The ubiquitin hydrolase activating factor Bro1 enhances ESCRT-III stability by inhibiting Vps4-mediated disassembly.
Abstract
Endosomal sorting complexes required for transport (ESCRTs) promote the invagination of vesicles into the lumen of endosomes, the budding of enveloped viruses, and the separation of cells during cytokinesis. These processes share a topologically similar membrane scission event facilitated by ESCRT-III assembly at the cytosolic surface of the membrane. The Snf7 subunit of ESCRT-III in yeast binds directly to an auxiliary protein, Bro1. Like ESCRT-III, Bro1 is required for the formation of intralumenal vesicles at endosomes, but its role in membrane scission is unknown. We show that overexpression of Bro1 or its N-terminal Bro1 domain that binds Snf7 enhances the stability of ESCRT-III by inhibiting Vps4-mediated disassembly in vivo and in vitro. This stabilization effect correlates with a reduced frequency in the detachment of intralumenal vesicles as observed by electron tomography, implicating Bro1 as a regulator of ESCRT-III disassembly and membrane scission activity.
Introduction
The endosomal sorting complexes required for transport (ESCRTs) are recruited to the cytosolic surface of endosomes, where they selectively package ubiquitinated transmembrane protein cargoes into the intralumenal vesicles (ILVs) of multivesicular bodies (MVBs). The ILVs and their cargoes are subsequently degraded upon fusion of MVBs with lysosomes. ESCRT-0, -I, and -II are stable heteromeric complexes, each containing subunits that bind directly to ubiquitin (Ub) linkages on the cytosolic domains of cargoes (Piper and Katzmann, 2007; Raiborg and Stenmark, 2009). Recent studies indicate that ESCRT-I and -II can also induce membrane invaginations of nascent ILVs in vitro (Wollert and Hurley, 2010) and that ESCRT-II can stimulate assembly of ESCRT-III (Im et al., 2009; Saksena et al., 2009).
The polymerization of ESCRT-III subunits is required for completion of ILV budding. Genetic and biochemical studies of ESCRT-III proteins from Saccharomyces cerevisiae have contributed much to our understanding about assembly of the complex, principles that are likely to be generally conserved. ESCRT-III in yeast is comprised of four core subunits (Snf7, Vps20, Vps24, and Vps2; Babst et al., 2002), the most abundant of which is Snf7 (Teis et al., 2008). Polymerization of Snf7 on membranes is stimulated upon binding Vps20, which relieves an autoinhibitory intramolecular interaction between the N- and C-terminal regions of Snf7 (Teis et al., 2008; Saksena et al., 2009). Membrane-associated Snf7 polymers are thought to be capped by Vps24, which recruits Vps2 (Babst et al., 2002; Teis et al., 2008). In turn, Vps2 promotes activation of the AAA-ATPase Vps4, which catalyzes disassembly of ESCRT-III and recycling of its subunits from membranes to the cytosol (Babst et al., 1998; Babst et al., 2002). Did2, Ist1, and Vps60 are three accessory ESCRT-III subunits that appear to serve primarily as regulators of Vps4 activity (Nickerson et al., 2006; Azmi et al., 2008; Dimaano et al., 2008; Rue et al., 2008).
In vitro reconstitution of ESCRT-III assembly on synthetic membranes revealed that excess amounts of the yeast core ESCRT-III subunits deform liposomes (Saksena et al., 2009) and drive both the formation and detachment of lumenal membrane invaginations (Wollert et al., 2009). In limiting amounts, however, ESCRT-III subunits lack the ability to deform the membrane and, instead, their activity is restricted to facilitating the severing of membrane invaginations generated by ESCRT-I and -II (Wollert and Hurley, 2010). These observations suggest that ESCRT-III is responsible for execution of the membrane scission step during ILV formation. Consistent with these in vitro findings, mammalian ESCRT-III is required during cytokinesis for midbody abscission and for the detachment of fully assembled retroviruses from the plasma membrane (McDonald and Martin-Serrano, 2009). Both abscission and retroviral budding require membranes to be severed in a manner that is topologically equivalent to ILV formation at MVBs, and all three processes are disabled by inhibiting Vps4 function (Garrus et al., 2001; Carlton et al., 2008). Overexpression of catalytically inactive Vps4 in mammalian cells dramatically increases the frequency of ILV budding profiles and reduces the number of freely detached ILVs at MVBs (Sachse et al., 2004). However, based on studies of reconstituted ILV budding in vitro (Wollert et al., 2009; Wollert and Hurley, 2010), Vps4 is thought to function indirectly in membrane scission by recycling ESCRT-III subunits for repeated rounds of assembly.
In addition to its membrane scission activity, ESCRT-III is required for coordinating the deubiquitination of transmembrane protein cargoes before their enclosure within ILVs. In humans, two distinct Ub hydrolases bind directly to ESCRT-III via microtubule-interacting and trafficking (MIT) domains (Agromayor and Martin-Serrano, 2006; Tsang et al., 2006; Row et al., 2007), and in yeast, recruitment of the Ub hydrolase Doa4 to ESCRT-III is promoted by Bro1, which binds both to Snf7 and to the Doa4 catalytic domain (Luhtala and Odorizzi, 2004; Kim et al., 2005; Richter et al., 2007). Bro1 and its closest human orthologue, Alix, each have a conserved N-terminal Bro1 domain that binds directly to Snf7 and CHMP4 proteins, respectively (Kim et al., 2005; Fisher et al., 2007). Although this interaction is essential for Alix to promote retrovirus budding (Fisher et al., 2007), the means by which it contributes to ESCRT-III function in membrane scission remain unclear. In yeast, Bro1 likely has a role in ILV budding independent of its activity in regulating deubiquitination because ILV formation is strongly impaired by deletion of BRO1 but not DOA4 (Richter et al., 2007). In this study, we report that binding of Bro1 to Snf7 enhances the stability of ESCRT-III in vivo and inhibits Vps4-mediated disassembly of the complex in vitro. This effect correlates with a reduced efficiency in detachment of ILVs from the limiting endosomal membrane. Conversely, mutation of the Bro1-binding site in Snf7 strongly inhibits vesicle formation and cargo sorting. These results implicate Bro1 as a regulator of ESCRT-III disassembly and membrane scission activity.
Results
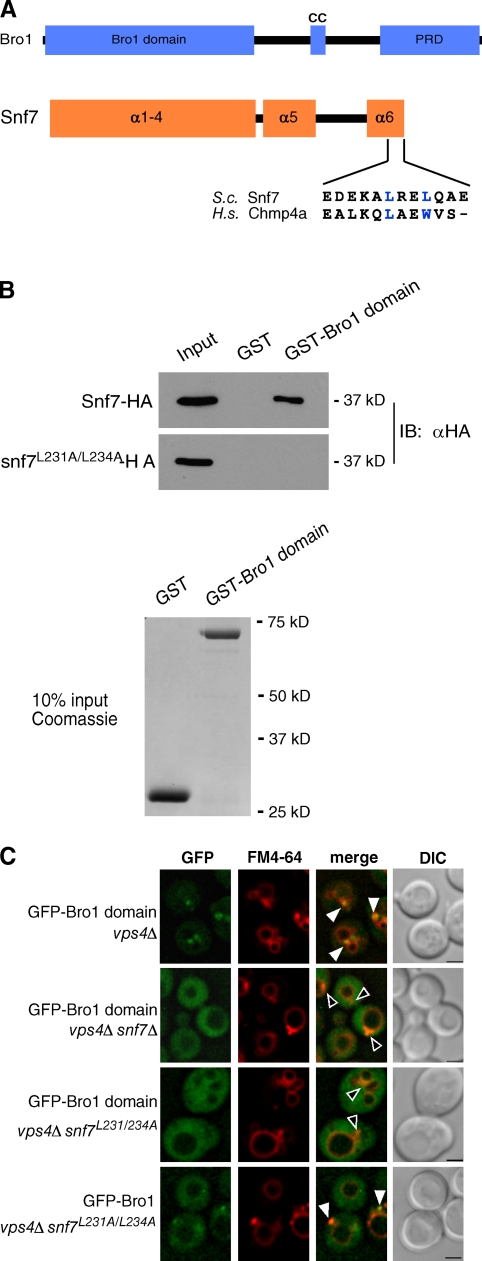
The Bro1 domain binds a MIM1-like motif in the Snf7 subunit of ESCRT-III
Yeast two-hybrid analysis (unpublished data) was used to map the amino acids in Snf7 required for its interaction with the Bro1 domain of yeast Bro1 (Fig. 1 A). This site is centered around residues Leu-231 and Leu-234 at the C terminus of Snf7, which resembles the site in CHMP4 proteins that binds the Bro1 domain of Alix (McCullough et al., 2008) and is similar to the MIM1 motif that was identified in Vps2/CHMP2 and Did2/CHMP1 proteins (Obita et al., 2007; Stuchell-Brereton et al., 2007). The MIT domain of Vps4 has relatively strong affinity for MIM1 of Vps2/CHMP2 and Did2/CHMP1 but binds poorly to the MIM1-like site in CHMP4 proteins (Kieffer et al., 2008), which is consistent with the lack of in vitro binding between purified Snf7 and Vps4 (Shestakova et al., 2010; unpublished data).
