The dystrophin protein complex, an important regulator of muscle membrane integrity, also maintains neural organization through interactions with the L1CAM family member SAX-7.
Abstract
The dystrophin protein complex (DPC), composed of dystrophin and associated proteins, is essential for maintaining muscle membrane integrity. The link between mutations in dystrophin and the devastating muscle failure of Duchenne’s muscular dystrophy (DMD) has been well established. Less well appreciated are the accompanying cognitive impairment and neuropsychiatric disorders also presented in many DMD patients, which suggest a wider role for dystrophin in membrane–cytoskeleton function. This study provides genetic evidence of a novel role for DYS-1/dystrophin in maintaining neural organization in Caenorhabditis elegans. This neuronal function is distinct from the established role of DYS-1/dystrophin in maintaining muscle integrity and regulating locomotion. SAX-7, an L1 cell adhesion molecule (CAM) homologue, and STN-2/γ-syntrophin also function to maintain neural integrity in C. elegans. This study provides biochemical data that show that SAX-7 associates with DYS-1 in an STN-2/γ-syntrophin–dependent manner. These results reveal a recruitment of L1CAMs to the DPC to ensure neural integrity is maintained.
Introduction
It has been widely established that the actin-binding protein dystrophin is critical for muscle membrane cytoskeletal integrity. Loss-of-function mutations in dystrophin results in the X-linked catastrophic muscle-wasting disorder Duchenne’s muscle dystrophy (DMD). Dystrophin is localized to the sarcolemma in skeletal muscles, where it associates with multiple transmembrane proteins, which include dystroglycan, to form the large membrane-spanning specialized adhesion complex known as the dystrophin protein complex (DPC; Hoffman et al., 1987; Koenig et al., 1988; Zubrzycka-Gaarn et al., 1988; Ervasti and Campbell, 1991; Ibraghimov-Beskrovnaya et al., 1992). Dystrophin binds F-actin inside the cell. Through extracellular interactions with laminin by dystroglycan, the DPC connects the muscle actin cytoskeleton to the ECM and, as such, is hypothesized to provide mechanical protection against the physical stresses of repetitive muscle contractions. Interruption of this linkage results in impaired membrane integrity and degeneration of muscle tissue (Mokri and Engel, 1975; Deconinck and Dan, 2007; Ervasti, 2007). Mutations in other DPC components, such as the sarcoglycans, also result in progressive muscle dystrophies, highlighting the importance of the DPC in maintaining muscle membrane integrity (Sandonà and Betto, 2009). Interestingly, a significant number of DMD patients also exhibit cognitive deficits, the underlying pathology of which is not clear (Bresolin et al., 1994; Mehler, 2000; Wicksell et al., 2004). The prevalence of neural defects accompanying many dystrophin mutations suggests a wider role for dystrophin in humans, thus underscoring the importance of identifying their functions in the nervous system.
Like muscles, the nervous system is also subjected to mechanical forces exerted externally as well as within the organism itself. Neuronal circuitries established during development must withstand the physical stresses of brain growth and synaptic activity-dependent neuronal remodeling as well as the mechanical impacts of body movements and environmental insults (Bénard and Hobert, 2009). Adhesion protein complexes are known to participate in maintaining neural integrity against these physical forces. For example, impaired N-cadherin function causes defects in maintaining the positions of neurons in zebrafish (Lele et al., 2002; Masai et al., 2003). The identification of molecules that participate in maintaining neural architecture has been relatively successful in Caenorhabditis elegans because of the simplicity of its nervous system and the ease of performing genetic manipulations. One protein that is required to maintain the C. elegans neural organization is SAX-7, a homologue of the L1 family of cell adhesion molecules (CAMs; Bénard and Hobert, 2009; Chen and Zhou, 2010).
Vertebrate L1CAMs, which include L1, neuronal CAM (NrCAM), neurofascin, and CHL1, are known to function in nervous system development and function (Hortsch, 2003). Mutations in human NrCAM can result in autism, whereas impaired L1 function causes several different X-linked neural disorders, which include corpus callosum hyperplasia, mental retardation, adducted thumbs, spastic paraplegia, and hydrocephalus (Rosenthal et al., 1992; Van Camp et al., 1993; Jouet et al., 1994; Fransen et al., 1995; Marui et al., 2009). As transmembrane proteins, L1CAMs have been shown to form cell–cell and cell–ECM adhesion via diverse extracellular interactions as well as associate with the membrane actin cytoskeleton by binding cytoskeletal linkers, such as ankyrin. This cytoskeletal anchorage via ankyrin has been shown to be important for L1CAM function (Needham et al., 2001; Buhusi et al., 2008). Ankyrin interaction is similarly important for SAX-7 function (Zhou et al., 2008). Additional binding sites to cytoskeletal linkers that are conserved in L1CAM cytoplasmic tails also contribute to SAX-7 function, underscoring the importance of cytoskeletal anchorage to L1CAMs. One of these sites, a PDZ (PSD95, Dlg, Zo1)-binding (PB) motif, was shown to mediate interaction with STN-2/γ-syntrophin (Zhou et al., 2008), a putative component of the DPC. This finding raises the possibility for dystrophin to function in conjunction with L1CAMs in the nervous system.
In this study, we present genetic data that the sole C. elegans dystrophin gene, dys-1, plays a role in maintaining neural organization. This novel role, which requires dys-1 function in neurons, is distinguishable from its requirement in body-wall muscle to maintain muscle integrity and regulate membrane excitability for proper locomotion (Bessou et al., 1998; Gieseler et al., 2000). We provide biochemical data that SAX-7 associates with DYS-1/dystrophin in an STN-2/γ-syntrophin–dependent fashion. Linkage to DYS-1/dystrophin likely provides SAX-7 anchorage to the actin cytoskeleton, thereby regulating SAX-7–mediated cell adhesions. This study provides insight into the common molecular mechanisms that may underlie diverse cognitive conditions, particularly those of the DMD and L1CAM disorders.
Results
dys-1 participates in positional maintenance of neurons
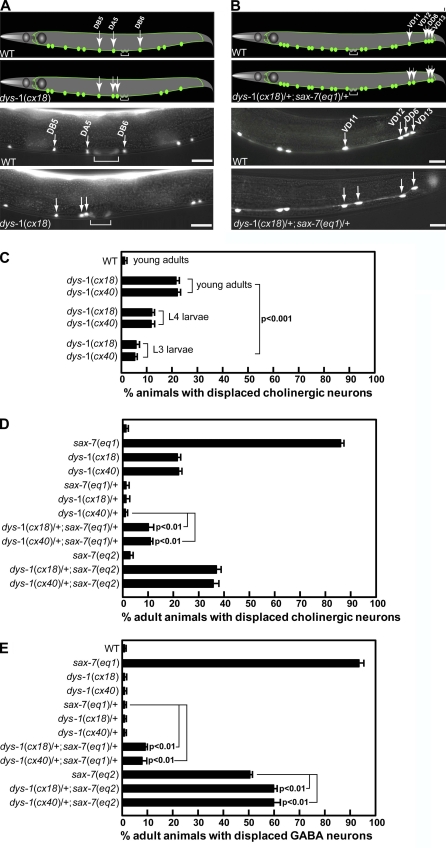
We previously identified STN-2/γ-syntrophin as a SAX-7 interactor that regulates SAX-7 activity in maintaining placement of neurons and their axons (Zhou et al., 2008). Although the role of STN-2/γ-syntrophin has not been characterized, mammalian syntrophins are known components of the DPC that link diverse cytoplasmic and membrane proteins, including signaling proteins as well as water and sodium channels, to the DPC (Brenman et al., 1996; Hasegawa et al., 1999; Lumeng et al., 1999; Adams et al., 2001; Gavillet et al., 2006; Hirn et al., 2008). Consistent with STN-2/γ-syntrophin functioning together with DYS-1/dystrophin, stn-2(tm1869) mutant animals exhibit similar abnormal movements with exaggerated head bending (Fig. 1), which was previously described for dys-1 mutant animals (Bessou et al., 1998). Indeed, dys-1 and stn-2 mutant animals show the characteristic head bend (Fig. 1 A), and they move with a reduced number of body bends per minute as compared with wild-type animals (Fig. 1 B). As a putative DPC component, we hypothesized that STN-2/γ-syntrophin may link SAX-7 to the DPC and that this association may be important for SAX-7 function. To test this hypothesis, we assayed whether DYS-1/dystrophin participates in the positional maintenance of neurons (see Materials and methods). Analysis of dys-1 mutant animals revealed displaced cholinergic neurons in >20% of dys-1(cx18) and dys-1(cx40) adult animals (Fig. 2). Displacement of the affected neurons, which are embryonically derived (Sulston et al., 1983), is less prevalent in younger dys-1 animals and is first detected in a small proportion of L3-staged larvae and an increasing proportion of L4-staged larvae (Fig. 2 C). This apparent progressive displacement is similarly observed in sax-7 mutant animals (Wang et al., 2005) and is consistent with a role for dys-1 in maintaining the position of neurons.
Figure 1.
dys-1 and stn-2 are required in body-wall muscles, but not neurons, to regulate locomotion behavior. (A) Quantification of young adult animals that exhibit exaggerated head bending during locomotion. Two sample sets were analyzed for wild type (WT), dys-1, and stn-2, in which n = 50 for each set. This phenotype is suppressed in dys-1 and stn-2 animals when the respective gene is expressed in the muscle (driven by the myo-3 promoter) but not in neurons (driven by the unc-119 promoter). Three independent transgenic lines were analyzed for each construct. The error bar of each transgenic line shows the standard error of the proportions of four sample sets in which n = 50 in each set. (B) Quantification of body bends per minute for the different strains (see Materials and methods). Three independent transgenic lines were analyzed for each construct. The error bars show the standard error of proportions of four sample sets in which n = 15 in each sample set. Statistical significance was assessed by Student’s t test as compared with wild-type animals.
Figure 2.
Mutations in dys-1 cause defects in maintaining neuronal positions. (A and B) Schematics and corresponding micrographs of VNC cholinergic neurons, as visualized in a young adult wild-type (WT) and dys-1(cx18) animal expressing the Punc-129::gfp marker (A) and VNC GABA neurons, as visualized in a young adult wild-type and a dys-1(cx18)/+;sax-7(eq1)/+ animal expressing UNC-47::GFP (B). The brackets in A mark the vulval muscles, which also express unc-129::gfp. The green dots and lines represent the neuronal cell bodies and axonal processes, respectively. (C–E) The cholinergic neurons DB5, DA5, and DB6 and GABA neurons VD11, VD12, DD6, and VD13 are stereotypically positioned in wild-type animals (see Materials and methods; Fig. S4). The relative positions of the cholinergic neurons are altered in >20% dys-1 adult animals, a phenotype that is less prevalent in dys-1 larvae (C), suggesting a positional maintenance role for dys-1. The quantification of young adult animals exhibiting displaced cholinergic (D) and GABA (E) neurons in dys-1 and sax-7 mutant backgrounds reveals a genetic interaction between dys-1 and sax-7. Error bars show the standard error of the proportions of three sample sets in which n = 100 in each set. The p-values in C–E show the statistical significance as assessed by the Z test between the indicated strains. Bars, 10 µm.
