Abstract
WT1 is essential for normal kidney development, and genetic alterations are associated with Wilms’ tumor, Denys Drash (DDS), and Frasier syndromes. Although generally considered a transcription factor this study has revealed that WT1 interacts with an essential splicing factor, U2AF65, and associates with the splicing machinery. WT1 is alternatively spliced and isoforms that include three amino acids, KTS, show stronger interaction with U2AF65 in vitro and better colocalization with splicing factors in vivo. Interestingly a mutation associated with DDS enhanced both −KTS WT1 binding to U2AF65 and splicing-factor colocalization. These data illustrate the functional importance of WT1 isoforms and suggest that WT1 playsa role in pre-mRNA splicing.
Keywords: WT1, U2AF65, spliceosome, alternative splicing
Defects in the WT1 gene predispose to childhood kidney tumors and can also result in Denys Drash (DDS) or Frasier syndromes, in which symptoms include genital malformation and nephropathy (Barbaux et al. 1997; Little and Wells 1997). WT1 is essential for urogenital development, and is expressed in cells that undergo a mesenchyme to epithelial transition (Pritchard-Jones et al. 1990; Kreidberg et al. 1993). The gene product has hallmarks of a transcription factor; four ‘Krüppel-like’ zinc fingers at its carboxy terminus, and an amino-terminal transcriptional regulatory domain. The zinc fingers are related to those of the transcription factor EGR1, and WT1 can bind to the GC-rich EGR1 DNA-binding site (Rauscher et al. 1990). WT1 both activates and represses GC-rich promoters, depending on context (Wang et al. 1993), generating the hypothesis that the protein activates genes of the epithelial lineage, and represses those that promote mesenchymal cell proliferation (Hastie 1994). To date, however, no clear in vivo targets for regulation by WT1 have been identified.
The WT1 gene encodes numerous protein isoforms. Two translational start sites have been identified, a standard methionine and a non-AUG leucine, which adds 68 amino-terminal residues (Breuning and Pelletier 1996). A specific nucleotide in the pre-mRNA is apparently edited, resulting in proteins differing by a single amino acid (Sharma et al. 1994). The pre-mRNA is also alternatively spliced (Haber et al. 1991). A first alternative exon encodes 17 amino acids that can be inserted towards the center of the protein, and a second 3 amino acids, KTS, which can be included between the third and fourth zinc fingers. The strong conservation of this latter motif throughout evolution suggests that it plays an important role (Kent et al. 1995). The KTS insertion reduces both DNA binding affinity and specificity (Bickmore et al. 1992). DNA binding by WT1 is known to play an important function as many DDS patients have mutations that specifically alter this activity (Little and Wells 1997). Significantly, Frasier syndrome is associated with WT1 mutations at splice-donor sites that prevent generation of +KTS isoforms (Barbaux et al. 1997). The isoform ratio appears constant throughout development (Harber et al. 1991), and the severe phenotype that arises from isoform imbalance in Frasier syndrome patients indicates that maintaining appropriate proportions of each protein is critical for function.
Some surprising clues to potential WT1 function came from careful examination of cell localization. The protein has a distinct nuclear pattern, being found in ‘speckles’ and coiled bodies (Larson et al. 1995). This pattern is characteristic of splicing, rather than transcription, factors. Indeed staining WT1-expressing cells for both WT1 and splice factors revealed good colocalization (Larson et al. 1995). Intriguingly more detailed analysis following transfection of individual WT1 forms revealed a marked, though not absolute, correlation between isoform and subnuclear distribution. Isoforms including the KTS motif colocalized dominantly with splicing factors, whereas −KTS proteins colocalized better with transcription factors (Larsson et al. 1995). This illustrates the differences between the isoforms, and suggests WT1 might not function exclusively as a transcription factor.
The major components of nuclear speckles and coiled bodies are splicing proteins. Coiled bodies were described originally as nucleolar accessory bodies and may play a role in splicing-complex assembly, maturation, or recycling. One coiled-body-specific protein, p80-coilin, may be central to this process (Bohmann et al. 1995). Recent evidence has suggested that speckles are dynamic structures, perhaps representing storage sites from which splicing factors are recruited (Misteli et al. 1997). Splicing itself is a complex two-step process, involving multiple RNA and protein species. Initially U1 snRNP, of which U170K is a major component, binds to the 5′ splice site, and U2AF65 to the 3′ splice site. These two complexes are bridged by SR protein family members such as SRp20, SC35, and ASF/SF2 (Wu and Maniatis 1993). U2AF65, with its partner U2AF35, promotes annealing of the U2 snRNA to the polypyrimidine tract adjacent to the 3′ splice site, facilitating splicing itself (Zamore et al. 1992). These proteins display a nuclear speckled pattern and have several common motifs, including arginine/serine-rich RS domains, and RNA recognition motifs (RRM). There is some evidence from molecular modeling that WT1 may have an amino-terminal RRM (Kennedy et al. 1996). Interestingly RNA binding by WT1 has been demonstrated in vitro, although this activity mapped to the first zinc finger, which has a slightly different structure then the remaining three (Caricasole et al. 1996). As WT1 colocalizes and indeed coimmunoprecipitates with splicing factors (Larsson et al. 1995), and has RNA-binding potential, it was clearly important to test whether WT1 associates directly with the splicing machinery.