Figure 1.
The Bro1 domain binds a MIM1-like motif in Snf7. (A) Schematic diagrams of Bro1 and Snf7, including the amino acid sequence alignment of MIM1-like motifs from yeast Snf7 and human CHMP4a. (B) In vitro pull-downs of purified GST or GST-Bro1 domain incubated with bacterial lysates expressing Snf7-HA or Snf7L231A/L234A-HA. GST and GST-Bro1 domain inputs were loaded at 10% of their total used in each pull-down. IB, immunoblotting. (C) Fluorescence and DIC microscopy of vps4Δ, vps4Δ snf7Δ, and vps4Δ snf7L231/234A cells expressing GFP-Bro1 domain or GFP-Bro1. Arrowheads indicate where GFP does (closed) or does not (open) colocalize with FM4-64–labeled class E compartments. Bars, 2 µm.
Leu-231 and Leu-234 in the MIM1-like site of Snf7 were essential for its in vitro binding to the Bro1 domain of yeast Bro1 (Fig. 1 B). In vivo, these residues were also required for Snf7 to mediate endosomal localization of the Bro1 domain fused to GFP (Fig. 1 C). However, full-length Bro1-GFP weakly localized to endosomes despite disruption of the Bro1 domain–binding site in Snf7 (snf7L231A/L234A; Fig. 1 C), which matches our previous observation that full-length Bro1 weakly localizes to endosomes despite mutation of the Snf7 interaction surface in the Bro1 domain, whereas the same mutation completely disables endosomal localization of the Bro1 domain alone (Kim et al., 2005).
Bro1 binding to Snf7 enhances the stability of ESCRT-III polymers in vivo
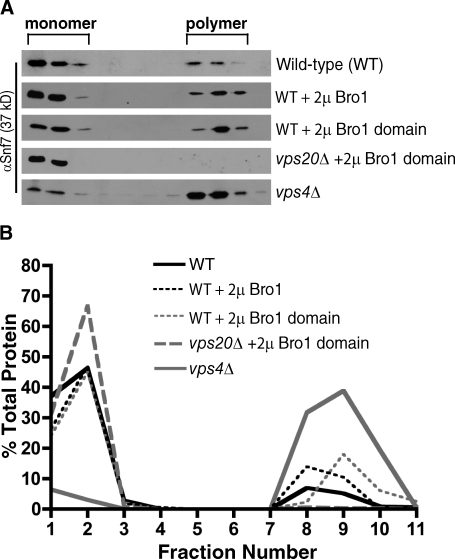
The position of the Bro1 domain–binding site at the C terminus of Snf7 (Fig. 1) suggested that Bro1 might act like Vps20 to relieve Snf7 autoinhibition (Teis et al., 2008; Saksena et al., 2009), thereby promoting Snf7 polymerization and incorporation into ESCRT-III. However, it was previously shown that Bro1 is not essential for Snf7 polymerization based on the analysis of cells in which ESCRT-III disassembly by the Vps4 ATPase had been disabled by deletion of the VPS4 gene (Teis et al. 2008). Therefore, we investigated whether Bro1 might influence Snf7 polymerization in the context of functional Vps4 by examining the distribution of Snf7 in detergent-solubilized yeast membranes separated by isopycnic density gradient centrifugation. In extracts from wild-type cells, the majority of Snf7 was seen in low density fractions, but a small amount (19%) was detected in higher density fractions (Fig. 2, A and B). This high density pool of Snf7 likely corresponds to the transient ESCRT-III complexes that were reported to exist at steady-state (Teis et al., 2008) because the same gradient fractions contained ∼90% of Snf7 in extracts from vps4Δ cells (Fig. 2, A and B) in which ESCRT-III is irreversibly trapped as an assembled complex (Babst et al., 1998; Teis et al., 2008). The polymeric Snf7 we observed had the same properties that were reported previously (Teis et al., 2008) because its stabilization upon deletion of VPS4 was largely dependent on Vps20 but not Bro1 (Fig. 2, A and B). Moreover, recombinant Snf7 that was expressed in bacteria and isolated as a soluble polymer by size-exclusion chromatography before being subjected to isopycnic density gradient centrifugation also migrated to high density fractions (Fig. S1 A).
Figure 2.
Overexpression of Bro1 or the Bro1 domain stabilizes ESCRT-III polymers at endosomes. (A) Western blot analysis of detergent-solubilized yeast membranes resolved by isopycnic density gradient centrifugation. Inclusion of 1% Triton X-100 throughout the gradients yielded identical sedimentation profiles. (B) Quantitation of triplicate gradients from A.
Although a relatively minor percentage of membrane-associated Snf7 was polymeric in extracts from wild-type cells (19%), the percentage of polymeric Snf7 increased upon overexpression of either full-length Bro1 or the Bro1 domain (∼29% and ∼27%, respectively; Fig. 2, A and B). This effect required that Snf7 first undergo activation by Vps20 because overexpression of the Bro1 domain in vps20Δ cells failed to enhance detection of polymeric Snf7 (Fig. 2, A and B). Bro1 domain overexpression also did not cause an increase in the amount of mutant snf7L231A/L234A that could be detected as a polymer, and wild-type Snf7 was similarly unaffected by overexpression of a mutant Bro1 domain that lacks the Snf7-binding site (Fig. S1 B), indicating that the effect of Bro1 domain overexpression toward stabilizing ESCRT-III depends on its ability to bind Snf7. Bro1 domain overexpression caused a similar shift toward high density gradient fractions for another core ESCRT-III subunit, Vps24 (Fig. S2), although concomitant shifts in ESCRT-I or ESCRT-II were not observed (not depicted). However, the total amounts of membrane-associated Snf7 and Vps24 were not significantly altered by overexpression of the Bro1 domain (Fig. S3), indicating that the membrane recruitment step preceding ESCRT-III assembly was not enhanced. Moreover, we did not detect an increase in the amount of Vps4 in high density gradient fractions upon overexpression of the Bro1 domain (unpublished data), indicating that binding of ESCRT-III to Vps4 was not dramatically increased under these conditions. Together, these observations suggest that driving the interaction between Bro1 and Snf7 by overexpression of Bro1 or the Bro1 domain enhances the stability of ESCRT-III assembled at endosomal membranes despite Vps4 being present.
