching morphogenesis. Arrows, elongating cell branches; arrowheads, retracting cell branch. A color key for MT growth dynamics classifications is shown below. Bar, 10 µm. (C) Summary table depicting data trends (red triangles/squares) for cell branching frequency (fold change) compared with myosin II activity and MT assembly dynamics (mean values) on substrates of varying compliance in 2D and 3D ECMs. (*, Straight et al., 2003; **, Fischer et al., 2009).
To determine if local modulation of MT dynamics mediates cell branching in real time, we analyzed MT dynamics during cell branch formation and/or elongation by time-lapse imaging of EB3-GFP in cells plated on compliant 3D ECMs to promote frequent branching. Because branch formation occurs infrequently (branch initiations at ∼10-min intervals) relative to the time scales of MT dynamic instability (growth excursion initiations at ∼10-s intervals) and GFP photobleaching (minutes), it was rare to capture high-quality movies suitable for accurate tracking of MT dynamics during new branch formation (Video 8). However, analysis of EB3-GFP tracks in the example shown in Fig. 7 B revealed that MTs displayed locally faster and shorter-lived growth (yellow tracks) at a site in the peripheral cell body where a cell branch was initiating (Fig. 7 B, left arrow) and slower, longer-lived growth (green tracks) in a branch that later retracted before continuing to elongate (Fig. 7 B, arrowhead). During elongation of existing branches, qualitative examination of MT tracks suggested that MTs grew more slowly and persistently toward elongating branch tips (Fig. 7 B, arrows) than at the base of the same branch. Together, these results suggest that local modulation of MT growth speed in the cell body may locally induce branch formation, whereas slower, more persistent MT growth in cell branches may promote branch elongation.
Discussion
Mechanosensing of the ECM is gaining importance as a potential physiological regulator of EC branching morphogenesis and motility during formation of vasculature (Ingber, 2002; Ghosh et al., 2008). We recently showed that 3D ECM dimensionality mechanosensing by ECs synergizes with ECM compliance mechanosensing to enhance cell branching, which suggests that compliance and dimensionality regulate distinct molecular pathways of cell branch formation (Fischer et al., 2009). Here, we tested the hypothesis that compliance and dimensionality mechanosensing in ECs may affect cell branching morphogenesis differentially through myosin II– and MT-dependent pathways. This work produced three main advances. First, we used a recently developed MT plus end–tracking program to show that specific parameters of MT assembly dynamics, growth speed and growth persistence, are globally and regionally modified by and contribute to ECM compliance and dimensionality mechanosensing. Second, we demonstrated that engagement of compliant 2D or 3D ECMs induces local differences in MT growth speed that require myosin II contractility. Finally, we showed that MT growth persistence is modulated by myosin II–mediated compliance mechanosensing when ECs are cultured on 2D ECMs, whereas 3D ECM engagement makes MT growth persistence insensitive to changes in ECM compliance. Thus, compliance and dimensionality ECM mechanosensing pathways independently regulate specific and distinct MT dynamics parameters in ECs to guide branching morphogenesis in physically complex ECMs.
Our results reveal new relationships between ECM compliance, topology mechanosensing, and MT dynamic instability, which are summarized in Fig. 7 C. On the stiffest 2D ECMs (glass), ECs extend few branches, myosin II–mediated cortical tension is uniformly high (Fischer et al., 2009), MT growth is uniformly slow, and MT growth persistence is highest within the cell body. On more compliant 2D ECMs (Fig. 7 C, 2D ECM, 0.7 kPa), reduction of myosin II contractility and spatial inhomogeneity in cortical tension promotes branch initiation (Fischer et al., 2009). This reduction in myosin II contractility also promotes faster, less persistent MT growth in the peripheral cell body, which likely further modulates cortical contractility to enhance cell branching. Additional inhibition of myosin II contractility with blebbistatin (Fig. 7 C, 2D ECM, Glass + Blebb) further promotes branch frequency while at the same time reducing MT growth persistence. This revealed that both cell branching morphology and MT dynamics are responsive to a range of myosin II contractility. MT dynamics are also required for cell branching morphology in response to compliance mechanosensing, which suggests a potential feedback mechanism between myosin II contractility and the regulation of MT dynamics in this process.