To further investigate a role for dys-1 in neuronal position maintenance, we assessed whether dys-1(cx18) or dys-1(cx40) genetically interacts with sax-7. Genetic or pharmacological suppression of movements in sax-7 animals was previously shown to suppress the neuronal displacement phenotype (Sasakura et al., 2005; Pocock et al., 2008). Thus, in these genetic assays, we intentionally used dys-1/+ heterozygosity to circumvent the possibility that the prolonged muscle excitation displayed by dys-1 homozygous animals (Bessou et al., 1998; Kim et al., 2004, 2009) may nonspecifically enhance neuronal displacement in sax-7 mutant backgrounds. Moreover, because homozygous sax-7(eq1)-null animals exhibit >95% penetrance of displaced neurons (Fig. 2, D and E; Wang et al., 2005), we assessed the consequences of dys-1/+ heterozygosity on neuronal displacement in sax-7(eq1)/+ animals. Displaced cholinergic and γ-aminobutyric acid (GABA) neurons located along the ventral nerve cord (VNC) were observed in 10% dys-1/+;sax-7(eq1)/+ trans-heterozygous animals, whereas neuronal displacement was not detected in either dys-1/+ or sax-7(eq1)/+ single heterozygous mutant animals (Fig. 2, D and E). This genetic interaction between dys-1 and sax-7 is significant and is also illustrated by an enhancement of displaced neurons by dys-1/+ in animals that are homozygous for the hypomorphic sax-7 allele eq2 (Wang et al., 2005). Indeed, displaced cholinergic neurons were detected in 37% of dys-1/+;sax-7(eq2) as compared with 4% of sax-7(eq2) single mutant animals (Fig. 2 D). The penetrance of GABA neuron displacement is also increased in dys-1/+;sax-7(eq2) animals as compared with sax-7(eq2) single mutant animals (Fig. 2 E). Collectively, these results reveal a role for dys-1 in maintaining neuron positions.
Aging dys-1 animals are reported to exhibit some muscle degeneration, particularly in a sensitized hlh-1 mutant background (Gieseler et al., 2000; Grisoni et al., 2003). Thus, it is possible that dys-1/+ in a sensitized sax-7 background could result in muscle degeneration or abnormal muscle morphology, which may indirectly cause altered positioning of the adjacent VNC neurons and tissues along the ventral midline. Immunostaining of β-integrin and phalloidin staining of actin did not reveal the presence of muscle degeneration or obvious defects in muscle morphology and sarcomeric organization in young adult dys-1 or sax-7 animals or in dys-1/+ animals in sax-7 mutant backgrounds (Fig. S1). Furthermore, young adult dys-1 or sax-7 animals do not exhibit vulval displacement (Fig. S2), suggesting that this positional maintenance role for dys-1 and sax-7 is specific for neurons.
dys-1 and stn-2 are required in neurons, but not body-wall muscles, to maintain neuronal positioning
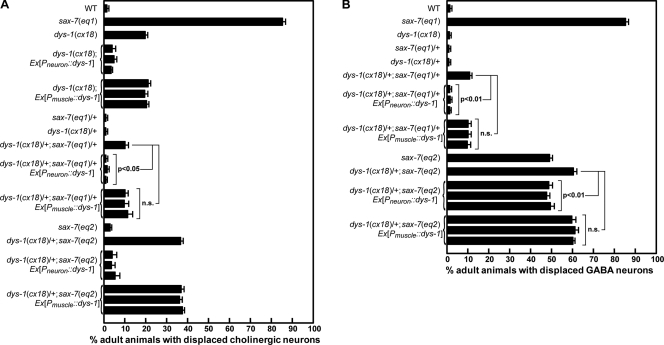
We previously showed that sax-7 is required in neurons as well as the adjacent hypodermis and body-wall muscles for positional maintenance of the VNC cholinergic and GABA neurons (Wang et al., 2005). dys-1 is predominantly expressed in the VNC neurons as well as body-wall and vulval muscles, as determined by Pdys-1::gfp transcriptional reporters (Bessou et al., 1998; Dupuy et al., 2007; Hunt-Newbury et al., 2007). To determine the site of function for dys-1, we assayed for rescue of the displaced neuron phenotype in dys-1 animals expressing full-length DYS-1/dystrophin in either neurons or muscles using tissue-specific promoters. Only DYS-1/dystrophin expressed in neurons rescued the neuronal displacement phenotype in dys-1 homozygous and dys-1/+;sax-7/+ animals (Fig. 3). Similarly, only neuronally expressed DYS-1/dystrophin suppressed the genetic enhancement displayed in dys-1/+;sax-7(eq2) animals, whereas DYS-1/dystrophin expressed in body-wall muscles did not (Fig. 3).
Figure 3.
dys-1 is required in neurons, but not muscle, to maintain neuronal position. (A and B) Tissue-specific expression assays reveal that dys-1 functions in neurons, but not body-wall muscles, to maintain the position of cholinergic (A) and GABA (B) neurons. Neuronal and muscle expressions of dys-1 were driven by the unc-119 and myo-3 promoters, respectively. Three independent transgenic lines were analyzed for each construct. The error bars show the standard error of the proportions of three sample sets in which n = 100 in each set. Statistical significance was assessed by the Z test. WT, wild type.
DYS-1/dystrophin expressed in body-wall muscles did, however, rescue the abnormal movement and head-bending phenotype, which is characteristic of dys-1 animals (Fig. 1), indicating that the expressed DYS-1/dystrophin in muscles is functional. In fact, transgenic dys-1 animals that expressed DYS-1/dystrophin in body-wall muscles are indistinguishable from wild-type animals, exhibiting no obvious phenotype other than displaced neurons. In contrast, dys-1 animals expressing DYS-1/dystrophin in neurons still displayed the characteristic locomotory and head-bending phenotype (Fig. 1). This result is consistent with a previous study that revealed a requirement for DYS-1 in muscles, but not in neurons, for wild-type movements (Bessou et al., 1998). Thus, dys-1 has two separable roles; i.e., dys-1 functions in neurons to maintain neuronal positioning and acts in body-wall muscles to control locomotory behavior.
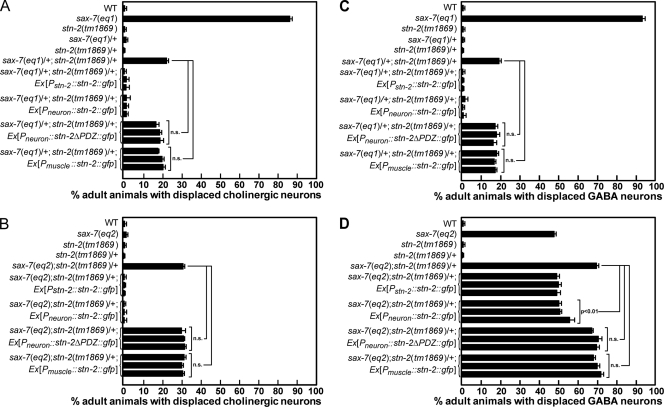
stn-2 is also expressed in VNC motor neurons and body-wall muscles (Zhou et al., 2008). Because dys-1 is required only in neurons to maintain neuronal positioning, we anticipated that stn-2 would also be required in neurons, but not muscles, for this function. Indeed, neuronal expression of STN-2/γ-syntrophin rescues the displaced VNC cholinergic and GABA neurons in stn-2/+ animals in either sax-7(eq1)/+ or a homozygous sax-7(eq2) sensitized background (Fig. 4). In contrast, muscle expression of STN-2/γ-syntrophin failed to rescue the neuronal displacement (Fig. 4). Like dys-1, expression of STN-2/γ-syntrophin in muscles, but not in neurons, rescues the abnormal locomotion and head bending exhibited by stn-2 animals (Fig. 1). Similar to dys-1, both stn-2 animals and stn-2/+ animals in either a sax-7(eq1)/+ or a homozygous sax-7(eq2) sensitized background do not exhibit muscle degeneration or obvious muscle abnormalities (Fig. S1), which may indirectly cause displacement of neurons. Thus, both dys-1 and stn-2 are required in neurons to maintain neuronal positioning, a function that is independent of their role in body-wall muscles.
Figure 4.
stn-2 is required in neurons, but not muscle, to maintain neuronal position. (A–D) Tissue-specific expression assays reveal that stn-2 functions in neurons, but not body-wall muscles, to maintain the position of cholinergic (A and B) and GABA (C and D) neurons. Moreover, this function requires the STN-2 PDZ domain. Three independent transgenic lines were analyzed for each construct in which neuronal and muscle expressions of STN-2 were driven by the unc-119 and myo-3 promoters, respectively. The error bars show the standard error of the proportions of three sample sets in which n = 100 in each set. Statistical significance was assessed by the Z test. WT, wild type.
STN-2 interacts with DYS-1 by yeast two-hybrid (Y2H) assays
Vertebrate syntrophins, including γ-syntrophins, can bind dystrophins via their C-terminal syntrophin unique (SU) region (Ahn and Kunkel, 1995; Ahn et al., 1996; Castelló et al., 1996). The 62–amino acid SU region of STN-2 shares 55 and 62% similarity with the SU region of human γ1- and γ2-syntrophin, respectively. This conservation of the SU region in STN-2 indicates a possible interaction between STN-2 and DYS-1.