Bro1 binding to Snf7 inhibits Vps4-mediated disassembly of ESCRT-III in vitro
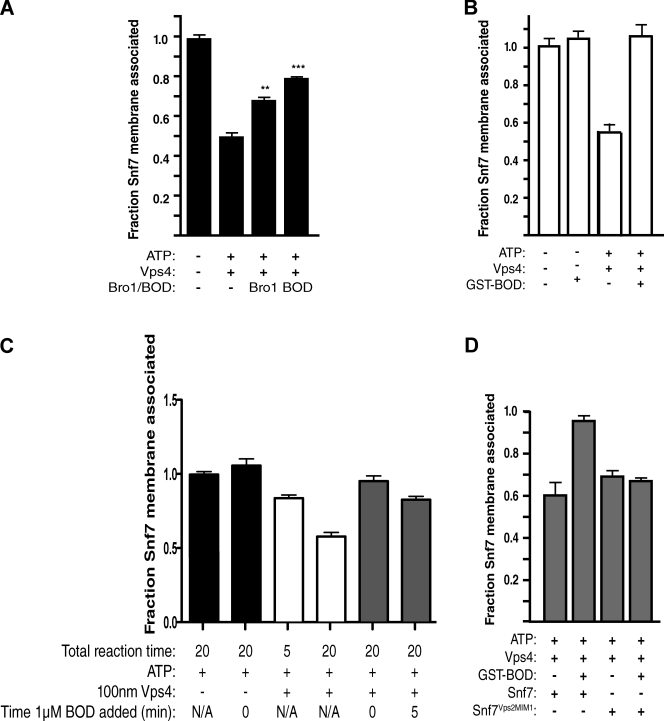
The stabilizing effect exerted by Bro1 toward ESCRT-III was examined further using an in vitro system that reconstitutes Vps4-mediated disassembly of the complex. In this assay, membrane-associated ESCRT-III generated from extracts of vps4Δ cells were incubated with 1 mM ATP and 10 nM purified Vps4 for 10 min at 30°C, and the extent of ESCRT-III disassembly was determined by probing the amount of Snf7 that remains membrane associated after centrifugation. As shown previously (Davies et al., 2010), this reaction condition resulted in a 50% reduction in membrane-associated Snf7 (Fig. 3 A), indicating that exogenous Vps4 has potent activity in mediating in vitro disassembly of ESCRT-III that had been preassembled in vivo. However, this activity was inhibited by ∼50% if ESCRT-III was generated from vps4Δ cells overexpressing the Bro1 domain and, to a lesser degree, if the source of ESCRT-III was vps4Δ cells overexpressing full-length Bro1 (Fig. 3 A).
Figure 3.
Bro1 binding to Snf7 inhibits in vitro disassembly of ESCRT-III. (A) 0.1 µM purified Vps4, ATP, and an ATP regeneration system were incubated with membranes isolated from vps4Δ yeast cells; overexpression of full-length Bro1 or the Bro1 domain (BOD) is indicated. The amount of membrane-associated Snf7 remaining after incubation at 30°C for 10 min was determined by quantitative Western blot analysis. P < 0.001 for both samples. (B) In vitro ESCRT-III release performed in the presence or absence of ATP, 0.1 µM Vps4, and 0.1 µM GST-Bro1 domain (GST-BOD). (C) In vitro ESCRT-III release performed as in B except that the reaction was allowed to proceed for 5 min in the absence of GST-BOD or for 20 min in the absence or presence of 1 µM GST-BOD that was added either at the same time as purified Vps4 or 5 min after the addition of Vps4. (D) In vitro ESCRT-III release performed with membranes from yeast expressing wild-type Snf7 or the Snf7Vps2MIM1 chimera in the presence or absence of ATP, 0.1 µM Vps4, and 0.1 µM GST-BOD. Error bars show the standard deviation.
The inhibitory effect of Bro1 on in vitro disassembly of ESCRT-III (Fig. 3 A) was similar to the in vivo results seen in Fig. 2, wherein the stability of membrane-associated ESCRT-III was enhanced upon overexpression of Bro1 or the Bro1 domain. Although these observations support a model in which Bro1 binding to Snf7 interferes with Vps4-mediated disassembly of ESCRT-III, an alternative possibility was that excess Bro1 or Bro1 domain compromises the ESCRT-III assembly process and results in aberrant complexes refractory to Vps4. However, the addition of purified Bro1 domain (fused to GST) effectively negated in vitro Vps4-mediated disassembly of preassembled ESCRT-III generated from vps4Δ cells (Fig. 3 B). Moreover, this inhibitory effect was observed even if Vps4 was allowed to initiate disassembly of ESCRT-III for 5 min before the addition of GST-Bro1 domain (Fig. 3 C). Thus, Bro1 inhibition of Vps4-mediated disassembly of ESCRT-III is manifested downstream of complex assembly.
We also tested whether the inhibitory effect of the Bro1 domain relied exclusively on its ability to bind Snf7 by generating ESCRT-III from vps4Δ cells in which a Snf7Vps2MIM1 chimera was expressed in place of wild-type Snf7. In this chimera, the MIM1-like Bro1 domain–binding site is replaced with the bona fide MIM1 from Vps2 that binds the MIT domain of Vps4 (Obita et al., 2007). The Snf7Vps2MIM1 chimera rescues in vitro Vps4-mediated disassembly of ESCRT-III formed in the absence of Vps2 expression, demonstrating that it effectively serves as a substrate for Vps4 (Davies et al., 2010). Similarly, ESCRT-III assembled with Snf7Vps2MIM1 in place of wild-type Snf7 was susceptible to disassembly by Vps4 even upon addition of purified Bro1 domain (Fig. 3 D). We conclude that direct binding of the Bro1 domain to Snf7 inhibits Vps4-mediated disassembly of ESCRT-III.
Overexpression of Bro1 or the Bro1 domain reduces ILV budding efficiency at MVBs
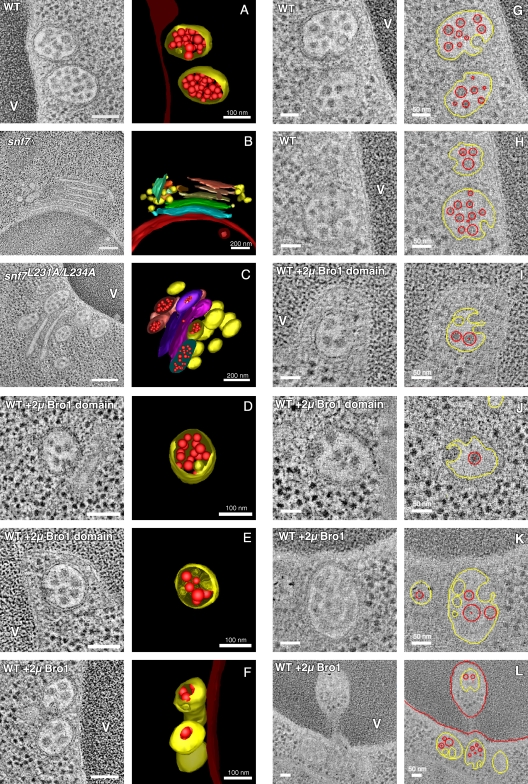
To evaluate whether binding of the Bro1 domain to Snf7 influences the membrane scission activity of ESCRT-III, we examined the morphology of yeast endosomes by EM and 3D tomography. Deletion of SNF7 or genes encoding the other core ESCRT-III subunits inhibits the biogenesis of MVBs like those seen in wild-type cells (Fig. 4 A and Video 1). Instead, snf7Δ cells (and other ESCRT-III deletion strains; not depicted) have abnormal endosomal structures known as class E compartments, which consist of stacks of aberrantly flattened endosomes that generally lack ILVs (Fig. 4 B and Video 2), which is consistent with ESCRT-III promoting membrane scission. Expression of the snf7L231A/L234A allele in place of wild-type SNF7 did not result in class E compartments like those seen in snf7Δ cells. Nevertheless, this mutation caused a sharp reduction in the number of ILVs and a gross distortion of endosomes in a manner reminiscent of the class E compartment morphology (Fig. 4 C, Fig. 5 C [quantification], and Video 3). This reduction in the amount of ILVs is consistent with the interpretation that Bro1 binding to Snf7 is critical for proper ESCRT-III function in vivo.
Figure 4.
Endosome morphologies in cells overexpressing Bro1 or the Bro1 domain. (A–F) 2D cross sections and 3D models from 200-nm-thick section electron tomograms. Spherical endosomal limiting membranes are depicted in yellow, whereas flattened class E compartments are depicted in various colors to discriminate discrete membranes. Lumenal vesicles are red. V, vacuole. Bars, 100 nm or 200 nm as noted. (G–L) Gallery of tomographic slices of wild-type cells with and without overexpression of Bro1 or the Bro1 domain. Freely detached ILVs are traced in red, whereas ILV budding profiles and the limiting endosomal membrane are traced in yellow. In some cases, the continuity of ILV budding profiles with the limiting endosomal membrane is out of plane in the tomographic slice but evident in the 3D reconstruction. Bars, 50 nm.
Figure 5.