When cells engage stiff 3D ECMs, myosin II–mediated compliance mechanosensing is additionally enhanced (Beningo et al., 2004; Fischer et al., 2009), further augmenting MT growth speeds compared with 2D ECMs (Fig. 7 C). As compliance is increased in 3D ECMs, both cell branching and MT growth speeds further increase. Indeed, over a range of experimental conditions in 2D and 3D ECMs, MT growth speeds and cell branching are strongly correlated (Fig. 7 A). However, when cells engage 3D ECMs, MT growth persistence no longer responds to changes in substrate compliance, but still responds to direct myosin II inhibition. This suggests that 3D ECM engagement uncouples compliance mechanosensing from myosin II–mediated regulation of MT growth persistence. One possible interpretation of this is that 3D ECM engagement may specifically regulate catastrophe factors, such as MCAK or Op18/stathmin (van der Vaart et al., 2009), to regulate MT dynamics. Although it is known that integrin engagement can regulate MT dynamics (Palazzo et al., 2004), our results show that different topologies of integrin engagement can differentially regulate MT catastrophe.
Our findings also demonstrate that unlike MT growth persistence, MT growth speeds are regionally regulated in branches and bodies of cells on compliant substrates with 3D ECM engagement. This suggests that the regional regulation of MT assembly/disassembly factors, such as XMAP215 or Op18/stathmin (van der Vaart et al., 2009), may be downstream of myosin II contractility. Regional regulation of MT growth speed was lost in either very stiff (55 kPa) 3D sandwich cultures, or when myosin II activity was directly inhibited by blebbistatin. Because we have previously shown that myosin II is more dynamic in the cortex of cells in compliant 3D ECMs (Fischer et al., 2009), this suggests that local differences in MT growth speed are likely caused by local stochastic fluctuations of myosin II activity at the cell cortex. We hypothesize that these stochastic fluctuations of myosin II at the cell cortex may alter the local activity of MT assembly factors, although the mechanism for this remains unclear. We suggest that local differences in myosin II contractility promote local differences in MT dynamics, which in turn feed back to further modulate actomyosin through Rho and Rac signaling (Rodriguez et al., 2003). This feedback may initiate the spatially confined breaking of cortical tension that permits branch formation during EC branching morphogenesis.
Materials and methods
Cells and DNA expression constructs
HUVECs were maintained in endothelial cell basal medium (EBM) supplemented with EGM-MV Single Quots (Lonza) at 37°C in 5% CO2. For live imaging, medium was supplemented with 25 µg Hepes, pH 7.2, and 30 U/ml Oxyrase. Transfection of GFP- or mApple-EB3 cDNAs (courtesy of M. Davidson, Florida State University, Tallahassee, FL) was performed using a nucleofector (Amaxa Biosystems) with solution kit IV (Lonza), setting A-034, and experiments were performed 6–10 h later to allow time for EB3 expression. Cells were treated with 0.1% DMSO (vehicle control), 20 µM taxol, 20 µM nocodazole, or 20 µM blebbistatin for 60 min before imaging. All blebbistatin experiments were performed using mApple-EB3 to avoid photo-inactivation of blebbistatin and phototoxicity from GFP (Sakamoto et al., 2005). For immunolabeling experiments, cells were fixed 90 min after drug treatment.