Results
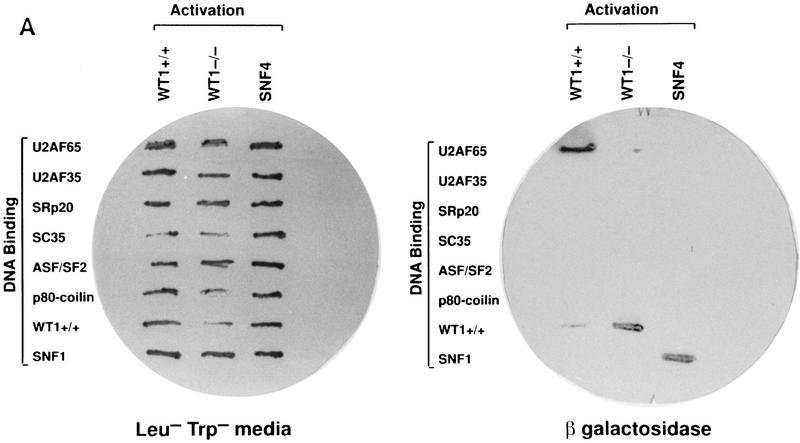
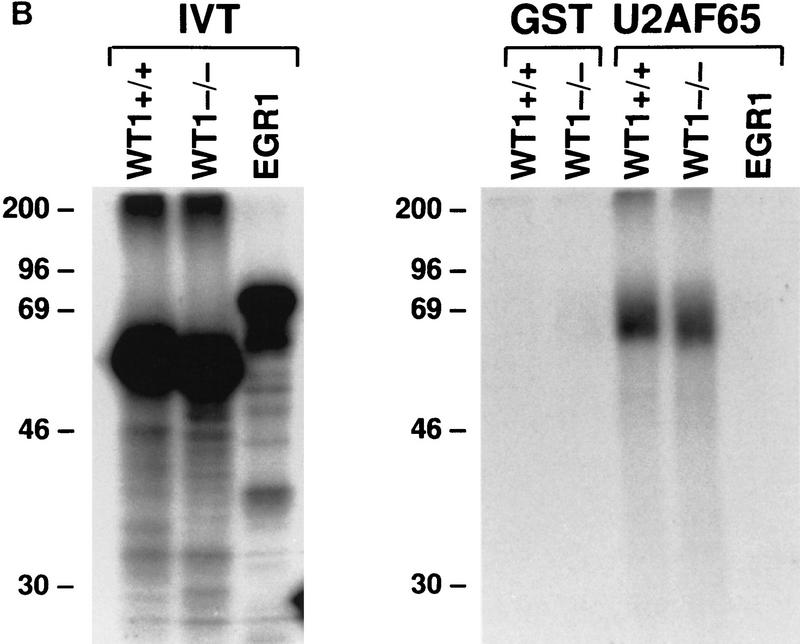
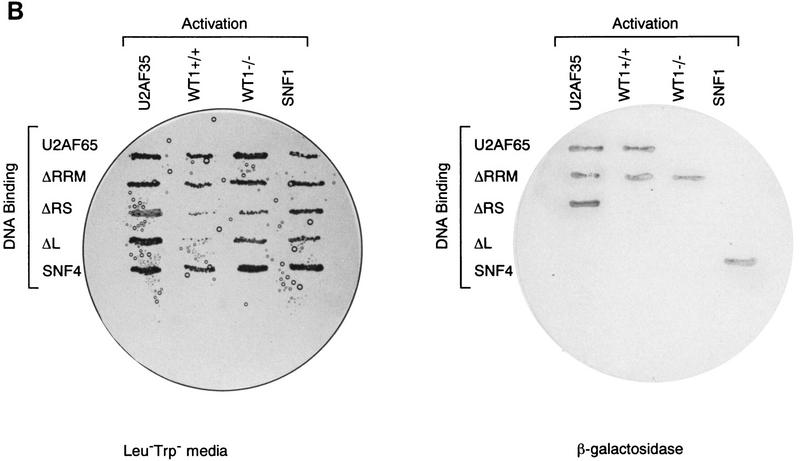
The ability of WT1 to interact directly with components of the splicing machinery was examined in a yeast two-hybrid system. Seven proteins were tested, including the splicing factors U170K, ASF/SF2, SC35, U2AF35, U2AF65, SRp20, and the coiled-body protein p80-coilin (Fig. 1A; data not shown). Only U2AF65 was identified as a potential binding partner, stimulating both growth without histidine and β-galactosidase activity in a dual-reporter system. The yeast proteins SNF1 and SNF4 were used as internal positive and negative controls (Harper et al. 1993). The observation that WT1 and U2AF65 interact was supported by the finding that in vitro-translated WT1, but not the related protein EGR1, could complex with bacterially expressed GST-U2AF65 (Fig. 1B).
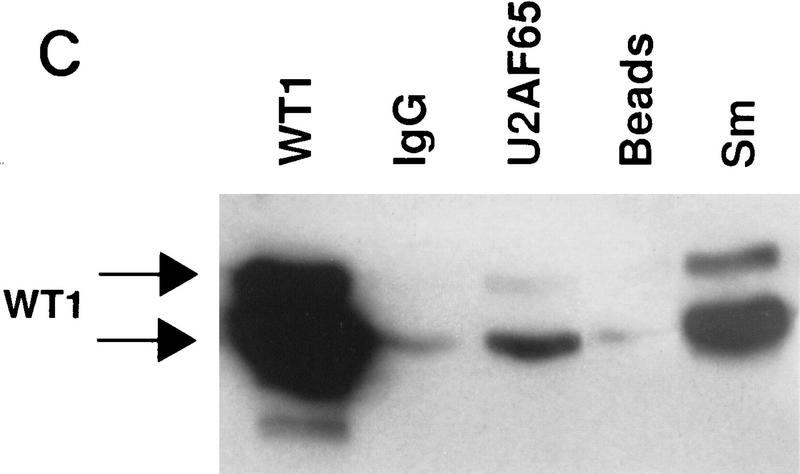
Figure 1.
WT1 interacts with U2AF65. (A) Yeast two-hybrid analysis. Yeast was transformed with plasmids encoding the indicated proteins, and individual colonies grown on Leu−Trp− (left) prior to the assessment of β-galactosidase activity (right). (B) In vitro binding assay. The indicated in vitro-translated proteins (left) were mixed with GST or GST–U2AF65 purified from bacteria, bound protein resolved by SDS-PAGE, and detected by fluorography (right). (C) WT1 coimmunoprecipitates with U2AF65. WT1-expressing M15 cells were immunoprecipitated with antibodies against the indicated proteins or normal mouse IgG, the complexes resolved by SDS-PAGE, and the presence of WT1 determined by Western blotting.
To establish whether WT1 and U2AF65 interact in vivo coimmunoprecipitations were performed from a mesonephric WT1-expressing cell line, M15 (Larsson et al. 1995). WT1 was present in U2AF65 immunoprecipitates, above background levels of control precipitations (Fig. 1C). The two WT1 immunoreactive bands correspond to isoforms initiated at the methionine and leucine starts (lower and higher band, respectively). These can be further resolved to show proteins containing/lacking the 17-amino-acid insert. Proteins with/without the KTS motif cannot be separated by size. WT1 is also a component of Sm immunoprecipitates (Larsson et al. 1995; Fig. 1C), which comprise a number of different splicing antigens, U2AF65 coprecipitating among them (data not shown). Therefore WT1 might form part of a multimeric complex including U2AF65 and other splicing factors. The ability to detect WT1:U2AF65 interaction by immunoprecipitation was variable and appeared to be dependent on the method used to prepare the extracts. This might suggest that only a small proportion of these proteins are complexed and/or the complex is transitory. From the relative intensity of the signals, only a fraction of WT1 appears associated with U2AF65. Significantly, the WT1 signal from U2AF65 immunoprecipitates is weaker than that from Sm (Fig. 1C), suggesting that WT1 might interact with other splicing proteins.