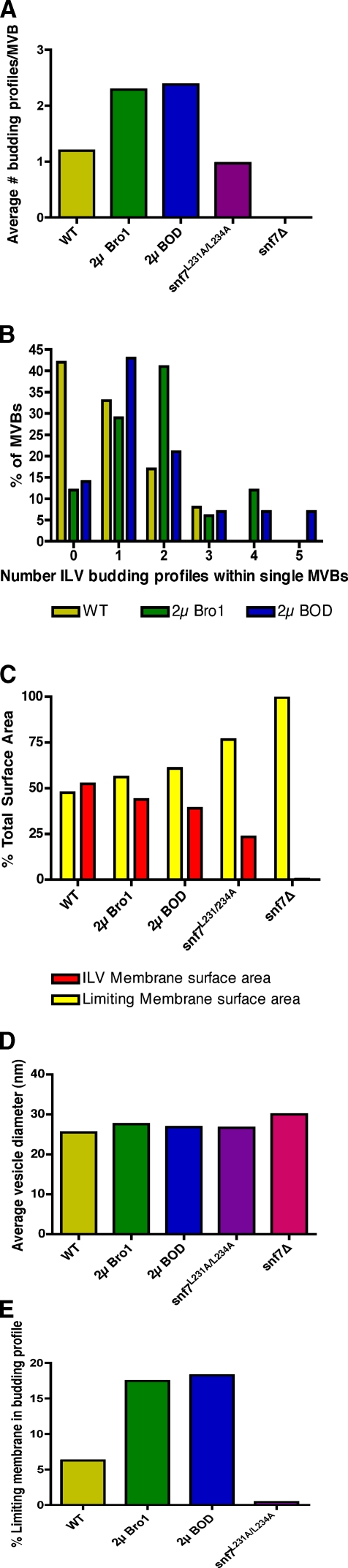
Quantitation of endosome morphology and vesicle budding profiles. Quantitation of the mean number of ILV budding profiles observed per MVB (A) versus the frequency distribution of the number of ILV budding profiles observed per MVB (B) observed in tomograms. Quantitation of the relative percentage of limiting versus lumenal membrane surface areas (C) and the mean ILV diameters (D) observed in tomograms. (E) The percentage of the limiting membrane incorporated into ILV budding profiles was measured for each strain. Note that no ILV budding profiles were seen for snf7Δ cells, but five freely detached ILVs were observed at the periphery of a class E compartment in a single tomogram of snf7Δ cells (Fig. S5); the rarity of this occurrence is reflected by the observation that the membrane surface area of these ILVs comprised 0.27% of the total endosomal membrane surface area in this strain. Tomograms for 12, 14, and 17 MVBs were prepared and modeled for wild-type cells, cells overexpressing the Bro1 domain, and cells overexpressing full-length Bro1, respectively. Four individual tomograms were prepared and modeled for snf7L231A/L234A cells, which contained a total of 33 MVB, VTE, and cisternal structures. Three individual tomograms were generated and modeled for snf7Δ cells, each containing E compartments that were composed of multiple cisternal stacks and occasionally also spherical endosomal membranes. Methods used for the preparation of the tomograms that were used to generate the measurements shown in this experiment are described in detail in the Materials and methods section.
Unexpectedly, we discovered that Bro1 domain overexpression in wild-type cells increased the frequency of intralumenal invaginations that had not detached from the limiting endosomal membrane (Fig. 4, D and E; and Videos 4 and 5). Such ILV budding profiles are not common, with a mean of 1.2 observed per MVB in wild-type cells, but 2.4 budding profiles per MVB on average were seen upon overexpression of the Bro1 domain (Fig. 5 A). Plotting the frequency distribution of ILV budding profiles (Fig. 5 B) showed that overexpression of the Bro1 domain resulted in more instances of multiple budding profiles at individual MVBs (five budding profiles at the MVB shown in Fig. 4 E; Video 5). Overexpression of full-length Bro1 had the same effect as the Bro1 domain (Fig. 4 F; Fig. 5, A and B; and Video 6), demonstrating that the increase in ILV budding profiles was not simply caused by the absence of an activity normally provided by the C-terminal region of Bro1. A gallery of tomograms highlighting ILV budding profiles versus detached ILVs at individual MVBs in wild-type cells with and without overexpression of Bro1 or the Bro1 domain is shown in Fig. 4 (G–L).
The low frequency of ILV budding profiles in wild-type yeast presumably reflects the rapid rate at which membrane scission normally occurs. This process is most likely delayed rather than accelerated in response to Bro1 or Bro1 domain overexpression because quantitation of membrane surface areas showed a decrease in the lumenal membrane content and a corresponding increase in the amount of limiting membrane (Fig. 5 C), although the mean vesicle diameter did not change (Fig. 5 D). In contrast, an acceleration of ILV formation would be expected to show an increase in the amount of lumenal membrane content within endosomes, as is observed in yeast lacking Ist1 (Nickerson et al., 2010), an accessory ESCRT-III protein that inhibits Vps4 activity (Dimaano et al., 2008). Compared with wild-type cells, the ratio of lumenal to limiting membrane surface area was lower upon mutation of the Bro1 domain–binding site in Snf7 (snf7L231A/L234A; 23.3% of total endosomal membrane surface area was lumenal; Fig. 5 C), whereas snf7Δ cells were almost exclusively devoid of lumenal membranes (0.27% of the total endosomal membrane surface area; Fig. 5 C), which is consistent with the stronger effects on endosomal morphology caused by these mutations (Fig. 4).
Measurements of tomograms further revealed that the amount of membrane incorporated into ILV budding profiles in wild-type cells accounted for only 6.3% of the limiting endosomal membrane surface area. In contrast, overexpression of Bro1 or the Bro1 domain resulted in the incorporation of 18.6% and 17.5%, respectively (Fig. 5 E), which is proportional to the increased frequency of ILV budding profiles observed under these conditions (Fig. 5 A). Despite the increase in the number of ILV budding profiles, their overall morphology was not altered significantly, as neither the diameter of the vesicles attached to the limiting endosomal membrane (Fig. S4 A) nor the diameter of the membrane neck at ILV budding profiles (Fig. S4 B) were statistically different in wild-type cells with or without overexpression of Bro1 or the Bro1 domain.
Cargo sorting and deubiquitination are not affected by Bro1 or Bro1 domain overexpression in wild-type cells
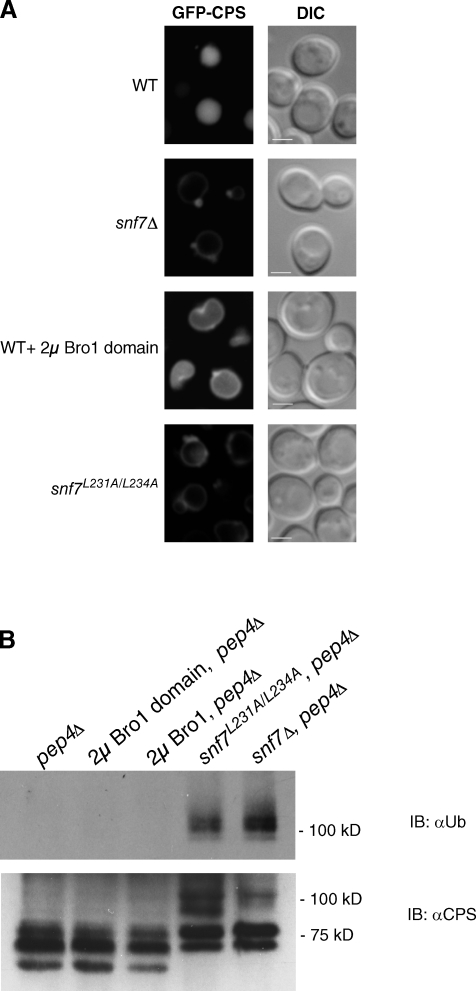
Transmembrane proteins sorted into the ILVs of MVBs are ultimately delivered to the vacuole lumen upon MVB–vacuole fusion, but disrupting the activity of any of the ESCRTs results in mislocalization of these cargoes to the vacuole membrane and their accumulation at class E compartments (Odorizzi et al., 1998). Therefore, we evaluated how the effects on ILV budding revealed by EM tomography (Fig. 4) impacted the efficiency of cargo sorting using live cell fluorescence microscopy of yeast expressing GFP fused to the cytoplasmic domain of carboxypeptidase S (GFP-CPS). We previously reported mislocalization of GFP-CPS upon mutation of the Snf7-binding site in the Bro1 domain (Kim et al., 2005). Similarly, mislocalization of GFP-CPS occurred both in snf7Δ cells and in snf7L231A/L234A cells lacking the Bro1-binding site within Snf7 (Fig. 6 A), which was consistent with the strong reduction in ILV formation caused by these mutations (Fig. 5). Delivery of GFP-CPS into the vacuole lumen in wild-type cells was only mildly affected upon overexpression of the Bro1 domain (Fig. 6 A), whereas full-length Bro1 overexpression did not appear to cause significant cargo mislocalization (Luhtala and Odorizzi, 2004).