2D and 3D cell culture
PA gels were cross-linked to 22 × 22 mm No. 1.5 coverslips (Corning) that had been preactivated. Preactivation involved first incubating coverslips in 0.5% 3-aminopropyltrimethyoxysilane diluted in water (10 min on a stir plate), followed by 6 × 5 min water washes. Coverslips were dried at 37°C, cooled to room temperature, and then immersed in 0.5% glutaraldehyde solution in PBS on a stir plate for 30 min. Coverslips were then washed three times for 10 min in water and dried at room temperature. For the experiments in this manuscript, PA gels of varying stiffness (55 kPa, 8.7 kPa, or 0.7 kPa; shear stress modulus) were prepared by varying concentrations of 40% acrylamide and 2% bis-acrylamide before tetramethylethylenediamine polymerization (Gardel et al., 2008).
To generate 3D ECMs of controlled compliance (sandwich gels; Fischer et al., 2009), coverslips with adhered PA gels were activated with 2 mM sulfo-SANPAH by exposure to 7,500 J of UV light, rinsed, and covalently cross-linked to a thin layer of unpolymerized rat tail type I collagen (1.6 mg/ml; BD). The covalently linked collagen was then polymerized for 4 h at 37°C and rinsed overnight in PBS, and the coverslip with bound collagen was assembled into a Rose chamber. HUVECs expressing fluorescent EB3 were cultured on the collagen and allowed to adhere (60 min) before being overlaid with an additional layer of 1.6 mg/ml collagen, which was subsequently polymerized as described. For live-cell imaging, chambers were filled with imaging media and sealed with a second coverslip.
2D compliant collagen-coupled PA substrates were assembled as described for 3D, except that the sulfo-SANPAH activated, PA-coated coverslips were covalently cross-linked to collagen at a concentration lower than required for polymerization (90 µg/ml for 8 h at 4°C). After overnight rinsing, cells were cultured on the collagen and prepared for imaging as above.
Microscopy
Live-cell imaging of fluorescent EB3.
GFP- or mApple-EB3 was imaged on a spinning disk confocal microscope using a 60× 1.2 NA water immersion objective lens on an epifluorescence microscope (TE2000; Nikon) equipped with Perfect Focus System, an electronic shutter (Smart shutter; Sutter Instrument Co.) for transmitted illumination, a linear-encoded x, y, z robotic stage with a piezo-driven z-axis top plate (ASI Technologies, Inc.) and a spinning disk confocal scan head (CSU-X; Yokogawa) equipped with a multi-bandpass dichromatic mirror (Semrock) and bandpass filters (Chroma Technology Corp.) in an electronic filter wheel for selection of GFP or Texas red emission. 561 and 488 nm laser illumination was provided by a custom-built laser combiner module (modification of LMM-3; Spectral Applied Research). This contained 500-mW solid-state lasers (488 nm [Coherent] and 561 nm [MPB Communications]) that were shuttered with electronic shutters and attenuated and/or directed to a fiber-coupled output port with an acoustic-optic tunable filter (Neos Technologies), and directed to the confocal scan head via a single-mode optical fiber (Oz Optics). Movies of EB3 dynamics were acquired using a cooled charge-coupled device (CoolSNAP HQ2; Photometrics) operated in the 14-bit mode for 2 min at 2-s image intervals using a 300-ms exposure time. Microscope system automation was controlled with MetaMorph software (MDS Analytical Technologies). For long-term imaging of fluorescent EB3 dynamics during cell branch formation and elongation, 1-min time-lapse image series (2-s intervals) were captured every 5 min for 30–60 min.
Immunofluorescence.
Fixation and processing of samples for immunofluorescence labeling was performed as described previously (Fischer et al., 2009), with the following modifications. Cells in 3D sandwich gels were fixed and immunolabeled without disruption of the collagen gel in 3% paraformaldehyde in CB buffer (10 mM MES, pH 6.1, 138 mM KCl, 3 mM MgCl2, and 2 mM EGTA) with 0.1% Triton X-100 for 30 min at room temperature, then rinsed in CB with 0.2% Triton X-100 for 30 min. After fixation, cells in PA-collagen gels were blocked with 4% BSA in CB (3 h, 37°C) and incubated in primary rat anti–α-tubulin antibody (overnight at 4°C, 1:1,000 dilution; AbD Serotec), then rinsed in CB three times for 15 min each before secondary antibodies Cy3-donkey anti–Rat (1:1,000 dilution; Jackson ImmunoResearch Laboratories, Inc.) and Alexa Fluor 488–phalloidin (1:200 dilution; Invitrogen) were applied simultaneously (2 h, 37°C). After extensive rinsing in CB, PA-collagen gels containing the cells were imaged directly in the Rose chambers to prevent disruption of the sandwich gels.