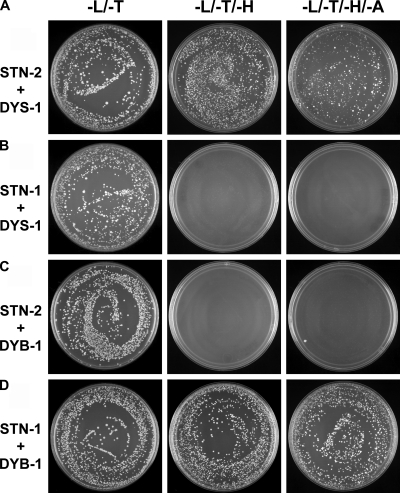
To investigate whether STN-2 interacts with DYS-1, a Y2H assay was performed. In this assay, rather than using full-length DYS-1, which is predicted to be a 417-kD protein, we tested the DYS-1 C-terminal end, which contains homologous sequences to mammalian dystrophins that are required for interacting with syntrophins (Bessou et al., 1998; Gieseler et al., 1999a). Yeast transformed with the full-length stn-2 and C-terminal dys-1 clones showed robust cell growth on the L/T/H and L/T/H/A selective media, indicating a positive interaction between STN-2 and DYS-1 (Fig. 5 A). An interaction between STN-1/αβ-syntrophin and DYS-1 was assessed as a negative control. As expected, STN-1/αβ-syntrophin and DYS-1 did not interact (Fig. 5 B), which was similar to results of a previous Y2H assay (Grisoni et al., 2003). We also did not observe an interaction between STN-2 and the dystrophin-like molecule DYB-1/dystrobrevin (Fig. 5 C). To determine that the negative controls STN-1 and DYB-1 were expressed and functional in the Y2H assays, a known interaction between them (Grisoni et al., 2003) was tested, which was positive as expected (Fig. 5 D). These negative controls underscore the specificity of the interaction between STN-2 and DYS-1. Thus, the two C. elegans syntrophins differentially interact with the dystrophin family of proteins; STN-2/γ-syntrophin interacts specifically with DYS-1/dystrophin, whereas STN-1/αβ-syntrophin interacts with DYB-1/dystrobrevin. Consistent with these molecular interactions, STN-1/αβ-syntrophin and DYB-1/dystrobrevin do not participate in neuronal positional maintenance (Fig. S3; Zhou et al., 2008).
Figure 5.
STN-2, but not STN-1, molecularly interacts with DYS-1, as determined by Y2H assays. The prey is comprised of full-length STN-2 or STN-1, whereas the bait is comprised of the C terminus of DYS-1 (amino acid residues 3,402–3,674) or full-length DYB-1. Cell growth on L/T selective media shows yeast cells that are successfully transformed with both the prey and bait constructs. Cell growth on L/T/H or L/T/H/A selective media reveals positive interactions between STN-2 and DYS-1 and between STN-1 and DYB-1.
STN-2 acts as an adaptor to link SAX-7 to the DYS-1 cytoskeleton
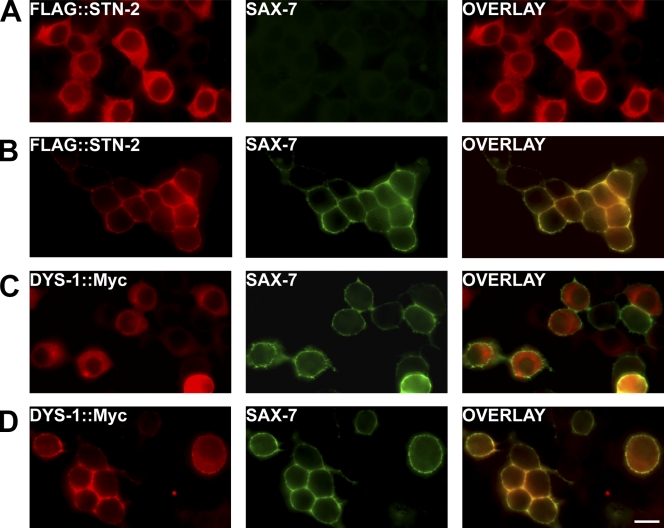
STN-2/γ-syntrophin can interact with both SAX-7 (Zhou et al., 2008) and DYS-1/dystrophin (Fig. 5 A), which was consistent with our hypothesis that STN-2/γ-syntrophin couples SAX-7 to DYS-1/dystrophin. To further test this hypothesis, we performed a protein recruitment assay using cultured cells, which was previously used to evaluate interactions between ankyrin and L1CAMs (Zhang et al., 1998; Chen et al., 2001; Zhou et al., 2008). FLAG::STN-2 is primarily localized in the cytoplasm when expressed in HEK293 cells (Fig. 6 A). However, cotransfection with a SAX-7 construct, which is composed of rat neurofascin extracellular and transmembrane domains fused to the SAX-7 cytoplasmic tail (SAX-7CT), results in the redistribution of STN-2 to the cell cortex where the neurofascin::SAX-7 chimera is localized (Fig. 6 B). This result is consistent with the previously described interaction of STN-2/γ-syntrophin with the SAX-7CT (Zhou et al., 2008). If STN-2/γ-syntrophin functions as an adaptor to link SAX-7 to DYS-1/dystrophin, SAX-7 should similarly recruit DYS-1/dystrophin to the cell cortex in an STN-2/γ-syntrophin–dependent manner. To test this prediction, HEK293T cells were transfected with dys-1::myc and neurofascin::sax-7 clones. As with the Y2H assay, the dys-1::myc clone encodes only the DYS-1 C-terminal region, which carries the syntrophin-interacting domain. DYS-1::Myc is cytoplasmic and does not colocalize with neurofascin::SAX-7 at the plasma membrane (Fig. 6 C). However, when FLAG::STN-2 is coexpressed together with DYS-1::Myc and neurofascin::SAX-7, DYS-1::Myc is largely redistributed from the cytoplasm to the cell cortex (Fig. 6 D). These results strongly suggest that DYS-1/dystrophin associates with SAX-7 only when STN-2/γ-syntrophin is present.
Figure 6.
Protein recruitment assays in HEK293 cells suggest that SAX-7 associates with DYS-1 only in the presence of STN-2. (A) FLAG::STN-2 is primarily localized in the cytoplasm, as detected by immunostaining with an anti-FLAG antibody. (B) Cotransfection with neurofascin::SAX-7CT (labeled SAX-7) causes FLAG::STN-2 to be redistributed to the cell cortex where SAX-7 is localized. (C) DYS-1::Myc, which contains the DYS-1 C-terminal amino acid residues 3,402–3,674, is primarily localized in the cytoplasm, as detected by anti-Myc antibody in cells transfected with both DYS-1::Myc and neurofascin::SAX-7CT. (D) When STN-2 is cotransfected, DYS-1::Myc is redistributed to the cell cortex where SAX-7 is localized, thus suggesting that SAX-7 can associate with DYS-1 via STN-2. Bar, 15 µm.
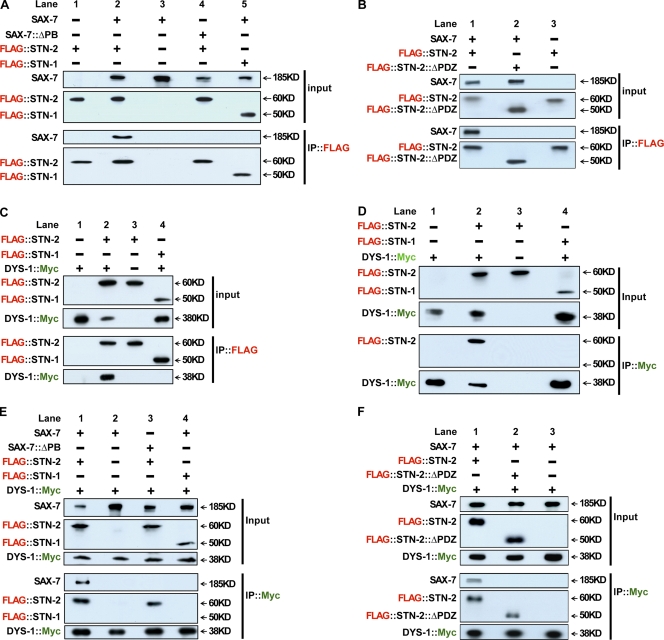
To further confirm that DYS-1/dystrophin and STN-2/γ-syntrophin form a complex with SAX-7, the biochemical interactions of the three proteins were tested in HEK293T cells via coimmunoprecipitation (IP [co-IP]) assays. Neurofascin::SAX-7 was detected in anti-FLAG IPs of HEK293T cell lysates containing FLAG::STN-2 and neurofascin::SAX-7 (Fig. 7, A [lane 2] and B [lane 1]). In contrast, neurofascin::SAX-7 was not detected in anti-FLAG IPs on cell lysates containing FLAG::STN-1 and neurofascin::SAX-7 (Fig. 7 A, lane 5) or FLAG::STN-2 and neurofascin::SAX-7CT lacking the PB sequence (Fig. 7 A, lane 4). These results were expected based on our previous results that STN-2/γ-syntrophin, but not STN-1/αβ-syntrophin, interacts with SAX-7 via the PB domain (Zhou et al., 2008). Neurofascin::SAX-7 was also not detected in IPs of cell lysates containing a mutated FLAG::STN-2 that lacks the PDZ domain (STN-2::ΔPDZ) and neurofascin::SAX-7 (Fig. 7 B, lane 2), suggesting that the PDZ domain is required for STN-2/γ-syntrophin to interact with SAX-7. The importance of the STN-2 PDZ domain is underscored by the result that expression of STN-2::ΔPDZ cannot rescue the neuronal displacement phenotype in sax-7(eq1)/+;stn-2/+ animals or suppress the phenotypic enhancement in sax-7(eq2)/sax-7(eq2);stn-2/+ animals (Fig. 4).
Figure 7.
STN-2 acts as a linker protein that bridges SAX-7 to DYS-1, as determined by co-IP assays in HEK293 cells. (A and B) Neurofascin::SAX-7CT (labeled SAX-7) interacts with STN-2, as shown in A (lane 2) and B (lane 1). This interaction depends on the SAX-7 PB sequence (A, lane 4) and the STN-2 PDZ domain (B, lane 2). This interaction between SAX-7 and STN-2 is specific because SAX-7 does not interact with STN-1 (A, lane 5). (C and D) The C-terminal end of DYS-1 (amino acid residues 3,402–3,674) interacts with STN-2, but not STN-1, as DYS-1 coimmunoprecipitates with STN-2 (C, lane 2) but not with STN-1 (C, lane 4). The reverse IP shows that STN-2 (D, lane 2), but not STN-1 (D, lane 4), coimmunoprecipitates with DYS-1. (E and F) Although SAX-7 does not coimmunoprecipitate with DYS-1 (E, lane 2; and F, lane 3), SAX-7 does so when STN-2 is present (E, lane 1; and F, lane 1), suggesting this tripartite complex requires STN-2 as a linking molecule. Co-IP of SAX-7 with STN-2 and DYS-1 requires the PB sequence of SAX-7 (E, lane 3) and the PDZ domain of STN-2 (F, lane 2).