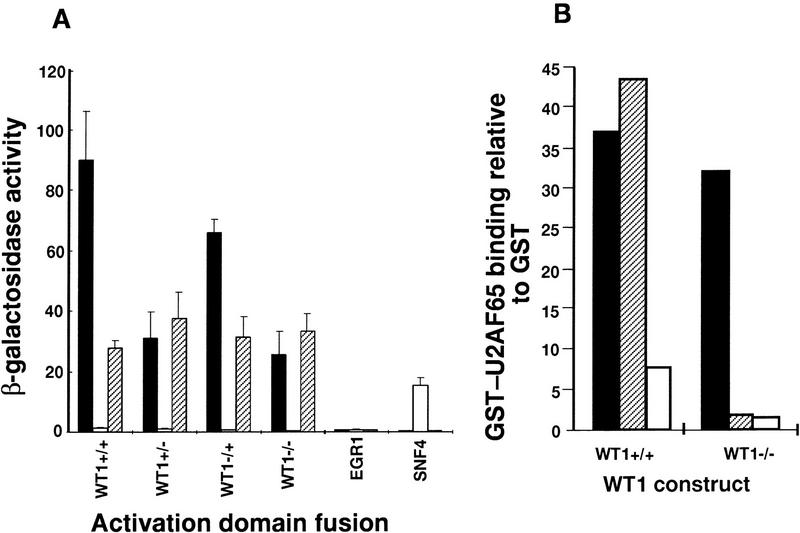
WT1 is a complex gene that encodes many proteins. To investigate whether U2AF65 binding is isoform dependent, the set of major WT1 proteins were tested. The first plus or minus symbol denotes the presence or absence of the 17-amino-acid insert, and the second represents KTS status. The initial yeast two-hybrid assays suggested that +KTS forms interacted more strongly with U2AF65 than forms lacking the KTS motif (Fig. 1A; data not shown). To examine this more closely β-galactosidase activity was measured in independent yeast colonies. This quantitative analysis indicates the relative strength of a particular interaction (Estojak et al. 1995). One described previously property of WT1 is homodimerization (Reddy et al. 1995), and this ability was used as an internal control. Whereas all isoforms showed similar ability to dimerize, the +KTS proteins showed stronger binding to U2AF65 than −KTS forms, although all isoforms showed at least some binding (Fig. 2A). This effect was not caused by differences in protein levels (data not shown). The differences in affinity of the WT1 isoforms for U2AF65 also became evident following titration of the GST binding experiments. At 100 mm salt, all forms of in vitro-translated WT1 bound broadly equally to GST–U2AF65 (Fig. 1B). However, at 250 mm salt, the binding of −KTS protein decreased (Fig. 2B). The enhanced binding of +KTS isoforms to U2AF65 correlates with their better colocalization with splicing factors (Larsson et al. 1995).
Figure 2.
U2AF65 binding is influenced by isoform. (A) Quantitation of β-galactosidase activity following transformation of yeast with the indicated constructs. (Solid bars) U2AF65; (open bars) SNF1; (hatched bars) WT1+/+ (WT1+/+). (B) WT1 binding to GST–U2AF65 compared to GST alone under different salt concentrations. (Solid bars) 100 mm; (hatched bars) 250 mm; (open bars) 500 mm.
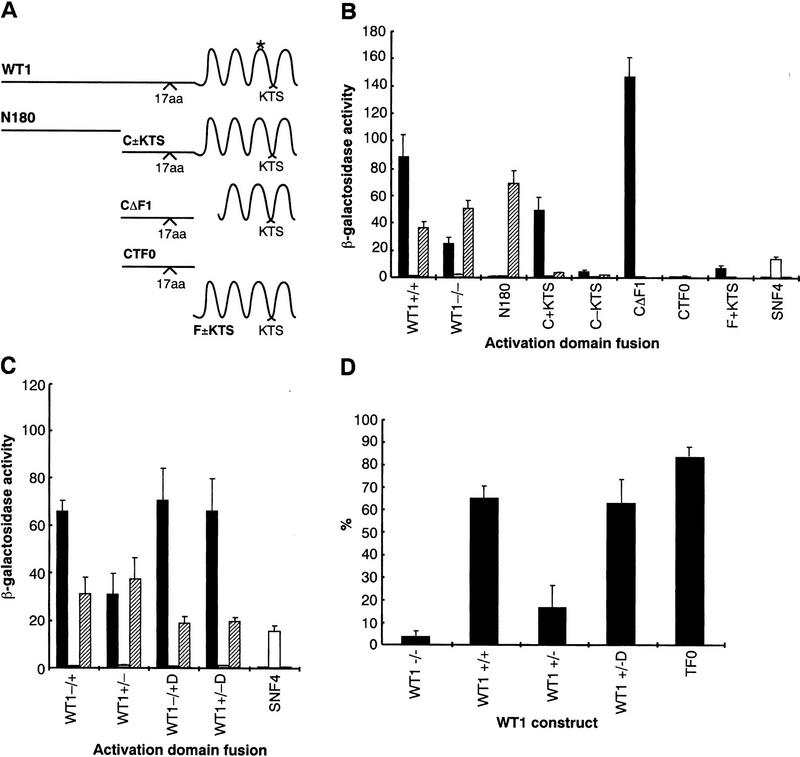
To map the region of WT1 that interacts with U2AF65, the protein was divided initially into two, N180 and C±KTS (Fig. 3A). The abilities of WT1 to dimerize and bind U2AF65 were found to be separable. N180, comprising the first 180 amino acids of WT1, could dimerize (Reddy et al. 1995), but did not bind U2AF65 (Fig. 3B). Conversely, the remainder of the protein was defective for dimerization but retained KTS-dependent U2AF65 binding (albeit at slightly reduced levels). Molecular modeling has revealed a potential RNA recognition motif within the amino terminus of WT1 (Kennedy et al. 1996). As WT1 binding to U2AF65 is independent of the amino terminus, it follows that RNA binding through this motif would not be essential for U2AF65 interaction. Within the remaining carboxy-terminal portion, deletion of finger 1 (CΔF1), another proposed RNA-interacting region (Caricasole et al. 1996), did not impair U2AF65 binding, displaying, if anything, increased association. Significantly, removal of all the zinc finger motifs (CTF0) resulted in loss of binding, whereas the finger region alone (F + KTS), displayed only a weak interaction. These results suggest that the zinc fingers provide elements critical for binding, but it seems likely that the interaction is stabilized by other residues. This view is supported by additional in vitro binding experiments (data not shown).
Figure 3.
U2AF65 binding is independent of dimerization, and affected by point mutation. (A) Schematic representation of WT1 and deletion constructs. Four major isoforms are generated by inclusion or exclusion of 17 amino acids and KTS, respectively. (*) Arg-to-Trp substitution associated with DDS. (B,C). Quantitation of β-galactosidase activity following transformation of yeast with U2AF65 (solid bars), SNF1 (open bars), and WT1+/+ (hatched bars, WT1+/+). Constructs carrying the DDS mutation are marked D. The ability of CΔF1, CTF0, and F+KTS to dimerize was not determined. (D) The percentage of WT1-transfected Cos7 cells showing good colocalization with splice factors.