Figure 6.
Cargo sorting and deubiquitination. (A) Fluorescence and DIC microscopy of cells expressing GFP-CPS. Bars, 2 µm. (B) Western blot analysis of anti-CPS immunoprecipitates. IB, immunoblotting.
Doa4 is the Ub hydrolase that deubiquitinates CPS and other MVB vesicle cargoes (Dupré and Haguenauer-Tsapis, 2001; Katzmann et al., 2001; Losko et al., 2001), and its activity is stimulated upon binding to Bro1 (Richter et al., 2007). This process occurs efficiently in wild-type cells, resulting in detection of very little ubiquitinated CPS (Ub-CPS; Fig. 6 B). In contrast, Ub-CPS accumulated upon deletion of SNF7 or upon mutation of its Bro1 domain–binding site (snf7L231A/L234A; Fig. 6 B), demonstrating that its interaction with Snf7 is important for Bro1 to promote Doa4 activity at endosomes. Conversely, overexpression of Bro1 or the Bro1 domain had no observable effects on CPS deubiquitination (Fig. 6 B), indicating that the enhancement of ESCRT-III stability and reduced efficiency in membrane scission is not a consequence of altered Doa4 function.
Discussion
In vitro studies have pointed to the assembly of ESCRT-III being crucial to the membrane scission event that results in detachment of ILVs (Wollert et al., 2009; Wollert and Hurley, 2010). Although the specific mechanism by which ESCRT-III executes this activity is unclear, its role in severing membrane connections is supported by in vivo studies of mammalian cells, showing a requirement for ESCRT-III in cytokinesis and retroviral budding, both of which are membrane scission events that are topologically equivalent to ILV formation at MVBs (McDonald and Martin-Serrano, 2009). Genetic dissection of the mechanism of action for ESCRT-III at endosomes in S. cerevisiae has been elusive because of the formation of aberrant class E compartments rather than MVBs in mutant yeast that generally lack functional ESCRT complexes (Rieder et al., 1996; Odorizzi et al., 1998; Luhtala and Odorizzi, 2004; Nickerson et al., 2006). In this study, we describe a potential role for Bro1 in regulating ESCRT-III by demonstrating that overexpression of either full-length Bro1 or its N-terminal Bro1 domain stabilizes ESCRT-III polymers and reduces the efficiency of ILV detachment from the limiting endosomal membrane in yeast that contain an otherwise wild-type ESCRT machinery.
ILV budding profiles in yeast are infrequently observed, even in thick sections imaged by 3D EM tomography, presumably because of the rapid rate at which membrane deformation and scission occur. The increased frequency of budding profiles caused by overexpression of Bro1 or the Bro1 domain must be interpreted as a reduction in ILV budding efficiency rather than an acceleration because these conditions also resulted in significantly less lumenal membrane content within MVBs. In the absence of monitoring this process in real time, it is impossible to exclude whether at least some of the ILV budding profiles are dead-end structures that fail to complete membrane scission. However, the ILVs seen in cells overexpressing Bro1 or the Bro1 domain appeared structurally normal. Neither their overall mean diameter nor the dimensions at the necks of the budding profiles were significantly different from the same measurements in wild-type cells, suggesting that these ILVs will eventually detach from the limiting endosomal membrane.
The specific mechanism underlying the reduced efficiency of ILV budding resulting from overexpression of Bro1 or the Bro1 domain is unclear, but a clue comes from our finding that this condition correlates with increased detection of Snf7 and Vps24, both core ESCRT-III subunits, in a high molecular mass polymer despite wild-type Vps4 expression. Vps4 is the AAA-ATPase that catalyzes disassembly of ESCRT-III (Babst et al., 1998), which accounts for very little of the assembled complex existing at steady-state (Teis et al., 2008). Based on in vitro reconstitution of ILV budding, Vps4 is thought to function indirectly in membrane scission by recycling ESCRT-III subunits for repeated rounds of assembly (Wollert et al., 2009; Wollert and Hurley, 2010). Given the intermediate degree to which Snf7 polymers are stabilized upon Bro1 or Bro1 domain overexpression (only ∼30% of the total amount of Snf7 that is trapped as a polymer in vps4Δ cells), it seems unlikely that a deficiency of free ESCRT-III subunits exists for complex assembly and subsequent execution of membrane scission. However, we presently cannot determine whether Bro1-mediated stabilization of ESCRT-III is the direct cause of the reduced efficiency in ILV budding, which would suggest an active role for Vps4 during membrane scission in vivo. Nevertheless, the possibility that membrane scission might be coupled to Vps4 activity is consistent with our previous discovery by EM tomography that MVBs have significantly more ILVs in the absence of Ist1 (Nickerson et al., 2010), an accessory ESCRT-III protein that negatively regulates Vps4 by inhibiting its binding to ESCRT-III (Dimaano et al., 2008). An active role for Vps4 in membrane scission might also explain why overexpression of catalytically inactive Vps4 in mammalian cells results in an increased frequency of ILV budding profiles at endosomes (Sachse et al., 2004).
The membrane scission activity of ESCRT-III in vitro is potently stimulated by ESCRT-II (Im et al., 2009), which is thought to trigger ESCRT-III polymerization through its direct binding to Vps20 (Saksena et al., 2009). Unlike ESCRT-III, however, ESCRT-II is not required for the budding of human immunodeficiency virus-1 (HIV-1; Langelier et al., 2006), raising the question of how ESCRT-III assembly is stimulated at the site of viral budding. The p6 domain of the Gag polyprotein of HIV-1 binds both to Alix (Strack et al., 2003) and to TSG101, a subunit of ESCRT-I (Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001). The yeast orthologue of TSG101 (Vps23) also binds Vps20 (Bowers et al., 2004; Teo et al., 2004), but an interaction between TSG101 and ESCRT-III has not been reported. Binding of Alix alone to p6 is sufficient to promote HIV-1 budding (Fisher et al., 2007; Usami et al., 2007), but whether this interaction facilitates ESCRT-III assembly is unknown. Our results suggest that Bro1 in yeast lacks this activity because its ability to stabilize ESCRT-III when overexpressed was dependent on the presence of Vps20.
Bro1 exerts its stabilization effect toward ESCRT-III downstream of complex assembly because the addition of exogenous Bro1 domain to preassembled ESCRT-III was sufficient to protect the complex from Vps4-mediated disassembly in vitro. Direct binding to Snf7 was required for Bro1 to inhibit ESCRT-III disassembly both in vivo and in vitro. The site at the C terminus of Snf7 to which the Bro1 domain binds is conserved, as a similar motif was identified at the C terminus of human CHMP4 proteins and shown to be required for binding to the Bro1 domain of Alix (McCullough et al., 2008). The amino acid sequence of this motif and its position near the C terminus of Snf7 resemble the MIM1 sites in Vps2/CHMP2 and Did2/CHMP1 that bind directly to the MIT domain of Vps4, but this site in Snf7 and CHMP4 proteins lacks residues critical for the MIT interaction (Obita et al., 2007; Stuchell-Brereton et al., 2007). Thus, it is unlikely that Bro1 stabilizes ESCRT-III by directly masking a site at which Vps4 might bind Snf7. Located upstream of the MIM1-like site in Snf7 and CHMP4 proteins is a second motif (MIM2) that binds weakly to the Vps4 MIT domain in vitro (Kieffer et al., 2008). Although it is possible that the Bro1 domain interaction sterically shields Vps4 recognition of the MIM2 site in Snf7, testing this hypothesis has been problematic because the Snf7–Vps4 interaction is so weak that we cannot detect binding of either full-length Snf7 or a fragment encompassing MIM2 either to full-length Vps4 or to the Vps4 MIT domain in isolation (unpublished data).