The interaction between STN-2/γ-syntrophin and DYS-1/dystrophin was tested next. Reciprocal co-IPs were observed with FLAG::STN-2 and DYS-1::Myc (Fig. 7, C and D, lane 2) but not with FLAG::STN-1 and DYS-1::Myc (Fig. 7, C and D, lane 4). It is interesting that both our Y2H and co-IP experiments, as well as an independent Y2H assay (Grisoni et al., 2003), consistently showed a lack of interaction between STN-1/αβ-syntrophin and the C-terminal end of DYS-1, whereas a previously published in vitro binding assay using a purified GST–STN-1 fusion protein suggested otherwise (Gieseler et al., 1999a). This difference may reflect dissimilar experimental approaches and conditions that could suggest a weak interaction between STN-1/αβ-syntrophin and DYS-1/dystrophin. For example, the intrinsic ability for GST to dimerize (Walker et al., 1993; Dirr et al., 1994; Lim et al., 1994) could have resulted in the oligomerization of the GST–STN-1 fusion protein that was used in the in vitro binding assay, which, in turn, may have enhanced a weak interaction between STN-1 and DYS-1. Similar oligomerization and influence of activity on diverse fusion proteins induced by GST have been reported (Haldeman et al., 1997; Mernagh et al., 1997; Niedziela-Majka et al., 1998).
To investigate whether SAX-7 could interact with DYS-1/dystrophin, anti-Myc IPs were performed on cell lysates containing both DYS-1::Myc and neurofascin::SAX-7. Neurofascin::SAX-7 did not coimmunoprecipitate (Fig. 7, E [lane 2] and F [lane 3]), which was consistent with a lack of association between SAX-7 and DYS-1/dystrophin, as suggested in the protein recruitment assay. However, in anti-Myc IPs on lysates of cells in which FLAG::STN-2 was cotransfected with both DYS-1::Myc and neurofascin::SAX-7CT, neurofascin::SAX-7CT coimmunoprecipitated along with FLAG::STN-2 (Fig. 7, E and F, lanes 1), which was consistent with SAX-7 being present in a complex with DYS-1 and STN-2. Collectively, these results strongly suggest a role for STN-2/γ-syntrophin as an adaptor protein that couples SAX-7 and DYS-1/dystrophin together in a protein complex.
Discussion
This study uncovers a novel neuronal role for DYS-1/dystrophin in maintaining the structure and organization of the C. elegans nervous system in collaboration with the SAX-7/L1CAM. This role is distinct from its previously established muscle function in maintaining muscle integrity and regulating locomotion. The presented genetic and biochemical data provide evidence that SAX-7 is linked to DYS-1/dystrophin via STN-2/γ-syntrophin and that this tripartite protein complex is required in neurons (Fig. 8). Dystrophin and the associated actin cytoskeleton likely provide anchorage to SAX-7, thereby regulating SAX-7 activity in maintaining neuronal positions.
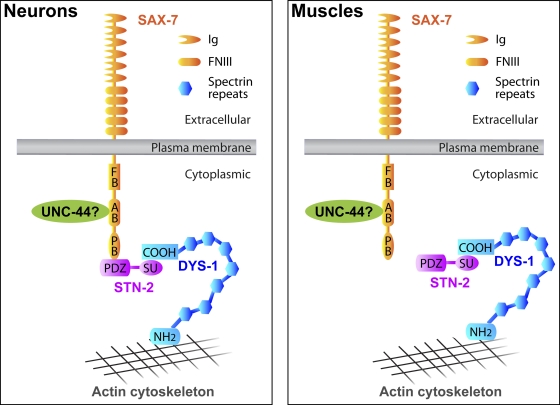
Figure 8.
A model depicting how STN-2/γ-syntrophin may regulate SAX-7 activity in maintaining neuronal positions, as based on genetic and biochemical data. The model speculates that STN-2/γ-syntrophin links SAX-7 to the DYS-1/dystrophin-based actin cytoskeleton in neurons but not muscles. This interaction is dependent on the STN-2 PDZ domain to interact with the SAX-7 PB motif and requires the STN-2 syntrophin unique (SU) domain to interact with the C-terminal end of DYS-1. DYS-1 is predicted to bind to the actin cytoskeleton by its N-terminal actin-binding domain. This linkage to the actin cytoskeleton may provide SAX-7 anchorage, thereby regulating SAX-7–mediated cell adhesion. Interruption of this anchorage leads to a defect in maintaining neuronal positions. UNC-44/ankyrin also interacts molecularly and genetically with SAX-7 (Zhou et al., 2008), but it is not known whether UNC-44/ankyrin is required in neurons and/or body-wall muscles; this unknown is depicted as “UNC-44?” in the schematic. Ig, Ig-like repeats; FNIII, fibronectin type III repeats; FB, FERM-binding sequence; and AB, ankyrin-binding sequence.
A distinct neuronal DPC functions to maintain neural architecture
Mutations in dys-1 cause progressive muscle degeneration in sensitized genetic backgrounds (Gieseler et al., 2000; Mariol et al., 2007), indicating a conserved role for dystrophin in maintaining muscle integrity. dys-1 mutant animals also exhibit a distinctive movement phenotype (i.e., exaggerated bending of the head and anterior body and prolonged muscle contraction), which is shown, in part, to be caused by abnormal cholinergic transmission (Bessou et al., 1998; Giugia et al., 1999). This phenotype can be rescued by wild-type DYS-1 expressed in muscle but not neurons (Bessou et al., 1998). Mutations in genes encoding other components of the C. elegans DPC, such as stn-2/γ-syntrophin, stn-1/αβ-syntrophin, and dyb-1/dystrobrevin, also lead to locomotory phenotypes that are indistinguishable from that of dys-1, suggesting that these DPC components function in muscles for proper cholinergic transmission (Fig. 1; Gieseler et al., 1999b, 2001; Grisoni et al., 2003; Kim et al., 2004). Indeed, stn-1 and stn-2 expression in muscles, but not neurons, can rescue the locomotory phenotype in the respective mutant animals (Fig. 1; Grisoni et al., 2003).
In addition to a role for the DPC in muscles, we identified a novel neuronal role of DYS-1/dystrophin and STN-2/γ-syntrophin in the positional maintenance of neurons, which is independent of their function in muscles. This neuronal role is not shared by STN-1/αβ-syntrophin or DYB-1/dystrobrevin (Fig. S3; Zhou et al., 2008), although both proteins are also extensively expressed in the nervous system (Gieseler et al., 2001; Grisoni et al., 2003). It is not known what function STN-1/αβ-syntrophin or DYB-1/dystrobrevin mediates in neurons or whether these roles involve DYS-1/dystrophin. Thus, at least in neurons, the DPCs are likely variable in composition and function.
It is not clear how neuronal displacement impacts neural functions in dys-1 mutant animals or whether dys-1 mediates additional functions in neurons other than maintaining neuronal positioning. Animals that lack dys-1 in neurons (i.e., dys-1 mutant animals expressing dys-1 only in muscles) have no obvious phenotype other than displaced neurons. Assays that assess behavior and higher order neural function (e.g., learning and memory) still need to be performed to address these questions.
STN-2/γ-syntrophin and DYS-1/dytrophin provide cytoskeletal anchorage to SAX-7
The protein recruitment assay and co-IP experiments suggest that STN-2/γ-syntrophin can act as an adaptor to link SAX-7 to DYS-1/dystrophin. As dystrophin is an actin-binding protein, the association of SAX-7 with DYS-1/dystrophin raises the possibility that SAX-7 is linked to the cortical actin cytoskeleton. Linkage of CAMs, including L1CAMs, cadherins, and integrins, to the cortical cytoskeleton has been shown to be required for robust cell adhesion (Bökel and Brown, 2002; Nagaraj and Hortsch, 2006; Hartsock and Nelson, 2008). Indeed, cultured cells expressing neurofascin that cannot bind ankyrin exhibit reduced cell–cell adhesion, presumably because of a poor association with the spectrin-based actin cytoskeleton (Garver et al., 1997; Tuvia et al., 1997). In C. elegans, UNC-44/ankyrin has been shown to function with SAX-7 in neuronal positional maintenance, which was consistent with the importance of linkage to the spectrin-actin cytoskeleton (Zhou et al., 2008). We thus speculate that STN-2/γ-syntrophin and DYS-1/dystrophin participate in neuronal positional maintenance by coupling SAX-7 to the cortical actin cytoskeleton. Interruption of this cytoskeleton linkage by either deletion of the SAX-7 PB sequence or the STN-2 PDZ domain or by knocking out stn-2 or dys-1 function leads to defects in maintaining neuronal position, probably caused by reduced SAX-7 activity in mediating cell adhesion.
It is thus puzzling that displaced cholinergic neurons are detected in dys-1, but not in stn-2, mutant animals. This difference suggests that DYS-1/dystrophin can also mediate SAX-7 function in an STN-2–independent fashion. Proteins that may compensate for the loss of STN-2/γ-syntrophin are not known, but one candidate protein may be UNC-44/ankyrin. A recent study uncovered the ability of mammalian ankyrins to bind dystrophin via a cysteine-rich domain, which is located close to, but distinct from, the syntrophin-interacting domain in the C terminus of dystrophin (Ayalon et al., 2008). Interaction with dystrophin has been shown for ankyrin-G and ankyrin-B, which are required for the proper localization of dystrophin at the costameres and sarcolemma, respectively, in murine skeletal muscle (Ayalon et al., 2008). Thus, UNC-44/ankyrin may compensate for the loss of STN-2 to couple SAX-7 to the dystrophin-based cytoskeleton, thus reflecting the lack of phenotype in stn-2 mutant animals. This speculation would require that the regulation of SAX-7 activity by UNC-44/ankyrin occurs in neurons, which is currently not known.
The expression of UNC-44/ankyrin in neurons, body-wall muscles, and the hypodermis (Chen et al., 2001) suggests that UNC-44/ankyrin could regulate SAX-7 activity in one or more of these tissues; SAX-7 is required in all three tissues to ensure that neural integrity is maintained (Wang et al., 2005). Consistent with UNC-44/ankyrin acting as an adaptor to link SAX-7 to the spectrin-actin cytoskeleton, SPC-1/α-spectrin and UNC-70/β-spectrin are similarly expressed in multiple tissues, including neurons, body-wall muscles, and the hypodermis (Hammarlund et al., 2000; Moorthy et al., 2000; Norman and Moerman, 2002). That SAX-7 may be coupled to both the dystrophin- and spectrin-based actin cytoskeletons raises the question of how cytoskeletal linkage of SAX-7 is coordinated. Are both the dystrophin- and spectrin-based cytoskeletons required in neurons for SAX-7 activity? Is SAX-7 associated with both cytoskeletons simultaneously, or are distinct subpopulations of SAX-7 linked to each cytoskeleton? If only the dystrophin-based cytoskeleton is required in neurons, how is linkage to the spectrin-based cytoskeleton prevented? In addition to the cortical actin cytoskeleton, dystrophin also binds intermediate filaments and microtubules (Prins et al., 2009; Le Rumeur et al., 2010). Thus, SAX-7 may be simultaneously anchored to one of these alternative cytoskeletons in addition to the spectrin-actin cytoskeleton.