DDS patients often have a missense mutation affecting a single residue within the WT1 zinc-finger region that impairs DNA binding (Little and Wells 1997). As the zinc-finger region of WT1 makes a significant contribution to U2AF65 binding, it was important to determine whether this mutation affected interaction. Comparison of +KTS and −KTS DDS mutant proteins revealed that the alteration increased affinity of the −KTS form for U2AF65. Dimerization ability, protein levels, and the properties of the +KTS protein appeared unaffected (Fig. 3C; other data not shown).
The relationship between nuclear distribution and affinity for U2AF65 is striking; strong U2AF65-binding WT1 proteins show good colocalization with splicing factors. It was therefore important to examine the staining pattern of mutant proteins. Interestingly the −KTS Drash mutant, which has increased U2AF65-binding potential, showed an apparent shift in nuclear distribution, displaying enhanced colocalization with splicing factors like +KTS isoforms, independently of expression level (Fig. 3D; data not shown). U2AF65 binding is not a prerequisite for this speckled distribution of WT1, because TF0 lost U2AF65 binding but still largely colocalizes with splicing factors (Fig. 3B and D).
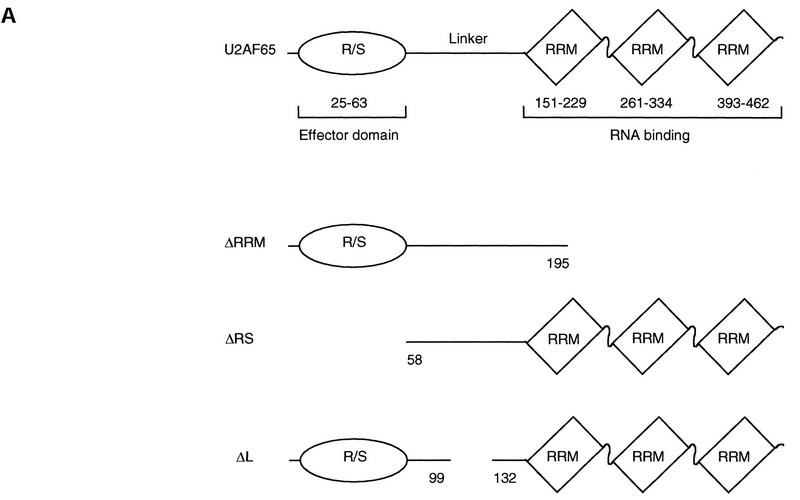
Having examined the region of WT1 responsible for interaction with U2AF65, attention shifted to the reciprocal domain. U2AF65 has three RRMs at its carboxy terminus (Fig. 4A). Removal of this region (ΔRRM) did not impair either the ability to interact with WT1 or U2AF35 (Fig. 4B). U2AF65 also has an essential amino-terminal R/S rich ‘effector’ domain separated from the RRM by a ‘linker’ region that binds partner proteins such as U2AF35 (Fig. 4A). Deleting either the RS-rich domain at the amino terminus (ΔRS) or sequences within the linker (ΔL) abrogated the ability of U2AF65 to bind to WT1, whereas U2AF35 binding was only lost following deletion of linker sequences (Fig. 4B). This suggests that both regions contribute to WT1 binding, and is supported by the finding that neither domain alone is sufficient for WT1 association (data not shown).
Figure 4.