Snf7 is the predominant subunit of the ESCRT-III polymer (Teis et al., 2008) and is critical for the membrane scission activity of the complex in vitro (Wollert et al., 2009). EM tomography suggests the membrane scission activity of ESCRT-III at endosomes in yeast relies on Snf7 binding to the Bro1 domain because snf7L231A/L234A cells, which have otherwise wild-type ESCRT protein activity, exhibit a strong reduction in the number of ILVs and a concomitant expansion of the limiting endosomal membrane, a morphology reminiscent of class E compartments seen in snf7Δ cells and other ESCRT mutant strains. The residual localization of full-length Bro1 to endosomes in snf7L231A/L234A cells might account for the few ILVs observed because complete removal of Bro1 function by deletion of the BRO1 gene results in class E compartments rather than MVBs (Luhtala and Odorizzi, 2004; Richter et al., 2007). Bro1 stimulates transmembrane protein cargo deubiquitination at endosomes by promoting both recruitment and activation of the Doa4 Ub hydrolase (Luhtala and Odorizzi, 2004; Richter et al., 2007). However, this activity alone cannot account for the importance of the Bro1–Snf7 interaction during membrane scission because mutations that specifically disable Bro1 from stimulating Doa4 do not impair ILV formation, nor does replacement of wild-type DOA4 with nonfunctional alleles (Richter et al., 2007).
The strong reduction in ILV formation seen in snf7L231A/L234A cells is consistent with the failure in delivery of CPS to the vacuole lumen via the MVB pathway. Given the central role Bro1 has in regulating deubiquitination at endosomes, it is not surprising that disruption of the Bro1 domain–binding site in Snf7 caused a strong accumulation of Ub-CPS. However, the efficiency of cargo deubiquitination controlled by endogenous full-length Bro1 in wild-type cells suffered virtually no reduction upon overexpression of the Bro1 domain. The C terminus of Bro1 stimulates Doa4 Ub hydrolase activity (Richter et al., 2007), and this region is absent from the Bro1 domain (Fig. 1 A), which would compete with endogenous Bro1 for binding to Snf7. Although its interaction with Snf7 comprises the primary mechanism for endosomal recruitment of Bro1, a small amount of Bro1 is detectable at endosomes if this interaction has been disabled (Fig. 1 C; Kim et al., 2005). The fact that cargo deubiquitination is not significantly compromised by Bro1 domain overexpression suggests that the Snf7-independent mechanism of Bro1 recruitment to endosomes (which has yet to be defined) might be sufficient to maintain Doa4 function.
The inhibition of ESCRT-III disassembly resulting from Bro1 binding to Snf7 might be critical for ESCRT-III reaching a state of assembly necessary to execute membrane scission, explaining the strong reduction in the efficiency of ILV formation observed upon disruption of the Bro1-binding site in Snf7. A similar stabilizing role for Alix would be consistent with observations that disruption of its interaction with CHMP4 proteins impairs cytokinesis and reduces the efficiency of retrovirus budding (Carlton and Martin-Serrano, 2007; Fisher et al., 2007; Morita et al., 2007; Usami et al., 2007; Carlton et al., 2008). At endosomes, a regulatory function in promoting ESCRT-III assembly would place Bro1 at the appropriate time and place to coordinate deubiquitination of transmembrane protein cargoes with the detachment of ILVs. However, a mechanism for coupling these seemingly separate activities of Bro1 has yet to be discovered.
Materials and methods
Yeast strains and plasmid constructions
Standard protocols were used to construct all yeast strains and plasmids described in Tables I and II (Longtine et al., 1998). The bacterial expression plasmid encoding SNF7-HA (pGO619) was constructed in vector pST39 (Tan, 2001) using a PCR product created from the wild-type SNF7 coding sequence and a reverse primer that includes one copy of the HA epitope. The L231A/L234A point mutations were created by site-directed mutagenesis of pGO619, yielding pGO621. The 2-µm plasmid encoding the Bro1 domain (pMWM3) was constructed by homologous recombination of linearized pRS426 with a PCR product containing the Bro1 promoter and Bro1 codons 1–387.
Table I.
S. cerevisiae strains used in this study
| Name | Genotype | Reference |
| SEY6210 | Mata leu2-3,112 ura3-52 his3Δ200 trp1-Δ901 lys2-Δ801 suc2-Δ9 | Robinson et al.,1988 |
| MBY3 | SEY6210; vps4Δ::TRP1 | Babst et al., 1997 |
| TVY1 | SEY6210; pep4Δ::LEU2 | Gerhardt et al., 1998 |
| MWY22 | SEY6210; pep4Δ::LEU2 snf7Δ::HIS3 | This study |
| MWY24 | SEY6210; snf7Δ::HIS3 | This study |
| MWY25 | SEY6210; snf7L231A/L234A::KAN | This study |
| MWY26 | SEY6210; snf7L231A/L234A-KAN vps4::TRP1 | This study |
| CRY181 | SEY6210; snf7L231A/L234A-KAN vps4::TRP1 pep4Δ::LEU2 | This study |
| EEY12 | SEY6210; snf7Δ::HIS3 vps4Δ::TRP1 | Babst et al., 2002 |
| MBY41 | SEY6210; vps20Δ::HIS3 vps4Δ::TRP1 | Babst et al., 2002 |
| GOY66 | SEY6210; bro1Δ::HIS3 vps4Δ::TRP1 | Odorizzi et al., 2003 |
| GOY65 | SEY6210; bro1Δ::HIS3 | Odorizzi et al., 2003 |
| EEY2-1 | SEY6210; vps20Δ::HIS3 | Babst et al., 2002 |
| MBY52 | SEY6210; vps4Δ::TRP1 pep4Δ::LEU2 prb1Δ::LEU2 | Katzmann et al., 2003 |
| JPY140 | SEY6210; vps4Δ::TRP1 pep4Δ::LEU2 snf7Δ::HIS3 | Davies et al., 2010 |
| CRY39 | SEY6210; Vps23-GFP::KAN | Nickerson et al., 2010 |
| CRY40 | SEY6210; Vps23-GFP::KAN vps4Δ::TRP1 | Nickerson et al., 2010 |
| GOY150 | SEY6210; Vps36-GFP::HIS3 | Nickerson et al., 2010 |
| GOY151 | SEY6210; Vps36-GFP::HIS3 vps4Δ::TRP1 | Nickerson et al., 2010 |
| GOY74 | SEY6210; Doa4-GFP::HIS3 | Luhtala and Odorizzi, 2004 |
| GOY75 | SEY6210; Doa4-GFP::HIS3 vps4Δ::TRP1 | Luhtala and Odorizzi, 2004 |
Table II.
Plasmids used in this study
| Plasmid | Protein expressed | Description |
| pGO337 | GFP-Bro1 1-387 | GFP-Bro11-387 URA3 ApR (pRS416) |
| pGO339 | GFP-Bro16 | GFP-Bro1 URA3 ApR (pRS41) |
| pGO47 | GFP-CPS | Odorizzi et al., 1998 |
| pCR180 | GFP-Sna3 | URA3 ApR (pRS415) GFP-Sna3 |
| pGO547 | Snf7 | pST39 Shine-Delgarno Snf7 |
| pGO619 | Snf7-HA | pST39 Shine-Delgarno Snf7-HA |
| pGO621 | Snf7L231A/L234A-HA | pST39 Shine-Delgarno Snf7 L231A/L234A-HA |
| pGEX-4T1 | GST | GE Healthcare |
| pGO626 | GST-Bro11-387-HA | pGEX-4T1 GST-Bro1 1-387-HA |
| pMB54 | GST-Vps4 | Azmi et al., 2006 |
| pGO216 | Bro1 | URA3 ApR (pRS426) Bro1 |
| pMWM3 | Bro11-387 | URA3 ApR (pRS426) Bro11-387 |
| pRS426 Snf7 | Snf7 | Davies et al., 2010 |
| pRS426 Snf7Vps2MIM1 | Snf7Vps2MIM1 | Davies et al., 2010 |
Protein interaction experiments
Lysates of bacteria expressing Snf7-HA or Snf7L231A/L234A-HA were prepared from cultures of BL21 (DE3; Agilent Technologies) transformed with pGO619 or pGO621; protein expression was induced at 20°C for 20 h after the addition of 0.5 mM isopropyl-β-d-thiogalactoside. Purified GST-Bro1 domain was obtained on glutathione Sepharose resin (GE Healthcare) from lysates of BL21 (DE3) that had been transformed with a plasmid encoding this gene fusion (Kim et al., 2005) and induced to express recombinant protein. For in vitro pull-down experiments, glutathione Sepharose and 5 µg GST-Bro1 domain (or GST alone) were incubated for 1 h at 4°C with lysates prepared from bacteria expressing Snf7-HA or Snf7L231/L234A-HA. Bound proteins were recovered by centrifugation of the resin, which was washed thrice with ice-cold PBS + 0.5% Triton X-100, then washed twice with ice-cold PBS. Bound proteins were eluted from the resin by boiling in Laemmli sample buffer, resolved by SDS-PAGE, and transferred to nitrocellulose for Western blot analysis using anti-HA monoclonal antiserum (Roche). 10% of the amount of GST or GST-Bro1 used in each pull-down was resolved by SDS-PAGE and analyzed by Coomassie staining.