Cell migration assay.
Cell migration assays were performed on 22-mm round No. 1.5 coverslips prepared with PA-collagen substrates as described for 2D and 3D culture, except they were assembled and mounted in a Gupton chamber, a custom-built multiposition stage insert that allowed us to image up to 16 coverslips per experiment. Phase-contrast images were acquired at 15-min intervals for 15 h on the microscope described above using a 20× 0.45 NA phase objective and an 0.52 NA LWD condenser using MetaMorph’s Multi-dimensional Acquisition (MDA) software module.
Quantification of cell branching and migration.
For analysis of cell branching, the “trace region” tool in MetaMorph was used to obtain cell outlines and cell areas. Branches were defined as protrusions that extended from the cell >10 µm in length. We defined the “branch origin” by locating the position on each side of the branch where the membrane displayed the greatest curvature and then connected those two points with a straight line. The “trace region” tool was then used to trace the outlines of the branches for each cell to calculate branch area and branch number. Branch length was the distance from the branch origin to the most distal point of the branch tip. Cell migration was quantified by hand-tracking the nucleolus in successive images from a time-lapse phase-contrast image series using the “track points” application in MetaMorph to determine instantaneous velocity and distance to origin. Statistical analysis was performed using the Analyze-It plug-in (Analyze-It Software Ltd.) for Excel (Microsoft), and branching and migration data were compared using a Bonferroni-corrected, one-way analysis of variance (ANOVA) test, with >95% confidence as the threshold for statistical significance.
MT dynamics analysis
MT dynamics were analyzed from EB3 movies using plusTipTracker (Matov et al., 2010), a Matlab-based, open-source software package that combines automated detection, tracking, analysis, and visualization tools for movies of fluorescently labeled MT plus end binding proteins (+TIPs). The +TIP comet detection algorithm relies on a watershed-based approach to estimate locally optimal thresholds. The track reconstruction algorithm uses the spatially and temporally globally optimized tracking framework described in Jaqaman et al. (2008), with cost functions modified to reflect MT track geometry. In brief, tracking occurs in two steps: frame-to-frame linking of comets into growth sub-tracks, and the linking of collinear, sequential growth sub-tracks into compound tracks. The cost of joining two candidate growth sub-tracks into a compound track is calculated from three spatial parameters and one temporal parameter. After calculating the cost of linking all pairs of candidate growth tracks, the links are chosen by minimizing the global cost, which is achieved by solving the Linear Assignment Problem (Jaqaman et al., 2008).
Determination of image quality.
Detection, tracking, and postprocessing analysis were performed on the first 30 frames only for each movie, as photo-bleaching was a limiting feature in some conditions and standardizing movie length was required for the MT tracking comparison. The quality of the movies was assessed by examining comet detection performance; movies were discarded from further analysis if EB3 expression was too high and led to too many false positives, or if focus drift or photobleaching led to a high standard deviation in mean comet number per frame over the course of the movie.
Tracking parameters.
Tracking control parameters were optimized based on a parameter sweep using the plusTipParamSweepGUI tool of plusTipTracker and verified by visual inspection of track overlays on movies. The same parameter set was used for all movies in the dataset: maximum gap length, 12 frames; minimum track length, 3 frames; search radius range, 5–10 pixels; maximum forward angle, 25°; maximum backward angle, 8°; maximum shrinkage factor, 1.0; fluctuation radius, 2 pixels. For this study, only growth excursions were of interest, so MT shrinkage or pause events were not analyzed. However, sub-track linking was still performed to correct for the many occurrences when comets cross over one another or disappear momentarily from the field of view by focal drift, which breaks the trajectories prematurely.