Both the amino-terminal RS domain and linker region of U2AF65 are required for WT1 binding. (A) Schematic representation of U2AF65 and deletion constructs. (B) Yeast was transformed with plasmids encoding the indicated proteins, and individual colonies grown on Leu−Trp− media (left) prior to the assessment of β-galactosidase activity (right).
To investigate whether WT1 plays an active role in splicing a model system was set up. Nuclear extracts were prepared from WT1-expressing M15 cells, and shown to be competent for the splicing of a pre-mRNA template, albeit less efficiently than HeLa extract (Fig. 5A). Large-scale splicing reactions containing biotinylated, 32P-labeled sense and antisense pre-mRNAs were performed and fractionated, with the presence of specific splicing complexes forming exclusively on the sense and not antisense pre-mRNA as indicated by the shoulder on the graph (Fig. 5B). This was confirmed by native gel electrophoresis, a representative example of which is shown in Figure 5C. Fractions were analyzed to detect proteins present on the biotinylated pre-mRNA. WT1 was detected, like an established splicing factor, U2-B″, specifically associated with the sense transcript (Fig. 5B), providing evidence that WT1 can become incorporated into spliceosomes.
Figure 5.
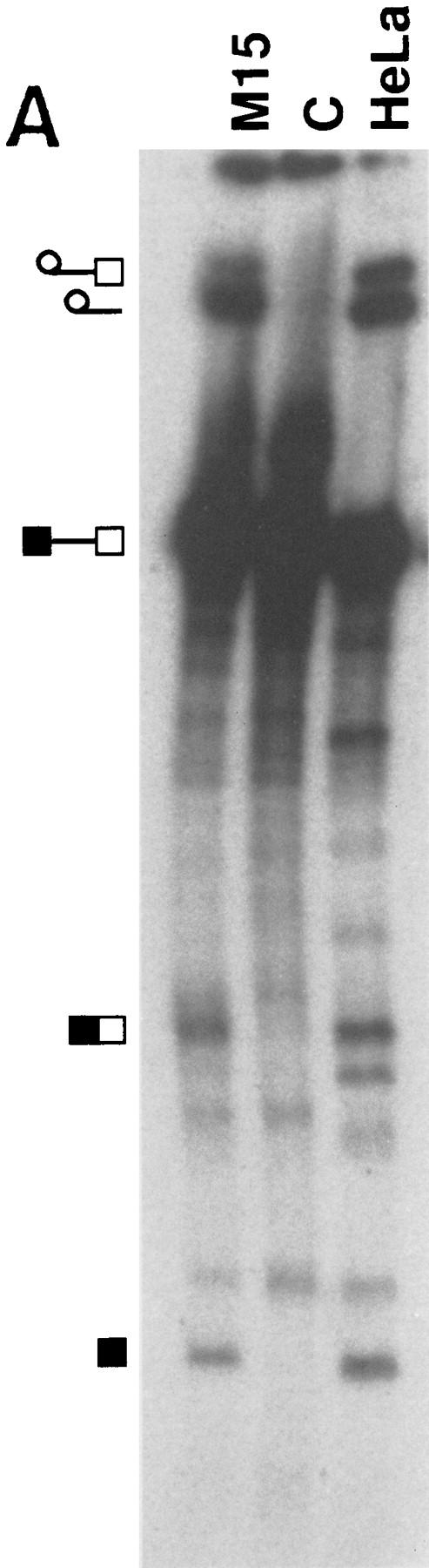
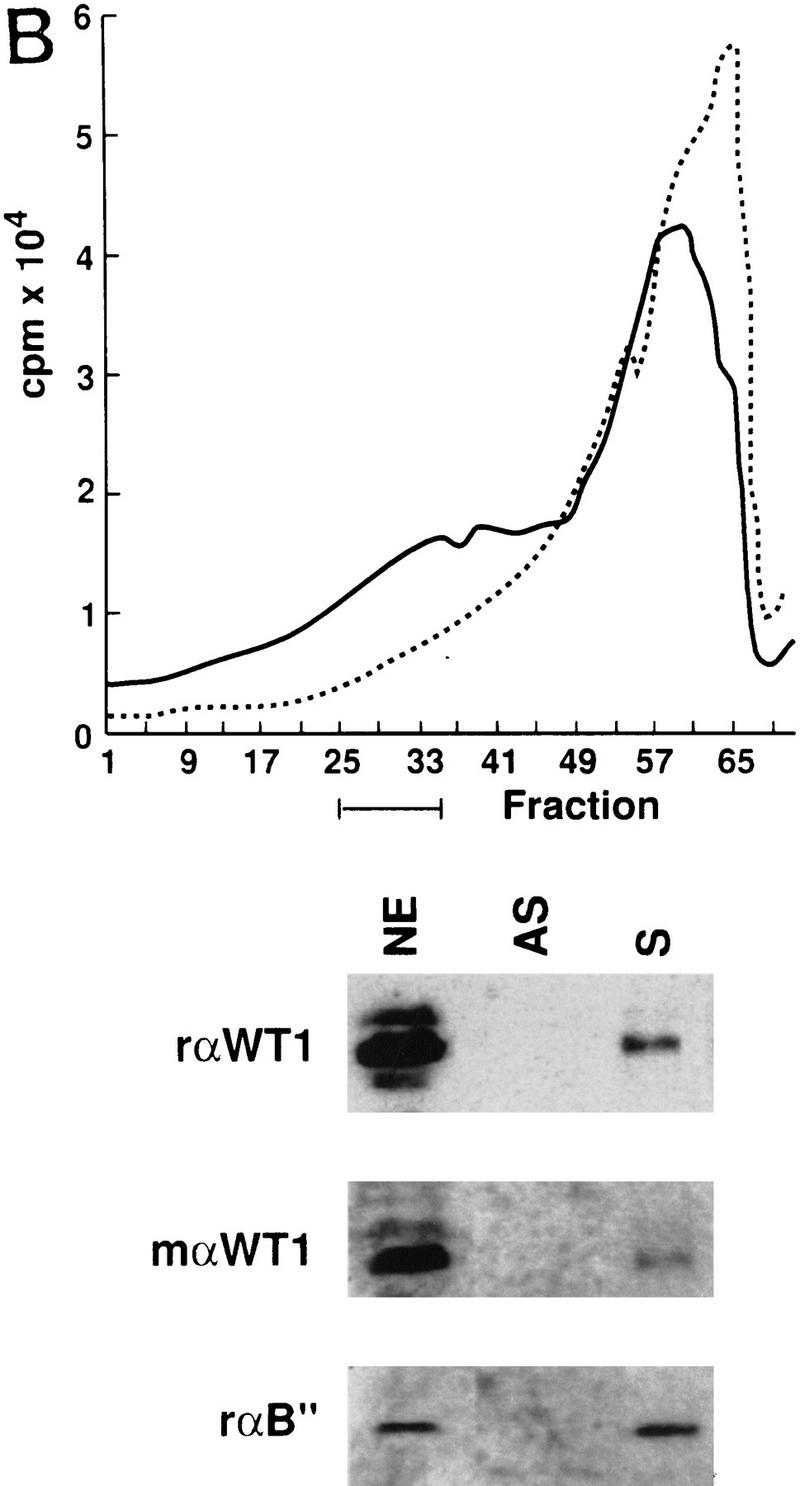
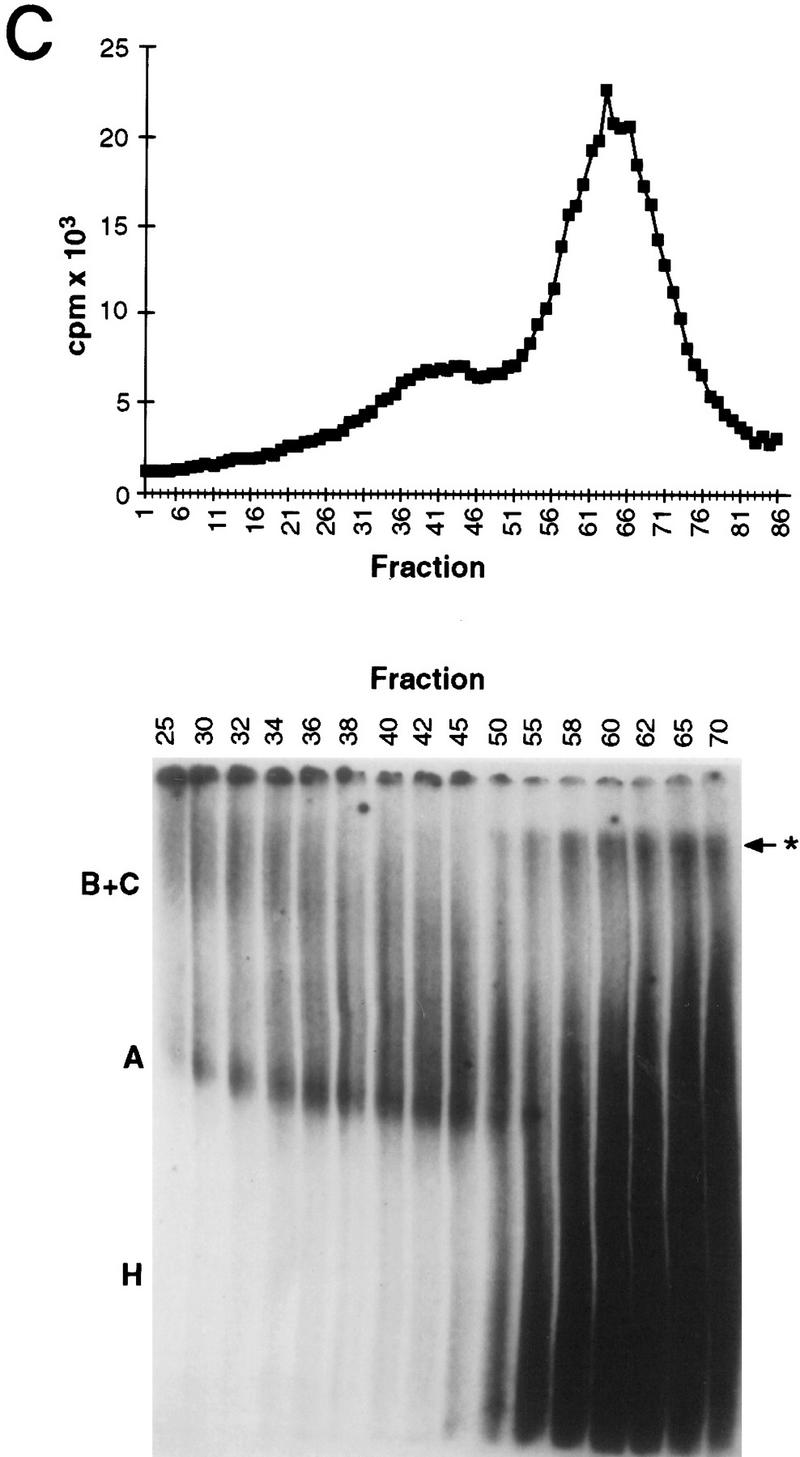
WT1 is incorporated into spliceosomes. (A) Nuclear extracts were prepared from WT1-expressing M15 cells, and their ability to splice a model pre-mRNA compared to that of HeLa extract. Control reaction C was set up with inactive M15 extract. Splicing intermediates were resolved by PAGE and detected by autoradiography. (B) Radioactive tracing was used to follow the fractionation of large-scale splicing reactions set up with sense (S) and anti-sense (AS) pre-mRNAs. The indicated fractions were pooled and streptavidin beads used to capture proteins associated with the 32P-labeled biotinylated RNA. Immunoblotting with the indicated antibodies against WT1 or U2-B″ were used to determine protein distribution. Nuclear extract (NE) was loaded as a positive control. (C) Native gel analysis of a fractionated splicing reaction. A, B+C, and H complexes are indicated. (*) Nonspecific aggregates.