Microscopy
Fluorescence microscopy of cells labeled with N-[3-triethylammoniumpro-pyl]-4-[p-diethylaminophenylhexatrienyl] pyridinium dibromide (FM4-64) and/or expressing GFP was performed as described previously (Luhtala and Odorizzi, 2004) using a microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with an NA 1.40 oil immersion 100× objective (Carl Zeiss, Inc.) at room temperature in minimal media. Differential interference contrast (DIC) and fluorescence microscopy images were acquired with a digital camera (Cooke Sensicam; Applied Scientific Instruments, Inc.) and processed using Slidebook (Intelligent Imaging Innovations) and Photoshop software (version CS2; Adobe).
For EM, yeast cells were high pressure frozen and freeze substituted as previously described (Nickerson et al., 2006; Richter et al., 2007) and embedded at −60°C in Lowicryl HM20 (Polysciences). Plastic blocks were trimmed and cut in 80-nm-thin sections and 250-nm-thick sections with a microtome (Leica) and placed on rhodium-plated copper slot grids. Tomographic samples were stained en bloc with 0.1% uranyl acetate and 0.25% glutaraldehyde as a minimal fixative with no additional after staining except for snf7Δ, which required 0.1% uranyl acetate and 2% glutaraldehyde for sufficient preservation for tomography. Thick sections were mapped on a transmission EM (Phillips CM10) at 80 kv, then a Tecnai 30 (FEI) at 300 kv was used for dual-tilt series images collected from 60 to −60° with 1° increments. Tilt series were shot at 31,000× magnification with a 0.764-nm working pixel (binning 2) and repeated at a 90° rotation for dual-axis tomography. 15-nm fiducial gold was used to coat sections on both sides for reconstruction of back projections using IMOD software. Manually assigned contours of the endosomal limiting membrane at the inner leaflet were used to measure the surface of the bilayers periodically every 3.85 nm and calculated using imodmesh. Best-fit sphere models were used to measure the diameters of nearly spherical lumenal vesicles from the outer leaflet of the membrane bilayers. Videos 1–6 were made from IMOD images and completed in QuickTime. Quantitation of ILV budding profiles/MVB were normalized to the mean percent MVB accounted for in the tomograms to more accurately represent the number of ILV budding profiles per MVB.
Isopycnic density gradient centrifugation
For the resolution of native ESCRT complexes, 20 A600 equivalents of cells were converted to spheroplasts, resuspended in 1 ml ice-cold lysis buffer (200 mM sorbitol, 50 mM potassium acetate, 20 mM Hepes, pH 7.2, and 2 mM EDTA) supplemented with protease inhibitors, and homogenized on ice. Soluble and membrane fractions were separated by spinning the lysates at 13,000 g for 15 min at 4°C. The resulting membrane pellet was resuspended in 950 µl lysis buffer, then solubilized by the addition of 0.5% Triton X-100 and rotation at 4°C for 30 min. Solubilized membranes were loaded onto the top of a 10–70% sucrose gradient prepared with the Tris gradient system (Biocomp Instruments) using 10% and 70% sucrose stocks prepared in 200 mM sorbitol, 50 mM potassium acetate, 20 mM Hepes, pH 7.2, and 2 mM EDTA. The gradient was subjected to centrifugation in an SW41 rotor (Beckman Coulter) at 100,000 g for 16–20 h at 4°C, and fractions were collected from the top. Sodium deoxycholate (0.015% final) and trichloroacetic acid (10% final) were added to each fraction, and proteins were precipitated on ice for at least 30 min, followed by centrifugation at 13,000 g for 10 min. Insoluble material was reprecipitated twice by sonication into ice-cold acetone and centrifugation, then sonicated into Laemmli sample buffer and resolved by SDS-PAGE, transferred to nitrocellulose, and examined by Western blotting using antisera against Snf7, Vps24, or GFP (Roche). Quantitation was performed on triplicate experiments by incubating membranes with Alexa Fluor 680 secondary antibody (Invitrogen) and visualizing with an infrared imager (Odyssey; LI-COR Biosciences). Bands were quantitated with Odyssey software (version 2.1). For the snf7L231A/L234A strain, 40 A600 equivalents were used because of decreased detection of the mutant version of the protein. The presence of 1% Triton X-100 throughout the sucrose gradient yielded identical results to gradients performed where Triton X-100 was omitted from the gradient.
In vitro ESCRT-III release assay
The in vitro ESCRT-III release assay was performed as previously described (Davies et al., 2010). Purified Vps4 and an ATP regeneration system (10 mM phosphocreatine, 10 U/ml creatine phosphokinase, and 1 mM ATP) were added to yeast membrane fractions containing ESCRT-III complexes (Babst et al.,1997; Azmi et al., 2006). Membranes were generated from sheroplasted cells that were lysed under native conditions and homogenized, then resuspended in ATPase reaction buffer (100 mM KOAc, 20 mM Hepes, pH 7.4, and 5 mM MgOAc) and passed through an 18-G needle three times followed by a 30-G needle five times to homogenize the membranes. Reactions were allowed to incubate for 10 min at 30°C, then subjected to centrifugation at 13,000 g to separate the membrane-associated and soluble ESCRT-III. SDS-PAGE followed by Western blot analyses was performed with anti-Snf7 antiserum and quantitated using ImageQuant software.
Ub-CPS immunoprecipitation
Detection of Ub-CPS was performed as described previously (Katzmann et al., 2001). The vacuolar protease gene PEP4 was deleted in all strains to reduce the nonspecific cleavage of Ub from CPS after cellular lysis. 10 A600 equivalents of cells were harvested and precipitated by the addition of trichloroacetic acid (10% final concentration). Whole cell lysates were generated by glass bead disruption in urea buffer (6 M urea, 1% SDS, 50 mM, Tris pH 7.5, 1 mM EDTA, and 5 mM NEM), spun at 13,000 g to clear, the supernatant was transferred to a new tube, and CPS was immunoprecipitated with anti-CPS polyclonal antiserum and protein A–Sepharose. Immunoprecipitates were spun down and washed four times, and the beads dried. Samples were resuspended in sample buffer, and 0.5 OD600 equivalents were subjected to SDS-PAGE, transferred to nitrocellulose, which was autoclaved twice, blocked in 20% fetal calf serum, and probed with antibodies against Ub (Invitrogen) or CPS.