Implications for mammalian L1CAMs
Mammalian dystrophin and DPC components are also expressed in neurons (Waite et al., 2009). In contrast to their functions in muscles, dystrophin and the DPC components in the mammalian nervous system have not been as extensively characterized. These neuronal DPC complexes apparently are molecularly heterogeneous (Blake and Kröger, 2000; Waite et al., 2009), suggesting multiple neuronal roles for the DPC.
The importance of dystrophin in the brain is directly indicated by cognitive impairments and neuropsychiatric disorders (e.g., autism and schizophrenia) that can be presented in DMD patients (Bresolin et al., 1994; Anderson et al., 2002). Autopsy studies on DMD patients revealed brain abnormalities that include disordered connections and architectural changes (Rosman, 1970; Jagadha and Becker, 1988; Moriuchi et al., 1993; Uchino et al., 1994; Kim et al., 1995). The mdx dystrophin mutant mice similarly exhibit an abnormal architecture of the brain that includes altered distribution of populations of neurons (Carretta et al., 2004; Del Tongo et al., 2009; Minciacchi et al., 2010). They also display altered spontaneous inhibitory postsynaptic currents in the cerebellar Purkinje cells, which may reflect the need for dystrophin to cluster GABAA receptors in hippocampal pyramidal neurons and inhibitory synapses of cerebellar Purkinje cells (Knuesel et al., 1999; Anderson et al., 2003; Kueh et al., 2008). Based on the role of C. elegans dystrophin in maintaining neural organization, impaired mammalian dystrophin may similarly affect neural integrity, thus accounting for some of the functional and architectural abnormalities present in the brains of DMD patients and mdx mice.
The association of DYS-1/dystrophin with SAX-7 via STN-2 suggests that mammalian L1CAMs could similarly function with dystrophin in the nervous system. Of the mammalian L1CAMs, NrCAM has a PB sequence (amino acids NSFV) that is most similar to that of SAX-7 (amino acids STFV). The similar expressions of NrCAM, γ-syntrophins, and dystrophin in the cerebral cortex, hippocampal pyramidal neurons, and the cerebellar cortex Purkinje cells of the mammalian brain are consistent with their ability to interact (Lidov et al., 1993; Piluso et al., 2000; Hogan et al., 2001; Backer et al., 2002; Alessi et al., 2006; Ishiguro et al., 2006; Heyden et al., 2008; Minciacchi et al., 2010). Although it is not known whether NrCAM functions in a similar capacity as SAX-7 for proper neural organization, the importance of NrCAM in the brain is underscored by the impaired cognitive function and social behavior exhibited by NrCAM-null mice (Moy et al., 2009). Moreover, mutations in human NrCAM can lead to autism (Marui et al., 2009), a disorder that is also presented in significant numbers of DMD patients (Wu et al., 2005; Hendriksen and Vles, 2008). The apparent overlap of neural symptoms of DMD and L1CAM disorders, together with our data that DYS-1/dystrophin and SAX-7 function in a complex, suggests the mammalian counterparts may act in a homologous fashion for proper neural architecture and nervous system function.
Materials and methods
Strains
C. elegans strains, provided by the Caenorhabditis Genetics Center, were grown on nematode growth medium plates at 21°C as described by Brenner (1974). N2 Bristol served as the wild-type strain. The alleles used in this study are listed by linkage groups as follows: LGI, dys-1(cx18), dys-1(cx40) (Bessou et al., 1998), and dyb-1(cx36) (Gieseler et al., 1999b); LGIV, sax-7(eq1) (Zhou et al., 2008), and sax-7(eq2) (Wang et al., 2005); and LGX and stn-2(tm1869) (Zhou et al., 2008).
C. elegans expression vectors and generation of transgenic animals
Transgenic animals were generated according to standard procedures (Mello et al., 1991). To generate Punc-119::dys-1 (pLC587), a 9.0-kb dys-1 genomic sequence from WRM0611bE10 (Geneservice) was pieced together at exon 30 with 3.9-kb dys-1 cDNA and 169-bp dys-1 3′ untranslated region (obtained from yk1473g12 and yk1434h02; Y. Kohara, National Institute of Genetics, Mishima, Japan) and subcloned into the pBluescript II KS vector between NotI and XmaI. The pan-neuronal unc-119 promoter (Maduro and Pilgrim, 1995, Maduro et al., 2000) was inserted at NotI to drive dys-1 expression in neurons. The resulting construct was injected at 190 ng/µl along with 2 ng/µl of the coinjection marker Pmyo-2::tdTomato (a gift from S. Panowski and A. Dillin, Salk Institute, La Jolla, CA) into dys-1(cx18).
To generate Pmyo-3::dys-1 (pLC586), a pLC587 derivative replacing the unc-119 promoter with the muscle-specific myo-3 promoter (Fire et al., 1998) was used to drive dys-1 expression in muscles. 160 ng/µl of the resulting construct was injected into dys-1(cx18) along with 40 ng/µl of the coinjection marker sur-5::dsRed (Yochem et al., 1998).
To generate Pstn-2::stn-2::gfp (pLC551), the stn-2 promoter (3.82-kb sequences upstream of the start codon) and 1.5-kb stn-2 cDNA (obtained from yk788e09; Y. Kohara) were placed in frame with gfp coding sequences and the unc-54 3′ untranslated region (Fire et al., 1990), all of which were subcloned into the pBluescript II KS vector between NotI and ApaI. 50 ng/µl of the resulting construct was injected into stn-2(tm1869) animals along with 50 ng/µl of the coinjection marker sur-5::dsRed (Yochem et al., 1998).
To generate Punc-119::stn-2::gfp (pLC553), a pLC551 derivative with the stn-2 promoter was replaced with the unc-119 promoter. 50 ng/µl of this construct was injected into stn-2(tm1869) animals along with 70 ng/µl of the coinjection marker Pstr-1::gfp (Troemel et al., 1995). Punc-119::stn-2::gfp::ΔPDZ (pLC590) was generated from a pLC553 derivative lacking the STN-2 PDZ domain (amino acids 74–150), which was injected at 50 ng/µl into stn-2(tm1869) animals along with 70 ng/µl of the coinjection marker Pstr-1::gfp (Troemel et al., 1995).
Pmyo-3::stn-2::gfp (pLC554) was generated from a pLC553 derivative that replaced the stn-2 promoter with the myo-3 promoter. This construct was injected at 50 ng/µl into stn-2(tm1869) animals along with 70 ng/µl of the coinjection marker Pstr-1::gfp (Troemel et al., 1995).
Live animal microscopy—scoring for displaced neurons
The oxIs12 (unc-47::gfp; McIntire et al., 1997) and evIs78 (Punc-129::gfp; Colavita et al., 1998) integrated transgenes were crossed into respective strains to visualize GABA or cholinergic neurons, respectively. Synchronized young adult animals of all these strains as well as synchronized dys-1 larvae of various stages were mounted on 2% agarose pads and scored for neuronal displacement with the 40× 1.3 NA Neofluar objective using a microscope (Axioplan 2 IE) with the image captured with a camera (AxioCam MRm) and imaging software (AxioVision 4.5; Carl Zeiss, Inc.).
Of the VNC cholinergic neurons, the relative positions of the DB5, DA5, and DB6 neurons are most consistently altered in the examined genetic strains as compared with wild type (Fig. 2 A) and, thus, were the focus of our analysis. The ratio of the distance between DB5 and DA5 (designated D1) to the distance between DA5 and DB6 (designated D2) was calculated (Fig. S4 A). D1 and D2 are similar in wild-type animals so that the mean D1/D2 ratio is slightly more than one (Fig. S4 B). Neurons are considered displaced when the D1/D2 ratio in an animal is two or greater, as a result of the D2 value being less than half that of D1 because DA5 and DB6 are closer to each other (Fig. 2 A).
Of the VNC GABA neurons, the relative positions of VD11, VD12, DD6, and VD13 neurons are most consistently altered in the examined genetic strains as compared with wild type (Fig. 2 B) and, thus, were the focus of our analysis. The ratio of the distance between VD11 and VD12 (designated D1) to the distance between VD12 and VD13 (designated D2) was calculated (Fig. S4 C). In wild-type animals, D1 is generally more than three times larger than D2 so that the mean D1/D2 ratio is 3.5 (Fig. S4 D). Neurons are considered displaced when the D1/D2 ratio in an animal is one or less, as a result of the D1 value being equal to or less than that of D2 because VD12 or DD6 is closer to VD11 (Fig. 2 B).
Live animal microscopy—determining vulval position
Synchronized L4 wild-type, sax-7, and dys-1 animals were mounted on 2% agarose pads and were examined under a microscope (Axioplan 2 IE) to determine the position of the vulva relative to the pharynx and the anus. Based on the calculation PV/(PV + VA), in which PV is the distance between the base of the pharyngeal terminal bulb and the center of the vulva and VA is the distance between the center of the vulva and the anal opening (Fig. S3 A), the vulva in sax-7 and dys-1 animals appears to be positioned similarly to that of wild type (Fig. S3 B).
Locomotion assay
A single young adult animal (8–12 h after the L4 stage) was transferred to a nematode growth medium plate with a fresh lawn of OP50 Escherichia coli. After 10 s of recovery, the number of body bends was measured during a subsequent 1-min period. A body bend is defined as a period of one sinusoidal track left by the animal on the bacteria lawn. Only animals without backward movement in that 1-min period were quantified.
Immunofluorescence analysis
Animals were fixed in methanol and stained for indirect immunofluorescence using the freeze-crack methanol fixation method (Miller and Shakes, 1995). Rhodamine-conjugated phalloidin (1:25 concentration; Invitrogen) against polymerized actin, the MH25 monoclonal antibody (1:300 concentration) against β-integrin (Francis and Waterston, 1985; Gettner et al., 1995), and the Alexa Fluor 488 secondary antibody (1:500 concentration; Invitrogen) were used. Samples were examined with a 100× 1.45 NA PlanFLUAR objective (Carl Zeiss, Inc.) using a microscope (Axioplan 2 IE) with the image captured with a camera (AxioCam MRm) and imaging software (AxioVision 4.5).
Y2H constructs
All Y2H bait and prey constructs were generated by subcloning coding sequences of stn-1, stn-2, dys-1, and dyb-1 into the respective vectors (Matchmaker GAL4 Two-Hybrid System 3; Takara Bio Inc.). The baits, pLC569 and pLC570, respectively contain stn-1 and stn-2 cDNA, which were subcloned into the pGBKT7 vector (Trp selective marker) in frame with the GAL4 DNA-binding domain. The preys, pLC571 and pLC572, respectively contain a dys-1 cDNA fragment encoding amino acids 3,402–3,674 and dyb-1 cDNA, which were subcloned into the pGADT7 vector (Leu selective marker) in frame with the GAL4 DNA-activating domain.