Region of interest (ROI) selection.
Binary masks of whole cells, individual branches, and cell peripheries were generated in two steps using plusTipTracker’s sub-ROI selection tool. First, whole cell and branch masks were manually selected based on the whole cell outlines and branch definitions were produced in MetaMorph as described above. In the second step, cell periphery masks were automatically generated from whole cell masks by including all pixels within 10 µm of the cell edge but not including pixels defined within branches. The subtraction of branch pixels was accomplished by choosing the all-branch mask generated in the first step as an exclusion mask in the second. Regional areas were calculated by summing the area of the mask and converting to square micrometers. The set of all MT growth excursions that spent ≥6s (three frames) within an ROI (branch or periphery) were extracted and stored within a subproject of the original cell.
Data grouping.
The plusTipPickGroups function of plusTipTracker was used to create groups of data defined by experimental condition (glass [2D only], 55 kPa [2D or 3D], 8.7 kPa [2D or 3D], or 0.7 kPa [2D or 3D]; drug or vehicle) and cellular region (whole cell, branch, or peripheral cell body). There were thus 42 groups to be compared, 14 each from the three regions. Groups of whole cells and peripheries ranged from 8 to 17 projects, whereas groups of branches ranged from 2 to 57 projects.
MT growth sub-track subpopulation analysis.
Tracks from within ROIs from all movies in the dataset were pooled using the plusTipPoolGroupData function to find the mean growth speed and mean growth lifetime. These values were used to split data for quadrant plot analysis. Quadrant plot analysis was performed as separate batch processes for the three regions, thus defining for each of the groups the total number of tracks in each of four subpopulations: slow and short-lived, slow and long-lived, fast and short-lived, and fast and long-lived. The relative proportions of these four subpopulations were used to generate percentage bar graphs for comparison. Statistical comparison of MT growth speeds and growth lifetimes were performed using a permutation test for means using a threshold for statistical significance of P < 0.001.
Online supplemental material
Fig. S1 shows whole cell and regional MT track density in either branches or peripheral cell bodies as detected by plusTipTracker software. Fig. S2 shows total track numbers for each category in every experimental condition, and the total correlated area associated with each experimental condition. Fig. S3 is the summary of all MT growth excursion class proportions for all MT populations and experimental conditions analyzed. Fig. S4 shows correlation analyses for MT growth lifetimes and EC branch frequency or branch length under all conditions analyzed. Table S1 lists all MT growth speeds and growth lifetimes under all experimental conditions, with associated means and standard errors. Video 1 shows HUVEC migration on glass with DMSO, Taxol, or nocodazole treatments. Video 2 shows HUVEC migration on 8.7 kPa 2D substrates with DMSO, Taxol, or nocodazole treatments. Video 3 shows HUVEC migration on 0.7 kPa 2D substrates with DMSO, Taxol, or nocodazole treatments. Videos 4 and 5 show HUVEC migration on 8.7 kPa and 0.7 kPa 3D sandwich gels with DMSO, Taxol, or nocodazole treatments. Video 6 compares GFP-EB3 dynamics in HUVECs on 8.7 kPa 2D and 8.7 kPa 3D sandwich gel substrates. Video 7 compares GFP-EB3 dynamics in HUVECs on 0.7 kPa 3D sandwich gels with EB3-GFP dynamics in HUVECs treated with blebbistatin on 8.7 kPa 3D sandwich gels. Video 8 shows long-term imaging of EC branching morphogenesis and GFP-EB3 dynamics. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006009/DC1.
Acknowledgments
We thank Mike Davidson for fluorescent EB3, William Shin for maintaining the microscopes, Dorothy Honemond for administrative assistance, and members of the Waterman laboratory and Orna Cohen-Fix (National Institute of Diabetes and Digestive and Kidney Diseases) for helpful discussions.