Discussion
It has been demonstrated previously that WT1 colocalizes in the nucleus with splicing factors, and is present in complexes with some spliceosomal components (Larsson et al. 1995). It has now been found that WT1 associates specifically with U2AF65, in a manner enhanced by three amino acids, KTS, inserted following alternative pre-mRNA splicing. In addition WT1 can be incorporated directly into spliceosomes. These findings re-emphasize individual isoform properties, and provide evidence of a new role for WT1 in pre-mRNA processing.
U2AF65 was identified from a series of splicing proteins as a potential WT1-binding partner. Significantly, U2AF65 was already a strong candidate for WT1 interaction following careful analysis of cell localization. Both U2AF65 and WT1 show a nuclear ‘speckled’ pattern characteristic of splicing factors. Unlike most splicing factors U2AF65 localizes in ring-like structures around nucleolar remnants when RNA synthesis is inhibited with actinomycin D, and it was found that WT1 behaves in a similar manner (Larsson et al.1995; Gama-Carvalho e al. 1997). Interestingly +KTS forms of WT1, which show best nuclear colocalization with splicing factors, appear to interact more strongly with U2AF65, providing an intriguing correlation between descriptive biology and molecular properties. However, U2AF65 binding is not essential for such a nuclear distribution, for example, TF0 is unable to bind U2AF65 but displays a speckled pattern. Nuclear ‘speckles’ contain splicing factors that can apparently be recruited for active splicing (Misteli et al. 1997), but can also contain splicing-defective factors (Gama-Carvalho et al. 1997). This suggests that adoption of a speckled distribution need not imply a functional role, as speckles might reflect storage sites for both active and inactive splicing proteins. It is therefore possible that WT1 proteins showing a speckled pattern fall into two categories. One category may represent proteins simply targeted to the speckles (for example, defective mutants such as TF0), whereas a second category could include functional proteins that can be recruited subsequently to participate in pre-mRNA processing. Experiments are planned to investigate potential WT1 recruitment from speckles and any correlation with U2AF65 binding.
Mapping the WT1 site to which U2AF65 binds has identified the zinc-finger motif as essential. EGR1 with its highly related zinc-finger structure does not appear to interact with U2AF65, whereas binding of WT1 apparently increased following KTS insertion. Interestingly a reciprocal finding was presented recently, in which −KTS WT1 had greatest affinity for the orphan nuclear receptor SF-1 (Nachtigal et al. 1998). These results suggest strongly that the +KTS and −KTS proteins adopt different conformations. It is of particular interest that a mutation found commonly in DDS patients affecting a zinc-finger residue caused an apparent increase in U2AF65 binding by the −KTS isoform. DDS is associated generally with WT1 mutations in only one allele, the disease seemingly arising in a dominant-negative manner (Little and Wells). It has been envisaged that DDS mutant proteins lose DNA-binding activity, and through association with wild-type proteins could functionally inactivate them. The loss of DNA binding by the −KTS DDS protein, coupled with increased affinity for U2AF65 (an apparent gain of function) might better explain the molecular basis for the disease. Frasier syndrome patients have mutations at splice-donor sites that specifically perturb the isoform ratio (Barbaux et al. 1997). The current data suggests that the isoform ratio can be disturbed phenotypically following dual loss/gain of function. It is interesting to note that in Frasier syndrome the ratio is skewed so there becomes excess −KTS protein, whereas in DDS there could be a phenotypic skew in the opposite direction. This might be important when considering the major difference between the two; both DDS and Frasier syndrome patients suffer genital abnormalities and nephropathy but Frasier syndrome patients are not apparently at risk of developing Wilms’ tumor per se. Some DDS patients have WT1 truncations (Little and Wells 1997) that can be shorter than TF0, however, and would be anticipated to lose U2AF65 binding. More than one mechanism may thus generate DDS.
The demonstration that WT1 could be incorporated into spliceosomes assembled on a model template connects the tissue-specific and developmentally regulated protein not only with the splicing machinery, but with splicing itself. U2AF65 is an essential splice factor that binds to the polypyrimidine tract upstream of the 3′ splice site and promotes U2 snRNA annealing to the branch point (Zamore et al. 1992). Through interaction with U2AF65, WT1 could therefore influence 3′ splice-site selection in a cell-type- and/or temporal-dependent manner. The observations that WT1 potentially binds RNA makes the prospect that WT1 might select a particular 3′ splice site and present this directly to the RRM of U2AF65 especially intriguing. U2AF65 contacts other proteins, such as U2AF35, Urp, and SF1, which are implicated in splice-site selection (Zamore et al. 1992; Abovich and Rosbash 1997; Tronchére et al. 1997). WT1 could compete with these proteins for binding, although this seems unlikely for SF1, in which the binding site has been mapped to the third RRM of U2AF65 (Berglund et al. 1998), a region not required for WT1 binding. Alternatively (or additionally), WT1 could affect the U2-annealing activity of U2AF65, which occurs via recruitment of UAP56 to the linker region (Fleckner et al. 1997). It is of interest that WT1 binding to U2AF65 appears to require sequences within the RS domain. The RS domain of U2AF65 has different properties from a classical RS domain (Valcárcel et al. 1996) and WT1 does not interact with other RS proteins, such as U2AF35. The U2AF65 RS domain promotes RNA–RNA interaction between the U2 snRNA and the branch point (Valcárcel et al. 1996) and it is possible that WT1 affects this function of U2AF65. From the relative strength of WT1 signal in the U2AF65 and Sm immunoprecipitates, it is clear that WT1 might interact with more than one component of the splicing machinery. So far the analysis has focused on specific proteins involved in the early stages of spliceosome assembly, but it is possible that WT1 acts at multiple stages during the splicing process. As U2AF65 is specifically associated with early spliceosomes (Bennett et al. 1992), which are difficult to detect by fractionation, perhaps WT1 is recruited initially to the spliceosome via U2AF65, but then retained in later complexes through interaction with other proteins. This is an area of continued investigation.