Y2H assays
All yeast assays were performed in the AH109 yeast strain, which were cultured on the following synthetic defined (SD) minimal media lacking specified amino acids according to the manufacturer’s instructions: SD/−Leu, SD/−Trp, SD/−Leu/−Trp, SD/−Leu/−Trp/−His + 2.5 mM 3-AT, SD/−Leu/−Trp/−His/−Ade + 12.5 mM 3-AT, SD/−Leu/−His + 2.5 mM 3-AT, and SD/−Trp/−His + 2.5 mM 3-AT (Takara Bio Inc.). 2.5 mM 3-AT was added to reduce the number of background colonies caused by leaky His expression. We confirmed that both bait and prey constructs (a) did not activate readout reporters on their own, e.g., grow on media lacking His or His/Ade, and (b) did not interact nonspecifically with the provided negative control pGADT7-T (for baits) or pGBKT7-lam (for preys). A positive interaction was considered robust if there were numerous large nonred (white) colonies growing on media lacking Leu/Trp/His/Ade. Positive interactions were further assayed by performing X-α- and β-galactosidase tests on these nonred colonies, which should turn strongly blue if there is robust interaction between bait and prey.
Mammalian cell expression vectors
Mammalian cell expression vectors used in this study were neurofascin::SAX-7CT (pLC101; Chen et al., 2001); neurofascin::SAX-7::ΔPB (pLC573), a pLC101 derivative lacking the PB motif STFV; Pcmv::FLAG::stn-1 (pLC565), the stn-1 cDNA (obtained from yk109h11) that was subcloned into p3×FLAG-CMV-7.1 (Sigma-Aldrich) between NotI and KpnI; Pcmv::FLAG::stn-2 (pLC566), the stn-2 cDNA that was subcloned into p3×FLAG-CMV-7.1 (Sigma-Aldrich) between SalI and BamHI; and Pcmv::FLAG::stn-2::ΔPDZ (pLC588), a pLC566 derivative lacking the PDZ domain (amino acids 74–150). Pcmv::dys-1::Myc (pLC574), a dys-1 cDNA fragment encoding amino acids 3,402–3,674, was cloned into pcDNA3.1/Myc–His B (Invitrogen) between KpnI and BamHI.
Cell culture and transfection
HEK293T cells were cultured in DME (Mediatech, Inc.) with 10% newborn bovine serum and treated with transfection reagent (Lipofectamine 2000; Invitrogen) according to the standard procedure provided by the manufacturer during DNA transfection. 40 h after transfection, cells were collected and washed with PBS in preparation for either immunofluorescence or co-IP assays.
Protein recruitment assay in HEK293 cells
The assay was performed as previously described (Zhang et al., 1998). Cells were fixed with formaldehyde, washed, and incubated with anti-FLAG (M2; Sigma-Aldrich), anti-Myc (9E10; Covance), or anti-SAX-7 (6991; Chen et al., 2001) antibodies to visualize FLAG::STN-2, DYS-1::Myc, or neurofascin::SAX-7CT, respectively. Images were acquired using Axioplan 2 IE, AxioCam MRm, and AxioVision 4.5.
Co-IP assays
Cell lysates were prepared in NETN buffer containing 1 mM NaF, 2.5 mM β-glycerophosphate, and a protease inhibitor cocktail (Roche). 200–500 µl of cell lysate was incubated with NETN buffer (0.05-M Tris, pH 7.8, 0.1-M NaCl, 1 mM EDTA, and 0.5% NP-40) containing the FLAG or Myc antibody at 4°C for 1 h. Immune complexes were precipitated with 20 µl of protein A/G beads (Santa Cruz Biotechnology, Inc.) at 4°C for 4 h. The beads were washed three times with NETN lysis buffer, and then the immune complexes were eluted by heating the beads to 95°C for 5 min in SDS sample buffer containing DTT. Blots were probed as described in the next section.
Western blot analysis and reagents
Cell lysates were prepared in NETN buffer containing 1 mM NaF, 2.5 mM β-glycerophosphate, and a protease inhibitor cocktail. Cell lysates were resolved by SDS-PAGE and electrophoretically transferred to the nitrocellulose membrane. Membranes were blocked in TBS-T (0.15-M NaCl, 20 mM Tris, pH 8, and 0.05% Tween 20) containing 5% bovine albumin (Sigma-Aldrich) for 1 h. Blots were probed with primary antibodies followed by horseradish peroxidase–conjugated anti–mouse (Mouse TrueBlot Ulture; eBioscience) or anti–rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). Blots were then developed on film using an enhanced chemiluminescence kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The primary antibodies used in this study included anti-FLAG (M2; Sigma-Aldrich), anti-Myc (9E10; Covance), and anti–SAX-7 (6991; Chen et al., 2001).
Online supplemental material
Fig. S1 shows that immunofluorescence experiments reveal normal body-wall muscles in animals of sax-7, stn-2, or dys-1 genetic backgrounds. Fig. S2 shows that the relative position of the vulva is normal in sax-7 and dys-1 animals. Fig. S3 demonstrates that dyb-1 mutant animals do not display neuronal position defects. Fig. S4 shows the positions of cholinergic and GABAergic neurons in wild-type animals. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006109/DC1.
Acknowledgments
We thank Karla Opperman and Wei Zhang for technical advice on the Y2H and co-IP assays, Ann Rougvie and Meg Titus for editorial advice, and the Caenorhabditis Genetics Center for providing genetic strains.
This study was supported in part by the University of Minnesota Paul and Sheila Wellstone Muscular Dystrophy Center Marzolf Award awarded to S. Zhou and by grant NS045873 from the National Institute of Neurological Disorders and Stroke.
Footnotes
Abbreviations used in this paper:
- CAM
- cell adhesion molecule
- DMD
- Duchenne’s muscular dystrophy
- DPC
- dystrophin protein complex
- GABA
- γ-aminobutyric acid
- IP
- immunoprecipitation
- NrCAM
- neuronal CAM
- PB
- PDZ-binding
- SAX-7CT
- SAX-7 cytoplasmic tail
- SU
- syntrophin unique
- VNC
- ventral nerve cord
- Y2H
- yeast two-hybrid
References
- Adams M.E., Mueller H.A., Froehner S.C. 2001. In vivo requirement of the α-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J. Cell Biol. 155:113–122 10.1083/jcb.200106158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn A.H., Kunkel L.M. 1995. Syntrophin binds to an alternatively spliced exon of dystrophin. J. Cell Biol. 128:363–371 10.1083/jcb.128.3.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn A.H., Freener C.A., Gussoni E., Yoshida M., Ozawa E., Kunkel L.M. 1996. The three human syntrophin genes are expressed in diverse tissues, have distinct chromosomal locations, and each bind to dystrophin and its relatives. J. Biol. Chem. 271:124–2730 10.1074/jbc.271.5.2724 [DOI] [PubMed] [Google Scholar]
- Alessi A., Bragg A.D., Percival J.M., Yoo J., Albrecht D.E., Froehner S.C., Adams M.E. 2006. gamma-Syntrophin scaffolding is spatially and functionally distinct from that of the alpha/beta syntrophins. Exp. Cell Res. 312:3084–3095 10.1016/j.yexcr.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Anderson J.L., Head S.I., Rae C., Morley J.W. 2002. Brain function in Duchenne muscular dystrophy. Brain. 125:4–13 10.1093/brain/awf012 [DOI] [PubMed] [Google Scholar]
- Anderson J.L., Head S.I., Morley J.W. 2003. Altered inhibitory input to Purkinje cells of dystrophin-deficient mice. Brain Res. 982:280–283 10.1016/S0006-8993(03)03018-X [DOI] [PubMed] [Google Scholar]
- Ayalon G., Davis J.Q., Scotland P.B., Bennett V. 2008. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell. 135:1189–1200 10.1016/j.cell.2008.10.018 [DOI] [PubMed] [Google Scholar]
- Backer S., Sakurai T., Grumet M., Sotelo C., Bloch-Gallego E. 2002. Nr-CAM and TAG-1 are expressed in distinct populations of developing precerebellar and cerebellar neurons. Neuroscience. 113:743–748 10.1016/S0306-4522(02)00221-X [DOI] [PubMed] [Google Scholar]
- Bénard C., Hobert O. 2009. Looking beyond development: maintaining nervous system architecture. Curr. Top. Dev. Biol. 87:175–194 10.1016/S0070-2153(09)01206-X [DOI] [PubMed] [Google Scholar]
- Bessou C., Giugia J.B., Franks C.J., Holden-Dye L., Ségalat L. 1998. Mutations in the Caenorhabditis elegans dystrophin-like gene dys-1 lead to hyperactivity and suggest a link with cholinergic transmission. Neurogenetics. 2:61–72 10.1007/s100480050053 [DOI] [PubMed] [Google Scholar]
- Blake D.J., Kröger S. 2000. The neurobiology of duchenne muscular dystrophy: learning lessons from muscle? Trends Neurosci. 23:92–99 10.1016/S0166-2236(99)01510-6 [DOI] [PubMed] [Google Scholar]
- Bökel C., Brown N.H. 2002. Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell. 3:311–321 10.1016/S1534-5807(02)00265-4 [DOI] [PubMed] [Google Scholar]
- Brenman J.E., Chao D.S., Gee S.H., McGee A.W., Craven S.E., Santillano D.R., Wu Z., Huang F., Xia H., Peters M.F., et al. 1996. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 84:757–767 10.1016/S0092-8674(00)81053-3 [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresolin N., Castelli E., Comi G.P., Felisari G., Bardoni A., Perani D., Grassi F., Turconi A., Mazzucchelli F., Gallotti D., et al. 1994. Cognitive impairment in Duchenne muscular dystrophy. Neuromuscul. Disord. 4:359–369 10.1016/0960-8966(94)90072-8 [DOI] [PubMed] [Google Scholar]
- Buhusi M., Schlatter M.C., Demyanenko G.P., Thresher R., Maness P.F. 2008. L1 interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. J. Neurosci. 28:177–188 10.1523/JNEUROSCI.3573-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretta D., Santarelli M., Sbriccoli A., Pinto F., Catini C., Minciacchi D. 2004. Spatial analysis reveals alterations of parvalbumin- and calbindin-positive local circuit neurons in the cerebral cortex of mutant mdx mice. Brain Res. 1016:1–11 10.1016/j.brainres.2004.04.021 [DOI] [PubMed] [Google Scholar]
- Castelló A., Brochériou V., Chafey P., Kahn A., Gilgenkrantz H. 1996. Characterization of the dystrophin-syntrophin interaction using the two-hybrid system in yeast. FEBS Lett. 383:124–128 10.1016/0014-5793(96)00214-1 [DOI] [PubMed] [Google Scholar]