This work was initiated as a project in the Physiology Course at the Marine Biological Laboratory in Woods Hole, MA, and we thank student Julie Janvore (Institute Curie, Paris, France) for her work at early stages of the study. C.M. Waterman, R.S. Fischer, and K.A. Myers are supported by the National Heart, Lung and Blood Institute, and G. Danuser and K.T. Applegate are supported by National Institute of General Medical Sciences (U01 GM067230).
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- EC
- endothelial cell
- HUVEC
- human umbilical vein EC
- MT
- microtubule
- PA
- polyacrylamide
- ROI
- region of interest
References
- Beningo K.A., Dembo M., Wang Y.L. 2004. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc. Natl. Acad. Sci. USA. 101:18024–18029 10.1073/pnas.0405747102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Yamada K.M. 2002. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 14:633–639 10.1016/S0955-0674(02)00364-2 [DOI] [PubMed] [Google Scholar]
- Dehmelt L., Halpain S. 2004. Actin and microtubules in neurite initiation: are MAPs the missing link? J. Neurobiol. 58:18–33 10.1002/neu.10284 [DOI] [PubMed] [Google Scholar]
- Dehmelt L., Smart F.M., Ozer R.S., Halpain S. 2003. The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J. Neurosci. 23:9479–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent E.W., Gertler F.B. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 40:209–227 10.1016/S0896-6273(03)00633-0 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Kalil K. 2001. Axon branching requires interactions between dynamic microtubules and actin filaments. J. Neurosci. 21:9757–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B.J. 2002. Molecular mechanisms of axon guidance. Science. 298:1959–1964 10.1126/science.1072165 [DOI] [PubMed] [Google Scholar]
- Discher D.E., Janmey P., Wang Y.L. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science. 310:1139–1143 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- Doyle A.D., Wang F.W., Matsumoto K., Yamada K.M. 2009. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 184:481–490 10.1083/jcb.200810041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Ram S., Yamada K.M. 2005. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 17:524–532 (Review) 10.1016/j.ceb.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Fischer R.S., Gardel M., Ma X., Adelstein R.S., Waterman C.M. 2009. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr. Biol. 19:260–265 10.1016/j.cub.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan L.A., Ju Y.E., Marg B., Osterfield M., Janmey P.A. 2002. Neurite branching on deformable substrates. Neuroreport. 13:2411–2415 10.1097/00001756-200212200-00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel M.L., Sabass B., Ji L., Danuser G., Schwarz U.S., Waterman C.M. 2008. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183:999–1005 10.1083/jcb.200810060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H., Betsholtz C. 2005. How do endothelial cells orientate? EXS. 94:3–15 10.1007/3-7643-7311-3_1 [DOI] [PubMed] [Google Scholar]
- Gerhardt H., Golding M., Fruttiger M., Ruhrberg C., Lundkvist A., Abramsson A., Jeltsch M., Mitchell C., Alitalo K., Shima D., Betsholtz C. 2003. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 161:1163–1177 10.1083/jcb.200302047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt H., Ruhrberg C., Abramsson A., Fujisawa H., Shima D., Betsholtz C. 2004. Neuropilin-1 is required for endothelial tip cell guidance in the developing central nervous system. Dev. Dyn. 231:503–509 10.1002/dvdy.20148 [DOI] [PubMed] [Google Scholar]
- Ghosh K., Thodeti C.K., Dudley A.C., Mammoto A., Klagsbrun M., Ingber D.E. 2008. Tumor-derived endothelial cells exhibit aberrant Rho-mediated mechanosensing and abnormal angiogenesis in vitro. Proc. Natl. Acad. Sci. USA. 105:11305–11310 10.1073/pnas.0800835105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.-H., Frey M.T., Burnham N.A., Wang Y.-L. 2006. Substrate rigidity regulates the formation and maintenance of tissues. Biophys. J. 90:2213–2220 10.1529/biophysj.105.070144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D.E. 2002. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ. Res. 91:877–887 10.1161/01.RES.0000039537.73816.E5 [DOI] [PubMed] [Google Scholar]