An increasing number of connections have been made between transcription and mRNA processing (Neugebauer and Roth 1997), and it is particularly intriguing to note the reciprocal properties of the WT1 isoforms. −KTS proteins have stronger DNA-binding potential, and weaker U2AF65-binding ability, whereas +KTS proteins display less DNA binding but greater U2AF65 binding. This emphasizes the effects a few amino acids can have on the properties of a protein, but may also be important when considering broader aspects of WT1 function. As WT1 can dimerize (Reddy et al. 1995) one molecule with strong DNA-binding activity could be coupled to one with high U2AF65 affinity. Although the mechanism by which WT1 represses or activates transcription is unclear, it is tempting to speculate that +KTS and −KTS WT1 might cooperate to ensure not only that the correct amount of a transcript is synthesized, but also that it is spliced correctly. Although WT1 was incorporated into spliceosomes on a model template, nothing is known about genuine RNA targets of WT1. Nevertheless it is easy to envisage how a protein that is able to influence both transcription and splicing would play a critical role in the control of proliferation and differentiation. The precise effects of WT1 and the nature of its targets are future questions, but the results presented here provide direct evidence that WT1 plays a role in post-transcriptional processing, changing the view that WT1 is specifically a transcription factor.
Materials and methods
Plasmids
Splice-factor cDNAs were cloned into pAS2 and WT1 into both pAS2 and pACT2 (Harper et al. 1993) following PCR. Specific WT1 isoforms are denoted by plus or minus symbols; the first indicating the presence or absence of 17 amino acids and the second KTS status. An NcoI site internal to the WT1 cDNA was used to generate N180 and C ± KTS, encompassing or deleting the first 180 amino acids, respectively. Other specific WT1 constructs were made by site directed mutagenesis or PCR. The DDS mutation results in Arg-to-Trp substitution at amino acid 394. ΔF1 deletes residues 314–352, whereas TF0 is truncated at amino acid 314. F + KTS was subcloned directly from GST + KTS (Bickmore et al. 1992). EGR1 cDNA was provided by V. Sukhatme (Beth Israel Hospital and Harvard Medical School) and cloned in-frame using an NcoI site within the cDNA (resulting in the loss of the first 28 residues). U2AF65 ΔN contains amino acids 58–462, ΔRRM contains amino acids 1–195, whereas residues 100–131 were deleted to create ΔL. For in vitro translation, cDNAs were subcloned directly from the yeast two-hybrid vector pACTII into pET11d (Novagen). For transfection experiments, the appropriate constructs were cloned into Rc/CMV (Invitrogen).
Antibodies
For immunohistochemistry, splicing proteins were detected with 3C5 (Turner and Franchi 1987) followed by FITC anti-mouse IgM, whereas rαWT1 C19 (Santa Cruz) followed by rhodamine anti-rabbit was used to detect WT1 [except TF0, which was detected by mαWT1 H2 (DAKO) followed by rhodamine anti-mouse IgG]. For immunoprecipitation or Western blotting, the following antibodies were used: H2 mαWT1 (DAKO), C19 rαWT1 (Santa Cruz), normal mouse IgG (Sigma), MC3 mαU2AF65 (Gama-Carvalho et al. 1997), Y12 mαSm proteins (Lerner et al. 1981), 4g3 rαU2-B′′ (Habets et al. 1989).
Cell lines
M15 (Larsson et al. 1995) and Cos7 cells (ECACC) were cultured at 37°C in DMEM supplemented with 10% FCS and in an atmosphere of 5% CO2.
Yeast two-hybrid analysis
Plasmids were introduced into yeast strain Y190 by lithium acetate transformation, and interaction tested by growth on leu−trp−his− media supplemented with 50 mm 3-amino triazole (Harper et al. 1993). To assess β-galactosidase activity filter lifts (from leu−trp− plates) and liquid assays (in leu−trp− media) were performed as described (Harshman et al. 1988). β-galactosidase activity was calculated: 1000 × A420/(volume × time × protein concentration). At least three independent colonies from three transformations were tested.
In vitro binding assays
GST–U2AF65 protein was prepared from bacteria according to Lin and Green (1991). In vitro translated (IVT) protein was prepared in a T7-driven rabbit reticulocyte-coupled transcription and translation system (Promega). Five micrograms of bacterially expressed GST–U2AF65 was mixed with [35S]methionine-labeled IVT protein in 200 μl of LSAB (100 mm NaCl, 100 mm Tris at pH 8, 0.1% NP40) at 4°C for 2 hr. Complexes were precipitated onto glutathione beads, washed extensively in LSAB, resolved by SDS-PAGE and detected by fluorography. To titrate the salt sensitivity of the GST–U2AF65:WT1 complex, reactions were performed as described above, increasing the salt concentration in LSAB to 250 and 500 mm. The amount of binding to GST–U2AF65 was compared to binding to GST alone following PhosphorImage analysis.
Immunofluorescence
Cos7 cells were electroporated (0.8 kV, 25 μF) with 5 μg of the appropriate cytomegalovirus (CMV) driven construct. Cells were fixed and stained 48 hr after transfection as described (Larsson et al. 1995). Distribution of WT1 was superimposed on the 3C5-staining pattern by dual-channel confocal microscopy. Individual transfected cells were categorized by eye as displaying good, intermediate, or poor colocalization with splice factors. Thirty cells were scored per transfection, and the percentage of cells falling into each group calculated. Following at least three independent transfections these proportions were averaged.
Immunoprecipitation
Nuclear extracts from ∼5 x 106 M15 cells were prepared according to Kim et al. (1997), diluted 1:4 with IPB100 (100 mm NaCl, 10 mm Tris, 1 mm EDTA at pH 8), and precleared with protein A-Sepharose before addition of the antibody cross-linked to protein A–Sepharose (Harlow and Lane 1988). Immune complexes were washed extensively in IPB100 + 0.05% NP-40, and resolved by 8% SDS-PAGE. Proteins were transferred to nitrocellulose by semidry transfer. Membranes were blocked with 5% nonfat milk, and probed. Bound antibody was coupled to goat anti-mouse/rabbit HRP as appropriate (Sigma), and detected by ECL-plus (Amersham) according to the manufacturer’s instructions.