- Chen L., Zhou S. 2010. “CRASH”ing with the worm: insights into L1CAM functions and mechanisms. Dev. Dyn. 239:1490–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ong B., Bennett V. 2001. LAD-1, the Caenorhabditis elegans L1CAM homologue, participates in embryonic and gonadal morphogenesis and is a substrate for fibroblast growth factor receptor pathway-dependent phosphotyrosine-based signaling. J. Cell Biol. 154:841–855 10.1083/jcb.200009004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita A., Krishna S., Zheng H., Padgett R.W., Culotti J.G. 1998. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 281:706–709 10.1126/science.281.5377.706 [DOI] [PubMed] [Google Scholar]
- Deconinck N., Dan B. 2007. Pathophysiology of duchenne muscular dystrophy: current hypotheses. Pediatr. Neurol. 36:1–7 10.1016/j.pediatrneurol.2006.09.016 [DOI] [PubMed] [Google Scholar]
- Del Tongo C., Carretta D., Fulgenzi G., Catini C., Minciacchi D. 2009. Parvalbumin-positive GABAergic interneurons are increased in the dorsal hippocampus of the dystrophic mdx mouse. Acta Neuropathol. 118:803–812 10.1007/s00401-009-0567-3 [DOI] [PubMed] [Google Scholar]
- Dirr H., Reinemer P., Huber R. 1994. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur. J. Biochem. 220:645–661 10.1111/j.1432-1033.1994.tb18666.x [DOI] [PubMed] [Google Scholar]
- Dupuy D., Bertin N., Hidalgo C.A., Venkatesan K., Tu D., Lee D., Rosenberg J., Svrzikapa N., Blanc A., Carnec A., et al. 2007. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25:663–668 10.1038/nbt1305 [DOI] [PubMed] [Google Scholar]
- Ervasti J.M. 2007. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim. Biophys. Acta. 1772:108–117 [DOI] [PubMed] [Google Scholar]
- Ervasti J.M., Campbell K.P. 1991. Membrane organization of the dystrophin-glycoprotein complex. Cell. 66:1121–1131 10.1016/0092-8674(91)90035-W [DOI] [PubMed] [Google Scholar]
- Fire A., Harrison S.W., Dixon D. 1990. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 93:189–198 10.1016/0378-1119(90)90224-F [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Francis G.R., Waterston R.H. 1985. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101:1532–1549 10.1083/jcb.101.4.1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E., Lemmon V., Van Camp G., Vits L., Coucke P., Willems P.J. 1995. CRASH syndrome: clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1. Eur. J. Hum. Genet. 3:273–284 [DOI] [PubMed] [Google Scholar]
- Garver T.D., Ren Q., Tuvia S., Bennett V. 1997. Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol. 137:703–714 10.1083/jcb.137.3.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavillet B., Rougier J.S., Domenighetti A.A., Behar R., Boixel C., Ruchat P., Lehr H.A., Pedrazzini T., Abriel H. 2006. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ. Res. 99:407–414 10.1161/01.RES.0000237466.13252.5e [DOI] [PubMed] [Google Scholar]
- Gettner S.N., Kenyon C., Reichardt L.F. 1995. Characterization of β pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J. Cell Biol. 129:1127–1141 10.1083/jcb.129.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler K., Abdel-Dayem M., Ségalat L. 1999a. In vitro interactions of Caenorhabditis elegans dystrophin with dystrobrevin and syntrophin. FEBS Lett. 461:59–62 10.1016/S0014-5793(99)01421-0 [DOI] [PubMed] [Google Scholar]
- Gieseler K., Bessou C., Ségalat L. 1999b. Dystrobrevin- and dystrophin-like mutants display similar phenotypes in the nematode Caenorhabditis elegans. Neurogenetics. 2:87–90 10.1007/s100480050057 [DOI] [PubMed] [Google Scholar]
- Gieseler K., Grisoni K., Ségalat L. 2000. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr. Biol. 10:1092–1097 10.1016/S0960-9822(00)00691-6 [DOI] [PubMed] [Google Scholar]
- Gieseler K., Mariol M.C., Bessou C., Migaud M., Franks C.J., Holden-Dye L., Ségalat L. 2001. Molecular, genetic and physiological characterisation of dystrobrevin-like (dyb-1) mutants of Caenorhabditis elegans. J. Mol. Biol. 307:107–117 10.1006/jmbi.2000.4480 [DOI] [PubMed] [Google Scholar]
- Giugia J., Gieseler K., Arpagaus M., Ségalat L. 1999. Mutations in the dystrophin-like dys-1 gene of Caenorhabditis elegans result in reduced acetylcholinesterase activity. FEBS Lett. 463:270–272 10.1016/S0014-5793(99)01651-8 [DOI] [PubMed] [Google Scholar]
- Grisoni K., Gieseler K., Mariol M.C., Martin E., Carre-Pierrat M., Moulder G., Barstead R., Ségalat L. 2003. The stn-1 syntrophin gene of C. elegans is functionally related to dystrophin and dystrobrevin. J. Mol. Biol. 332:1037–1046 10.1016/j.jmb.2003.08.021 [DOI] [PubMed] [Google Scholar]
- Haldeman M.T., Xia G., Kasperek E.M., Pickart C.M. 1997. Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry. 36:10526–10537 10.1021/bi970750u [DOI] [PubMed] [Google Scholar]
- Hammarlund M., Davis W.S., Jorgensen E.M. 2000. Mutations in β-spectrin disrupt axon outgrowth and sarcomere structure. J. Cell Biol. 149:931–942 10.1083/jcb.149.4.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock A., Nelson W.J. 2008. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta. 1778:660–669 10.1016/j.bbamem.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Cuenda A., Spillantini M.G., Thomas G.M., Buée-Scherrer V., Cohen P., Goedert M. 1999. Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J. Biol. Chem. 274:12626–12631 10.1074/jbc.274.18.12626 [DOI] [PubMed] [Google Scholar]
- Hendriksen J.G., Vles J.S. 2008. Neuropsychiatric disorders in males with duchenne muscular dystrophy: frequency rate of attention-deficit hyperactivity disorder (ADHD), autism spectrum disorder, and obsessive—compulsive disorder. J. Child Neurol. 23:477–481 10.1177/0883073807309775 [DOI] [PubMed] [Google Scholar]
- Heyden A., Angenstein F., Sallaz M., Seidenbecher C., Montag D. 2008. Abnormal axonal guidance and brain anatomy in mouse mutants for the cell recognition molecules close homolog of L1 and NgCAM-related cell adhesion molecule. Neuroscience. 155:221–233 10.1016/j.neuroscience.2008.04.080 [DOI] [PubMed] [Google Scholar]
- Hirn C., Shapovalov G., Petermann O., Roulet E., Ruegg U.T. 2008. Nav1.4 deregulation in dystrophic skeletal muscle leads to Na+ overload and enhanced cell death. J. Gen. Physiol. 132:199–208 10.1085/jgp.200810024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman E.P., Brown R.H., Jr, Kunkel L.M. 1987. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 51:919–928 10.1016/0092-8674(87)90579-4 [DOI] [PubMed] [Google Scholar]
- Hogan A., Shepherd L., Chabot J., Quenneville S., Prescott S.M., Topham M.K., Gee S.H. 2001. Interaction of gamma 1-syntrophin with diacylglycerol kinase-zeta. Regulation of nuclear localization by PDZ interactions. J. Biol. Chem. 276:26526–26533 10.1074/jbc.M104156200 [DOI] [PubMed] [Google Scholar]
- Hortsch M. 2003. Neural cell adhesion molecules—brain glue and much more! Front. Biosci. 8:d357–d359 10.2741/1006 [DOI] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A., et al. 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5:e237 10.1371/journal.pbio.0050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O., Ervasti J.M., Leveille C.J., Slaughter C.A., Sernett S.W., Campbell K.P. 1992. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 355:696–702 10.1038/355696a0 [DOI] [PubMed] [Google Scholar]
- Ishiguro H., Liu Q.R., Gong J.P., Hall F.S., Ujike H., Morales M., Sakurai T., Grumet M., Uhl G.R. 2006. NrCAM in addiction vulnerability: positional cloning, drug-regulation, haplotype-specific expression, and altered drug reward in knockout mice. Neuropsychopharmacology. 31:572–584 10.1038/sj.npp.1300855 [DOI] [PubMed] [Google Scholar]
- Jagadha V., Becker L.E. 1988. Brain morphology in Duchenne muscular dystrophy: a Golgi study. Pediatr. Neurol. 4:87–92 10.1016/0887-8994(88)90047-1 [DOI] [PubMed] [Google Scholar]
- Jouet M., Rosenthal A., Armstrong G., MacFarlane J., Stevenson R., Paterson J., Metzenberg A., Ionasescu V., Temple K., Kenwrick S. 1994. X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat. Genet. 7:402–407 10.1038/ng0794-402 [DOI] [PubMed] [Google Scholar]
- Kim H., Rogers M.J., Richmond J.E., McIntire S.L. 2004. SNF-6 is an acetylcholine transporter interacting with the dystrophin complex in Caenorhabditis elegans. Nature. 430:891–896 10.1038/nature02798 [DOI] [PubMed] [Google Scholar]
- Kim H., Pierce-Shimomura J.T., Oh H.J., Johnson B.E., Goodman M.B., McIntire S.L. 2009. The dystrophin complex controls bk channel localization and muscle activity in Caenorhabditis elegans. PLoS Genet. 5:e1000780 10.1371/journal.pgen.1000780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wu K., Black I.B. 1995. Deficiency of brain synaptic dystrophin in human Duchenne muscular dystrophy. Ann. Neurol. 38:446–449 10.1002/ana.410380315 [DOI] [PubMed] [Google Scholar]
- Knuesel I., Mastrocola M., Zuellig R.A., Bornhauser B., Schaub M.C., Fritschy J.M. 1999. Short communication: altered synaptic clustering of GABAA receptors in mice lacking dystrophin (mdx mice). Eur. J. Neurosci. 11:4457–4462 10.1046/j.1460-9568.1999.00887.x [DOI] [PubMed] [Google Scholar]
- Koenig M., Monaco A.P., Kunkel L.M. 1988. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 53:219–228 10.1016/0092-8674(88)90383-2 [DOI] [PubMed] [Google Scholar]
- Kueh S.L., Head S.I., Morley J.W. 2008. GABA(A) receptor expression and inhibitory post-synaptic currents in cerebellar Purkinje cells in dystrophin-deficient mdx mice. Clin. Exp. Pharmacol. Physiol. 35:207–210 [DOI] [PubMed] [Google Scholar]
- Lele Z., Folchert A., Concha M., Rauch G.J., Geisler R., Rosa F., Wilson S.W., Hammerschmidt M., Bally-Cuif L. 2002. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development. 129:3281–3294 [DOI] [PubMed] [Google Scholar]