- Jaqaman K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., Danuser G. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil K., Dent E.W. 2005. Touch and go: guidance cues signal to the growth cone cytoskeleton. Curr. Opin. Neurobiol. 15:521–526 10.1016/j.conb.2005.08.005 [DOI] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Beningo K., Anderson K., Wang Y.L., Small J.V. 2002. Tensile stress stimulates microtubule outgrowth in living cells. J. Cell Sci. 115:2283–2291 [DOI] [PubMed] [Google Scholar]
- Kollins K.M., Hu J., Bridgman P.C., Huang Y.Q., Gallo G. 2009. Myosin-II negatively regulates minor process extension and the temporal development of neuronal polarity. Dev. Neurobiol. 69:279–298 10.1002/dneu.20704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y.A., Vorobjev I.A., Borisy G.G. 2002. Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J. Cell Sci. 115:3527–3539 [DOI] [PubMed] [Google Scholar]
- Lo C.M., Wang H.B., Dembo M., Wang Y.L. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto A., Connor K.M., Mammoto T., Yung C.W., Huh D., Aderman C.M., Mostoslavsky G., Smith L.E., Ingber D.E. 2009. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 457:1103–1108 10.1038/nature07765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matov A., Applegate K., Kumar P., Thoma C., Krek W., Danuser G., Wittmann T. 2010. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods. 7:761–768 10.1038/nmeth.1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.F. 2004. Contraction reaction: mechanical regulation of Rho GTPase. Trends Cell Biol. 14:111–114 10.1016/j.tcb.2004.01.005 [DOI] [PubMed] [Google Scholar]
- Palazzo A.F., Eng C.H., Schlaepfer D.D., Marcantonio E.E., Gundersen G.G. 2004. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 303:836–839 10.1126/science.1091325 [DOI] [PubMed] [Google Scholar]
- Pelham R.J., Jr, Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 94:13661–13665 10.1073/pnas.94.25.13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., Doyle A.D., Yamada K.M. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10:538–549 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S., Grinnell F. 2007. Fibroblast mechanics in 3D collagen matrices. Adv. Drug Deliv. Rev. 59:1299–1305 10.1016/j.addr.2007.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S., Jiang H., Ho C.H., Grinnell F. 2007. Microtubule function in fibroblast spreading is modulated according to the tension state of cell-matrix interactions. Proc. Natl. Acad. Sci. USA. 104:5425–5430 10.1073/pnas.0608030104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez O.C., Schaefer A.W., Mandato C.A., Forscher P., Bement W.M., Waterman-Storer C.M. 2003. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat. Cell Biol. 5:599–609 10.1038/ncb0703-599 [DOI] [PubMed] [Google Scholar]
- Rösner H., Möller W., Wassermann T., Mihatsch J., Blum M. 2007. Attenuation of actinomyosinII contractile activity in growth cones accelerates filopodia-guided and microtubule-based neurite elongation. Brain Res. 1176:1–10 10.1016/j.brainres.2007.07.081 [DOI] [PubMed] [Google Scholar]
- Saez A., Buguin A., Silberzan P., Ladoux B. 2005. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys. J. 89:L52–L54 10.1529/biophysj.105.071217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Limouze J., Combs C.A., Straight A.F., Sellers J.R. 2005. Blebbistatin, a myosin II inhibitor, is photoinactivated by blue light. Biochemistry. 44:584–588 10.1021/bi0483357 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., Mitchison T.J. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- van der Vaart B., Akhmanova A., Straube A. 2009. Regulation of microtubule dynamic instability. Biochem. Soc. Trans. 37:1007–1013 10.1042/BST0371007 [DOI] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C.M. 2005. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J. Cell Biol. 169:929–939 10.1083/jcb.200412114 [DOI] [PMC free article] [PubMed] [Google Scholar]