Online supplemental material
Fig. S1 shows that Bro1 binding to Snf7 is required for the stabilization of ESCRT-III polymers at endosomes. Fig. S2 shows that overexpression of the Bro1 domain stabilizes ESCRT-III subunit Vps24 at the endosomal membrane. Fig. S3 reveals that the subcellular distribution of ESCRT-III proteins is not altered by overexpression of full-length Bro1 or the Bro1 domain. Fig. S4 demonstrates that the diameters of the membrane necks and vesicles contained within ILV budding profiles do not significantly between wild-type cells and cells overexpressing Bro1 or the Bro1 domain. Fig. S5 shows a tomographic model of an snf7Δ class E compartment that contains five intralumenal vesicles. Video 1 shows a 3D reconstruction of MVBs from a wild-type cell, the top MVB contains a vesicle budding profile. Video 2 contains a 3D reconstruction of an snf7Δ class E compartment. Video 3 shows endosomal structures from snf7L231A/L234A cells in which the endosomes appear to be intermediates between wild-type MVBs and E compartments. Video 4 highlights an MVB from cells overexpressing the Bro1 domain. The MVB contains a single ILV budding profile and several intralumenal vesicles. Video 5 contains a tomographic model of an MVB from cells overexpressing the Bro1 domain in which five simultaneous ILV budding profiles can be observed. Video 6 shows a cluster of three endosomes from cells overexpressing full-length Bro1. Two ILV budding profiles can be seen within the tubular endosome. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007018/DC1.
Acknowledgments
We thank Caleb Richter for yeast strain and plasmid constructions and Sylvie Urbé for helpful discussions.
This work was funded by the National Institutes of Health (grants GM-065505 to G. Odorizzi and GM-73024 D. Katzmann). M. Wemmer was supported by a National Institutes of Health training grant (GM-07135), I. Azmi was supported by an American Heart Association predoctoral grant (AHA07-155882), and B. Davies was supported by a grant from the Fraternal Order of Eagles.
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- ESCRT
- endosomal sorting complex required for transport
- ILV
- intralumenal vesicle
- MIT
- microtubule interacting and trafficking
- MVB
- multivesicular body
- Ub
- ubiquitin
References
- Agromayor M., Martin-Serrano J. 2006. Interaction of AMSH with ESCRT-III and deubiquitination of endosomal cargo. J. Biol. Chem. 281:23083–23091 10.1074/jbc.M513803200 [DOI] [PubMed] [Google Scholar]
- Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., Katzmann D.J. 2006. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172:705–717 10.1083/jcb.200508166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I.F., Davies B.A., Xiao J., Babst M., Xu Z., Katzmann D.J. 2008. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 14:50–61 10.1016/j.devcel.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., Emr S.D. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., Emr S.D. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Snyder W.B., Wendland B., Emr S.D. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289 10.1016/S1534-5807(02)00219-8 [DOI] [PubMed] [Google Scholar]
- Bowers K., Lottridge J., Helliwell S.B., Goldthwaite L.M., Luzio J.P., Stevens T.H. 2004. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 5:194–210 10.1111/j.1600-0854.2004.00169.x [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Martin-Serrano J. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Agromayor M., Martin-Serrano J. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 105:10541–10546 10.1073/pnas.0802008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B.A., Azmi I.F., Payne J., Shestakova A., Horazdovsky B.F., Babst M., Katzmann D.J. 2010. Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol. Biol. Cell. 21:3396–3408 10.1091/mbc.E10-06-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C., Jones C.B., Hanono A., Curtiss M., Babst M. 2008. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 19:465–474 10.1091/mbc.E07-08-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré S., Haguenauer-Tsapis R. 2001. Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol. Cell. Biol. 21:4482–4494 10.1128/MCB.21.14.4482-4494.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.D., Chung H.Y., Zhai Q., Robinson H., Sundquist W.I., Hill C.P. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 128:841–852 10.1016/j.cell.2007.01.035 [DOI] [PubMed] [Google Scholar]
- Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Côté M., Rich R.L., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:55–65 10.1016/S0092-8674(01)00506-2 [DOI] [PubMed] [Google Scholar]
- Gerhardt B., Kordas T.J., Thompson C.M., Patel P., Vida T. 1998. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 273:15818–15829 10.1074/jbc.273.25.15818 [DOI] [PubMed] [Google Scholar]
- Im Y.J., Wollert T., Boura E., Hurley J.H. 2009. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell. 17:234–243 10.1016/j.devcel.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Babst M., Emr S.D. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Stefan C.J., Babst M., Emr S.D. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423 10.1083/jcb.200302136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J.J., Morita E., De Domenico I., Ward D.M., Kaplan J., Sundquist W.I. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 15:62–73 10.1016/j.devcel.2008.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kim J., Sitaraman S., Hierro A., Beach B.M., Odorizzi G., Hurley J.H. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 8:937–947 10.1016/j.devcel.2005.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier C., von Schwedler U.K., Fisher R.D., De Domenico I., White P.L., Hill C.P., Kaplan J., Ward D., Sundquist W.I. 2006. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80:9465–9480 10.1128/JVI.01049-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Losko S., Kopp F., Kranz A., Kölling R. 2001. Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 Mutant. Mol. Biol. Cell. 12:1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N., Odorizzi G. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729 10.1083/jcb.200403139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319 10.1038/nm1201-1313 [DOI] [PubMed] [Google Scholar]
- McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., Hill C.P. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA. 105:7687–7691 10.1073/pnas.0801567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., Martin-Serrano J. 2009. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 122:2167–2177 10.1242/jcs.028308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Alam S.L., Eckert D.M., Gygi S.P., Sundquist W.I. 2007. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe. 2:41–53 10.1016/j.chom.2007.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D.P., West M., Odorizzi G. 2006. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175:715–720 10.1083/jcb.200606113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson D.P., West M., Henry R., Odorizzi G. 2010. Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol. Biol. Cell. 21:1023–1032 10.1091/mbc.E09-09-0776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D.J., Perisic O., Emr S.D., Williams R.L. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 449:735–739 10.1038/nature06171 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S.D. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858 10.1016/S0092-8674(00)81707-9 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Katzmann D.J., Babst M., Audhya A., Emr S.D. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- Piper R.C., Katzmann D.J. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458:445–452 10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Richter C., West M., Odorizzi G. 2007. Dual mechanisms specify Doa4-mediated deubiquitination at multivesicular bodies. EMBO J. 26:2454–2464 10.1038/sj.emboj.7601692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S.E., Banta L.M., Köhrer K., McCaffery J.M., Emr S.D. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 7:985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.S., Klionsky D.J., Banta L.M., Emr S.D. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row P.E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M.J., Sanderson C.M., Urbé S. 2007. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 282:30929–30937 10.1074/jbc.M704009200 [DOI] [PubMed] [Google Scholar]
- Rue S.M., Mattei S., Saksena S., Emr S.D. 2008. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell. 19:475–484 10.1091/mbc.E07-07-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse M., Strous G.J., Klumperman J. 2004. ATPase-deficient hVPS4 impairs formation of internal endosomal vesicles and stabilizes bilayered clathrin coats on endosomal vacuoles. J. Cell Sci. 117:1699–1708 10.1242/jcs.00998 [DOI] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A.E., Emr S.D. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 136:97–109 10.1016/j.cell.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B.A., Katzmann D.J., Babst M. 2010. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 21:1059–1071 10.1091/mbc.E09-07-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B., Calistri A., Craig S., Popova E., Göttlinger H.G. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 114:689–699 10.1016/S0092-8674(03)00653-6 [DOI] [PubMed] [Google Scholar]
- Stuchell-Brereton M.D., Skalicky J.J., Kieffer C., Karren M.A., Ghaffarian S., Sundquist W.I. 2007. ESCRT-III recognition by VPS4 ATPases. Nature. 449:740–744 10.1038/nature06172 [DOI] [PubMed] [Google Scholar]
- Tan S. 2001. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr. Purif. 21:224–234 10.1006/prep.2000.1363 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Emr S.D. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 15:578–589 10.1016/j.devcel.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Teo H., Perisic O., González B., Williams R.L. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 7:559–569 10.1016/j.devcel.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Tsang H.T., Connell J.W., Brown S.E., Thompson A., Reid E., Sanderson C.M. 2006. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 88:333–346 10.1016/j.ygeno.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Usami Y., Popov S., Göttlinger H.G. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 81:6614–6622 10.1128/JVI.00314-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L., Bouamr F., LaGrassa T.J., Agresta B., Kikonyogo A., Leis J., Carter C.A. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA. 98:7724–7729 10.1073/pnas.131059198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Hurley J.H. 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 464:864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Yang D., Ren X., Lee H.H., Im Y.J., Hurley J.H. 2009. The ESCRT machinery at a glance. J. Cell Sci. 122:2163–2166 10.1242/jcs.029884 [DOI] [PMC free article] [PubMed] [Google Scholar]