Splicing assays
Nuclear extract was prepared from 1 × 109 M15 cells according to Dignam (1983). Adenovirus major late precursor (Konarska and Sharp 1987) was used as a template and both 32P-labeled and biotinylated during transcription. Sense pre-mRNA was made from Sau3A-digested pBSAd1 with T3 RNA polymerase, whereas antisense message used the same template, linearized with HindIII, and transcribed with T7 polymerase. Splicing reactions were set up with 50% (vol/vol) nuclear extract, 1.5 mm ATP, 2 mm MgCl2, 5 mm creatine phosphate, 2 mm DTT, 200 U/ml RNAsin (Promega), and 2.5 mg/ml pre-mRNA and left at 30°C for 2 hr. Splicing intermediates were separated on 10% polyacrylamide–urea gel. For the fractionation experiments (Reed et al. 1988), large-scale (10-ml) splicing reactions were set up and after 30 min at 30°C loaded onto a 2.5 × 75-cm Sephacryl S-500 column equilibrated with FSP (20 mm Tris at pH 7.8, 60 mm KCl, 2.5 mm EDTA, and 0.1% Triton X-100) at 4°C. Fractions of 1.6 ml were collected at a flow rate of 0.4 ml/min. For radioactive monitoring, 50 μl was taken from each and the assembly of intact splicing complexes verified by native gel analysis (3.5% polyacrylamide, 0.4% agarose). The appropriate fractions were pooled, and supplemented to 2 mm DTT, 0.7 U/ml RNasin, before mixing with streptavidin-coated beads (Boehringer) at 4°C. Bound complexes were washed extensively in 20 mm Tris at pH 7.9, 100 mm NaCl, prior to RNase A treatment. Proteins were eluted with 20 mm Tris at pH 7.6, 20 mm DTT, 2% SDS, and precipitated with acetone, before SDS-PAGE. Western blotting was performed as outlined above.
Acknowledgments
We thank T. Maniatis, G. Akusjärvi, V. Sukhatme, M. Green, J. Steitz, B. Turner, and M. Carmo-Fonseca who generously provided reagents. A.I.L. is a Wellcome Trust Principal Research Fellow. N.D.H. is an International Scholar of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Nick.Hastie@hgu.mrc.ac.uk; FAX 0131 343 2620.
References
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Barbaux S, Niaudet P, Gubler M-C, Grünfield J-P, Jaubert F, Kuttenn F, Fékété CN, Souleyreau-Therville N, Thibaud E, Fellous M, McElreavey K. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint recognition. Genes & Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes & Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Oghene K, Little MH, Seawright A, van Heyningen V, Hastie ND. Modulation of DNA binding specificity by alternative splicing of the Wilms’ tumour wt1 gene transcript. Science. 1992;257:235–237. doi: 10.1126/science.1321494. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuning W, Pelletier J. A non-AUG translational initiation event generates novel WT1 isoforms. J Biol Chem. 1996;271:8646–8654. doi: 10.1074/jbc.271.15.8646. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Duarte A, Larsson SH, Hastie ND, Little M, Holmes G, Todorov I, Ward A. RNA binding by the Wilms’ tumour suppressor zinc finger proteins. Proc Natl Acad Sci. 1996;93:7562–7566. doi: 10.1073/pnas.93.15.7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estojak J, Brent R, Golemis EA. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcárcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcárcel J, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Sohn RL, Buckler AJ, Pelletier J, Call KM, Housman DE. Alternative splicing and genomic structure of the Wilms tumour gene WT1. Proc Natl Acad Sci. 1991;88:9618–9622. doi: 10.1073/pnas.88.21.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets WJ, Hoet MH, de Jong BAW, van der Kemp A, van Venrooij WJ. Mapping of B cell epitopes on small nuclear riboproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Habor Laboratory; 1988. [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 cdk-interacting protein is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Harshman KD, Moye-Rowley WS, Parker CS. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell. 1988;53:321–330. doi: 10.1016/0092-8674(88)90393-5. [DOI] [PubMed] [Google Scholar]
- Hastie ND. The genetics of Wilms’ tumour—A case of disrupted development. Annu Rev Biochem. 1994;28:523–558. doi: 10.1146/annurev.ge.28.120194.002515. [DOI] [PubMed] [Google Scholar]
- Kennedy D, Ramsdale T, Mattick J, Little M. An RNA recognition motif in Wilms’ tumour protein (WT1) revealed by structural modelling. Nat Genet. 1996;12:329–332. doi: 10.1038/ng0396-329. [DOI] [PubMed] [Google Scholar]
- Kent J, Coriat A-M, Sharpe PT, Hastie ND, van Heyningen V. The evolution of WT1 sequence and expression pattern in the vertebrates. Oncogene. 1995;11:1781–1792. [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA. Interactions between small nuclear ribonuclear particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring J M, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND. Subnuclear localisation of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibody to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Green MR. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Little M, Wells C. A clinical overview of WT1 gene mutations. Hum Mut. 1997;9:209–225. doi: 10.1002/(SICI)1098-1004(1997)9:3<209::AID-HUMU2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Misteli T, Cáceres J, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA. Wilms’ tumour 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Transcription units as RNA processing units. Genes & Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- Pritchard-Jones K, Fleming S, Davidson D, Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J, Housman D, van Heyningen V, Hastie N. The candidate Wilms’ tumour gene is involved in genitourinary development. Nature. 1990;346:194–197. doi: 10.1038/346194a0. [DOI] [PubMed] [Google Scholar]
- Rauscher FJ, Morris JF, Tounay OE, Cook DM, Curran T. Binding of the Wilms’ tumour locus zinc finger protein to the EGR-1 consensus sequence. Science. 1990;250:1259–1262. doi: 10.1126/science.2244209. [DOI] [PubMed] [Google Scholar]
- Reddy JC, Morris JC, Wang J, English MA, Haber DA, Shi Y, Licht JD. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem. 1995;270:10878–10884. doi: 10.1074/jbc.270.18.10878. [DOI] [PubMed] [Google Scholar]
- Reed R, Griffith J, Maniatis T. Purification and visualisation of native spliceosomes. Cell. 1988;53:949–961. doi: 10.1016/s0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Sharma PM, Bowman M, Madden SL, Rauscher FJ, Sukumar S. RNA editing in the Wilms’ tumor susceptibility gene, WT1. Genes & Dev. 1994;8:720–731. doi: 10.1101/gad.8.6.720. [DOI] [PubMed] [Google Scholar]
- Tronchère H, Wang J, Fu X-D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- Turner BM, Franchi L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J Cell Sci. 1987;87:269–282. doi: 10.1242/jcs.87.2.269. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branchpoint and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Qiu QQ, Dueul TF. The Wilms’ tumour gene product WT1 activates or represses transcription through separate functional domains. J Biol Chem. 1993;268:9172–9175. [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]