- Le Rumeur E., Winder S.J., Hubert J.F. 2010. Dystrophin: more than just the sum of its parts. Biochim. Biophys. Acta. 1804:1713–1722 [DOI] [PubMed] [Google Scholar]
- Lidov H.G., Byers T.J., Kunkel L.M. 1993. The distribution of dystrophin in the murine central nervous system: an immunocytochemical study. Neuroscience. 54:167–187 10.1016/0306-4522(93)90392-S [DOI] [PubMed] [Google Scholar]
- Lim K., Ho J.X., Keeling K., Gilliland G.L., Ji X., Rüker F., Carter D.C. 1994. Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci. 3:2233–2244 10.1002/pro.5560031209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng C., Phelps S., Crawford G.E., Walden P.D., Barald K., Chamberlain J.S. 1999. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat. Neurosci. 2:611–617 10.1038/10165 [DOI] [PubMed] [Google Scholar]
- Maduro M., Pilgrim D. 1995. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 141:977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M.F., Gordon M., Jacobs R., Pilgrim D.B. 2000. The UNC-119 family of neural proteins is functionally conserved between humans, Drosophila and C. elegans. J. Neurogenet. 13:191–212 10.3109/01677060009084494 [DOI] [PubMed] [Google Scholar]
- Mariol M.C., Martin E., Chambonnier L., Ségalat L. 2007. Dystrophin-dependent muscle degeneration requires a fully functional contractile machinery to occur in C. elegans. Neuromuscul. Disord. 17:56–60 10.1016/j.nmd.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Marui T., Funatogawa I., Koishi S., Yamamoto K., Matsumoto H., Hashimoto O., Nanba E., Nishida H., Sugiyama T., Kasai K., et al. 2009. Association of the neuronal cell adhesion molecule (NRCAM) gene variants with autism. Int. J. Neuropsychopharmacol. 12:1–10 (published erratum appears in Int. J. Neuropsychopharmacol. 2009. 12:439) 10.1017/S1461145708009127 [DOI] [PubMed] [Google Scholar]
- Masai I., Lele Z., Yamaguchi M., Komori A., Nakata A., Nishiwaki Y., Wada H., Tanaka H., Nojima Y., Hammerschmidt M., et al. 2003. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development. 130:2479–2494 10.1242/dev.00465 [DOI] [PubMed] [Google Scholar]
- McIntire S.L., Reimer R.J., Schuske K., Edwards R.H., Jorgensen E.M. 1997. Identification and characterization of the vesicular GABA transporter. Nature. 389:870–876 10.1038/39908 [DOI] [PubMed] [Google Scholar]
- Mehler M.F. 2000. Brain dystrophin, neurogenetics and mental retardation. Brain Res. Brain Res. Rev. 32:277–307 10.1016/S0165-0173(99)00090-9 [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mernagh D.R., Reynolds L.A., Kneale G.G. 1997. DNA binding and subunit interactions in the type I methyltransferase M.EcoR124I. Nucleic Acids Res. 25:987–991 10.1093/nar/25.5.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.M., Shakes D.C. 1995. Immunofluorescence microscopy. Methods Cell Biol. 48:365–394 10.1016/S0091-679X(08)61396-5 [DOI] [PubMed] [Google Scholar]
- Minciacchi D., Del Tongo C., Carretta D., Nosi D., Granato A. 2010. Alterations of the cortico-cortical network in sensori-motor areas of dystrophin deficient mice. Neuroscience. 166:1129–1139 10.1016/j.neuroscience.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Mokri B., Engel A.G. 1975. Duchenne dystrophy: electron microscopic findings pointing to a basic or early abnormality in the plasma membrane of the muscle fiber. Neurology. 25:1111–1120 [DOI] [PubMed] [Google Scholar]
- Moorthy S., Chen L., Bennett V. 2000. Caenorhabditis elegans β-G spectrin is dispensable for establishment of epithelial polarity, but essential for muscular and neuronal function. J. Cell Biol. 149:915–930 10.1083/jcb.149.4.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriuchi T., Kagawa N., Mukoyama M., Hizawa K. 1993. Autopsy analyses of the muscular dystrophies. Tokushima J. Exp. Med. 40:83–93 [PubMed] [Google Scholar]
- Moy S.S., Nonneman R.J., Young N.B., Demyanenko G.P., Maness P.F. 2009. Impaired sociability and cognitive function in Nrcam-null mice. Behav. Brain Res. 205:123–131 10.1016/j.bbr.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj K., Hortsch M. 2006. Phosphorylation of L1-type cell-adhesion molecules—ankyrins away! Trends Biochem. Sci. 31:544–546 10.1016/j.tibs.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Needham L.K., Thelen K., Maness P.F. 2001. Cytoplasmic domain mutations of the L1 cell adhesion molecule reduce L1-ankyrin interactions. J. Neurosci. 21:1490–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedziela-Majka A., Rymarczyk G., Kochman M., Ozyhar A. 1998. GST-Induced dimerization of DNA-binding domains alters characteristics of their interaction with DNA. Protein Expr. Purif. 14:208–220 10.1006/prep.1998.0932 [DOI] [PubMed] [Google Scholar]
- Norman K.R., Moerman D.G. 2002. α spectrin is essential for morphogenesis and body wall muscle formation in Caenorhabditis elegans. J. Cell Biol. 157:665–677 10.1083/jcb.200111051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piluso G., Mirabella M., Ricci E., Belsito A., Abbondanza C., Servidei S., Puca A.A., Tonali P., Puca G.A., Nigro V. 2000. Gamma1- and gamma2-syntrophins, two novel dystrophin-binding proteins localized in neuronal cells. J. Biol. Chem. 275:15851–15860 10.1074/jbc.M000439200 [DOI] [PubMed] [Google Scholar]
- Pocock R., Bénard C.Y., Shapiro L., Hobert O. 2008. Functional dissection of the C. elegans cell adhesion molecule SAX-7, a homologue of human L1. Mol. Cell. Neurosci. 37:56–68 10.1016/j.mcn.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Prins K.W., Humston J.L., Mehta A., Tate V., Ralston E., Ervasti J.M. 2009. Dystrophin is a microtubule-associated protein. J. Cell Biol. 186:363–369 10.1083/jcb.200905048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal A., Jouet M., Kenwrick S. 1992. Aberrant splicing of neural cell adhesion molecule L1 mRNA in a family with X-linked hydrocephalus. Nat. Genet. 2:107–112 10.1038/ng1092-107 [DOI] [PubMed] [Google Scholar]
- Rosman N.P. 1970. The cerebral defect and myopathy in Duchenne muscular dystrophy. A comparative clinicopathological study. Neurology. 20:329–335 [DOI] [PubMed] [Google Scholar]
- Sandonà D., Betto R. 2009. Sarcoglycanopathies: molecular pathogenesis and therapeutic prospects. Expert Rev. Mol. Med. 11:e28 10.1017/S1462399409001203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura H., Inada H., Kuhara A., Fusaoka E., Takemoto D., Takeuchi K., Mori I. 2005. Maintenance of neuronal positions in organized ganglia by SAX-7, a Caenorhabditis elegans homologue of L1. EMBO J. 24:1477–1488 10.1038/sj.emboj.7600621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Troemel E.R., Chou J.H., Dwyer N.D., Colbert H.A., Bargmann C.I. 1995. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 83:207–218 10.1016/0092-8674(95)90162-0 [DOI] [PubMed] [Google Scholar]
- Tuvia S., Garver T.D., Bennett V. 1997. The phosphorylation state of the FIGQY tyrosine of neurofascin determines ankyrin-binding activity and patterns of cell segregation. Proc. Natl. Acad. Sci. USA. 94:12957–12962 10.1073/pnas.94.24.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchino M., Teramoto H., Naoe H., Yoshioka K., Miike T., Ando M. 1994. Localisation and characterisation of dystrophin in the central nervous system of controls and patients with Duchenne muscular dystrophy. J. Neurol. Neurosurg. Psychiatry. 57:426–429 10.1136/jnnp.57.4.426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp G., Vits L., Coucke P., Lyonnet S., Schrander-Stumpel C., Darby J., Holden J., Munnich A., Willems P.J. 1993. A duplication in the L1CAM gene associated with X-linked hydrocephalus. Nat. Genet. 4:421–425 10.1038/ng0893-421 [DOI] [PubMed] [Google Scholar]
- Waite A., Tinsley C.L., Locke M., Blake D.J. 2009. The neurobiology of the dystrophin-associated glycoprotein complex. Ann. Med. 41:344–359 10.1080/07853890802668522 [DOI] [PubMed] [Google Scholar]
- Walker J., Crowley P., Moreman A.D., Barrett J. 1993. Biochemical properties of cloned glutathione S-transferases from Schistosoma mansoni and Schistosoma japonicum. Mol. Biochem. Parasitol. 61:255–264 10.1016/0166-6851(93)90071-5 [DOI] [PubMed] [Google Scholar]
- Wang X., Kweon J., Larson S., Chen L. 2005. A role for the C. elegans L1CAM homologue lad-1/sax-7 in maintaining tissue attachment. Dev. Biol. 284:273–291 10.1016/j.ydbio.2005.05.020 [DOI] [PubMed] [Google Scholar]
- Wicksell R.K., Kihlgren M., Melin L., Eeg-Olofsson O. 2004. Specific cognitive deficits are common in children with Duchenne muscular dystrophy. Dev. Med. Child Neurol. 46:154–159 10.1111/j.1469-8749.2004.tb00466.x [DOI] [PubMed] [Google Scholar]
- Wu J.Y., Kuban K.C., Allred E., Shapiro F., Darras B.T. 2005. Association of Duchenne muscular dystrophy with autism spectrum disorder. J. Child Neurol. 20:790–795 10.1177/08830738050200100201 [DOI] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M. 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 149:1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Davis J.Q., Carpenter S., Bennett V. 1998. Structural requirements for association of neurofascin with ankyrin. J. Biol. Chem. 273:30785–30794 10.1074/jbc.273.46.30785 [DOI] [PubMed] [Google Scholar]
- Zhou S., Opperman K., Wang X., Chen L. 2008. unc-44 Ankyrin and stn-2 gamma-syntrophin regulate sax-7 L1CAM function in maintaining neuronal positioning in Caenorhabditis elegans. Genetics. 180:1429–1443 10.1534/genetics.108.091272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubrzycka-Gaarn E.E., Bulman D.E., Karpati G., Burghes A.H., Belfall B., Klamut H.J., Talbot J., Hodges R.S., Ray P.N., Worton R.G. 1988. The Duchenne muscular dystrophy gene product is localized in sarcolemma of human skeletal muscle. Nature. 333:466–469 10.1038/333466a0 [DOI] [PubMed] [Google Scholar]